Abstract
Background
Numerous reports have highlighted that pyroptosis is closely linked to tumorigenesis and drug resistance of tumors. However, the potential role of pyroptosis in regulating immune cell infiltration in tumor microenvironment (TME) remains unclear.
Methods
Here, we performed consensus clustering analysis based on the expression of 10 typical pyroptosis-related regulators (PRRs) to construct pyroptosis-mediated tumor pattern clusters and pyroptosis-related gene signature in breast cancer (BC). GSVA combined with ssGSEA methods were applied to evaluate the differences in biological pathway and immune cell infiltration level, respectively. The PCA method was employed to construct the pyro-score to quantify the pyroptosis pattern level of individual BC patient.
Results
We determined three distinct pyro-clusters among 1852 BC samples, which exhibited different survival outcomes and enriched biological processes. The TME features demonstrated that these three clusters corresponded to three established immune profiles: immune-desert, immune-excluded and immune-inflamed phenotype, respectively. Based on pyroptosis-related signature genes, we constructed the pyro-score and stratified BC patients into high and low pyro-score group. Patients with high pyro-score exhibited favorable outcome and increased infiltration of immune cells. Further investigation revealed that high pyro-score was also related to high expression of immunosuppressive molecules, decreased tumor mutation burden (TMB) and high rate of mutation in significantly mutated genes (SMGs) (eg, PIK3CA and CDH1).
Conclusion
This research emphasizes the indispensable role of pyroptosis in TME complexity and diversity. Assessing the pyroptosis pattern level of individual BC patient will assist us in better understanding TME features and directing more effective immunotherapeutic approaches.
Keywords: breast cancer, immune landscape, immunotherapy, pyroptosis, tumor microenvironment
Background
Pyroptosis (also named pyroptotic cell death), which is a caspase-dependent and inflammasome-mediated inflammatory form of cell death,1–3 is closely related to multiple pathological processes. As a novel procedural cell death, pyroptosis is controlled by specific genes encoding signals: pyroptosis-related regulators (PRRs).4 The expression and function of these PRRs exert important influence on pyroptosis and exploration of these PRRs could better understand the potential mechanisms of pyroptosis in diseases.5,6 Mounting evidence has revealed that genetic variation and expression perturbations of PRRs were correlated with the occurrence and development of malignant tumors and abnormal immunity.7–9 Comprehensive analysis of genetic change and dysregulated expression underlying tumor heterogeneity will be beneficial to identify potential therapeutic targets from the perspective of pyroptosis.
Breast cancer (BC) remains the most prevalent malignant tumor and the main cause of cancer-related mortality for women globally and its incidence and fatality rate were 24.2% and 15.0%, respectively.10,11 In recent years, with the deepening research on tumor microenvironment (TME) features (containing tumoral, immune and stromal compartment, and secreted cytokines), the role of immune cells in tumor development and progression has been emphasized.12–14 For instance, the simple immune score assessed by the density and location of CD3+ and cytotoxic CD8+ T cells could serve as an accurate predictor for BC prognosis and recurrence.15 Moreover, current immunotherapies have contributed to improve BC patients’ survival status and most notably, immune checkpoint inhibitors (ICIs): PD-1, PD-L1 and CTLA4 blockades have been approved for BC treatment by FDA.16,17 Assessment of immune infiltration from the perspective of TME constituents has become an essential method to predict the efficacy of ICIs.18,19 Currently, many researchers have proposed three basic immune profiles by parsing TME constituents: immune-inflamed/excluded/desert phenotype, characterized by distinct TME features and immunotherapeutic efficacy.20 Thus, identification of immune phenotypes from the perspective of TME constituents will be helpful to guide and predict immunotherapeutic responsiveness.21,22
Recent studies have identified the relationship between pyroptosis and immune cells in TME. Tang et al demonstrated that CD8+ T cells could release GZMA and GZMB to induce pyroptosis. Induced cell pyroptosis could activate macrophages-derived IL-1β to regulate anti-tumor immune response.23 Erkes et al found that BRAF and MEK inhibitors could activate CD4+ and CD8+ T cells infiltration and enhance anti-tumor immune response via GSDME gene-associated pyroptosis. Loss of GSDME gene will show defective HMGB1 release and decreased infiltration of T cells.24 However, due to technical limitations, current studies only focused on one or two PRRs and immune cells. Thus, comprehensive analysis of multiple PRRs-mediated TME cell-infiltrating features will be necessary and assist us in understanding anti-tumor immunity.
In our research, we incorporated genomic and transcriptomic data of 1852 BC samples from TCGA and GEO datasets to comprehensively evaluate the relationship between pyroptosis-mediated tumor pattern clusters and TME cell infiltration characteristics. Three distinct pyro-clusters using consensus clustering analysis were identified, which were characterized by three previously reported immune profiles: immune-inflamed/excluded/desert phenotype,20 indicating that pyroptosis takes on an indispensable role in the formation of diverse TME. Besides, to quantify the pattern level mediated by pyroptotic cell death in individual BC patient, we constructed a pyro-score using principal component analysis (PCA), which could predict ICIs response, suggesting that pyroptosis performed an essential role in guiding therapy for BC.
Methods
Obtainment and Preprocessing of BC Datasets
Transcriptome expression data and clinical information of BC patients were acquired from public repositories of the TCGA (https://portal.gdc.cancer.gov/) and GEO (https://www.ncbi.nlm.nih.gov/geo/) databases. The inclusion criteria for GEO datasets were based on: (1) datasets with adequate sample size greater than 80 were selected; (2) datasets were derived from the same platform: Affymetrix Human Genome U133 Plus 2.0 Array; (3) datasets without clinical characteristics and survival outcome were removed. A total of 6 eligible BC datasets (containing 1852 samples) were incorporated into this study, including TCGA-BRCA (N = 1109), GSE20685 (N = 327),25 GSE20711 (N = 88),26 GSE42568 (n = 104),27 GSE58812 (N = 107),28 GSE88770 (N = 117)29 datasets (Table 1). For TCGA datasets, RNA sequencing data with FPKM value were obtained and then transformed into transcripts per kilobase million (TPM), which are more similar to those deriving from GEO microarray datasets. Due to the fact that all GEO microarray data were derived from Affymetrix, we extracted expression matrix from the raw “CEL” and implemented robust multiarray averaging method provided by “affy” and “simpleaffy” packages to achieve background correction and quantile normalization. The “combat” algorithm of “sva” package was used to achieve the batch-effect removal,30 while merging the expression matrix of TCGA and GEO datasets. The genomic mutation data acquired from the UCSC Xena database was used for copy number variation (CNV) and somatic mutation analysis. The chromosomal location of CNV alterations in PRRs was plotted using “Rcircos” package.
Table 1.
Baseline Characteristics of Breast Cancer Patients in TCGA and GEO Datasets
| TCGA-BRCA | GSE20685 | GSE20711 | GSE42568 | GSE58812 | GSE88770 | |
|---|---|---|---|---|---|---|
| No. of patients | 1109 | 327 | 88 | 104 | 107 | 117 |
| Platform | Illumina RNAseq | Affymetrix U133 Plus 2.0 Array |
Affymetrix U133 Plus 2.0 Array |
Affymetrix U133 Plus 2.0 Array |
Affymetrix U133 Plus 2.0 Array |
Affymetrix U133 Plus 2.0 Array |
| Age | ||||||
| ≤65 | 775 (70.7%) | 305 (93.3%) | NA | 68 (65.4%) | 72 (67.3%) | NA |
| >65 | 321 (29.3%) | 22 (6.7%) | NA | 36 (34.6%) | 35 (32.7%) | NA |
| Gender | ||||||
| Male | 12 (1.1%) | 0 (0%) | NA | NA | NA | 0 (0%) |
| Female | 1084 (98.9%) | 327 (100%) | NA | NA | NA | 117 (100%) |
| Stage | ||||||
| I | 183 (17.1%) | NA | NA | NA | NA | NA |
| II | 621 (57.9%) | NA | NA | NA | NA | NA |
| III | 248 (23.1%) | NA | NA | NA | NA | NA |
| IV | 20 (1.9%) | NA | NA | NA | NA | NA |
| Unknown | 24 (2.2%) | NA | NA | NA | NA | NA |
| ER status | ||||||
| Positive | 789 (77.4%) | NA | 42 (48.3%) | 67 (66.3%) | 0 (0%) | 106 (90.6%) |
| Negative | 231 (22.6%) | NA | 45 (51.7%) | 34 (33.7%) | 107 (100%) | 11 (9.4%) |
| Unknown | 78 (7.1%) | NA | 1 (1.1%) | 3 (2.9%) | NA | NA |
| PR status | ||||||
| Positive | 685 (67.4%) | NA | NA | NA | 0 (0%) | 79 (68.1%) |
| Negative | 332 (32.6%) | NA | NA | NA | 107 (100%) | 37 (31.9%) |
| Unknown | 81 (7.4%) | NA | NA | NA | NA | 1 (0.9%) |
| HER-2 status | ||||||
| Positive | 162 (15.8%) | NA | 26 (29.5%) | NA | 0 (0%) | 7 (6.1%) |
| Negative | 865 (84.2%) | NA | 62 (70.5%) | NA | 107 (100%) | 108 (93.9%) |
| Unknown | 71 (6.5%) | NA | NA | NA | NA | 2 (1.7%) |
| Subtype | ||||||
| Luminal A | 305 (28.6%) | NA | 13 (14.8%) | NA | 0 (0%) | NA |
| Luminal B | 253 (23.7%) | NA | 22 (25.0%) | NA | 0 (0%) | NA |
| HER-2 | 145 (13.6%) | NA | 26 (29.5%) | NA | 0 (0%) | NA |
| Basal | 228 (21.4%) | NA | 27 (30.7%) | NA | 107 (100%) | NA |
| Normal | 136 (12.7%) | NA | 0 (0%) | NA | 0 (0%) | NA |
| Unknown | 31 (2.8%) | NA | NA | NA | NA | NA |
| Survival status | ||||||
| Alive | 947 (86.4%) | 244 (74.6%) | 63 (71.6%) | 69 (66.3%) | 78 (72.9%) | 89 (76.1%) |
| Deceased | 149 (13.6%) | 83 (25.4%) | 25 (28.4%) | 35 (33.7%) | 29 (27.1%) | 28 (23.9%) |
Definition of PRRs
Previous studies revealed that GSDMD could be cleaved by inflammasome-activated CASP1 and LPS-activated CASP4/5, thus executing the pyroptosis.31,32 Then, CASP8 was found to serve as a regulator of GSDMD cleavage.33 From then on, many researchers paid more attention to the role of gasdermins in cells. Mounting evidence revealed that cell apoptosis could be converted into pyroptosis when GSDME is cleaved by CASP3 and GZMB.34,35 Besides, GZMA are capable of cleaving GSDMB, thus converting apoptosis into pyroptosis.36 Accordingly, a total of 10 genes (GSDMD, CASP1, CASP4, CASP5, CASP8, GSDME, CASP3, GZMB, GZMA, GSDMB) were defined as PRRs in this study. Then, functional enrichment analysis of these PRRs was performed by using online database: Metascape (https://metascape.org/)37 and gene ontology (GO) provided by R “clusterProlifer” package.38
Consensus Clustering Analysis of PRRs
Based on the expression of these PRRs, we applied unsupervised clustering analysis to construct distinct tumor pattern clusters mediated by pyroptosis and stratified patients for further analysis. To determine the optimal number of clusters and guarantee their stability, the consensus clustering algorithm provided by “ConsensusClusterPlus” package was performed repeatedly for 100 times. Consensus clustering, as a highly useful technique in cancer research, could detect unknown groups in a dataset based on intrinsic biological features and no external information. Besides, this method could provide quantitative and visual stability evidence derived from repeated subsampling and clustering. Based on these merits, the expression of these PRRs was repeatedly factorized and the outputs aggregated to obtain consensus clustering of BC samples.
Gene Set Variation Analysis (GSVA) and Exploration of Tumor-Infiltrating Immune Cells
R “GSVA” package was utilized to perform GSVA enrichment analysis,39 which could explore the difference in biological processes between distinct pattern clusters mediated by pyroptosis. The gene set of “c2.cp.kegg.v7.4.symbols.gmt” downloaded from MsigDB database was used as the well-defined biological signature. Adjusted P-value less than 0.05 was regarded as statistically different. To investigate the immune infiltration landscape between distinct clusters, single-sample gene set enrichment analysis (ssGSEA) was implemented to calculate the infiltration levels of 23 different types of immune cells.40 Based on a Gaussian fitting model and multidimensional scaling, we estimated the bio-similarity of immune cells, calculated the enriched score of each immune cell and uniformly distributed the normalized score from 0 to 1.
Establishment of Differentially Expressed Genes (DEGs) Between Distinct Clusters Mediated by Pyroptosis
Distinct tumor clusters have been identified by the above consensus clustering analysis. Subsequently, to investigate DEGs between different clusters, we applied the empirical Bayesian approach of R “limma” package41 to analyze the expression of genes and screen out the DEGs using adjusted P-value <0.001 as the significance criteria.
Construction of the Pyro-Score
To quantify the pyroptosis pattern level of individual BC patient, we developed a scoring scheme by the following procedures. Firstly, the prognostic analysis of the overlapping DEGs identified between different tumor pattern clusters was performed by univariate Cox regression analysis and genes with prognostic impact were extracted. Then, feature selection of these genes with prognostic value was analyzed by recursive feature elimination with random forest and the 10-fold cross-validation method provided by “caret” package. Finally, the expression profile of the determined genes was employed to perform PCA analysis and we extracted principal components 1 and 2 as the signature score. The advantage of this method is focusing on the score on the set with the largest block of well correlated (or anticorrelated) genes in the set, while down-weighting contributions from genes that do not track with other set members. The pyro-score was defined using a formula similar to previous studies42–44 :∑(PC1i+PC2i), in which i represents the expression of PRRs.
Statistical Analysis
All data processing was generated in R-4.1.0. For quantitative data, we performed Student’s t-tests and the Wilcoxon rank-sum test to estimate the statistical significance for normally and nonnormally distributed variables, respectively. For comparisons of more than two groups, we used one-way analysis of variance and Kruskal–Wallis tests as parametric and nonparametric methods, respectively. To analyze the relationship between tumor pattern clusters mediated by pyroptosis and prognosis, we applied R “Survminer” package to perform Kaplan–Meier survival analysis and the Cox proportional hazards model. The surv-cutpoint function provided by R “survival” package was used to divide patients into high and low pyro-score and tumor mutation burden (TMB) group. All statistical P-values were two-side, with P-value <0.05 as statistical difference and adjusted P-value was calculated using the Benjamini–Hochberg correction.
Results
Landscape of Genetic Alterations of PRRs in BC
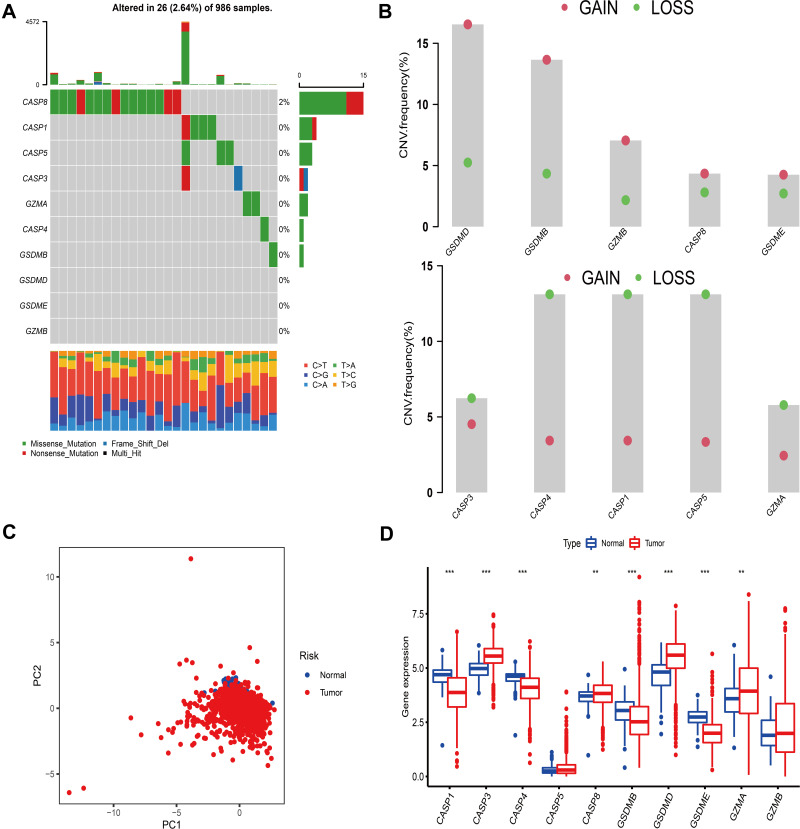
A total of 10 PRRs were finally investigated in this study. Metascape and GO enrichment analysis of these regulators were performed, and the results revealed that biological processes associated with pyroptosis were indeed significantly enriched (Figure S1A and B). Figure 1A depicts the prevalence of somatic mutations of 10 PRRs among 986 BC samples with available variant classification and variant type information, out of which 26 (2.64%) experienced genetic alterations of PRRs, mainly including missense mutation and nonsense mutation. Considering that CASP8 exhibited the highest mutation frequency, we investigated the difference in expression of PRRs between 965 CASP8-wild tumor samples and 15 mutant ones and a total of 3 PRRs (CASP1, CASP5 and GZMA) were found differentially expressed (Figure S2). Further exploration of 10 PRRs revealed a prevalent CNV. GSDMD, GSDMB, GZMB, CASP8, GSDME had prevalent CNV amplification, whose mean levels were 1.009, 1.436, 0.475, 0.384 and 0.509, respectively, while CASP4, CASP1, CASP5, CASP3 and GZMA showed widespread heterozygous deletions (Figure 1B and Table S1). The chromosomal locations of CNV alterations of PRRs are summarized in Table 2 and Figure S1C. Besides, principal component analysis based on these PRRs was performed and BC samples and normal ones could not be distinguished at all (Figure 1C). To explore whether the above genetic variations affected PRRs expression, we calculated the difference in gene expression values between BC and normal samples and observed that CASP3, CASP8, GSDMD, GZMA were significantly increased in BC tissues, whereas CASP1, CASP4, GSDMB, GSDME were markedly downregulated in tumor tissues (Figure 1D). Compared with normal samples, PRRs with CNV amplification exhibited remarkedly higher expression in BC samples, such as CASP8, GSDMD, while other RPGs with deleted CNV, such as CASP1, CASP4 were significantly decreased in tumor tissues (Figure 1B and D), indicating that there was high heterogeneity of genomic and transcriptomic landscape of PRRs between BC samples and normal ones and this expression imbalance of PRRs performed a vital role in the tumorigenesis and BC progression.
Figure 1.
Landscape of genetic and transcriptomic variation of pyroptosis-related regulators in breast cancer. (A) 26 of the 986 BC patients experienced genetic alterations of 10 PRRs, with a frequency of 2.64%, mostly including missense mutation and nonsense mutation. (B) The CNV frequency of 10 PRRs was prevalent. The column represented the alteration frequency. The deletion frequency, green dot; The amplification frequency, pink dot. (C) PCA of 10 PRRs to distinguish BC from normal samples. (D) The difference of mRNA expression levels of 10 PRRs between BC and normal samples. The asterisks represented the statistical P-value (**P < 0.01; ***P < 0.001).
Table 2.
The Chromosomal Locations of Copy Number Variation of 10 Pyroptosis-Related Regulators in Breast Cancer
| Chromosome | Start | End | bp | CNV Frequency | |
|---|---|---|---|---|---|
| CASP8 | Chr2 | 201,233,463 | 201,286,594 | 53,132 | Gain |
| CASP3 | Chr4 | 184,627,696 | 184,649,509 | 21,814 | Loss |
| GZMA | Chr5 | 55,102,646 | 55,110,252 | 7607 | Loss |
| GSDME | Chr7 | 24,698,355 | 24,757,939 | 59,585 | Gain |
| GSDMD | Chr8 | 143,557,913 | 143,563,062 | 5150 | Gain |
| CASP4 | Chr11 | 104,942,866 | 104,968,574 | 25,709 | Loss |
| CASP5 | Chr11 | 104,994,240 | 105,023,168 | 28,929 | Loss |
| CASP1 | Chr11 | 105,025,508 | 105,035,250 | 9743 | Loss |
| GZMB | Chr14 | 24,630,954 | 24,634,190 | 3237 | Gain |
| GSDMB | Chr17 | 39,904,597 | 39,918,650 | 14,054 | Gain |
Construction of Tumor Pattern Clusters Mediated by PRRs
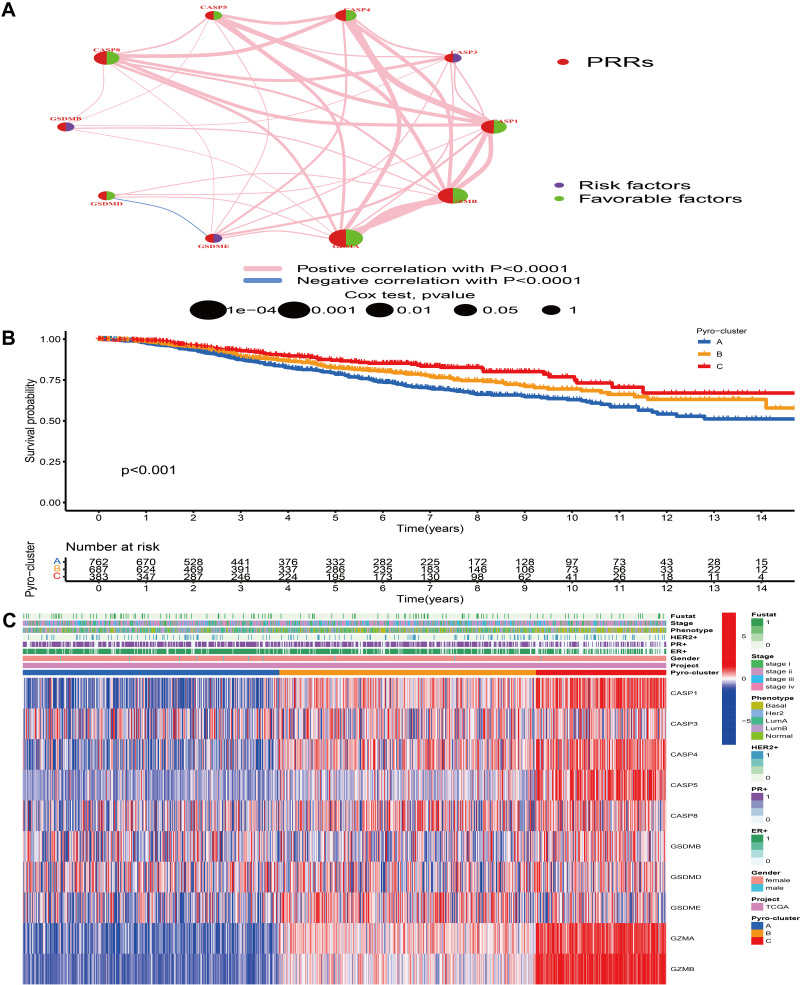
TCGA-BRCA and 5 GEO datasets (GSE20685, GSE20711, GSE42568, GSE58812, GSE88770), whose survival information was available, were merged into one meta-cohort. Univariate Cox regression and Kaplan–Meier analysis were performed to determine the prognostic value of PRRs in BC samples and P-value <0.05 was regarded as the criteria for infiltering. Figure 2A comprehensively illustrates the landscape of the interactions and connections of PRRs and their prognostic value in BC. This network suggested that the cross-talk between these genes was significantly associated with survival outcome of BC patients and performed a vital role in the development of diverse tumor pattern clusters mediated by PRRs. Based on the expression of these PRRs, the consensus clustering analysis was conducted to classify patients with qualitatively different pyroptosis-mediated patterns. We discovered that k = 3 appeared to be an optimal selection for sorting the whole BC samples and 3 distinct clusters were finally obtained (Figure S3). We defined these clusters as pyro-cluster A, B and C, which included 762, 688 and 384 samples, respectively. The predictive ability of these 3 pyro-clusters in survival outcome demonstrated that pyro-cluster C possessed the significant prognostic benefit and pyro-cluster A exhibited the worst survival outcome (Figure 2B). Figure 2C depicted the difference of gene expression profiles and clinical characteristics between distinct pyro-clusters, and we observed that CASP1, CASP4, CASP5, GZMA, GZMB were evidently elevated in pyro-cluster C.
Figure 2.
Tumor pattern clusters mediated by pyroptosis-related regulators. (A) The interaction of expression on 10 PRRs in BC. The lines connecting PRRs represented their interaction with each other. The size of each circle represented the prognosis effect of each regulator and scaled by P-value. Protective factors for patients’ survival were indicated by a green dot in the circle center and risk factors indicated by the purple dot in the circle center. (B) Kaplan–Meier curves of OS for 1834 BC patients in one meta cohort with different pyro-clusters. The numbers of patients in pyro-cluster A, B and C were 762, 688 and 384, respectively. (C) Unsupervised clustering of PRRs in one meta cohort. The pyro-cluster, gender, ER+, PR+, HER2+, phenotype, stage and survival status were used as patient annotations. Red represented the high expression of regulators and blue represented low expression.
The Pyro-Clusters Characterized by Distinct Immune Profiles
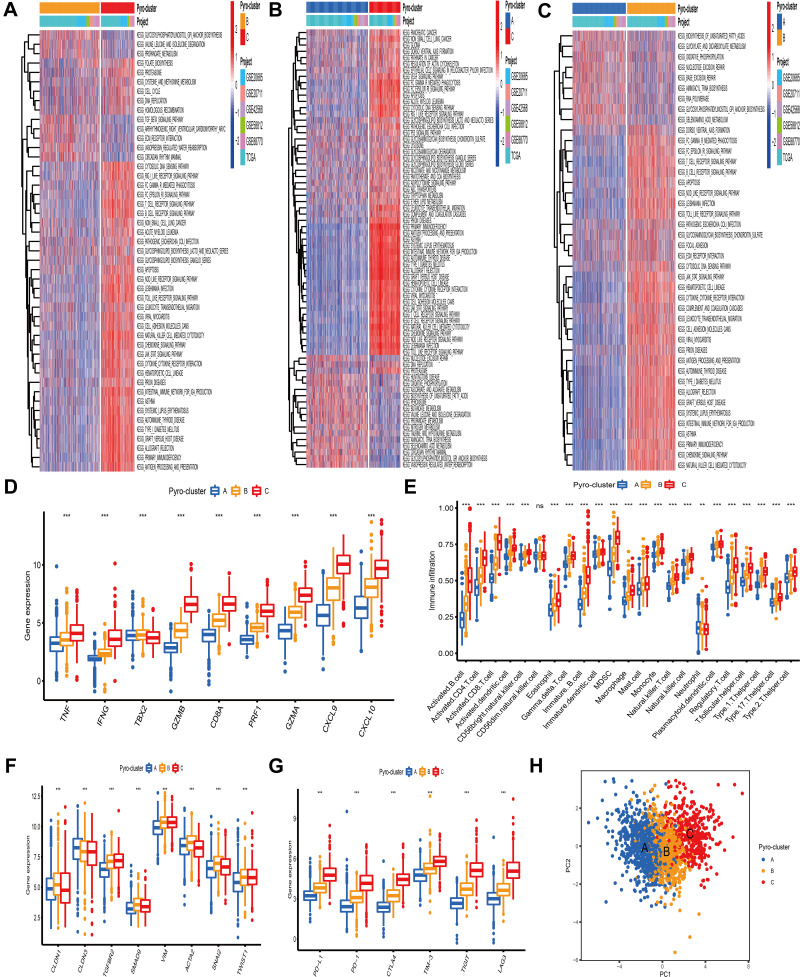
To understand the biological behaviors underlying these 3 distinct clusters mediated by PRRs, we conducted GSVA analysis and found that compared with pyro-cluster A and B, pyro-cluster C with favorable prognosis presented immune activation enrichment pathways, including allograft rejection, cytokine–cytokine receptor interaction, chemokine signaling pathway, T/B cell receptor signaling pathway, natural killer cell mediated cytotoxicity and Toll-like receptor signaling pathway (Figure 3A and C). Besides, some transcripts of immune activation, including TNF, IFNG, GZMB, CD8A, PRF1, GZMA, CXCL9, CXCL10 were also found highly expressed in pyro-cluster C (Figure 3D). Pyro-cluster B was prominently enriched in pathways associated with carcinogenic activation and stromal pathway, including TGF beta signaling pathway, ECM receptor interaction, while pyro-cluster A was remarkedly associated with biological process associated with immune suppression. Besides, we performed ssGSEA to evaluate the infiltration levels of 23 different types of immune cells in TME among distinct pyro-cluster. As shown in Figure 3E, with the exception of neutrophils (mainly elevated in pyro-cluster A), other immune cells were significantly elevated in pyro-cluster C and B, especially antitumor lymphocyte cell subpopulations, such as activated CD4+/CD8+ T cells, B cells and natural killer cells. However, pyro-cluster B did not exhibit favorable prognosis, compared with pyro-cluster C. Previous researches revealed that immune-excluded phenotype consisted of abundant immune cells. However, these immune cells did not penetrate the tumor parenchyma but instead were positioned in the stroma surrounding tumor cell nests. The abundant stromal elements were regarded as T-cell suppressive. The GSVA findings have revealed that pyro-cluster B was remarkedly correlated with stromal activation. Thus, we speculated that stromal activation in pyro-cluster B suppressed the antitumor activity of immune cells. Subsequent analysis revealed that pyro-cluster B definitely exhibited stromal activation, as some transcripts of TGF beta/EMT pathway, including CLDN1, TGFBR2, SMAD9, VIM, ACTA2, SNAI2, TWIST1 were highly expressed in pyro-cluster B (P-value <0.001) (Figure 3F). Considering that immunosuppressive molecule is an effective predictor of immunotherapeutic responsiveness, we also calculated PD-L1, PD-1, CTLA4, TIM-3, TIGIT and LAG3 expression level among distinct clusters and observed a prominent upregulation of pyro-cluster C (Figure 3G). Based on these findings, we determined that these 3 pyro-clusters were characterized by distinct immune infiltration landscapes. As expected, we considered that pyro-cluster B as immune-excluded phenotype, characterized by weakened immune cell infiltration and stromal activation; cluster C as immune-inflamed phenotype consisting of abundance of infiltrating immune cells and immune activation and cluster A as immune-desert phenotype, characterized by immune suppression. Moreover, we performed PCA to distinguish between 3 pyro-clusters based on the expression of PRRs and observed that they were generally separated with a relatively clear resolution (Figure 3H).
Figure 3.
Biological pathway and tumor microenvironment characteristics in distinct pyro-clusters. (A–C) Heatmap showed the GSVA score of representative Hallmark pathways in distinct pyro-clusters. The TCGA-BRCA and GEO cohort (GSE20685, GSE20711, GSE42568, GSE58812, GSE88770) compositions were used as sample annotations. (D) Difference in the immune-activation related gene expression among three pyro-clusters. (E) The fraction of tumor-infiltrating cells in three pyro-clusters using the ssGSEA. Within each group, the scattered dots represented TME cell expression values. The thick line represented the median value. The bottom and top of the boxes were the 25th and 75th percentiles (interquartile range). The statistical difference of three pyro-clusters was compared through the Kruskal–Wallis H-test. (F) Difference in the TGF/beta-EMT pathway-related gene expression among three pyro-clusters. (G) Difference in the immune-checkpoint related gene expression among three pyro-clusters. (H) PCA of PRRs to distinguish between pyro-cluster A, B and C. ***P < 0.001, ns, not significant.
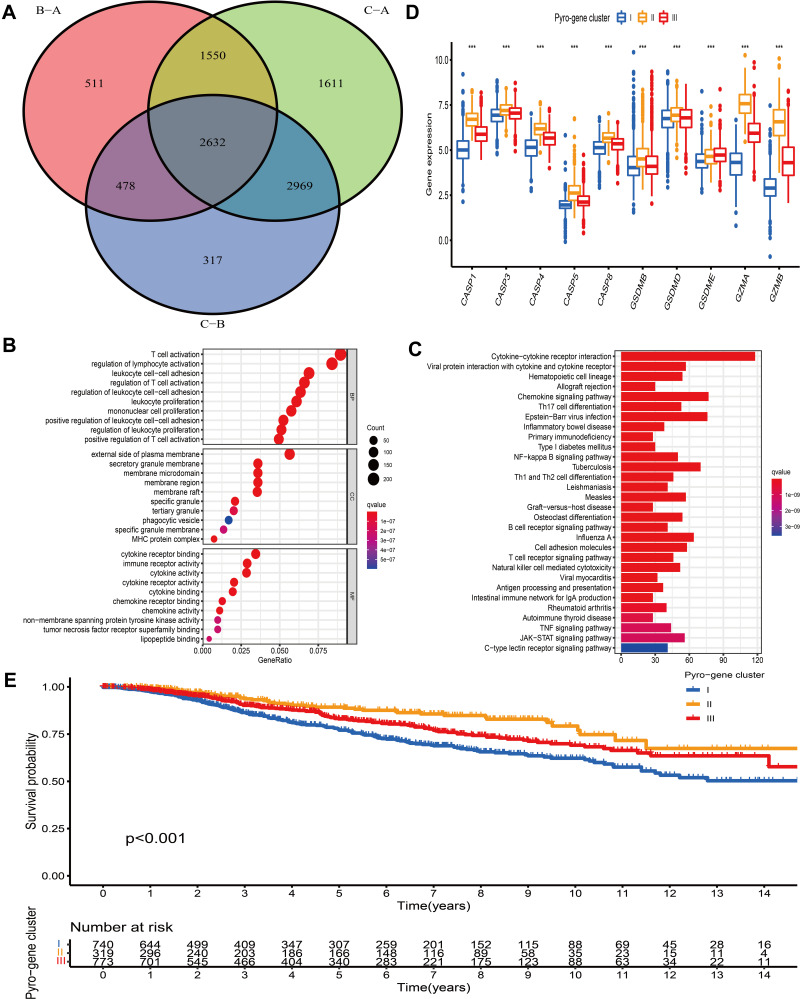
Identification of Pyro-Gene Clusters and Exploration of Their Functional Annotations
Based on the expression of PRRs, the consensus clustering analysis could stratify BC patients into 3 pyro-clusters. However, the transcriptional perturbations underlying these 3 clusters were not fully elucidated. Thus, the expression change of 16,437 genes between distinct pyro-clusters in BC was investigated by “limma” package and a total of 2632 overlapping DEGs among the 3 pyro-clusters were determined and regarded as the crucial pyroptosis-related gene signature distinguishing 3 pyro-clusters (Figure 4A). Somatic mutation analysis of these DEGs demonstrated that 922 out of 986 (93.51%) tumor samples carried genetic alterations. Further analysis of CNV alterations revealed that these genes exhibited either amplification or deletion. GO enrichment analysis of these DEGs revealed that biological processes associated with immune regulation and inflammatory response were prominently enriched (Figure 4B). KEGG enrichment analysis also demonstrated that these DEGs played an immunomodulatory role in TME (Figure 4C). Based on the 2632 pyroptosis-related signature genes, we conducted unsupervised consensus clustering analysis by using “ConsensusClusterPlus” package38 and identified 3 distinct pyroptosis-related signature subtypes (Figure S4). We termed these 3 subtypes as pyro-gene clusters I, II and III, among which the difference in expression of PRRs was significant (Figure 4D). Besides, analysis of clinicopathological features among these 3 pyro-gene clusters revealed that patients with basal phenotype, ER-, PR- and HER2+ were mainly concentrated in cluster I (Figure S5). Further prognostic analysis revealed that pyro-gene cluster I exhibited the worse survival outcome and pyro-gene cluster II was proven to be related to favorable prognosis (Figure 4E).
Figure 4.
Construction of pyro-gene signature and functional annotation. (A) 2632 pyroptosis-related DEGs between three pyro-clusters were shown in the Venn diagram. (B and C) Functional annotation for pyroptosis-related signature genes using GO and KEGG enrichment analysis. (D) The expression of PRRs in three pyro-gene clusters. The upper and lower ends of the boxes represented an interquartile range of values. The lines in the boxes represented the median value. The asterisks represented the statistical P-value The one-way ANOVA test was used to test the statistical differences among three pyro-gene clusters. (E) The survival curves of the pyro-gene clusters were estimated by the Kaplan-Meier plotter (P <0.001). ***P < 0.001.
Construction and Validation of the Pyroptosis-mediated Prognostic Model
To help clinicians predict the likelihood of survival for BC patients, we constructed a prognostic model from the perspective of pyroptosis. R “caret” package was utilized to randomly assigned (at a ratio of 1:1) 1832 BC patients into discovery (916 samples) and validation (916 ones) cohorts. We then applied prognostic pyroptosis-related signature genes into LASSO regression algorithm and determined a total of 357 overall survival (OS)-related genes, based on the optimum λ value and minimum partial likelihood deviance (Figure S6A and B), for the subsequent multivariate Cox regression analysis. According to the optimum AIC value, we incorporated 18 genes into the prognostic model, whose formula was presented as follows:
Risk score = (expression of BCL2A1 * −0.238470252350429) + (expression of TRDV3* −0.346925429019898) + (expression of SPNS2 * 0.185309943913867) + (expression of GNG8 * −0.189794329786803) + (expression of GP1BA * −0.405484788043643) + (expression of TMEM200C * −0.373334800071322) + (expression of DDX60L * −0.190514793988741) + (expression of GRHL2 * 0.240968659418045) + (expression of TBPL1 * 0.298873968952058) + (expression of GSTM3 * −0.116072513675859) + (expression of IL17RB * −0.191525854234791) + (expression of ESM1 * 0.165800870027823) + (expression of PCSK6 * −0.177033178053132) + (expression of NDUFA5 * −0.546965749404047) + (expression of CLDN7 * 0.219203629474111) + (expression of BACE1 * 0.439324243330006) + (expression of CARD14 * 0.219615672010187) + (expression of HEATR3 * 0.325749622674532).
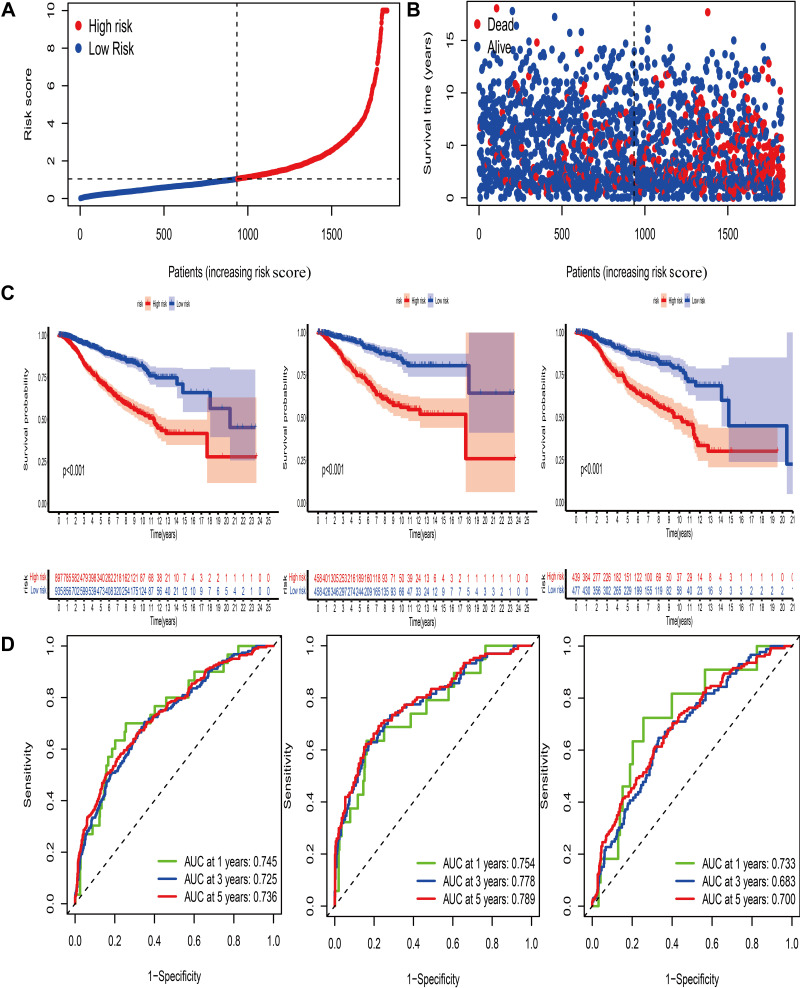
The median risk score calculated based on the formula was utilized to stratify BC patients into 2 groups (Figure 5A). The distribution plot of the risk score and survival analysis revealed that high-risk groups were more likely to suffer from earlier death and exhibited a dismal outcome, compared with low-risk group (Figure 5B–D). The AUC of 5-year ROC in the all, discovery and validation dataset was 0.736, 0.789 and 0.700, respectively (Figure 7D). To further assess the predictive ability of this signature in OS, we compared this pyroptosis-related gene signature with the previously established autophagy and ferroptosis-related models in BC and observed that our signature exhibited better predictive abilities (Figure S6C and D).
Figure 5.
Construction of the pyroptosis-mediated prognostic model. (A and B) Ranked dot and scatter plots showing the risk score distribution and patient survival status. (C) Kaplan–Meier analysis of the OS between the two groups in the all, discovery and validation dataset, respectively. (D) ROC curves to predict the sensitivity and specificity of 1-, 3- and 5-year survival according to the prognostic model in the all, discovery and validation dataset, respectively.
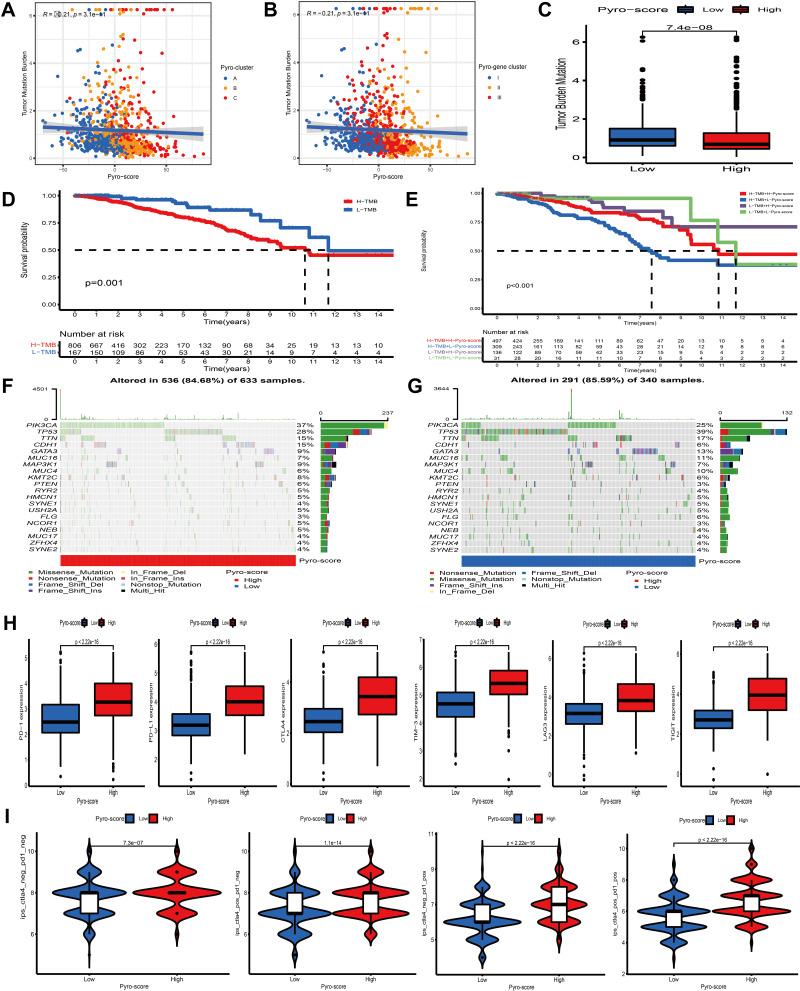
Figure 7.
Characteristics of pyro-score in tumor somatic mutation and immunotherapies. (A and B) Correlation analysis between pyro-score and TMB. (C) Relative distribution of TMB in high versus low pyro-score group. (D) Kaplan–Meier curves for high and low TMB patient groups (P= 0.001). (E) Kaplan–Meier curves for subgroup patients stratified by both pyro-score and TMB (P< 0.001). (F and G) Mutational landscape of SMGs in TCGA-BRCA stratified by high (left panel) versus low pyro-score (right panel) groups. Individual patients were represented in each column. The upper bar plot showed TMB, the right bar plot showed the mutation frequency of each gene in separate pyro-score groups. (H) Relative distribution of immunosuppressive molecules expression in high pyro-score versus low pyro-score groups. (I) Relative distribution of immunotherapeutic efficacy in high pyro-score versus low pyro-score groups.
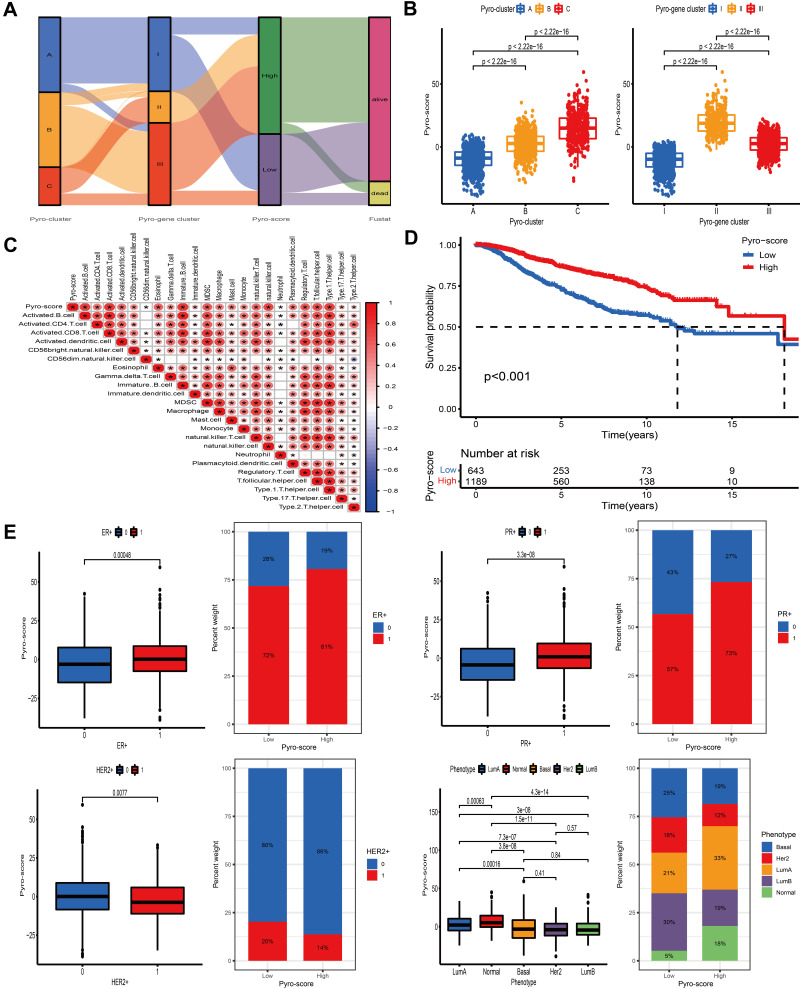
Development of the Pyro-Score and Evaluation of Its Clinical Significance
The above analyses have revealed that pyroptosis played an essential role in survival outcome and immune regulation of tumor-infiltrating cells. However, these findings were only based on the patient population and could not accurately predict the patterns of pyroptotic cell death in individual tumor. Consequently, we constructed a scoring system based on identified pyroptosis-related signature genes and defined it as pyro-score to quantify the pattern mediated by pyroptotic cell death in individual BC patient. Considering that the quantification of pyroptosis is complex, we used the alluvial diagram to illustrate the workflow of pyro-score construction (Figure 6A). The results revealed that pyro-gene clusters II and III were linked to high pyro-score, while pyro-gene cluster I was related with low pyro-score. Moreover, Kruskal–Wallis test demonstrated that pyro-cluster C exhibited the highest pyro-score, followed by pyro-cluster B and pyro-cluster A (Figure 6B). Spearman analysis was performed to examine the relationship between the pyro-score and immune landscape. The correlation matrix revealed that the pyro-score was positively associated with tumor-infiltrating lymphocytes, including activated CD4+/CD8+ T cells, activated B cells and natural killer T cells and negatively correlated with tumor-associated neutrophils, demonstrating the crosstalk between pyro-score and tumor-infiltrating immune cells (Figure 6C). Furthermore, the predictive ability of pyro-score for BC prognosis was estimated by stratifying patients into high- and low-score group, according to the cutoff value of −4.50714330196327. Unexpectedly, patients in high-score group exhibited a prominent survival benefit (Figure 6D). Considering the highly complex heterogeneity of BC, we further explored the prognostic impact of pyro-score in distinct classifications of BC and observed the similar results in clinical subtypes: luminal A, B and basal-like (Figure S7A). The relationship between the pyro-score and ER status, PR status, HER2+ status and molecular subtype was also analyzed. As shown in Figure 6E, ER+, PR+ were correlated with a high pyro-score, while HER2+ and Basal subtype were significantly associated with a low pyro-score.
Figure 6.
Construction of pyro-score and exploration of its clinical features. (A) Alluvial diagram of pyro-clusters in groups with different pyro-gene clusters, and pyro-score. (B) Distribution of pyro-score in the different pyro-clusters and pyro-gene clusters. (C) Correlations between pyro-score and the immune landscape using Spearman analysis. The negative correlation was marked with blue and positive correlation with red. The star signified the statistical difference (*P-value <0.05). (D) Kaplan–Meier curves for patients with high and low pyro-score subgroups. (E) Bar plot showing pyro-score in groups with ER+, PR+, HER2+, phenotypes. The differences between different groups were compared through the Kruskal–Wallis test.
Estimation of the Role of Pyro-Score in Tumor Somatic Mutation, Immunotherapy and Chemotherapeutic Efficacy
Accumulating evidence has revealed that cancer-specific antigens generated by somatic mutations could influence the responsiveness to immunotherapy. Thus, the distribution pattern of TMB was investigated between high and low pyro-score groups. The results demonstrated that pyro-score was negatively correlated with TMB and high-score group had lower TMB (Figure 7A–C). BC patients were categorized into high- or low-TMB group based on the cutoff value of 0.421052631578947. Survival analysis further demonstrated that high-TMB group exhibited an unfavorable survival outcome and, as expected, combined high-TMB and low pyro-score group had the worst prognosis (Figure 7D and E). Significantly mutant genes (SMGs) analysis was also conducted between different pyro-score group and the SMGs landscape revealed that PIK3CA (38% vs 25%) and CDH1 (16% vs 5%) exhibited higher somatic mutation rate in high-score group, whereas TP53 (38% vs 28%) had higher somatic mutation rate in low-score group (Figure 7F and G).
Immunotherapy based on immunosuppressive molecules, such as PD-1/CTLA-4 has achieved a considerable breakthrough in antitumor response. Many well-known predictors, such as PD-L1 were extensively used to assess immune response. Our analysis showed that the expression levels of common immunosuppressive molecules, including PD-1, PD-L1 and CTLA4 and novel immune checkpoint proteins, including TIM-3, LAG3, TIGIT, were pronouncedly elevated in the high pyro-score group (Figure 7H), indirectly demonstrating the essential role of pyro-score in mediating immune response. As a superior predictor of responsiveness of anti-PD-1 and CTLA-4 therapies, IPS could calculate the determinants of tumor immunogenicity and depict the cancer antigenomes and intra-tumoral immune profiles.45 This scoring scheme derived from a panel of immune-related genes, which belong to four classes: suppressor cells, effector cells, immunomodulators or checkpoints, and MHC-related molecules. By averaging the samplewise Z scores of the four classes within the respective category, the sum of the weighted averaged Z score was calculated as the IPS. We observed that patients in high pyro-score group exhibited significant therapeutic benefits from ICIs treatment represented by IPS (CTLA4-/PD-1-, CTLA4+/PD-1-, CTLA4-/PD-1+ and CTLA4+/PD-1+), indirectly suggesting that pyro-score played an essential role in predicting response to immunotherapies (Figure 7I).
To assess whether pyro-score was associated with the half inhibitory concentration (IC50) of common antitumor drugs and chemotherapeutic efficacy, we applied “pRRophetic” package in R. By constructing the ridge regression model based on Genomics of Drug Sensitivity in Cancer (GDSC) (www.cancerrxgene.org/) cell-line expression spectrum and TCGA gene expression profiles, the package could apply pRRophetic algorithm to predict drug IC50. We compared the difference of drug IC50 between different pyro-score group and determined that patients in high pyro-score group were negatively correlated with IC50 of cisplatin, doxorubicin, gemcitabine, methotrexate, roscovitine, vinblastine and vinorelbine, demonstrating that they were the potentially beneficial candidates from these commonly used chemotherapeutic drugs (Figure S7B).
Discussion
Accumulating evidence has revealed that regulators involved in pyroptosis perform an indispensable role in inflammation and antitumor immunity.46,47 As plenty of studies are restricted to single TME infiltrating immune cell or PRR, comprehensive analysis of the overall TME immune features mediated by multiple PRRs is not conducted. Thus, identification of distinct tumor pattern clusters mediated by pyroptosis in TME will deepen our understanding of antitumor immunity and direct more effective immunotherapeutic plans.
In this research, three pyro-clusters characterized by distinct immune profiles were identified. Pyro-cluster A was characterized by immune suppression, corresponding to an immune-desert phenotype. Pyro-cluster B was characterized by the infiltration of immune cells and activation of stroma, corresponding to an immune-excluded phenotype. Pyro-cluster C was characterized by immune activation and anti-tumor lymphocyte infiltration, corresponding to immune-inflamed phenotype. Recent researches revealed that TME components performed an essential role in tumor progression and immune response.19 Baseline infiltration level of CD4/8+ T cells, natural killer cells, M1 macrophages and secreted cytokines were prominently related to immunotherapeutic efficacy.19,48,49 We also found that pyro-cluster C was prominently related to anti-tumor lymphocyte infiltration and high expression level of immunosuppressive molecules, indicating its potential value in predicting immunotherapeutic benefits. A previous study found that activation of TGF beta and EMT-related signaling pathway could impede the lymphocytes from penetrating into the tumor parenchyma.50 TGF beta-targeted molecular agents have been found to take on an important role in reshaping TME and restoring anti-cancer immunity.22,51 Thus, we speculated that combination of ICIs and TGF beta blockade may be beneficial to pyro-cluster B patients.
Moreover, DEGs identified between three pyro-clusters were prominently elevated in biological pathways associated with inflammatory response and immune regulation, indicating that these DEGs were regarded as pyroptosis-related gene signature. Based on these pyroptosis-related genes, three transcriptomic clusters were constructed and characterized by different survival outcomes, which were in great accord with the results of pyro-clusters. Furthermore, a scoring scheme named as pyro-score was established to quantify the pyroptosis pattern level of individual BC patient and direct therapeutic interventions more precisely. As a result, pyro-cluster C characterized by immune-inflamed phenotype showed high pyro-score, while pyro-cluster B and A characterized by immune-excluded and desert phenotype, respectively, exhibited low pyro-score. In addition, we observed that pyro-score could serve as a prognostic biomarker in BC and significantly correlated with molecular phenotype, indicating that this pyro-score plays an important complementary role to clinical predictors. Besides, subgroup analysis of classical classifications of BC demonstrated that pyro-score performed a vital role in predicting clinical outcome in luminal A, B and basal-like subtypes. Further analyses demonstrated that pyro-score was markedly linked to immunosuppressive molecules and immunotherapy, implying that pyro-score could influence therapeutic efficacy. Based on these findings, we believed that pyroptosis could be used in clinical practice to identify immune profiles and direct therapeutic strategies.
Assessment of mutated genes capable of driving tumors is one milestone towards cancer detection and therapeutic approach selection. Here, we observed that compared with low pyro-score group, PIK3CA and CDH1 exhibited elevated mutation rate in high pyro-score group, while TP53 showed augmented mutation rate in low pyro-score group. Recent researches revealed that CDH1 and PIK3CA mutations in genetically modified mice could result in immune-related subtype for invasive lobular carcinoma of breast, which was characterized by enhanced immune infiltration and a strong signature of Treg cells and immunosuppressive molecule-based immune checkpoint activation.52 TP53 is a frequently mutated tumor suppressor gene, which could enhance immune activity in BC.53 These pyro-score mediated driver gene mutations remarkedly correlated with immune activity, highlighting the complicated connection between pyroptosis and tumor immunogenomic features.
Although we obtained ten typical regulators involved in pyroptosis in this research, a series of novel regulators would be identified. Future researches were required to optimize the accuracy of pyroptosis-mediated patterns by integrating more PRRs. Besides, we constructed tumor pattern clusters and pyro-score using retrospective cohorts; thus, prospective datasets of BC samples were required to verify our results.
Conclusions
In conclusion, we comprehensively analyzed pyroptosis-mediated tumor patterns of 1852 BC patients based on 10 typical PRRs, and systematically linked this pattern cluster with TME cell-infiltrating features. This integrated analysis demonstrated that pyroptosis takes on an indispensable role in regulation of antitumor immunity. More broadly, assessing the pyroptosis pattern level of individual tumor will assist us in better understanding TME characteristics and directing more effective immunotherapeutic approaches.
Funding Statement
This study was supported by National Scientific Foundation of China (No. 81870554) and Foundation from Renji Hospital, School of Medicine, Shanghai Jiaotong University (2019NYLYCP0102).
Abbreviations
PRRs, pyroptosis-related regulators; BC, breast cancer; TME, tumor microenvironment; ICIs, immune checkpoint inhibitors; TPM, transcripts per kilobase million; CNV, copy number variation; GSVA, gene set variation analysis; ssGSEA, single-sample gene set enrichment analysis; DEGs, differentially expressed genes; PCA, principal component analysis; TMB, tumor mutation burden; SMGs, significantly mutated genes.
Data Sharing Statement
The datasets analyzed during the current study are available in the TCGA (https://portal.gdc.cancer.gov/) and GEO (https://www.ncbi.nlm.nih.gov/geo/) databases.
Ethics Approval and Informed Consent
All datasets in the present research were downloaded from public databases, including TCGA and GEO. Thus, the present study was exempted from the approval of ethics review board in Renji Hospital, School of Medicine, Shanghai Jiao Tong University. The current study follows the TCGA and GEO data access policies and publication guidelines.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Fang Y, Tian S, Pan Y, et al. Pyroptosis: a new frontier in cancer. Biomed Pharmacother. 2020;121:109595. doi: 10.1016/j.biopha.2019.109595 [DOI] [PubMed] [Google Scholar]
- 2.Kovacs SB, Miao EA. Gasdermins: effectors of Pyroptosis. Trends Cell Biol. 2017;27(9):673–684. doi: 10.1016/j.tcb.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi J, Gao W, Shao F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem Sci. 2017;42(4):245–254. doi: 10.1016/j.tibs.2016.10.004 [DOI] [PubMed] [Google Scholar]
- 4.Xia X, Wang X, Cheng Z, et al. The role of pyroptosis in cancer: pro-cancer or pro-”host”?. Cell Death Dis. 2019;10(9):650. doi: 10.1038/s41419-019-1883-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malireddi RKS, Kesavardhana S, Kanneganti TD. ZBP1 and TAK1: master regulators of NLRP3 inflammasome/ Pyroptosis, Apoptosis, and Necroptosis (PAN-optosis). Front Cell Infect Microbiol. 2019;9:406. doi: 10.3389/fcimb.2019.00406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue Y, ENOSI TUIPULOTU D, Tan WH, et al. Emerging activators and regulators of inflammasomes and pyroptosis. Trends Immunol. 2019;40(11):1035–1052. doi: 10.1016/j.it.2019.09.005 [DOI] [PubMed] [Google Scholar]
- 7.Hou J, Zhao R, Xia W, et al. PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis. Nat Cell Biol. 2020;22(10):1264–1275. doi: 10.1038/s41556-020-0575-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu H, Zhang S, Wu J, et al. Molecular targeted therapies elicit concurrent apoptotic and GSDME-dependent pyroptotic tumor cell death. Clin Cancer Res. 2018;24(23):6066–6077. doi: 10.1158/1078-0432.CCR-18-1478 [DOI] [PubMed] [Google Scholar]
- 9.Zhao P, Wang M, Chen M, et al. Programming cell pyroptosis with biomimetic nanoparticles for solid tumor immunotherapy. Biomaterials. 2020;254:120142. doi: 10.1016/j.biomaterials.2020.120142 [DOI] [PubMed] [Google Scholar]
- 10.Kalimutho M, Nones K, Srihari S, et al. Patterns of genomic instability in breast cancer. Trends Pharmacol Sci. 2019;40(3):198–211. doi: 10.1016/j.tips.2019.01.005 [DOI] [PubMed] [Google Scholar]
- 11.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 12.Simonaggio A, Epaillard N, Pobel C, et al. Tumor microenvironment features as predictive biomarkers of response to Immune Checkpoint Inhibitors (ICI) in metastatic clear cell Renal Cell Carcinoma (mccRCC). Cancers. 2021;13(2):231. doi: 10.3390/cancers13020231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fridman WH, Zitvogel L, Sautès-fridman C, et al. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14(12):717–734. doi: 10.1038/nrclinonc.2017.101 [DOI] [PubMed] [Google Scholar]
- 14.Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. 2015;15(11):669–682. doi: 10.1038/nri3902 [DOI] [PubMed] [Google Scholar]
- 15.Deepak KGK, Vempati R, Nagaraju GP, et al. Tumor microenvironment: challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol Res. 2020;153:104683. doi: 10.1016/j.phrs.2020.104683. [DOI] [PubMed] [Google Scholar]
- 16.Barzaman K, Moradi-kalbolandi S, Hosseinzadeh A, et al. Breast cancer immunotherapy: current and novel approaches. Int Immunopharmacol. 2021;98:107886. doi: 10.1016/j.intimp.2021.107886 [DOI] [PubMed] [Google Scholar]
- 17.Gaynor N, Crown J, Collins DM. Immune checkpoint inhibitors: key trials and an emerging role in breast cancer. Semin Cancer Biol. 2020. doi: 10.1016/j.semcancer.2020.06.016 [DOI] [PubMed] [Google Scholar]
- 18.Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. doi: 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov. 2019;18(3):197–218. doi: 10.1038/s41573-018-0007-y [DOI] [PubMed] [Google Scholar]
- 20.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330. doi: 10.1038/nature21349 [DOI] [PubMed] [Google Scholar]
- 21.Bareche Y, Buisseret L, Gruosso T, et al. Unraveling triple-negative breast cancer tumor microenvironment heterogeneity: towards an optimized treatment approach. J Natl Cancer Inst. 2020;112(7):708–719. doi: 10.1093/jnci/djz208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang R, Xu J, Zhang B, et al. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J Hematol Oncol. 2020;13(1):110. doi: 10.1186/s13045-020-00946-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erkes DA, Cai W, Sanchez IM, et al. Mutant BRAF and MEK inhibitors regulate the tumor immune microenvironment via pyroptosis. Cancer Discov. 2020;10(2):254–269. doi: 10.1158/2159-8290.CD-19-0672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kao KJ, Chang KM, Hsu HC, et al. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer. 2011;11(143). doi: 10.1186/1471-2407-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dedeurwaerder S, Desmedt C, Calonne E, et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol Med. 2011;3(12):726–741. doi: 10.1002/emmm.201100801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clarke C, Madden SF, Doolan P, et al. Correlating transcriptional networks to breast cancer survival: a large-scale coexpression analysis. Carcinogenesis. 2013;34(10):2300–2308. doi: 10.1093/carcin/bgt208 [DOI] [PubMed] [Google Scholar]
- 28.Jézéquel P, Loussouarn D, Guérin-charbonnel C, et al. Gene-expression molecular subtyping of triple-negative breast cancer tumours: importance of immune response. Breast Cancer Res. 2015;17(1). doi: 10.1186/s13058-015-0550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metzger-filho O, Michiels S, Bertucci F, et al. Genomic grade adds prognostic value in invasive lobular carcinoma. Ann Oncol. 2013;24(2):377–384. doi: 10.1093/annonc/mds280 [DOI] [PubMed] [Google Scholar]
- 30.Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He WT, Wan H, Hu L, et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 2015;25(12):1285–1298. doi: 10.1038/cr.2015.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi J, Zhao Y, Wang K, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526(7575):660–665. doi: 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- 33.Orning P, Weng D, Starheim K, et al. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science. 2018;362(6418):1064–1069. doi: 10.1126/science.aau2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers C, Fernandes-alnemri T, Mayes L, et al. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun. 2017;8(1). doi: 10.1038/ncomms14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z, Zhang Y, Xia S, et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature. 2020;579(7799):415–420. doi: 10.1038/s41586-020-2071-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Z, He H, Wang K, et al. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science. 2020;368(6494). doi: 10.1126/science.aaz7548. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14(1). doi: 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren EH, Deng YJ, Yuan WH, et al. An immune-related gene signature for determining Ewing sarcoma prognosis based on machine learning. J Cancer Res Clin Oncol. 2021;147(1):153–165. doi: 10.1007/s00432-020-03396-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng D, Li M, Zhou R, et al. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol Res. 2019;7(5):737–750. doi: 10.1158/2326-6066.CIR-18-0436 [DOI] [PubMed] [Google Scholar]
- 43.Zhang B, Wu Q, Li B, et al. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. 2020;19(1):53. doi: 10.1186/s12943-020-01170-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. doi: 10.1093/jnci/djj052 [DOI] [PubMed] [Google Scholar]
- 45.Charoentong P, Finotello F, Angelova M, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019 [DOI] [PubMed] [Google Scholar]
- 46.Orning P, Lien E, Fitzgerald KA. Gasdermins and their role in immunity and inflammation. J Exp Med. 2019;216(11):2453–2465. doi: 10.1084/jem.20190545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsuchiya K. Switching from apoptosis to pyroptosis: gasdermin-elicited inflammation and antitumor immunity. Int J Mol Sci. 2021;22(1):426. doi: 10.3390/ijms22010426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Topalian SL, Taube JM, Anders RA, et al. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16(5):275–287. doi: 10.1038/nrc.2016.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng D, Ye Z, Wu J, et al. Macrophage correlates with immunophenotype and predicts anti-PD-L1 response of urothelial cancer. Theranostics. 2020;10(15):7002–7014. doi: 10.7150/thno.46176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tauriello DVF, Palomo-ponce S, Stork D, et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554(7693):538–543. doi: 10.1038/nature25492 [DOI] [PubMed] [Google Scholar]
- 51.Panagi M, Voutouri C, Mpekris F, et al. TGF-β inhibition combined with cytotoxic nanomedicine normalizes triple negative breast cancer microenvironment towards anti-tumor immunity. Theranostics. 2020;10(4):1910–1922. doi: 10.7150/thno.36936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.An Y, Adams JR, Hollern DP, et al. Cdh1 and Pik3ca mutations cooperate to induce immune-related invasive lobular carcinoma of the breast. Cell Rep. 2018;25(3):702–14.e6. doi: 10.1016/j.celrep.2018.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Z, Jiang Z, Gao Y, et al. TP53 mutations promote immunogenic activity in breast cancer. J Oncol. 2019;2019:5952836. doi: 10.1155/2019/5952836. [DOI] [PMC free article] [PubMed] [Google Scholar]