Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a highly pathogenic and contagious coronavirus that caused a global pandemic with 5.2 million fatalities to date. Questions concerning serologic features of long-term immunity, especially dominant epitopes mediating durable antibody responses after SARS-CoV-2 infection, remain to be elucidated.
Objective
We aimed to dissect the kinetics and longevity of immune responses in coronavirus disease 2019 (COVID-19) patients, as well as the epitopes responsible for sustained long-term humoral immunity against SARS-CoV-2.
Methods
We assessed SARS-CoV-2 immune dynamics up to 180 to 220 days after disease onset in 31 individuals who predominantly experienced moderate symptoms of COVID-19, then performed a proteome-wide profiling of dominant epitopes responsible for persistent humoral immune responses.
Results
Longitudinal analysis revealed sustained SARS-CoV-2 spike protein–specific antibodies and neutralizing antibodies in COVID-19 patients, along with activation of cytokine production at early stages after SARS-CoV-2 infection. Highly reactive epitopes that were capable of mediating long-term antibody responses were shown to be located at the spike and ORF1ab proteins. Key epitopes of the SARS-CoV-2 spike protein were mapped to the N-terminal domain of the S1 subunit and the S2 subunit, with varying degrees of sequence homology among endemic human coronaviruses and high sequence identity between the early SARS-CoV-2 (Wuhan-Hu-1) and current circulating variants.
Conclusion
SARS-CoV-2 infection induces persistent humoral immunity in COVID-19–convalescent individuals by targeting dominant epitopes located at the spike and ORF1ab proteins that mediate long-term immune responses. Our findings provide a path to aid rational vaccine design and diagnostic development.
Key words: SARS-CoV-2, COVID-19, long-term immune response, humoral immunity, proteome-wide peptide microarray, dominant epitope
Abbreviations used: BSA, Bovine serum albumin; CoV, Coronavirus; COVID-19, Coronavirus disease 2019; FP, Fusion peptide; M, Membrane protein; MERS, Middle East respiratory syndrome; N, Nucleocapsid protein; nsp, Nonstructural protein; NT50, 50% neutralization titer; NTD, N-terminal domain; OD, Optical density; ORF, Open reading frame; PBS, Phosphate-buffered saline; PBS-T, PBS–Tween 20; PDB, Protein Data Bank (http://www.wwpdb.org/); RBD, Receptor binding domain; S, Spike protein; SARS, Severe acute respiratory syndrome
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative pathogen of the ongoing coronavirus disease 2019 (COVID-19) pandemic, belongs to the Betacoronavirus genus, which includes 2 other known highly pathogenic human coronaviruses: the severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) coronaviruses.1 Since the onset of the first recorded case in late December 2019, SARS-CoV-2 infection has resulted in more than 263 million confirmed cases and 5.2 million deaths worldwide as of November 2021, affecting 220 countries and regions across 5 continents.2 After infection, clinical presentations range widely, including asymptomatic infections, mild or moderate symptoms, or even life-threatening severe pneumonia.3 Despite the availability of multiple types of vaccine approaches, many countries are still struggling to contain new waves of infections, with virus variants emerging that appear to exhibit increased transmissibility and resistance to antiviral immune responses.
An effective humoral immune response mediated by neutralizing antibodies should possess a powerful and indispensable adaptive immunity to block infection and eliminate viral pathogens. After SARS-CoV-2 infection, a high rate (above 90%) of robust seroconversion among infected individuals was observed at 7 to 14 days after disease onset.4, 5, 6 The production of antigen-specific IgA, IgG, and IgM recognizing the spike protein (S) and nucleocapsid protein (N) of SARS-CoV-2 was detectable during the disease’s acute phase and the early stage of convalescence.4 , 5 , 7 The magnitude of neutralizing antibodies appeared to be associated with age, symptomatic infection, and disease severity. Elderly patients and individuals exhibiting severe COVID-19 symptoms tend to develop higher levels of neutralizing antibodies.4 , 8, 9, 10 A number of studies concerning the longevity of antibody responses have revealed that despite a loss of serum/plasma reactivity to virus antigens and declining neutralizing antibody titers over time in some symptomatic COVID-19 cases, a sustained level of overall long-term humoral immunity was observed for up to 8 to 12 months after infection in COVID-19–convalescent individuals.10, 11, 12, 13 Furthermore, the number of SARS-CoV-2 antigen-specific memory B cells stayed stable for at least 6 to 12 months,12 , 13 along with ongoing selection and accumulation of B-cell clones expressing neutralizing antibodies,12 , 14 indicating the maintenance of persistent humoral immunity after SARS-CoV-2 infection.
Neutralizing antibodies play a fundamental role in host defenses against viral infection. A panel of highly potent monoclonal antibodies of SARS-CoV-2 has been identified,15, 16, 17 primarily targeting epitopes located at the receptor binding domain of the S protein that assembles into trimers on the virion surface and facilitates virus entry and fusion on engaging the angiotensin-converting enzyme 2 receptor.18 Other regions of S protein, including the N-terminal domain (NTD) of S1 subunit and the S2 subunit, also contain immunogenic epitopes that are capable of inducing neutralizing antibodies.16 , 19 , 20 Besides neutralization, antibodies of SARS-CoV-2 may confer protection in vivo via Fc-mediated effector functions, such as antibody-dependent phagocytosis triggered by natural killer cells and antibody-dependent cytotoxicity by monocytes or macrophages.21 , 22 In addition to combating viral infections, antibodies generated by natural infection or vaccination may facilitate virus pathogenesis, either by increased inflammation activation or enhanced virus infectivity through antigen/antibody immune complex formation or by Fc-dependent functions,23, 24, 25 although the enhanced viral infection has not been observed in the context of SARS-CoV-2 in vivo. Theses divergent roles of antibody responses necessitate a systematic characterization of SARS-CoV-2 epitopes, as well as properties of long-lasting antibodies targeting neutralizing or nonneutralizing epitopes.
Despite recent progress on the kinetics and duration of antibody responses after SARS-CoV-2 infection, longitudinal analysis regarding prominent epitopes that sustain long-term humoral immune responses is still limited. Previous studies have profiled epitopes of antibody response in COVID-19–infected individuals mainly by means of ELISA-based assays,26 , 27 phage- or bacterial-display–based approaches,28, 29, 30, 31 and microarray-based technologies.30 , 32, 33, 34, 35 A number of immunodominant epitopes of SARS-CoV-2 S protein have been identified; the most frequently detected regions were mapped close to or spanning the fusion peptide (FP) of the S2 subunit, the second heptad repeat within the S2 subunit and on the C-terminal domain of the S1 subunit.26 , 28, 29, 30 , 32 , 33 Additionally, serologic screening also revealed epitopes located at other proteins across the SARS-CoV-2 proteome, including N protein, membrane (M) protein, and ORF1ab, ORF3a, and ORF7a.28, 29, 30 , 34 , 35 Although previous studies have provided important insight into antigenic epitopes, these studies have largely focused on the epitope profiling of COVID-19 patients in the early convalescence phase. The exact epitopes mediating durable SARS-CoV-2–specific antibody responses as well as the dynamics of epitope recognition remain to be elucidated.
Here, with the aim of obtaining a deeper understanding of the kinetics and longevity of humoral immune responses in COVID-19 patients, especially epitopes responsible for sustained long-term immunity against SARS-CoV-2, we conducted a comprehensive longitudinal analysis of immune profiling in 31 COVID-19 patients up to 180 to 220 days after symptom onset. On the basis of the evaluation of antigen-binding and neutralizing antibodies as well as serum cytokine levels, we further identified a panel of epitopes located at ORF1ab, S, and N proteins of SARS-CoV-2 that mediates persistent humoral immune responses using a peptide microarray encompassing the complete proteome of SARS-CoV-2. Our results shed light on the features of long-term immunity to overcome viral infection and will help inform rational vaccine design and the development of improved serologic diagnostic tools.
Methods
Study participants and sample collection
To longitudinally evaluate the kinetics of SARS-CoV-2–directed immune responses, 101 serum samples were collected from 31 SARS-CoV-2–infected individuals who were admitted to Nantong Third Hospital Affiliated to Nantong University (Nantong, China) between January and March 2020. All patients were diagnosed with confirmed SARS-CoV-2 infection through reverse transcription quantitative PCR testing; disease severity was defined as mild to moderate (nonsevere) or severe COVID-19 according to version 7 the Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia released by the National Health Commission of the People’s Republic of China.36 Patients were followed up longitudinally for 4 to 8 months after recovery from acute infection. Blood samples were collected in a longitudinal manner from 4 to 219 days after symptom onset for 20 patients among 31 study participants (median 4.5 samples per individual, ranging from 2 to 8), while single-time-point sampling was performed for each of the other 11 individuals who were in a late convalescent disease phase (days 122 to 214 after disease onset). Accordingly, sera (n = 20) from age- and sex-matched healthy donors were also included as the control group. No study participant had a documented prior infection with SARS or MERS.
The study was approved by the ethical committee of Nantong Third Hospital Affiliated to Nantong University (approval EL2020006). Written informed consent was obtained from each of the study participants. Serum was separated from peripheral blood in serum gel tubes via centrifugation, formed into aliquots, and stored at −80°C before use.
Cell lines
HEK293T and Huh7 cells were cultured in Dulbecco modified Eagle medium (Gibco; Thermo Fisher Scientific, Waltham, Mass) supplemented with 10% heat-inactivated fetal bovine serum (Gibco), 100 U/mL penicillin (Gibco), and 100 μg/mL streptomycin (Gibco) at 37°C with 5% CO2.
Detection of IgG antibody by ELISA
The levels of antigen-binding IgG antibody in human serum samples were determined by ELISA. Ninety-six–well ELISA plates (Corning, Corning, NY) were precoated with SARS-CoV-2 nucleocapsid protein (Sino Biological, Beijing, China, catalog no. 40588-V08B, 50 ng per well) or spike protein (Sino Biological, 40589-V08B1, 100 ng per well), SARS-CoV spike protein (Sino Biological, 40634-V08B, 100 ng per well), or MERS-CoV spike protein (Sino Biological, 40069-V08B, 50 ng per well) overnight at 4°C. After blocking with 5% nonfat milk in phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBS-T) for 2 hours at 37°C, 3-fold serially diluted heat-inactivated serum samples at a starting dilution of 1:200 were added into precoated plates for 2 hours at 37°C. Wells were washed 3 times between each step with PBS-T. Reactions were visualized by incubation with horseradish peroxidase–conjugated anti-human IgG antibody (Abcam, Cambridge, United Kingdom, 1:100,000 dilution) for 1 hour at 37°C, followed by adding tetramethylbenzidine substrate (Life Technologies, Carlsbad, Calif). The developing reactions were stopped with 2 mol H2SO4, and absorbance was measured at 450 nm with the correction wavelength set at 630 nm (OD450 − OD630). For calculating the end point titer of binding antibodies, data were linearized by plotting the log10 of serum dilutions versus the corrected optical density (OD) values (OD450 − OD630) within the linear portion of the curve (y = kx + b, r 2 > 0.99), and the serum dilution at which the adjusted OD value (OD450 − OD630) = 0 was calculated to give an end point titer.
Antigen-specific IgG antibodies in peptide-immunized mice were assessed by peptide-based ELISA. Briefly, 96-well ELISA plates (Thermo Fisher Scientific) were coated with 1 μg per well of 4 mixed peptides (peptides 318, 356, 510, and 530) in carbonate buffer (pH 9.6) overnight at 4°C. After blocking with PBS-T containing 5% nonfat milk, wells were incubated with serum samples diluted at 1:40 for 1 hour at 37°C. Subsequently, plates were washed and incubated with horseradish peroxidase–conjugated anti-mouse IgG antibody (Abcam). Reactions were visualized with tetramethylbenzidine substrate (Life Technologies), and the absorbance at 450 nm was measured after stopping reactions with 2 mol H2SO4.
Pseudovirus production and titration
The codon-optimized gene encoding SARS-CoV-2 (NC_045512) or SARS-CoV (AY291315.1) spike protein with C-terminal 19 aa deletion, or MERS-CoV (JX869059) spike protein truncated with C-terminal 16 aa was cloned into pcDNA3.1(+) vector, respectively. HEK 293T cells cultured in a 100 mm tissue culture dish were cotransfected with 1 μg of a plasmid encoding spike protein and 15 μg of an env-deficient, luciferase-expressing backbone (pNL4-3.luc.RE) using polyethyleneimine (Polysciences, Warrington, Pa). Pseudovirus-containing cell culture supernatants were collected at 48 hours after transfection, filtered, and stored at −80°C in aliquots. For determination of virus titers (50% tissue culture infectious dose), serial dilutions of pseudoviruses were added to 1 × 104 Huh7 cells preseeded in 96-well plates. After 12 hours’ infection, the virus-containing medium was replaced by fresh growth medium, and cells were cultured for an additional 48 hours. Luciferase activity of cell lysis was measured with Steady-Glo Luciferase Assay System (Promega, Madison, Wisc) using a microplate reader (BioTek Instruments, Winooski, Vt), and wells producing relative luminescence units higher than 10 times of the average background value were considered positive.
Pseudovirus neutralization assay
Heat-inactivated serum samples from COVID-19 patients or healthy donors were 2-fold serially diluted and incubated with 200 pseudoviruses at 50% tissue culture infectious dose for 1 hour at 37°C. The mixtures were then applied to infect Huh7 cells preseeded in 96-well plates in duplicate. Wells were replenished with fresh growth medium at 12 hours after infection, and the luciferase activity of cells was determined 48 hours later. The 50% neutralization titers (NT50) against SARS-CoV-2 and SARS-CoV pseudoviruses were calculated by nonlinear regression using GraphPad Prism 8.0 software (GraphPad Software, La Jolla, Calif); and the NT50 of MERS-CoV pseudovirus was defined as the highest dilution of serum that resulted in a 50% reduction of relative luminescence units compared to the virus control without applying serum samples.
Protein microarray-based cytokine detection
The quantitative measurement of multiple cytokines (IL-1α, IL-1β, IL-4, IL-6, IL-8, IL-10, IL-13, MCP-1, IFN-γ, and TNF-α) in serum samples was performed using a multiplex ELISA array (RayBiotech, Peachtree Corners, Ga) according to the manufacturer’s protocol. Briefly, glass slides precoated with cytokine-specific antibodies were blocked with sample diluent at room temperature for 30 minutes, followed by the addition of 60 μL 2-fold diluted serum samples or cytokine standard dilutions. After overnight incubation at 4°C, slides were washed 5 times and stained with 80 μL biotinylated detection antibody cocktail and then Cy3 equivalent dye-conjugated streptavidin at room temperature for 1 hour. The fluorescence intensity was detected by the InnoScan 300 microarray scanner (Innopsys, Chicago, Ill) at 532 nm, and data were analyzed by Q-Analyzer software (RayBiotech).
Peptide synthesis and conjugation with bovine serum albumin
A total of 515 peptides (15 aa in length, overlapping by 11 aa) that cover the SARS-CoV-2 proteome were synthesized by GL Biochem (Shanghai, China) on the basis of the amino acid sequence of SARS-CoV-2 strain Wuhan-Hu-1. Peptides were conjugated with bovine serum albumin (BSA) using the cross-linker Sulfo-SMCC (Thermo Fisher Scientific) according to the manufacturer’s instruction. Briefly, Sulfo-SMCC was added at 30-fold molar excess to BSA, followed by dialysis to PBS. Subsequently, the peptide containing cysteine was then added in a ratio of 1:1 (wt/wt) and incubated for 2 hours, and further dialyzed with PBS to eliminate free peptides.
Peptide microarray fabrication
To prepare the peptide microarray, peptides of SARS-CoV-2, as well as the negative control (BSA) and positive controls (anti-human IgG and anti-human IgM antibodies; Sigma-Aldrich, St Louis, Mo) were immobilized on PATH substrate slides (Grace Bio-Labs, Bend, Ore) in triplicate using a Super Marathon printer (Arrayjet, Edinburgh, Scotland, United Kingdom). Then peptide microarrays were preserved at −80°C for further use.
Microarray-based epitope mapping
Microarray-based serum analysis was performed as Li et al,37 with minor modifications. In order to create individual chambers for the identical subarrays, a 14-chamber rubber gasket was mounted onto each peptide microarray slides. The slide arrays were warmed to room temperature before use, then blocked with 3% BSA in PBS-T for 3 hours. The serum samples from COVID-19 patients or pooled sera from 20 healthy donors (control group) were diluted at 1:200 in PBS-T for most samples and then incubated with each subarray for 2 hours at 4°C. The arrays were washed with PBS-T, and incubated with Cy3-conjugated goat anti-human IgG and Alexa Fluor 647–conjugated donkey anti-human IgM (Jackson ImmunoResearch Laboratories, West Grove, Pa) at 1:1000 dilution each for 1 hour at room temperature. After incubation, the arrays were washed with PBS-T, then dried fully by centrifugation at room temperature. Subsequently, the arrays were scanned using a LuxScan 10K-A microarray scanner (CapitalBio, Beijing, China), and the fluorescence intensity data was obtained and analyzed by GenePix Pro 6.0 software (Molecular Devices, Sunnyvale, Calif).
Peptide microarray data analysis
The IgG and IgM data were analyzed respectively. The signal intensity of each spot was defined as the foreground minus the background, and averaged the triple spots for each peptide. The cutoff value for positive peptide response of COVID-19 samples was set as twice the signal intensity of the healthy donor control. Peptides with positive rates above 80% among samples tested at all 3 sampling time points (days 10-60, 100-150, and 180-220 after disease onset) were defined as dominant and persistent peptides, and peptides with a positive response frequency above 60% at all 3 time points were considered as subdominant and persistent. The average signal intensity of each peptide in 3 sampling time point groups was calculated; peptides showing high signal intensity (above the mean + SD of signal intensities of all tested samples) were also selected. Data processing and analysis were performed by R v3.6.3 software (https://www.r-project.org/), and significant signal intensity changes among different sampling time point groups were evaluated based on Limma. Differences with P < .05 were considered statistically significant.
Mouse immunization
Female BALB/c mice aged 6 to 8 weeks were purchased from Beijing Vitalstar Biotechnology (Beijing, China). All mice were housed in specific-pathogen-free facilities, and the animal study was approved by the Institutional Animal Care and Use Committee of Tianjin Medical University (Tianjin, China).
For immunization, groups of mice (n = 5) were injected intramuscularly with 25 μg per dose of each peptide (peptides 318, 356, 510, and 530 respectively) mixed with 50 μg alum (InvivoGen, San Diego, Calif) and 10 μg CpG adjuvants, and boosted twice with 50 μg per dose of peptides in the presence of adjuvants at 2-week intervals. Vaccination with adjuvants alone served as the negative control. Sera were collected from immunized mice at day 10 or day 14 after the second and third immunizations.
Statistical and structural analyses
All graphs were plotted by GraphPad Prism. Statistical comparisons between groups in Fig 1, Fig 2, and 4 were performed by 1-way ANOVA with Tukey multiple comparison. Correlations shown in Fig 1 and in Fig E9 and Fig E10 in the Online Repository available at www.jacionline.org were determined by Pearson correlation analysis. Differences with P < .05 were considered statistically significant.
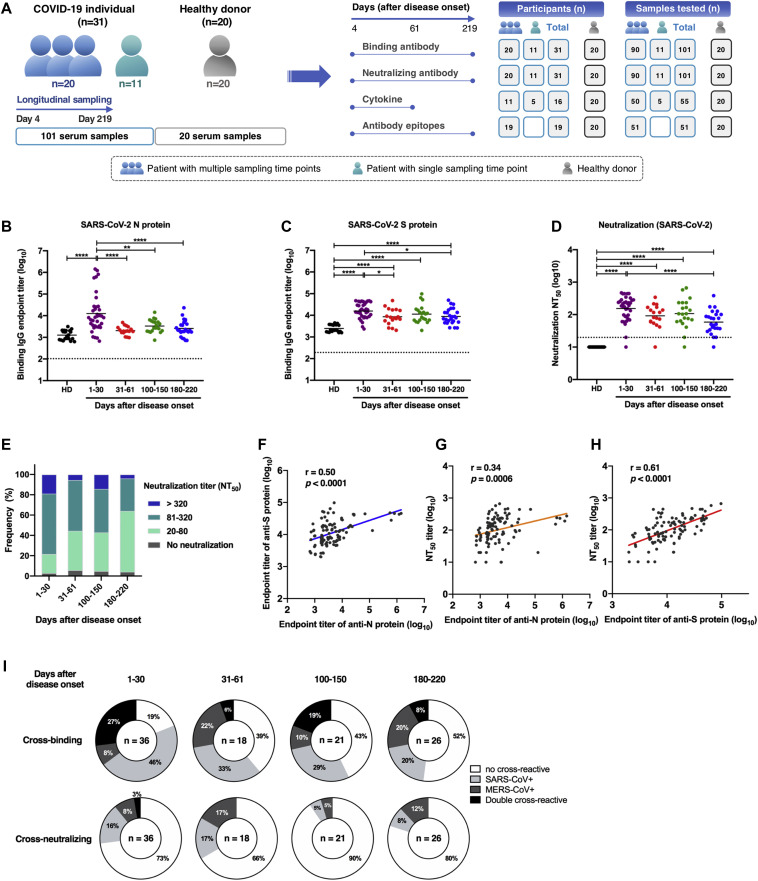
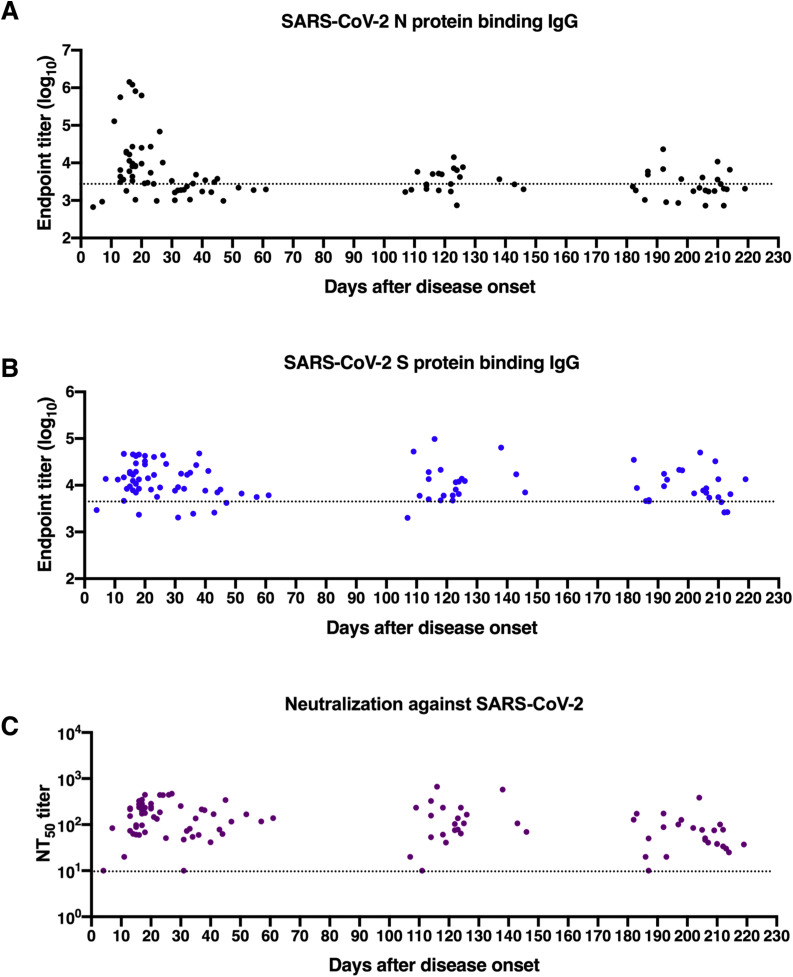
Fig 1.
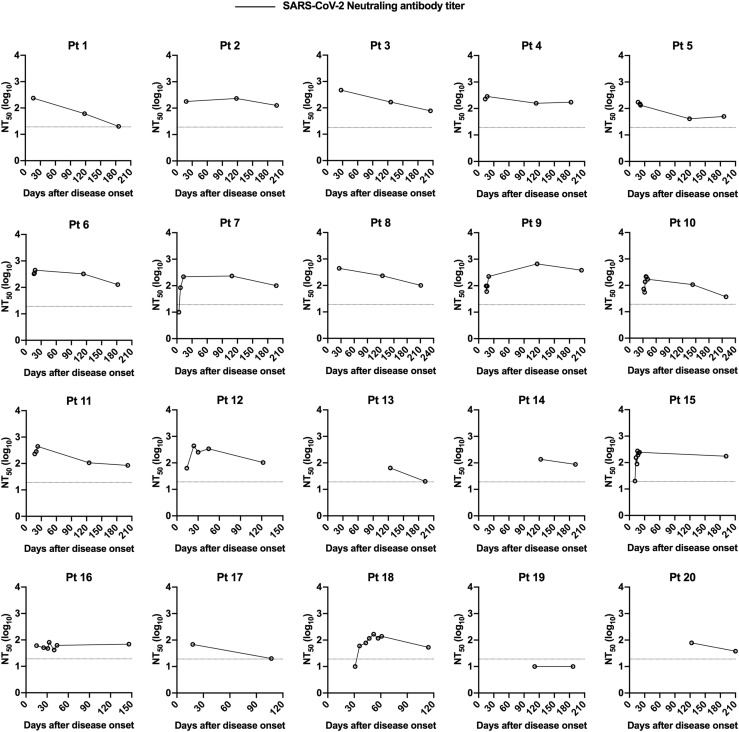
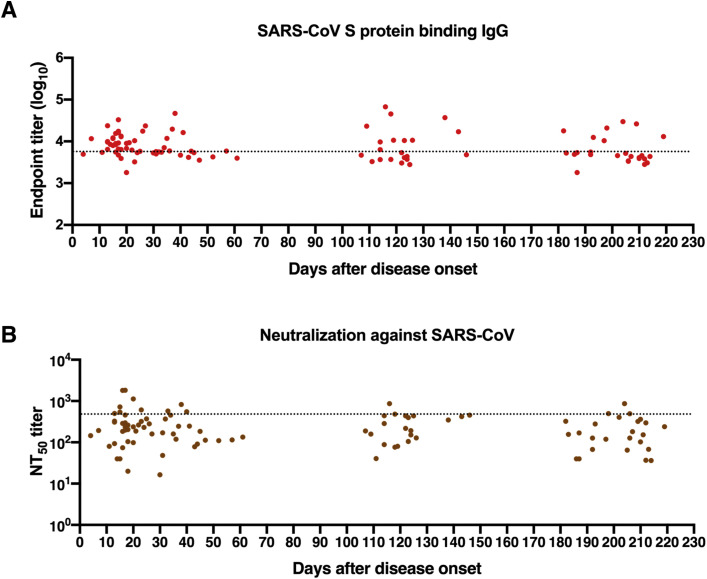
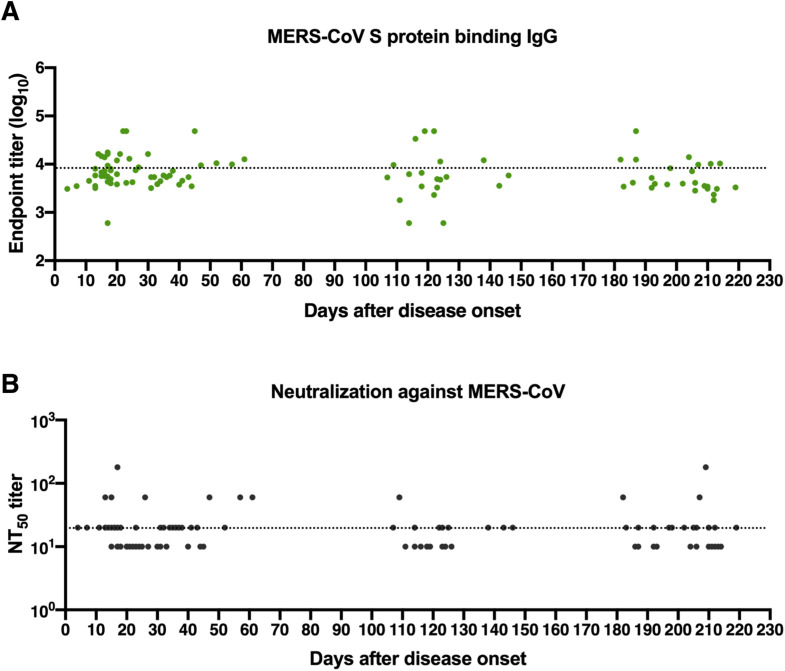
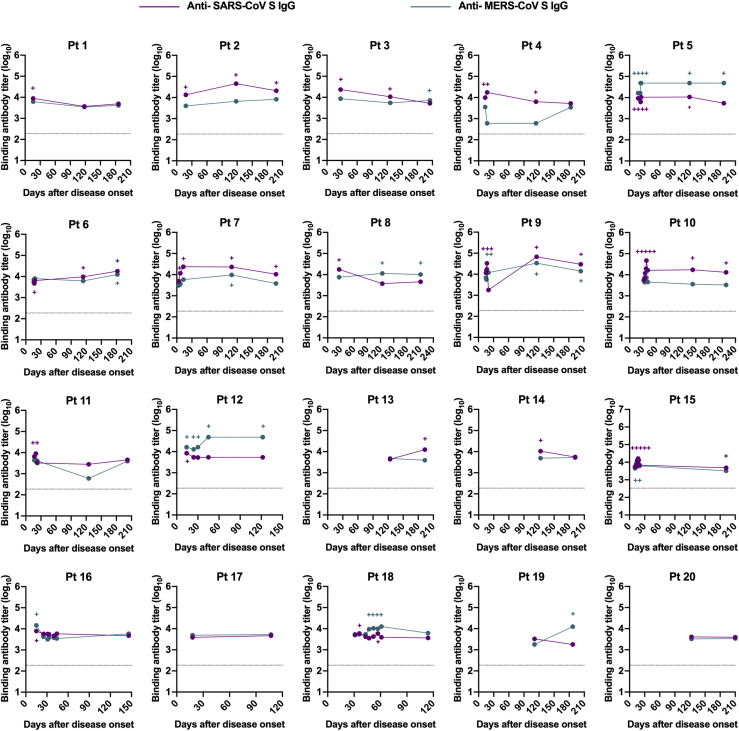
Longitudinal dynamics of antibody responses in COVID-19 patients. A, Schematic of the study design. A total of 31 SARS-CoV-2–infected individuals and 20 healthy donors were enrolled onto the study. Serum sample collection was performed longitudinally at multiple time points for 20 patients among 31 SARS-CoV-2–infected individuals, while sampling at a single time point was conducted for the other 11 patients. The number of participants and serum samples used in various assays are listed. B and C, A total of 101 serum samples from 31 COVID-19 patients were tested by ELISA for SARS-CoV-2 N protein (B) and S protein (C) binding IgG antibodies at different time points after symptom onset (days 1-30, n = 37; days 31-61, n = 18; days 100-150, n = 21; days 180-220, n = 25). Serum samples from 20 healthy donors were included as the control group. D, NT50 against SARS-CoV-2 pseudoviruses over time was calculated by nonlinear regression. E, Distribution of serum neutralizing activity in COVID-19 patients at indicated time points after disease onset are presented. Samples with a NT50 titer below 20 were defined as no neutralizing activity. F, Correlation analysis between log10 serum end point titers of SARS-CoV-2 S protein–specific IgG antibodies and N protein-binding IgG antibodies (n = 101). G and H, Correlations of serum neutralizing titers against SARS-CoV-2 with end point titers of N protein (G) or S protein binding (H) IgG antibodies. I, Serum samples obtained from COVID-19 patients at indicated time points after disease onset were evaluated for the cross-binding to SARS-CoV and MERS-CoV S proteins as well as the cross-neutralizing activity against SARS-CoV and MERS-CoV pseudoviruses. For cross-binding and SARS-CoV cross-neutralizing, the cutoff value for the positive response was set as the mean plus 2× SD of serum binding end point titers or NT50 values of 20 healthy donors. For MERS-CoV cross-neutralizing, samples with a NT50 of >20 (the lowest serum dilution) were considered positive. Small horizontal solid lines in (B-D) indicate the mean value of each group; dotted lines represent the lowest serum dilution in each test. Statistical significances were determined by 1-way ANOVA with Tukey multiple comparison. ∗P < .05, ∗∗P < .01, and ∗∗∗∗P < .0001. Correlation analyses in (F-H) were calculated by Pearson correlation test.
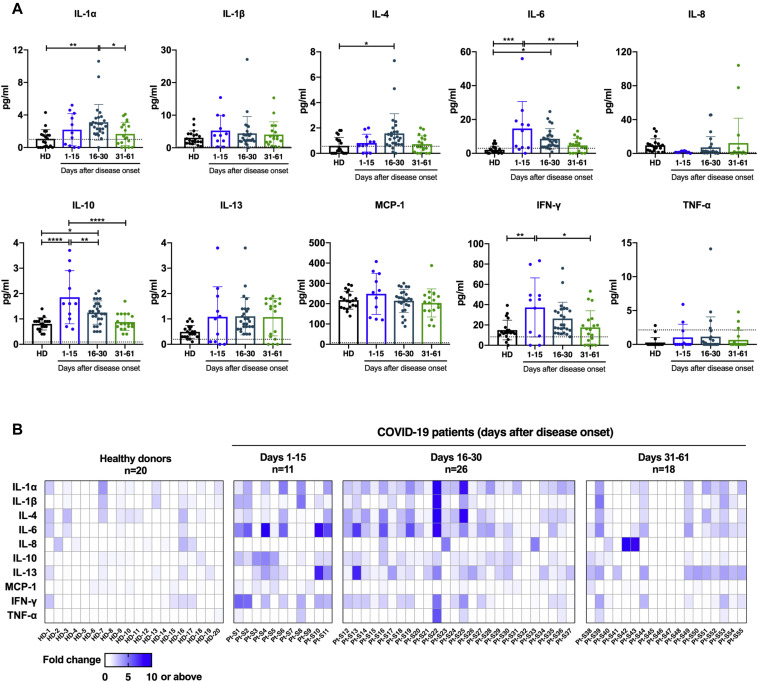
Fig 2.
Kinetics of cytokine production in COVID-19 patients. A, The cytokine production levels in 55 serum samples collected from 16 COVID-19 patients during the acute phase and an early stage of convalescence were detected by protein-microarray–based ELISA (days 1-15, n = 11; days 16-30, n = 26; days 31-61, n = 18; healthy donors, n = 20). Each dot represents an individual serum sample; dotted lines denote the limit of detection. Statistical significance was determined by 1-way ANOVA with Tukey multiple comparison. ∗P < .05, ∗∗P < .01, and ∗∗∗∗P < .0001. B, Fold change of each cytokine production level in COVID-19 patients shown in (A) compared to the mean values of 20 samples from healthy donors. Each column indicates a distinct serum sample collected from indicated time points after onset of symptoms; each row represents 1 individual cytokine tested.
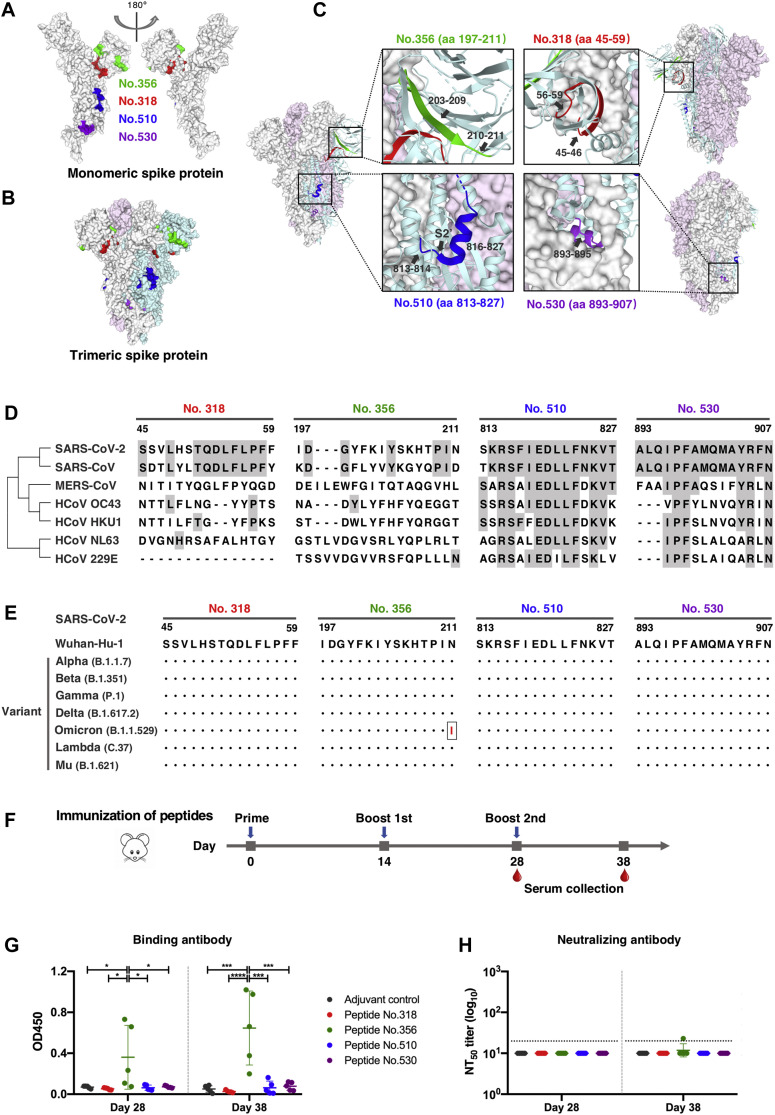
Fig 4.
Dominant epitopes of SARS-CoV-2 spike protein in terms of mediating durable humoral immune responses. A and B, Locations of the dominant and persistent epitopes on 3-D structures of the monomeric (A) and trimeric (B) S protein (PDB ID: 6VXX). Epitopes are highlighted in green (peptide 356, aa 197-211), red (peptide 318, aa 45-59), blue (peptide 510, aa 813-827), and purple (peptide 530, aa 893-907), respectively. The three S monomers in closed-conformation are depicted in gray, pink, and cyan, respectively. C, Detailed structure analysis of dominant epitopes in SARS-CoV-2 S protein on the closed state of the S trimer (PDB ID: 6VXX). D, Sequence alignment of identified epitopes among common human coronaviruses. Epitope residues that are conserved between SARS-CoV-2 and other human coronaviruses are shaded in gray.E, Epitope conservation analysis of the early SARS-CoV-2 (strain Wuhan-Hu-1) and 7 emerging variants. Black dots represent identical residues between the Wuhan-Hu-1 strain and the indicated variant. A single N211I substitution occurring in the Omicron variant is highlighted in red.F, BALB/c mice were immunized 3 times with each of the 4 selected S-protein peptides mixed with alum and CpG adjuvants. Sera were collected at day 14 after the second immunization and at day 10 after the third immunization. G, The levels of antigen-specific IgG antibody in immunized mouse sera (1:40 diluted) were tested by peptide-based ELISA. Statistical differences between groups were determined by 1-way ANOVA with Tukey multiple comparison. ∗P < .05, ∗∗∗P < .001, and ∗∗∗∗P < .0001. H, Neutralizing activity of mouse sera against pseudotyped SARS-CoV-2. Dotted horizontal lines indicate the lowest serum dilution in the assay (1:20).
The structures of SARS-CoV-2 spike protein (Protein Data Bank [PDB; http://www.wwpdb.org/] ID: 6VXX and PDB ID: 6ZGI) and the dimerization domain of SARS-CoV-2 nucleocapsid protein (PDB ID: 6YUN) were utilized to dissect the structural details of identified epitopes located on the spike protein and the dimerization domain of the nucleocapsid protein.
Sequence alignment and homology analysis among various human coronaviruses were performed by the Clustal W algorithm in MEGA v10.1.6 software.
Results
SARS-CoV-2 infection induces sustained antigen-specific binding and neutralizing antibodies
To longitudinally assess antibody responses after SARS-CoV-2 infection, 101 serum samples were collected from 31 individuals with PCR-confirmed SARS-CoV-2 infection (Fig 1 , A). Study participants included 26 patients with moderate COVID-19, 1 with asymptomatic presentation, 2 with mild illness, and 2 with severe symptoms (see Table E1 in the Online Repository available at www.jacionline.org). Patients enrolled onto this study ranged from 17 to 66 years of age (median age, 45 years), with an approximately equal distribution of male (51.6%) and female (48.4%) subjects. The most common symptoms among study participants included fever (83.9%), cough (67.7%), myalgia (22.6%), and chills (22.6%), with a median duration of illness of 15 days. A total of 90 serum samples from 20 COVID-19 patients were collected during hospitalization and discharge after recovery at multiple time points up to 219 days after symptom onset (Fig 1, A, and see Table E2 in the Online Repository). Sampling from the other 11 participants was performed at a single time point during late convalescence (days 122 to 214 after symptom onset). Additionally, samples from 20 healthy donors with matching age and sex distribution were included as a control group (Table E1).
Antigen-specific IgG antibodies in serum samples were quantified by ELISA precoated with SARS-CoV-2 N protein or S protein. Compared to healthy donors, both anti-N and anti-S IgG antibodies were developed in COVID-19 patients within 30 days after onset of symptoms, with a geometric mean end point titer of 4.10 (log10 anti-N IgG) and 4.20 (log10 anti-S IgG) (Fig 1, B and C; and see Table E3 in the Online Repository at www.jacionline.org; Fig E1, A and B; and Fig E2), in agreement with previous observations that seroconversion occurs at 1 to 2 weeks after SARS-CoV-2 infection.5 , 38 Afterward, antigen binding antibody titers declined to varying degrees over time. Rapidly and dramatically decreasing anti-N IgG antibody levels were observed in COVID-19 patients, whereas higher levels of anti-S IgG titers were maintained up to 180 to 220 days after disease onset compared to healthy donors (Fig 1, B and C). Correlation analysis revealed a significant correlation between log10 anti-N IgG titers and anti-S IgG titers (r = 0.50 and P < .001; Fig 1, F).
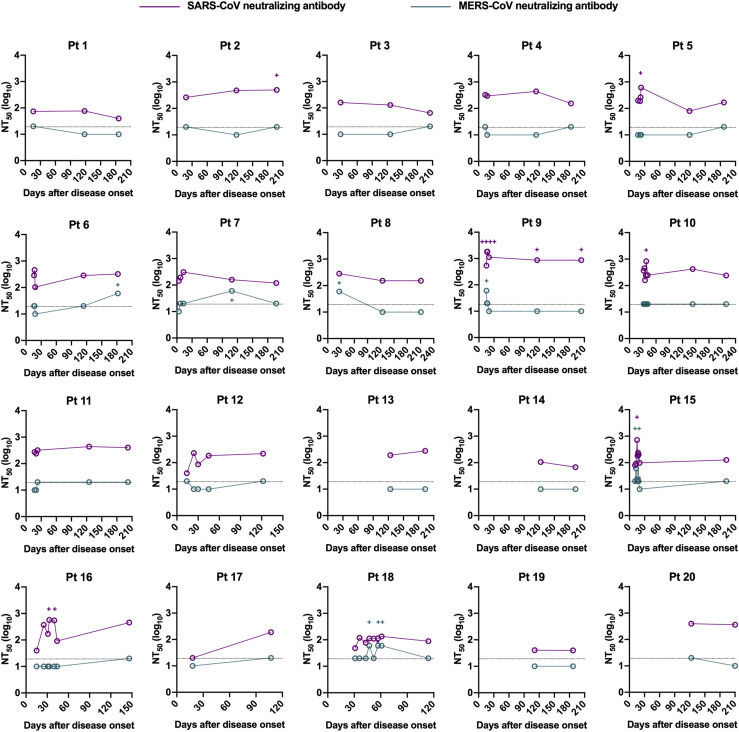
In addition to quantifying binding antibodies, the dynamics of functional neutralizing antibodies in individuals with COVID-19 were further determined using pseudotyped SARS-CoV-2. Over 95% of patient serum samples (35/37) collected between 4 and 30 days after symptom onset had robust neutralizing activities against SARS-CoV-2, and a large proportion of samples exhibited moderate (NT50 80-320) to strong (NT50 > 320) neutralizing activity (Fig 1, D and E). Of note, 2 samples with no or low neutralizing titers (NT50 at a minimum serum dilution of 1:20) were collected at an early period after viral infection (at day 4 and day 11 after symptom onset) before eliciting potent neutralizing antibodies (Fig 1, D, and Fig E1, C). Despite declining levels of SARS-CoV-2 neutralizing antibodies in COVID-19 patients over time, most patient serum samples (24/25) obtained during 180 to 220 days after symptom onset sustained positivity for neutralization against SARS-CoV-2, with a 2.8-fold decrease in geometric mean NT50 titers (Fig 1, D and E; Table E3; Fig E3). As expected, compared to anti-N IgG antibodies, a stronger correlation between SARS-CoV-2 S binding IgG titers and neutralizing antibody titers was identified (r = 0.61 and P < .001; Fig 1, G and H), consistent with findings that SARS-CoV-2 S protein is the major target of neutralizing antibodies.
The S protein of SARS-CoV-2 shares a high sequence similarity with highly virulent SARS-CoV (76% sequence identity) and shows a low sequence homology to MERS-CoV (34% sequence identity). Serologic cross-reactivity among coronaviruses has been reported;8 , 10 , 28 , 29 however, data on longitudinal follow-up evaluation of cross-reactivity of antibodies against SARS-CoV-2 toward other coronaviruses remain limited. We determined cross-binding and cross-neutralizing antibodies in longitudinal sera from SARS-CoV-2–infected individuals. Results showed that more than 80% of patient serum samples bound to S proteins of SARS-CoV and/or MERS-CoV within 1 month after symptom onset, with 27% of samples exhibiting double cross-reactivity (Fig 1, I). Reactivity to SARS-CoV S protein possessed a larger proportion (73%) than MERS-CoV (35%) among samples tested in an early period (1-30 days after disease onset) (Fig 1, I). The double cross-binding and SARS-CoV cross-binding antibodies exhibited a gradual decline over time, with only 8% double cross-binding and 20% SARS-CoV single cross-binding samples at 180 to 220 days after disease onset (Fig 1, I; in the in the Online Repository available at www.jacionline.org, see Fig E4, A; Fig E5, A; and Fig E6 in this article's Online Repository at www.jacionline.org). Intriguingly, positivity of MERS-CoV S protein–reactive antibodies stayed relatively constant over time (Fig 1, I). Different from the high cross-binding capacity, only a small number of patient serum samples cross-neutralized SARS-CoV and/or MERS-CoV pseudoviruses (Fig 1, I; Fig E4, B; Fig E5, B; and see Fig E7 in the Online Repository). Approximately 27% samples collected at 1 to 30 days after symptom onset presented cross-neutralizing activity, and this number dropped to 20% at 180 to 220 days, with more dramatic changes in SARS-CoV cross-neutralizing activity (Fig 1, I). A higher proportion of samples cross-neutralized SARS-CoV than MERS-CoV within 30 days after disease onset. It is worth noting that despite similar proportions of samples showing SARS-CoV and MERS-CoV cross-neutralization, the NT50 values for MERS-CoV were much lower than that of SARS-CoV among serum samples that were positive for cross-neutralization (Fig E4, B; Fig E5, B; Fig E7).
SARS-CoV-2 infection results in activation of cytokine production
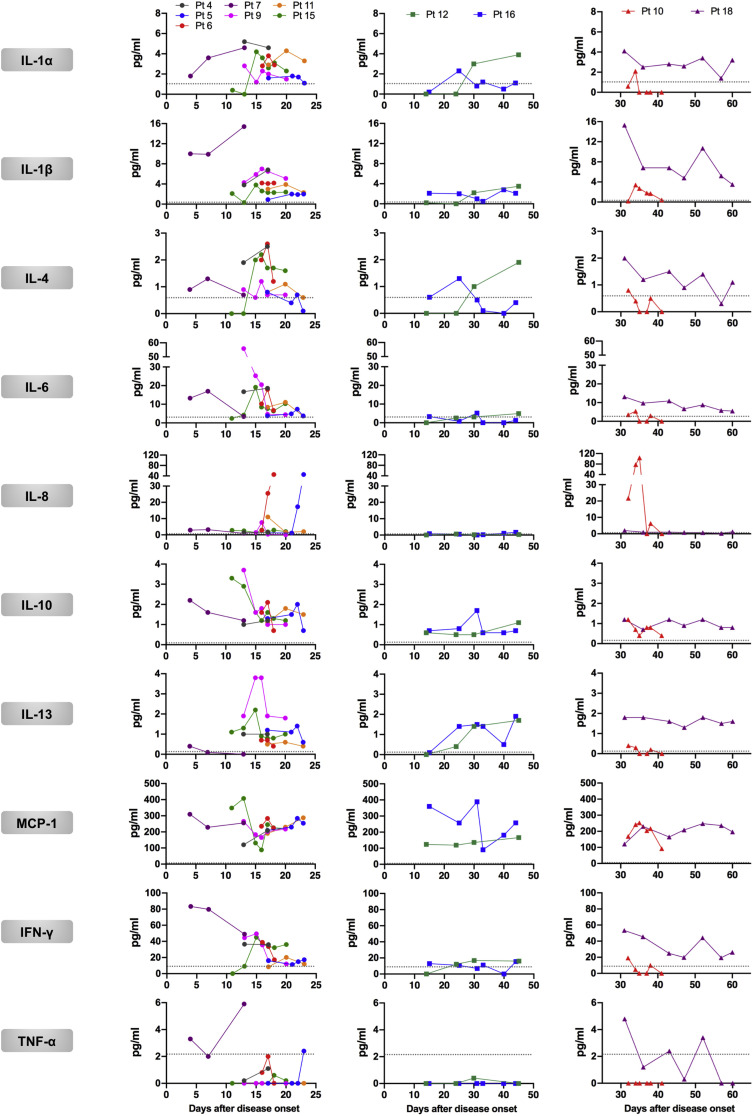
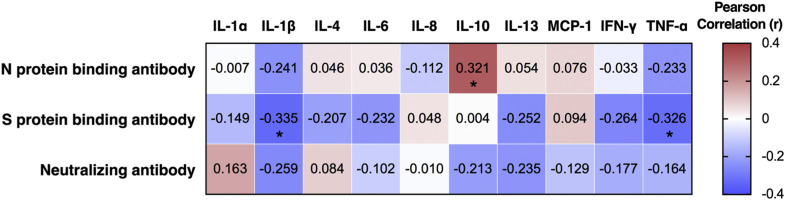
To comprehensively characterize the immunologic changes after SARS-CoV-2 infection, we further assessed changes in dynamics in cytokine levels in sera of COVID-19 patients. Fifty-five longitudinal serum samples that were collected within the first 2 months of symptom onset were used for the detection of cytokine production by protein-based microarray. Increased serum levels of multiple cytokines were observed during the first month of illness, including IL-1α (proinflammatory), IL-6 (proinflammatory), and IL-10 (anti-inflammatory), which have been linked to cytokine release syndrome in severe COVID-19 cases,39 , 40 as well as IFN-γ (TH1 type) and IL-4 (TH2 type) (Fig 2 , A, and Fig E8 in the Online Repository available at www.jacionline.org). Specifically, serum levels of IL-6, IL-10, and IFN-γ increased within 15 days after onset of symptoms in patients and decreased in later phases, whereas the release of IL-1α and IL-4 was shown to be remarkably increased during 16 to 30 days from disease onset, and rapidly dropped to normal afterward (Fig 2, A). Correlation analysis indicated a weak or no linear association between antigen-specific antibody responses and cytokine production after SARS-CoV-2 infection, although a statistical significance was observed between levels of IL-10 and SARS-CoV-2 N protein binding antibody (r = 0.321 and P < .05), between IL-1β production and S protein binding antibody (r = −0.335 and P < .05), and between TNF-α and S protein binding antibody levels (r = −0.335 and P < .05; see Fig E9 in the Online Repository).
To allow direct visualization and comparison among patient samples across multiple cytokine responses over time, we constructed a heat map showing fold changes of cytokine release relative to the healthy donor control group (Fig 2, B). Consistent with our findings presented above, elevated serum cytokine levels after SARS-CoV-2 infection were predominantly observed during the acute phase and an early period of convalescence (within 30 days after disease onset) (Fig 2, A and B). Among cytokines tested, proinflammatory IL-6 exhibited the most robust response, with a 4.9-fold increase and a 2.9-fold increase on average for samples collected during 1 to 15 days and 16 to 30 after onset of symptoms, respectively (Fig 2, B). Of note, 1 serum sample (sample Pt-S22, collected at day 18 after disease onset) obtained from a COVID-19–infected individual with moderate disease, exhibited a marked increase in the production of multiple proinflammatory cytokines, including IL-1α, IL-1β, IL-6, and TNF-α (Fig 2, B). Additionally, hyperproduction of cytokines including IL-1α, IL-4, IL-6, IL-10, IL-13, and IFN-γ was also detected in 1 sample (sample Pt-S11, collected at day 15 after disease onset) collected from a severe case of COVID-19; presumably these are associated with disease severity and outcome (Fig 2, B). These results indicate broad inflammatory activation and changes over time involving concomitant release of proinflammatory and anti-inflammatory cytokines as well as TH1-type and TH2-type cytokines in COVID-19 patients.
Proteome-wide epitope mapping identifies dominant epitopes mediating persistent humoral immune responses in COVID-19 patients
To better characterize the distinguishing features of humoral immunity to SARS-CoV-2 over time, we applied a proteome-wide epitope mapping using peptide-based microarrays. A peptide library covering the SARS-CoV-2 proteome was generated and immobilized onto slides, in which each peptide was 15 aa in length with an 11 aa overlap. A total of 51 longitudinal serum samples from 19 patients with either asymptomatic (n = 1) or mild (n = 2) to moderate (n = 15) or severe (n = 1) SARS-CoV-2 infections were tested (Table I ). To achieve relatively balanced sampling numbers, intervals, and time points, samples were collected sequentially at 2 or 3 time points from each COVID-19 participant ranging from 16 to 219 days after symptom onset. The majority of patients (18/19) generated neutralizing antibodies after viral infection, except for the single individual with asymptomatic infection (Table I). According to the different sampling time points, samples were divided into 3 groups: days 10-60 (n = 18), days 100-150 (n = 18), and days 180-220 (n = 15). Longitudinal assessment of virus epitope profiles in COVID-19 patients was performed for both serum IgG and IgM antibodies, with pooled sera from 20 healthy donors used as negative control.
Table I.
Characteristics of COVID-19 patients and sample cohorts in epitope mapping
| Patient ID | Age (years) | Sex | Disease severity | Duration of illness (days) | Serum samples |
|||
|---|---|---|---|---|---|---|---|---|
| Sample ID | Sampling days after disease onset | SARS-CoV-2 |
||||||
| S binding IgG titer (log10) | Neutralization (NT50) | |||||||
| 1 | 39 | F | Moderate | 14 | 1-1 | 16 | 4.10 | 237.32 |
| 1-2 | 118 | 3.68 | 60.71 | |||||
| 1-3 | 186 | 3.66 | 20.00 | |||||
| 2 | 54 | F | Moderate | 15 | 2-1 | 18 | 4.13 | 178.04 |
| 2-2 | 118 | 4.33 | 232.92 | |||||
| 2-3 | 198 | 4.32 | 126.74 | |||||
| 3 | 26 | M | Moderate | 17 | 3-1 | 27 | 4.46 | 468.97 |
| 3-2 | 126 | 4.09 | 165.02 | |||||
| 3-3 | 205 | 3.89 | 77.08 | |||||
| 4 | 52 | F | Moderate | 16 | 4-1 | 17 | 4.29 | 285.41 |
| 4-2 | 114 | 4.13 | 157.25 | |||||
| 4-3 | 183 | 3.94 | 172.55 | |||||
| 5 | 56 | F | Moderate | 14 | 5-1 | 23 | 4.22 | 186.14 |
| 5-2 | 119 | 3.78 | 40.95 | |||||
| 5-3 | 187 | 3.68 | 50.29 | |||||
| 6 | 50 | F | Moderate | 14 | 6-1 | 18 | 3.93 | 445.35 |
| 6-2 | 114 | 4.28 | 324.84 | |||||
| 6-3 | 182 | 4.55 | 127.41 | |||||
| 7 | 54 | M | Moderate | 20 | 7-1 | 13 | 4.67 | 216.59 |
| 7-2 | 109 | 4.72 | 233.08 | |||||
| 7-3 | 197 | 4.33 | 100.53 | |||||
| 8 | 44 | F | Moderate | 9 | 8-1 | 26 | 4.64 | 443.69 |
| 8-2 | 124 | 3.81 | 233.08 | |||||
| 8-3 | 211 | 3.64 | 100.53 | |||||
| 9 | 52 | F | Moderate | 19 | 9-1 | 20 | 4.63 | 222.13 |
| 9-2 | 116 | 4.99 | 664.89 | |||||
| 9-3 | 204 | 4.70 | 384.45 | |||||
| 10 | 41 | M | Moderate | 13 | 10-1 | 41 | 4.31 | 169.26 |
| 10-2 | 143 | 4.24 | 106.33 | |||||
| 10-3 | 219 | 4.13 | 37.05 | |||||
| 11 | 66 | F | Moderate | 10 | 11-1 | 23 | 4.61 | 441.91 |
| 11-2 | 125 | 4.14 | 106.40 | |||||
| 11-3 | 202 | 3.83 | 84.84 | |||||
| 12 | 30 | M | Moderate | 28 | 12-1 | 23 | 3.75 | 437.74 |
| 12-2 | 44 | 3.91 | 341.81 | |||||
| 12-3 | 121 | 3.67 | 103.08 | |||||
| 13 | 64 | M | Moderate | 19 | 13-1 | 124 | 4.08 | 64.01 |
| 13-2 | 193 | 4.12 | 20.00 | |||||
| 14 | 66 | M | Mild | 19 | 14-1 | 123 | 4.07 | 136.81 |
| 14-2 | 192 | 3.98 | 87.88 | |||||
| 15 | 38 | F | Severe | 22 | 15-1 | 20 | 4.45 | 243.80 |
| 15-2 | 192 | 4.25 | 174.08 | |||||
| 16 | 30 | M | Moderate | 45 | 16-1 | 15 | 3.98 | 60.65 |
| 16-2 | 44 | 3.85 | 62.50 | |||||
| 16-3 | 146 | 3.85 | 69.30 | |||||
| 17 | 20 | M | Moderate | 22 | 17-1 | 18 | 3.37 | 68.51 |
| 17-2 | 107 | 3.30 | 20.00 | |||||
| 18 | 17 | M | Mild | 30 | 18-1 | 52 | 3.82 | 167.14 |
| 18-2 | 114 | 3.70 | 53.26 | |||||
| 19 | 45 | F | Asymptomatic | — | 19-1 | 111 | 3.78 | <20 |
| 19-2 | 187 | 3.66 | <20 | |||||
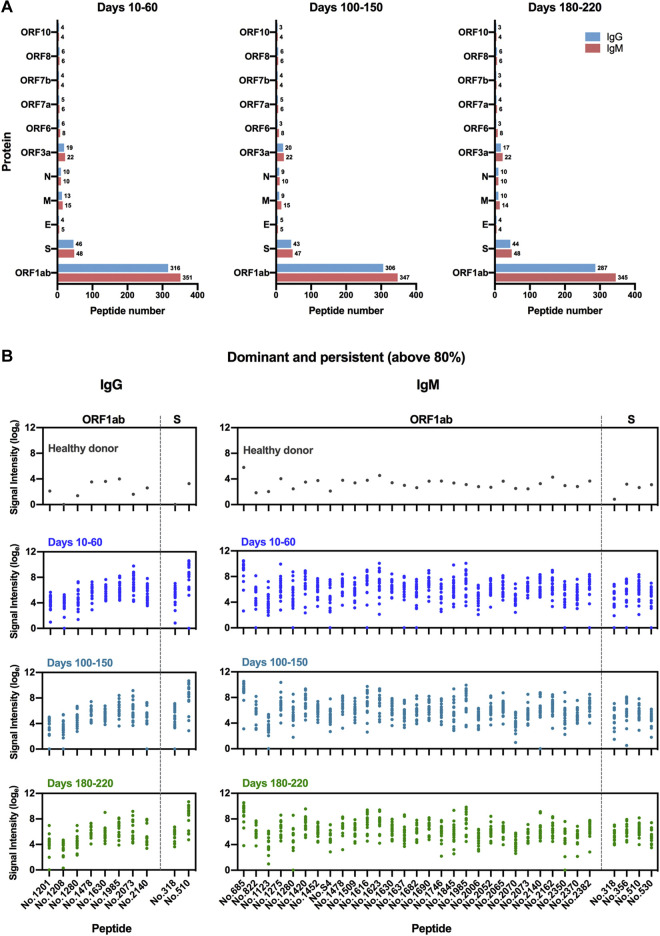
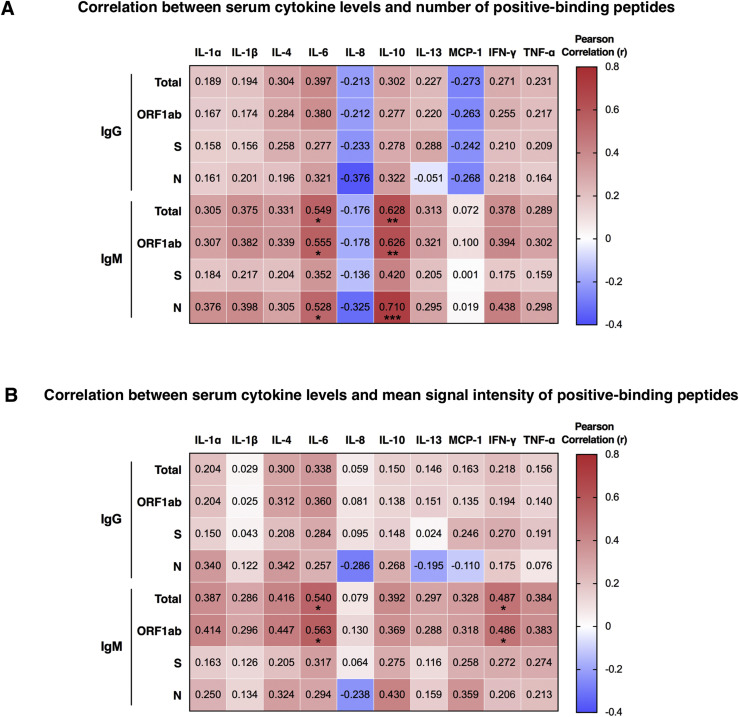
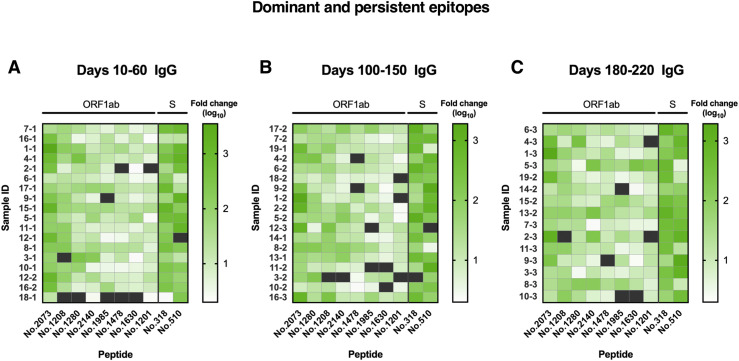
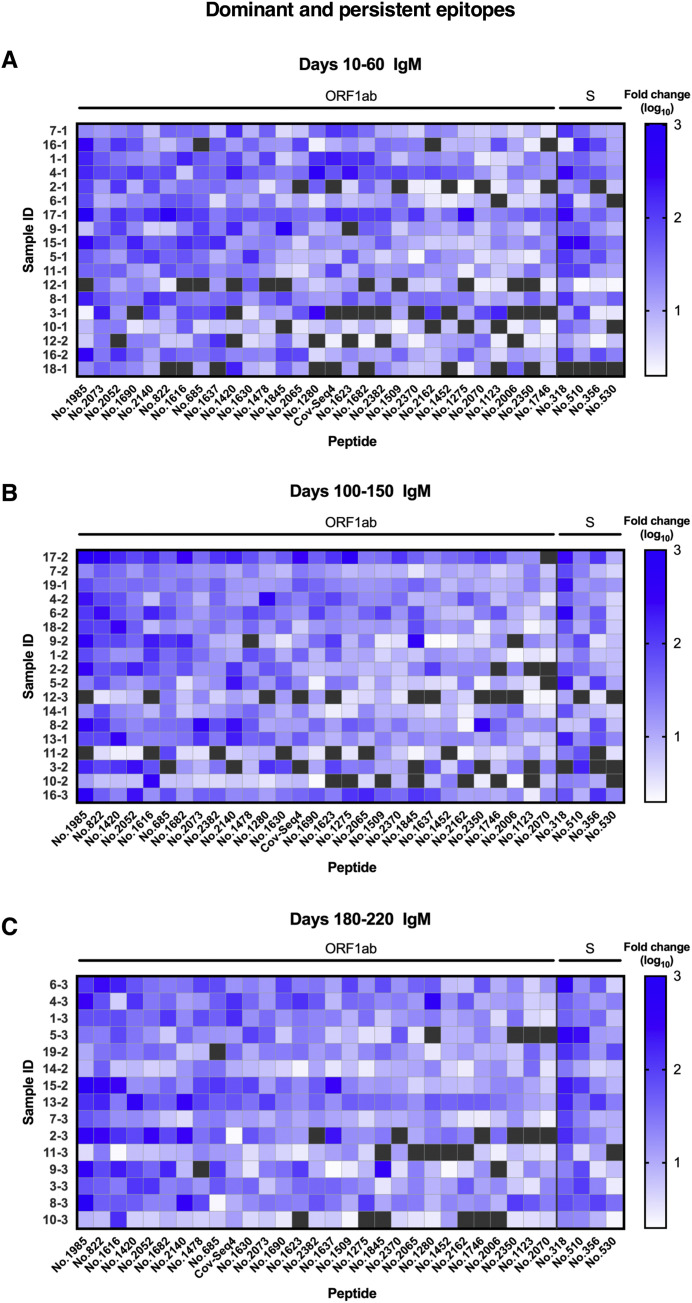
Using the SARS-CoV-2 proteome microarray, the kinetics of peptide-binding antibody responses was determined and analyzed for (1) binding signal intensity, and (2) percentage of positive-reactive samples (positive rate) for each peptide. On the basis of the cutoff value for positive peptide-binding response, which was set as twice of the signal intensity of the negative control, we identified a total of 460 positive peptides for IgG and 479 positive peptides for IgM that were reactive with at least 1 patient serum sample. Positive peptide numbers and the distribution of responses across different open reading frames (ORFs) of SARS-CoV-2 were relatively stable among different sampling groups, with a trend of slight decrease in positive peptide numbers over time (Fig 3 , A). The most reactivity was identified in the replicase polyprotein ORF1ab, which is the largest ORF, encompassing more than two thirds of the entire genome (Fig 3, A). Interestingly, we observed moderate to strong degrees of correlation between positive peptide-binding responses and serum cytokine levels early after SARS-CoV-2 infection (days 10-60 after disease onset; see Fig E10 in the Online Repository at www.jacionline.org). The number of positive binding peptides for IgM was shown to be associated with serum levels of IL-6 and IL-10; likewise, the average signal intensity of total reactive peptides and that of ORF1ab binding peptides for IgM presented positive associations with IL-6 and IFN-γ production in serum samples. These data suggest that changes in cytokine levels after SARS-CoV-2 infection may influence the magnitude and breadth of epitopes recognized by antigen-specific humoral responses.
Fig 3.
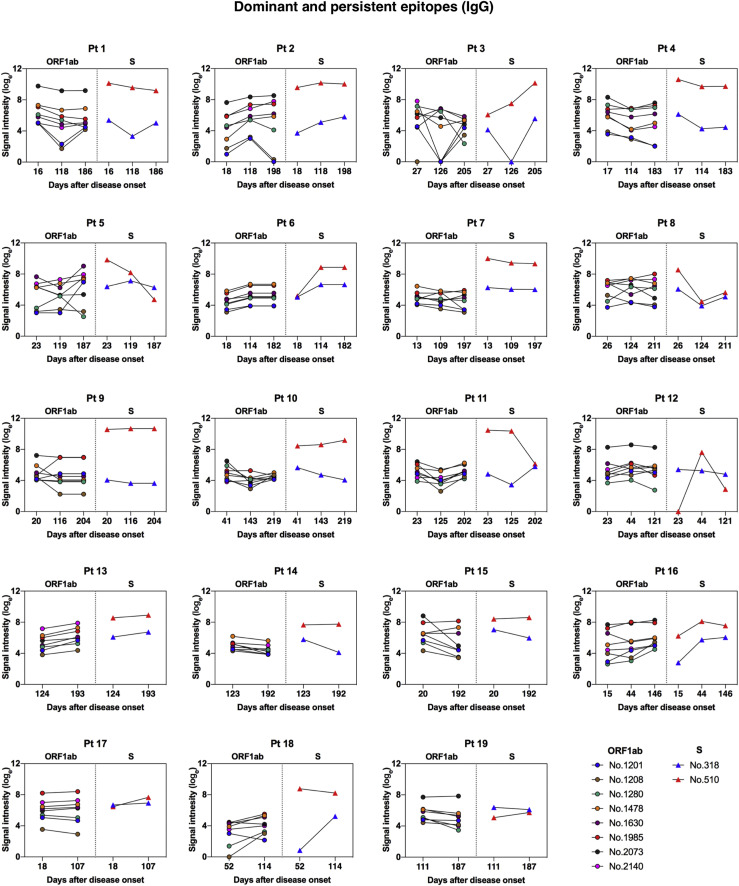
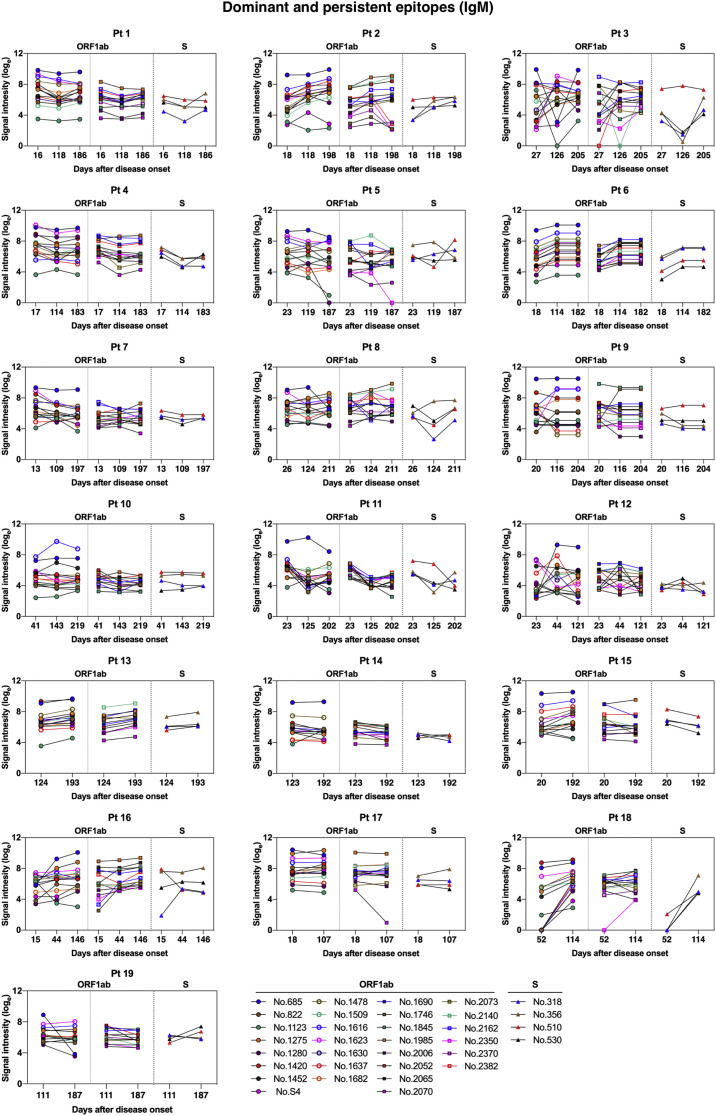
IgM and IgG recognition of dominant epitopes that contribute to long-term antibody responses in SARS-CoV-2–infected individuals. Longitudinal analysis of serum recognition of epitopes in 19 individuals with COVID-19 using a peptide microarray covering the proteome of SARS-CoV-2. The cutoff value for the positive response of peptide binding in patient samples (n = 51) was set as twice the signal intensity of the pooled sera from 20 healthy donors. A, Peptide counts and distribution of positive binding peptides that were detectable in 1 or more samples collected at 10-60 days (n = 18), 100-150 days (n = 18), and 180-220 days (n = 15) after disease onset. Numbers indicate the total identified IgG and IgM epitopes from each ORF. B, Signal intensities of dominant and persistent IgG and IgM epitopes (x-axis) that were sustainably reactive in more than 80% samples within all 3 sampling time points. Each dot indicates the pooled sera from healthy donors (top) or a distinct patient serum sample collected from indicated time points after symptom onset. E, Envelope protein.
On the basis of identified positive epitopes, we further selected the most common epitopes that consistently remained reactive in more than 80% of the COVID-19 samples among each of the 3 sampling groups (termed dominant and persistent epitopes). Results revealed that these highly dominant epitopes capable of mediating long-term humoral immune responses were located at the SARS-CoV-2 ORF1ab polyprotein and S protein, with more epitopes recognized by IgM antibodies (n = 33) than IgG antibodies (n = 10) (Fig 3, B, and Table II ; see Fig E11 and Fig E12 in the Online Repository at www.jacionline.org). The ORF1ab polyprotein possessed the maximal number of dominant epitopes mediating long-term responses, and epitopes were broadly distributed on the regions of nonstructural proteins (nsp) 2-5, nsp 8-10, nsp 12-14, and nsp 16 (Table II). Notably, we identified one immunodominant epitope, 2073 (ORF1ab, aa 5801-5815), that could be recognized by IgG and IgM antibodies from 100% of the COVID-19 patients, regardless of serum sampling time points (Fig 3, B, and Table II; see Fig E13 and Fig E14 in the Online Repository). This highly reactive peptide is located within the helicase (nsp 13) region of the ORF1ab polyprotein, which is essential for unwinding double-stranded RNA templates during SARS-CoV-2 replication.41 Among selected peptides of ORF1ab, 2 immunodominant peptides with the highest average signal intensities recognized by IgG antibodies, number 1985 (ORF1ab, aa 5449-5463) and number 2073 (ORF1ab, aa 5801-5815), are both located on nsp 13. For IgM responses, peptide 685 (ORF1ab, aa 249-263) on nsp 2 and peptide 1985 (ORF1ab, aa 5449-5463) on nsp 13 presented the most robust binding intensities (Fig 3, B). Considering the variation in baseline signals (pooled sera from healthy donors) for different peptides (Fig 3, B), we further calculated fold changes of signal intensities regarding each key peptide, relative to the healthy control group. Results showed that peptide 2073 (IgG binding) and peptide 1985 (IgM binding) located on nsp 13 region sustained the top peptide binding intensities (fold change) among patient sera collected up to 180 to 220 days (Fig E13 and Fig E14).
Table II.
Epitopes with >80% positive rate at all 3 sampling time points
| Protein | Subunit | Peptide ID | Peptide sequence | Positive response frequency |
||
|---|---|---|---|---|---|---|
| Days 10-60 | Days 100-150 | Days 180-220 | ||||
| IgG | ||||||
| ORF1ab | nsp3 | 1201 | 2313-SFKWDLTAFGLVAEW-2327 | 0.94 | 0.83 | 0.87 |
| nsp3 | 1208 | 2341-LGLAAIMQLFFSYFA-2355 | 0.89 | 0.94 | 0.93 | |
| nsp3 | 1280 | 2629-VLSTFISAARQGFVD-2643 | 0.94 | 1.00 | 1.00 | |
| nsp5 (3CL pro) | 1478 | 3421-SFCYMHHMELPTGVH-3435 | 0.89 | 0.89 | 0.93 | |
| nsp8 | 1630 | 4029-MQTMLFTMLRKLDND-4043 | 0.94 | 0.89 | 0.93 | |
| nsp13 (helicase) | 1985 | 5449-TCTERLKLFAAETLK-5463 | 0.89 | 0.89 | 0.87 | |
| nsp13 (helicase) | 2073 | 5801-KGVITHDVSSAINRP-5815 | 1.00 | 1.00 | 1.00 | |
| nsp14 | 2140 | 6069-DQFKHLIPLMYKGLP-6083 | 1.00 | 0.94 | 1.00 | |
| S | S1, NTD | 318 | 45-SSVLHSTQDLFLPFF-59 | 1.00 | 0.94 | 1.00 |
| S2, FP | 510 | 813-SKRSFIEDLLFNKVT-827 | 0.94 | 0.94 | 1.00 | |
| IgM | ||||||
| ORF1ab | nsp2 | 685 | 249-YELQTPFEIKLAKKF-263 | 0.89 | 0.94 | 0.93 |
| nsp2 | 822 | 797-EKYCALAPNMMVTNN-811 | 0.94 | 1.00 | 1.00 | |
| nsp3 | 1123 | 2001-ATYKPNTWCIRCLWS-2015 | 0.83 | 0.83 | 0.87 | |
| nsp3 | 1275 | 2609-LKTLVATAEAELAKN-2623 | 0.89 | 0.94 | 0.93 | |
| nsp3 | 1280 | 2629-VLSTFISAARQGFVD-2643 | 0.89 | 0.94 | 0.87 | |
| nsp4 | 1420 | 3189-MYLKLRSDVLLPLTQ-3203 | 0.83 | 1.00 | 1.00 | |
| nsp5 (3CL pro) | 1452 | 3317-YEDLLIRKSNHNFLV-3331 | 0.83 | 0.94 | 0.93 | |
| nsp5 (3CL pro) | S4 | 3353-KVDTANPKTPK-3363 | 0.83 | 0.89 | 1.00 | |
| nsp5 (3CL pro) | 1478 | 3421-SFCYMHHMELPTGVH-3435 | 0.94 | 0.94 | 0.93 | |
| nsp5 (3CL pro) | 1509 | 3545-LGSALLEDEFTPFDV-3559 | 0.89 | 0.94 | 1.00 | |
| nsp8 | 1616 | 3973-SEVVLKKLKKSLNVA-3987 | 0.89 | 0.89 | 1.00 | |
| nsp8 | 1623 | 4001-LEKMADQAMTQMYKQ-4015 | 0.83 | 0.83 | 0.93 | |
| nsp8 | 1630 | 4029-MQTMLFTMLRKLDND-4043 | 1.00 | 0.94 | 1.00 | |
| nsp8 | 1637 | 4057-VPLNIIPLTTAAKLM-4071 | 0.94 | 0.94 | 1.00 | |
| nsp9 | 1682 | 4237-LNRGMVLGSLAATVR-4251 | 0.83 | 1.00 | 1.00 | |
| nsp10 | 1690 | 4269-FCAFAVDAAKAYKDY-4283 | 0.94 | 1.00 | 1.00 | |
| nsp12 (RdRp) | 1746 | 4493-FFKFRIDGDMVPHIS-4507 | 0.83 | 0.83 | 0.87 | |
| nsp12 (RdRp) | 1845 | 4889-NLDKSAGFPFNKWGK-4903 | 0.89 | 0.83 | 0.87 | |
| nsp13 (helicase) | 1985 | 5449-TCTERLKLFAAETLK-5463 | 0.94 | 0.89 | 1.00 | |
| nsp13 (helicase) | 2006 | 5533-VVYRGTTTYKLNVGD-5547 | 0.83 | 0.89 | 0.87 | |
| nsp13 (helicase) | 2052 | 5717-AKHYVYIGDPAQLPA-5731 | 0.94 | 1.00 | 1.00 | |
| nsp13 (helicase) | 2065 | 5769-PAEIVDTVSALVYDN-5783 | 0.94 | 0.94 | 0.93 | |
| nsp13 (helicase) | 2070 | 5789-KDKSAQCFKMFYKGV-5803 | 0.94 | 0.83 | 0.87 | |
| nsp13 (helicase) | 2073 | 5801-KGVITHDVSSAINRP-5815 | 1.00 | 1.00 | 1.00 | |
| nsp14 | 2140 | 6069-DQFKHLIPLMYKGLP-6083 | 1.00 | 0.94 | 1.00 | |
| nsp14 | 2162 | 6157-GFDYVYNPFMIDVQQ-6171 | 0.89 | 0.94 | 0.87 | |
| nsp16 | 2350 | 6909-LIGDCATVHTANKWD-6923 | 0.83 | 0.89 | 0.87 | |
| nsp16 | 2370 | 6989-TAFVTNVNASSSEAF-7003 | 0.94 | 1.00 | 0.93 | |
| nsp16 | 2382 | 7037-LSSYSLFDMSKFPLK-7051 | 0.89 | 0.94 | 0.93 | |
| S | S1, NTD | 318 | 45-SSVLHSTQDLFLPFF-59 | 0.94 | 0.94 | 1.00 |
| S1, NTD | 356 | 197-IDGYFKIYSKHTPIN-211 | 0.89 | 0.89 | 1.00 | |
| S2, FP | 510 | 813-SKRSFIEDLLFNKVT-827 | 0.94 | 0.94 | 1.00 | |
| S2 | 530 | 893-ALQIPFAMQMAYRFN-907 | 0.83 | 0.83 | 0.93 | |
Dominant and persistent epitopes of SARS-CoV-2 S protein are located at NTD and S2 subunits
A total of 4 dominant and persistent epitopes from the SARS-CoV-2 S protein were identified: peptide 318 (S, aa 45-59) and peptide 356 (S, aa 197-211), which are located within the NTD region; peptide 510 (S, aa 813-827), which covers the S2’ cleavage site and parts of the FP of S2 subunit, and peptide 530 (S, aa 893-907), which is located at the connecting region between FP and the first heptad repeat region of the S2 subunit (Fig 3, B, and Table II). Among these key peptides of S protein that we selected, peptide 318, located at the NTD of the S protein, possessed the most robust binding intensity (fold change relative to the control; Fig E13 and Fig E14). Structural analyses revealed that these epitopes are fully exposed on the surface of monomeric S protein; however, some residues of epitopes for peptides 318, 356, and 530 are concealed under the surface of the trimeric S protein (Fig 4 , A and B), suggesting that both S monomer and trimer structures are recognized efficiently by host immune system under certain circumstances. To be specific, 2 loop segments of peptide 318 (aa 45-46 and aa 56-59) are exposed on the trimeric S protein, with the central β-strand buried inside; and most residues of peptide 356 are accessible on the surface, including a core β-strand (aa 203-209) and a loop segment (aa 210-211). Peptide 510 contains an S2’ cleavage site and a central helix of FP, both of which are fully presented on the surface of S protein; residues of peptide 530 are mostly cryptic, with only a small fraction of the loop (aa 893-895) exposed on the trimeric S protein (Fig 4, C).
Sequence homology analysis among 7 common human coronaviruses revealed that 2 epitopes located at the S2 subunit (peptides 510 and 530) share high sequence identity with other coronaviruses, suggesting serologic cross-reactivity targeting these epitopes among human coronaviruses (Fig 4, D). The sequence of peptide 318 exhibited high similarity with SARS-CoV, while a low level of sequence homology for peptide 356 was shown among coronaviruses, suggesting SARS-CoV-2–specific antibody responses targeting this region (Fig 4, D).
Considering new emerging and circulating SARS-CoV-2 variants globally, we further performed sequence alignment with regard to key epitopes of the S protein between the early SARS-CoV-2 strain (Wuhan-Hu-1) and 5 variants of concern (Alpha, Beta, Gamma, Delta, and Omicron), as well as 2 variants of interest (Lambda and Mu), according to the World Health Organization classification of variant viruses (updated on November 30, 2021). Results showed that sequences of these dominant and persistent epitopes are almost identical among variants analyzed, except for a single N211I mutation identified in the new Omicron variant (Fig 4, E). These data indicate that antibodies generated by early SARS-CoV-2 strains may consistently recognize current circulating variants, and that these dominant epitopes may be capable of mediating sustained long-term antibody responses on infection of SARS-CoV-2 variants.
To further determine the immunologic characteristics of identified epitopes on the S protein, and to further investigate the potential value of these S-protein epitopes as peptide vaccine candidates, we performed a mouse immunization study using selected peptides. BALB/c mice were inoculated 3 times with each peptide in the presence of alum and CpG adjuvants (Fig 4, F). Among the 4 peptides selected, vaccination with peptide 356 elicited antigen-specific antibodies after the second and third doses (Fig 4, G). The results of the serum neutralization assay revealed that immunization with linear peptides did not generate significant levels of neutralizing antibodies against SARS-CoV-2 (Fig 4, H), suggesting these linear peptides perform poorly in inducing robust neutralizing antibody responses.
N protein of SARS-CoV-2 lacks most reactive epitopes mediating durable antibody responses after viral infection
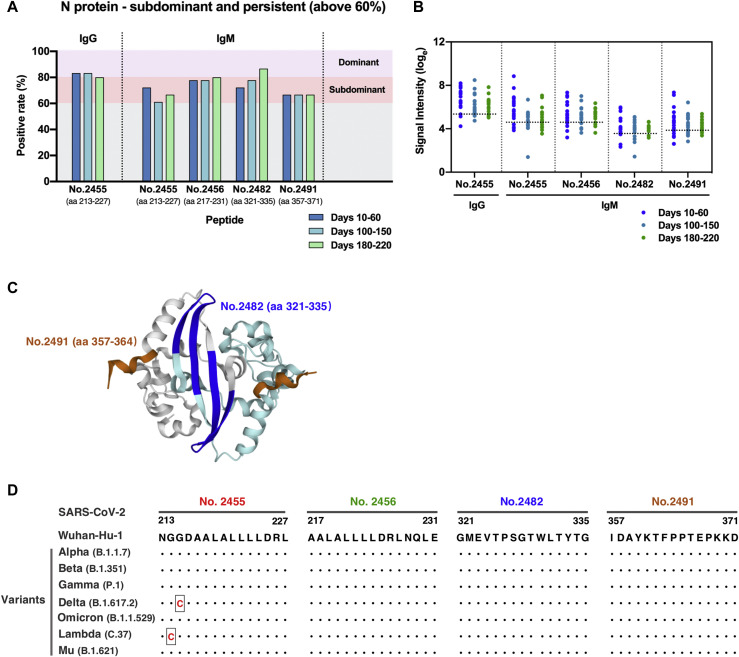
A number of studies have elucidated the potent antigenicity of the SARS-CoV-2 N protein.4 , 11 , 12 However, we failed to identify dominant and persistent epitopes located at the N protein on the basis of the current selection criteria (above 80% positive rate at all 3 sampling time points). For longitudinal evaluation of epitope profiles of the N protein in individuals with COVID-19, we carried out a second round of epitope screening that was based on data obtained from the peptide microarray for the selection of subdominant epitopes that consistently remain positive reactivity in more than 60% of the COVID-19 samples for each of the 3 sampling time points (termed subdominant and persistent epitopes). A total of 4 subdominant and persistent epitopes of the SARS-CoV-2 N protein were identified, among which peptide 2455 (N, aa 213-227) presented reactivity to both of IgG and IgM antibodies in COVID-19 patients, with relatively higher levels of signal intensity (Fig 5 , A and B, and see Table E4 in the Online Repository at www.jacionline.org). The 2 overlapping peptides, peptide 2455 (N, aa 213-227) and peptide 2456 (N, aa 217-231), are located at the Ser/Arg-rich linker region between the N-terminal RNA binding domain and C-terminal dimerization domain of the N protein. Peptide 2482 (N, aa 321-335) and peptide 2491 (N, aa 357-371) are fully or partially located within the dimerization domain of the N protein (Table E4). Because of the lack of 3-D structures regarding the intact conformation of the N protein, we only performed structural analyses of 2 identified epitopes on the dimeric structure of the C-terminal dimerization domain. Residues of peptide 2482 forms 2 β strands that are arranged in an antiparallel manner in the dimerization interfaces, whereas aa 357-364 of peptide 2491 form helix-based structures that are located at opposite ends of the dimer (Fig 5, C).
Fig 5.
Subdominant epitopes located at the SARS-CoV-2 nucleocapsid protein capable of mediating persistent antibody responses. A, IgG and IgM recognition frequencies of subdominant peptides (durably reactive in more than 60% samples) among patient serum samples collected at multiple time points after disease onset. B, Signal intensity kinetics of identified subdominant epitopes over time. Each dot represents a distinct patient serum sample obtained from COVID-19 patients at indicated time points after symptom onset. Dotted horizontal lines indicate cutoff values of positive response for each peptide. C, Detailed structure analysis of epitopes on the C-terminal dimerization domain of the N protein (PDB ID: 6YUN). Epitopes are labeled in blue (peptide 2482, aa 321-335) and brown (peptide 2491, aa 357-364). The 2 monomeric structures are depicted in gray and cyan, respectively. D, Sequence alignment analysis of the identified N-protein epitopes between the early SARS-CoV-2 (strain Wuhan-Hu-1) and 7 emerging variants. Black dots denote identical sequences between the Wuhan-Hu-1 strain and the indicated variant. Changes in amino acid sequence are highlighted in red.
Sequence alignment between early SARS-CoV-2 (strain Wuhan-Hu-1) and 7 emerging variants further revealed that sequences of these subdominant and persistent epitopes in N protein are almost identical among current circulating variants, with a single G215C substitution of peptide 2455 occurring in the Delta variant and a single G214C mutation of peptide 2455 identified in the Lambda variant, suggesting that antibodies targeting these peptides potentially recognize antigen of emerging SARS-CoV-2 variants (Fig 5, D).
Longitudinal serologic analysis identifies epitopes with high signal intensity but decreasing reactivity over time
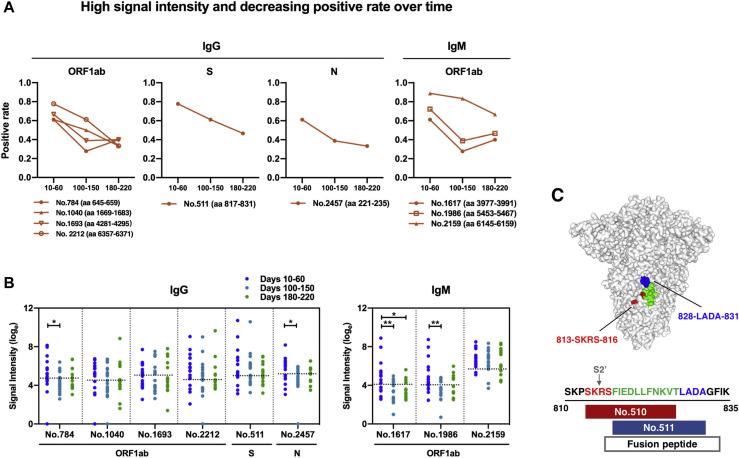
In addition to the selected highly reactive epitopes that are responsible for sustained humoral immune responses mentioned above, we further identified and characterized a panel of 9 positive peptides that showed robust binding intensities (above the mean + SD of signal intensities of all tested samples) with a trend of decreasing reactivity (positive rate changes above 20%) among serum samples from COVID-19 patients over time, in accordance with a general trend of waning in humoral immune responses. Theses epitopes are positioned within the 3 dominant antigens: ORF1ab, S, and N proteins (Fig 6 , A; and see Table E5 in the Online Repository at www.jacionline.org). Additionally, significant reduction in signal intensities of 4 peptides from ORF1ab (peptides 784-IgG, 1617-IgM, and 1986-IgM) and N (peptide 2457-IgG) proteins were observed between samples from an early period of sampling time points (days 10-60 after disease onset) and the later phase of sampling time points (days 100-150 or days 180-220 after symptom onset) (Fig 6, B). Notably, despite largely overlapping with each other, 2 peptides of the S protein peptide 510 (dominant and persistent; Fig 3, B) and peptide 511 (high signal intensity and decreasing positive rate over time; Fig 6, A) exhibited different patterns of reactivity among patient serum samples over time. Sequence and location analysis indicated that peptide 510 contains a S2’ cleavage site and additional amino acids of 813-SKRS-816, whereas peptide 511 is completely within the FP with extended 828-LADA-831 residues that are fully exposed on the surface of trimeric S protein (Fig 6, C). These results revealed new features of epitopes that may ultimately contribute to longer-lasting and stronger humoral immunity against SARS-CoV-2.
Fig 6.
Liner epitopes with high binding intensity and declining reactive frequencies over time, recognized by SARS-CoV-2–infected individuals. A and B, Longitudinal analysis and distribution of identified peptides exhibiting high binding signal intensity (above mean + SD of signal intensities of all tested samples) but decreasing positive rate over time. The recognition frequency (A) and signal intensity (B) of epitopes were plotted with 3 sampling time points after symptom onset. Each dot in (B) stands for an individual patient serum sample collected at indicated time points after symptom onset; dotted horizontal lines indicate cutoff values of positivity for each peptide. Statistical significance analysis was performed based on Limma of R v3.6.3 software. ∗P < .05 and ∗∗P < .01. C, Location and sequence comparisons of 2 adjacent epitopes, peptide 510 (dominant and persistent) and peptide 511 (high signal intensity and decreasing positivity over time), on SARS-CoV-2 S protein structure (PDB ID: 6ZGI). The overlapping regions between 2 epitopes are highlighted in green; unique sequences are labeled in red and blue.
Discussion
A systematic characterization of long-term immune responses on SARS-CoV-2 infection is critical for the development of improved diagnostics, effective therapeutic interventions, and vaccines. In the current study, we performed a comprehensive longitudinal analysis of COVID-19 patients over 180 to 220 days’ follow-up, showing persistent humoral immune responses and activated cytokine production after viral infection. Significantly, by taking advantage of the peptide-based microarray spanning the proteome of SARS-CoV-2, we further revealed kinetics of epitope recognition and identified a panel of dominant epitopes capable of mediating long-term humoral immunity. The findings we report here regarding the longevity of humoral immune responses after SARS-CoV-2 infection confirm some previously published data5 , 10 , 12 , 13 , 42 but extend them by performing deep serologic profiling and epitope screening using longitudinal serum samples from individuals with COVID-19 through the proteome-wide microarray approach.
In this study, we identified 4 dominant epitopes (peptides 318, 356, 510, and 530) within the SARS-CoV-2 S protein that were capable of persistently reactive with more than 80% COVID-19 patient samples tested, up to 180 to 220 days after symptom onset. Peptide 510 (S, aa 813-827), comprising the S2’ cleavage site and the FP of S2 subunit, has been commonly identified in previous studies of other groups.26 , 28 , 32, 33, 34 Functional analyses indicated antibodies targeting this region may exhibit limited neutralization potency against SARS-CoV-226 , 33 despite being highly exposed on the surface of the S protein (Fig 4, A-C). In the case of peptide 530 (S, aa 893-907), which is positioned between the FP and the first heptad repeat region of the S2 subunit, residues are generally buried inside the trimeric structure of the S protein, which makes it hard to access via robust neutralizing antibodies against SARS-CoV-2 (Fig 4, A-C). Additionally, 2 S1-NTD–directed peptides that could mediate long-term antibody responses of SARS-CoV-2 were also selected. Analyses regarding the peptide sequence and location indicated that residues of these 2 peptides—peptide 318 (S, aa 45-59) and peptide 356 (S, aa 197-211)—are in close proximity to the reported epitopes recognized by infection-enhancing antibodies23 , 25 but apart from the key sites of highly potent neutralizing antibodies targeting NTD of the S1 subunit,16 , 19 , 43 suggesting the possibility of epitope recognition by nonneutralizing antibodies toward these 2 identified peptides. In addition to the details mentioned above, mouse immunization with selected peptides further indicated the low efficacy of these non–receptor binding domain linear peptides in inducing robust neutralizing antibody responses (Fig 4, G). Collectively, these data suggest that dominant linear epitopes mediating long-term humoral immune responses on SARS-CoV-2 infection probably induce antibodies with no or limited neutralizing potency.
A fuller understanding the epitope landscape of SARS-CoV-2 antibodies, especially S protein–directed epitopes, provides new insight into the functional dissection of antibodies, which further facilitates innovative and rational vaccine design. Neutralizing antibodies confer protection to eliminate viral infections, whereas nonneutralizing antibodies may play a beneficial, neutral, or even harmful role during virus clearance. Nonneutralizing antibodies provide additional protection in vivo via a range of Fc-mediated effector functions in the context of antibody-dependent phagocytosis and antibody-dependent cytotoxicity. In contrast, some studies have proposed the possibility of a pathogenic role of nonneutralizing antibodies in coronavirus infection. Previous studies using multiple types of vaccine candidates for SARS-CoV and MERS-CoV observed enhanced immunopathology in vaccinated small animals and nonhuman primates after virus challenge.24 , 44, 45, 46, 47, 48, 49, 50 More recently, studies of 2 groups reported that nonneutralizing antibodies targeting NTD of SARS-CoV-2 S protein were capable of enhancing viral infection in vitro through Fcγ receptor–independent mechanisms,23 , 25 although passively administered infection-enhancing antibodies in animal models have been shown to be protective against SARS-CoV-2 infection in vivo. Considering the controversial roles of antibodies during coronavirus infection, further investigations are required to validate the potential role of antibodies recognizing these dominant and persistent epitopes in combating viral infection in vivo. The next stage of rational vaccine design for SARS-CoV-2 could be conceived by eliciting highly potent neutralizing antibodies and protective nonneutralizing antibodies, along with a reduction of the presentation of infection-enhancing epitopes or immunodominant epitopes that have no beneficial effects.
Although a number of previous studies have only focused on the S protein of SARS-CoV-2 aiming to delineate antibody functions regarding neutralization, we performed a comprehensive proteome-wide epitope mapping and identified a panel of epitopes within the ORF1ab polyprotein that were consistently recognized by a high proportion of patient serum samples over time. Although these ORF1ab-directed peptides distributed on the multiple nonstructural proteins may not elicit functional antibodies targeting the SARS-CoV-2 virion, they could be applicable as a diagnostic tool to help differentiate natural infection from vaccination. With the increasing number of vaccine recipients worldwide, current serologic tests based on the S protein and the N protein are facing challenges as an effective approach to aid molecular tests for detection of SARS-CoV-2 infection, and also for the determination of immune status after viral infection. By taking advantage of the most common and persistently reactive peptides within the ORF1ab among COVID-19–infected individuals, serologic diagnosis of natural infection will be carried out without taking into account of vaccination status involving receptor binding domain–based, S protein–based, and inactivated virus–based vaccine approaches. Future studies are needed to assess the reactivity, sensitivity, and specificity of identified ORF1ab peptides in larger cohorts comprising both SARS-CoV-2–infected individuals and vaccine recipients. Additionally, further evaluation of the detection efficiency through multiple peptide combination strategies will be required to overcome the lower sensitivity of peptides compared to the full-length protein and the possible cross-reactivity among common human coronaviruses.
The major limitations of our study are the relatively few samples obtained from a small patient cohort size, and the majority of participants experienced nonsevere COVID-19 diseases. These may limit some of our conclusions with respect to the positive response frequency and magnitude of immune responses, which may vary according to disease severity. Nevertheless, the data presented in this study provide valuable insights into the kinetics of immune responses over time after SARS-CoV-2 infection and features of dominant epitopes capable of mediating sustained humoral immunity in individuals with COVID-19. Together, these findings offer a deeper understanding of longevity of natural immunity induced by viral infection, and have broad implications for innovative vaccination strategies and improved diagnostic approaches.
Key messages.
-
•
The comprehensive longitudinal analysis of 31 COVID-19 patients over 180 to 220 days’ follow-up indicated persistent humoral immunity and activated cytokine production after SARS-CoV-2 infection.
-
•
Using the peptide-based microarray spanning the proteome of SARS-CoV-2, we revealed the kinetics of epitope recognition and identified dominant epitopes capable of mediating long-term humoral immune responses in individuals with COVID-19.
Footnotes
The first 5 authors contributed equally to this article, and all should be considered first author.
This work was supported by the following grants: National Natural Science Foundation of China (grant 31870922 to D.Z.), Natural Science Research Project of Nantong Science and Technology Bureau (XG202003-2 and XG202003-4 to R.L.), National Key Research and Development Program of China (2017YFA0103501 to X.K.), Science and Technology Service Network Initiative (KFJ-STS-QYZD-187 to X.K.), Key Program of the Chinese Academy of Sciences (QYZDJ-SSW-SM01 to X.K.), and General Program of National Natural Science Foundation of China (81570827 to L.H. and 31471224 to X.K.).
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Supplementary data
Fig E1.
Fig E2.
Fig E3.
Fig E4.
Fig E5.
Fig E6.
Fig E7.
Fig E8.
Fig E9.
Fig E10.
Fig E11.
Fig E12.
Fig E13.
Fig E14.
References
- 1.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO) WHO coronavirus (COVID-19) dashboard, 2021. https://COVID19.who.int/ Available at:
- 3.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 4.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J., Liang B., Chen C., Wang H., Fang Y., Shen S., et al. SARS-CoV-2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID-19. Nat Commun. 2021;12:1813. doi: 10.1038/s41467-021-22034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 7.Sterlin D., Mathian A., Miyara M., Mohr A., Anna F., Claer L., et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci Transl Med. 2021;13 doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J., Wu Q., Liu Z., Wang Q., Wu J., Hu Y., et al. Spike-specific circulating T follicular helper cell and cross-neutralizing antibody responses in COVID-19–convalescent individuals. Nat Microbiol. 2021;6:51–58. doi: 10.1038/s41564-020-00824-5. [DOI] [PubMed] [Google Scholar]
- 9.Wang P., Liu L., Nair M.S., Yin M.T., Luo Y., Wang Q., et al. SARS-CoV-2 neutralizing antibody responses are more robust in patients with severe disease. Emerg Microbes Infect. 2020;9:2091–2093. doi: 10.1080/22221751.2020.1823890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanshylla K., Di Cristanziano V., Kleipass F., Dewald F., Schommers P., Gieselmann L., et al. Kinetics and correlates of the neutralizing antibody response to SARS-CoV-2 infection in humans. Cell Host Microbe. 2021;29:917–929.e4. doi: 10.1016/j.chom.2021.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang T., Liang B., Fang Y., Lu S., Li S., Wang H., et al. Declining levels of neutralizing antibodies against SARS-CoV-2 in convalescent COVID-19 patients one year post symptom onset. Front Immunol. 2021;12:708523. doi: 10.3389/fimmu.2021.708523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z., Muecksch F., Schaefer-Babajew D., Finkin S., Viant C., Gaebler C., et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595:426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371 doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokal A., Chappert P., Barba-Spaeth G., Roeser A., Fourati S., Azzaoui I., et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell. 2021;184:1201–1213.e14. doi: 10.1016/j.cell.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 16.Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 17.Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu C., Wang Y., Liu C., Zhang C., Han W., Hong X., et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci Adv. 2021;7 doi: 10.1126/sciadv.abe5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauer M.M., Tortorici M.A., Park Y.J., Walls A.C., Homad L., Acton O.J., et al. Structural basis for broad coronavirus neutralization. Nat Struct Mol Biol. 2021;28:478–486. doi: 10.1038/s41594-021-00596-4. [DOI] [PubMed] [Google Scholar]
- 21.Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y., Soh W.T., Kishikawa J.I., Hirose M., Nakayama E.E., Li S., et al. An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies. Cell. 2021;184:3452–3466.e18. doi: 10.1016/j.cell.2021.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4 doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li D., Edwards R.J., Manne K., Martinez D.R., Schäfer A., Alam S.M., et al. In vitro and in vivo functions of SARS-CoV-2 infection–enhancing and neutralizing antibodies. Cell. 2021;184:4203–4219.e32. doi: 10.1016/j.cell.2021.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poh C.M., Carissimo G., Wang B., Amrun S.N., Lee C.Y., Chee R.S., et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat Commun. 2020;11:2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang B.Z., Hu Y.F., Chen L.L., Yau T., Tong Y.G., Hu J.C., et al. Mining of epitopes on spike protein of SARS-CoV-2 from COVID-19 patients. Cell Res. 2020;30:702–704. doi: 10.1038/s41422-020-0366-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoddard C.I., Galloway J., Chu H.Y., Shipley M.M., Sung K., Itell H.L., et al. Epitope profiling reveals binding signatures of SARS-CoV-2 immune response in natural infection and cross-reactivity with endemic human CoVs. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.H., Leng Y., et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370 doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zamecnik C.R., Rajan J.V., Yamauchi K.A., Mann S.A., Loudermilk R.P., Sowa G.M., et al. ReScan, a multiplex diagnostic pipeline, pans human sera for SARS-CoV-2 antigens. Cell Rep Med. 2020;1 doi: 10.1016/j.xcrm.2020.100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haynes W.A., Kamath K., Bozekowski J., Baum-Jones E., Campbell M., Casanovas-Massana A., et al. High-resolution epitope mapping and characterization of SARS-CoV-2 antibodies in large cohorts of subjects with COVID-19. Commun Biol. 2021;4:1317. doi: 10.1038/s42003-021-02835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Lai D.Y., Lei Q., Xu Z.W., Wang F., Hou H., et al. Systematic evaluation of IgG responses to SARS-CoV-2 spike protein–derived peptides for monitoring COVID-19 patients. Cell Mol Immunol. 2021;18:621–631. doi: 10.1038/s41423-020-00612-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y., Ma M.L., Lei Q., Wang F., Hong W., Lai D.Y., et al. Linear epitope landscape of the SARS-CoV-2 spike protein constructed from 1,051 COVID-19 patients. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H., Wu X., Zhang X., Hou X., Liang T., Wang D., et al. SARS-CoV-2 Proteome Microarray for Mapping COVID-19 Antibody Interactions at Amino Acid Resolution. ACS Cent Sci. 2020;6:2238–2249. doi: 10.1021/acscentsci.0c00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi Z., Ling Y., Zhang X., Chen J., Hu K., Wang Y., et al. Functional mapping of B-cell linear epitopes of SARS-CoV-2 in COVID-19 convalescent population. Emerg Microbes Infect. 2020;9:1988–1996. doi: 10.1080/22221751.2020.1815591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.National Health Commission & National Administration of Traditional Chinese Medicine Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7) Chin Med J (Engl) 2020;133:1087–1095. doi: 10.1097/CM9.0000000000000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y., Li C.Q., Guo S.J., Guo W., Jiang H.W., Li H.C., et al. Longitudinal serum autoantibody repertoire profiling identifies surgery-associated biomarkers in lung adenocarcinoma. EBioMedicine. 2020;53 doi: 10.1016/j.ebiom.2020.102674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., et al. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin Infect Dis. 2020;71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 41.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., et al. Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex. Cell. 2020;182:1560–1573.e13. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wajnberg A., Amanat F., Firpo A., Altman D.R., Bailey M.J., Mansour M., et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., et al. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tseng C.T., Sbrana E., Iwata-Yoshikawa N., Newman P.C., Garron T., Atmar R.L., et al. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M., et al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol. 2011;85:12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weingartl H., Czub M., Czub S., Neufeld J., Marszal P., Gren J., et al. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol. 2004;78:12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yasui F., Kai C., Kitabatake M., Inoue S., Yoneda M., Yokochi S., et al. Prior immunization with severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) nucleocapsid protein causes severe pneumonia in mice infected with SARS-CoV. J Immunol. 2008;181:6337–6348. doi: 10.4049/jimmunol.181.9.6337. [DOI] [PubMed] [Google Scholar]
- 48.Deming D., Sheahan T., Heise M., Yount B., Davis N., Sims A., et al. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 2006;3:e525. doi: 10.1371/journal.pmed.0030525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q., Zhang L., Kuwahara K., Li L., Liu Z., Li T., et al. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates. ACS Infect Dis. 2016;2:361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agrawal A.S., Tao X., Algaissi A., Garron T., Narayanan K., Peng B.H., et al. Immunization with inactivated Middle East respiratory syndrome coronavirus vaccine leads to lung immunopathology on challenge with live virus. Hum Vaccin Immunother. 2016;12:2351–2356. doi: 10.1080/21645515.2016.1177688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.