To the Editor,
We read with interest the paper of Li et al. reporting serum amyloid A (SAA) as a possible biomarker to evaluate the severity of coronavirus disease and progression. (1) They found that the ratio of SAA to lymphocyte (SAA/L) was significantly increased in critically severe patients. A recent meta-analysis also supports the use of this biomarker to assess the severity of the disease although a high heterogeneity among the studies was reported. (2) Other biomarkers have also been linked with disease severity and complications and we would like to share our data regarding the association between the level of SARS-CoV-2 nucleocapsid antigens and the severity of COVID-19.
Eighty-one unvaccinated patients with documented nasopharyngeal (NP) RT-PCR positive for SARS-CoV-2 (eSwab tubes; Copan Italia, Brescia, Italy) for which a serum sample (BD SST II Advance®; Becton Dickinson, NJ, USA) was obtained within 12 h of NP sampling were included. The median delta collection time was 1 min (10th - 90th percentiles: −74 to 183 min). Thirty-six patients (44.4%) were women (median age = 77 years, min-max = 20-97) and 45 (55.6%) were men (median age = 76.5 years, min-max = 33–94). Eighteen patients were categorized as asymptomatics (WHO score of 1), 6 as symptomatic ambulatory mild disease (WHO score of 2 and 3), 43 as symptomatic hospitalized moderate disease (WHO scores of 4 and 5) and 14 as symptomatic hospitalized severe disease patients (WHO score of 6 to 10). Eleven patients died (WHO score of 10). The mean time since symptoms was 5.5 days (standard deviation (SD): 4.4) and 7.1 days (SD: 7.2) in non-severe (WHO scores of 2 to 5) and severe patients (WHO score of 6 to 10), respectively. The SARS-CoV-2 nucleocapsid (N) antigen was quantified automatically in patient sera by a Single Molecular Array (Simoa) immunoassay using the Simoa HD-X analyzer (Quanterix, Massachusetts, USA), as described elsewhere. (3) The results are quantitative and expressed as pg/mL. Serum samples that provided results upper the calibration range were retested using a 1000-fold dilution. The LOD of the assay is 0.099 pg/mL and was considered as positivity cut-off. The within-run coefficient of variation (CV) is less than 10% according to the information of the manufacturer. RT-PCR was performed on the GeneXpert instrument (Cepheid, California, USA) using the Xpress SARS-CoV-2 assay targeting N2 and E genes for the majority of the samples and on a the LightCycler 480 Instrument II (Roche Diagnostics, Rotkreuz, Switzerland) using the LightMix Modular SARS-CoV E-gene set for very few samples obtained during day-care.
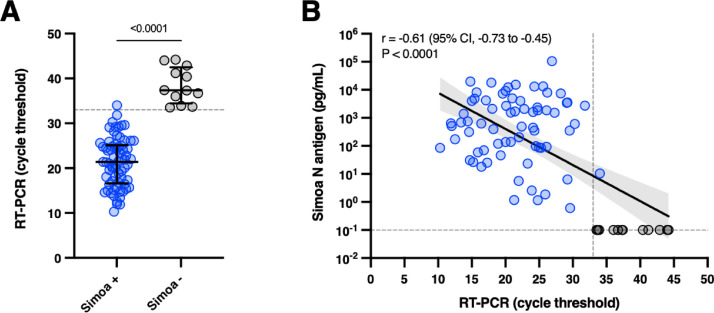
The mean Ct values obtained by RT-PCR were significantly lower in case of serum antigen positivity (p < 0.0001, unpaired t-test) (Fig. 1 A). Using a Ct threshold of 33 as a surrogate of contagiousness(4), the clinical sensitivity was 100% and the clinical specificity was 92.3%. Indeed, patients with low viral load (i.e. Ct > 33) have a very low risk of being infective and capable of transmitting the virus. Therefore, the NP RT-PCR positivity in these subjects may be attributable to residual low viral load(4). Wang et al. also found that negative plasma N antigen in COVID-19 patients corresponded to higher Ct values (38.2 = median; IQR = 37.4–39.2). (5) Highly significant correlations were found between RT-PCR results and antigen levels (Pearson r = −0.61; p < 0.0001) (Fig. 1 B). Other studies found a clinical sensitivity ranging from 85.2% to 93.0% (5, 6, 7) in patients that developed symptoms up to maximum two weeks. The clinical sensitivity increased to 94.2% to 100% considering samples collected within the first days since the onset of symptoms. After 2 weeks, the clinical sensitivity progressively decreased to values comprised between 43.2% to 74.5%. (3, 5, 6, 7) Given that the peak of N antigens is reached after 7 days, as for the viral load in NP samples, (8) and that a continuous decline is observed afterwards, the timing since symptoms is therefore essential for the evaluation of antigenemia.
Fig. 1.
[A] Positive and negative antigen results in serum according to RT-PCR cycle threshold values in nasopharyngeal samples. The gray dotted line corresponds to a cycle threshold of 33. [B] Linear regression of cycle threshold results obtained by RT-PCR versus antigenemia measured on the Simoa. The gray dotted line on the Y-axis represents positivity cut-off for N antigens. The gray dotted line on the X-axis corresponds to a cycle threshold of 33.
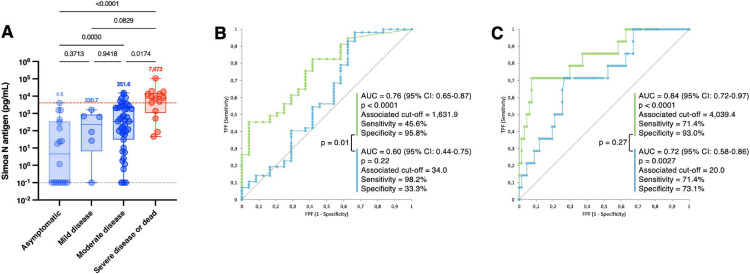
Interestingly, we found that N antigen levels were significantly increased in severe patients (median = 7673 pg/mL) compared to moderately affected (351.6 pg/mL, p = 0.0174) and asymptomatic patients (median = 4.6 pg/mL, p < 0.0001). Moderate patients also had significantly higher antigen levels compared to asymptomatic subjects (p = 0.0030) (Fig. 2 A). Using ROC curve analysis for the classification of hospitalized (WHO score of 4 to 10) versus non-hospitalized patients (WHO score of 1 to 3), a cut-off of 1631.9 pg/mL was identified (sensitivity = 45.6%, specificity = 95.8%; AUC = 0.76, p < 0.0001) (Fig. 2 B – green line). This AUC was similar to the one found by Li et al. using SAA/L for clinical classification (i.e. AUC = 0.75)(1). For the classification of severe (WHO score of 6 to 10) versus non-severe patients (WHO score of 1 to 5), a cut-off of 4039.4 pg/mL was identified with a sensitivity of 64.0% and a specificity of 89.0%. The calculated AUC was 0.84 (p < 0.0001) (Fig. 2 C – green line). Higher concentrations of N antigens were also observed in other studies in more severe patients (3, 5, 7, 9, 10) as well as positive correlations with inflammatory biomarker levels (i.e. CRP or IL-6). (9, 10)
Fig. 2.
[A]Antigenemia and RT-PCR results according to the WHO clinical progression scale on samples obtained on the day of diagnosis, i.e. within 12 h since the RT-PCR. The red dotted line corresponds to the severity cut-off, as determined by the ROC curve analyze (see panel C). The gray dotted lines correspond to the positivity cut-off of each antigen assay. Medians are represented on top of each whisker box. [B] ROC curve analysis for AUC determination according to the hospitalization status and [C] to the severity status. The green line corresponds to N antigen results and the blue line to the Ct results.
Our study confirms that severe patients exhibit higher N antigen levels compared to non-severe at the time of diagnosis. This may help in patients’ triage and monitoring of those more prone to develop a severe form of the disease. Compared to N antigen levels in serum, Ct values of RT-PCR were less associated to severity, especially considering the hospitalization status (Fig. 2 B and C – blue line). In our cohort, only asymptomatics had significantly higher Ct values compared to severe patients (28.4 versus 18.8, p = 0.037), an observation consistent with previous investigations. (5)
In conclusion, sensitive N antigen determination in serum provides a valuable marker for COVID-19 diagnosis and evaluation of severity. Further studies with more patients are needed to complement our data. A better discrimination of N antigen levels based on the days since symptoms as well as correlation with antibody seropositive are also needed.
Declaration of Competing Interest
None.
Author contributions
Conceptualization: JFA – JBA – JDO; Data curation: JFA – JBA – CDA – JDO; Formal analysis: JFA – JBA – CDA – JDO; Funding acquisition: JDO; Investigation: JFA – JDO; Methodology: JFA – JDO; Project administration: JMD – JDO; Resources: JDM – JDO; Supervision: JDO; Validation: JFA – JDO; Visualization: JFA – JDO; Writing – original draft: JFA – JDO; Writing – review & editing: JFA – JBA – CDA – JMD – JDO.
Data availability statement
The data that support the findings of this study are available from the corresponding author JDO, upon reasonable request.
Acknowledgment
The authors would like to thank the technical teams from the laboratories of QUALIblood, Clinique Saint-Luc Bouge and Clinique Saint-Pierre Ottignies for collecting the samples and performing the analyses.
References
- 1.Li H., Xiang X., Ren H., Xu L., Zhao L., Chen X., et al. Serum Amyloid A is a biomarker of severe Coronavirus Disease and poor prognosis. J Infect. Jun 2020;80(6):646–655. doi: 10.1016/j.jinf.2020.03.035. PubMed PMID: 32277967. Pubmed Central PMCID: PMC7141628. Epub 2020/04/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zinellu A., Paliogiannis P., Carru C., Mangoni A.A. Serum amyloid A concentrations, COVID-19 severity and mortality: an updated systematic review and meta-analysis. Int J Infect Dis. 2021;105:668–674. doi: 10.1016/j.ijid.2021.03.025. 2021-04-01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shan D., Johnson J.M., Fernandes S.C., Suib H., Hwang S., Wuelfing D., et al. N-protein presents early in blood, dried blood and saliva during asymptomatic and symptomatic SARS-CoV-2 infection. Nat Commun. 2021;12(1):1931. doi: 10.1038/s41467-021-22072-9. Mar 26PubMed PMID: 33771993. Pubmed Central PMCID: PMC7997897. Epub 2021/03/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basile K., McPhie K., Carter I., Alderson S., Rahman H., Donovan L., et al. Cell-based Culture Informs Infectivity and Safe De-Isolation Assessments in Patients with Coronavirus Disease 2019. Clin Infect Dis. 2021;73(9):e2952–e29e9. doi: 10.1093/cid/ciaa1579. Nov 2PubMed PMID: 33098412. Pubmed Central PMCID: PMC7665383. Epub 2020/10/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H., Hogan C.A., Verghese M., Solis D., Sibai M., Huang C., et al. SARS-CoV-2 Nucleocapsid Plasma Antigen for Diagnosis and Monitoring of COVID-19. Clin. Chem. 2021 doi: 10.1093/clinchem/hvab216. Oct 4PubMed PMID: 34605900. Pubmed Central PMCID: PMC8522398. Epub 2021/10/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hingrat Q.L., Visseaux B., Laouenan C., Tubiana S., Bouadma L., Yazdanpanah Y., et al. Detection of SARS-CoV-2N-antigen in blood during acute COVID-19 provides a sensitive new marker and new testing alternatives. Clin Microbiol Infect. 2020 Dec 8 doi: 10.1016/j.cmi.2020.11.025. PubMed PMID: 33307227. Pubmed Central PMCID: PMC7724284. Epub 2020/12/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., Ong C.M., Yun C., Mo W., Whitman J.D., Lynch K.L., et al. Diagnostic Value of Nucleocapsid Protein in Blood for SARS-CoV-2 Infection. Clinical chemistry. 2021 Aug 6. PubMed PMID: 34358289. Pubmed Central PMCID: PMC8436384. Epub 2021/08/07. [DOI] [PMC free article] [PubMed]
- 8.Lippi G., Simundic A.M., Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) Clin Chem Lab Med. 2020 Jun 25;58(7):1070–1076. doi: 10.1515/cclm-2020-0285. PubMed PMID: 32172228. Epub 2020/03/17. [DOI] [PubMed] [Google Scholar]
- 9.Brasen C.L., Christensen H., Olsen D.A., Kahns S., Andersen R.F., Madsen J.B., et al. Daily monitoring of viral load measured as SARS-CoV-2 antigen and RNA in blood, IL-6, CRP and complement C3d predicts outcome in patients hospitalized with COVID-19. Clin Chem Lab Med. 2021 Nov 25;59(12):1988–1997. doi: 10.1515/cclm-2021-0694. PubMed PMID: 34455731. Epub 2021/08/30. [DOI] [PubMed] [Google Scholar]
- 10.Perna F., Bruzzaniti S., Piemonte E., Maddaloni V., Atripaldi L., Sale S., et al. Serum levels of SARS-CoV-2 nucleocapsid antigen associate with inflammatory status and disease severity in COVID-19 patients. Clin Immunol. May 2021;226 doi: 10.1016/j.clim.2021.108720. PubMed PMID: 33819577. Pubmed Central PMCID: PMC8017913. Epub 2021/04/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author JDO, upon reasonable request.