Abstract
Chemical investigations of a marine sponge-associated Bacillus revealed six new imidazolium-containing compounds, bacillimidazoles A–F (1–6). Previous reports of related imidazolium-containing natural products are rare. Initially unveiled by timsTOF (trapped ion mobility spectrometry) MS data, extensive HRMS and 1D and 2D NMR analyses enabled the structural elucidation of 1–6. In addition, a plausible biosynthetic pathway to bacillimidazoles is proposed based on isotopic labeling experiments and invokes the highly reactive glycolytic adduct 2,3-butanedione. Combined, the results of structure elucidation efforts, isotopic labeling studies and bioinformatics suggest that 1–6 result from a fascinating intersection of primary and secondary metabolic pathways in Bacillus sp. WMMC1349. Antimicrobial assays revealed that, of 1–6, only compound six displayed discernible antibacterial activity, despite the close structural similarities shared by all six natural products.
Keywords: marine-derived Bacillus, antibacterial, biosynthetic gene cluster, isotopic enrichment, heterocycles, imidazolium
1. Introduction
Heterocyclic scaffolds are commonly encountered in natural products isolated from both terrestrial and marine organisms [1,2,3,4]. Their vast structural diversity, drug-like features, and biological properties have inspired both intensive efforts to discover new heterocyclic compounds, as well as imaginative total syntheses [4]. Nitrogen-containing heterocycles, such as pyrroles, imidazoles, oxazoles, pyridines, and quinolones, exhibit a diverse array of biological activities; these include, but are by no means limited to, antibacterial [5], antifungal [6], and anticancer [7] activities.
Marine-derived imidazole alkaloids have been one of the most fruitful families of bioactive compounds giving rise to many pharmaceutical leads [8]. Historically, marine-derived imidazole alkaloids have been most often isolated from sponges; imidazole alkaloids [9] featuring bromopyrrole-imidazoles [10], indole-containing imidazoles [11], and 2-aminoimidazoles are among the species most often identified from marine sponges [12]. Furthermore, in recent years, marine microorganisms have come to be viewed as sustainable and productive sources of new bioactive imidazole-containing natural products [9]. However, reports of positively charged imidazolium natural products remain relatively rare. Most reports of imidazolium-containing compounds feature 1,3-dimethyl-5-methylthiol [13,14,15] or 2-aminoimidazolium containing structures (guanidinium-like) [10,11]. A wide array of applications in ionic liquids [16], important biological activities [17], and their amenability to further structural modifications have made imidazolium salts an attractive target of contemporary research [18]. Therefore, discovering new imidazolium-based species and gaining further insight into their biosynthetic origins has become an interesting, yet challenging, task for natural product scientists.
2. Results & Discussion
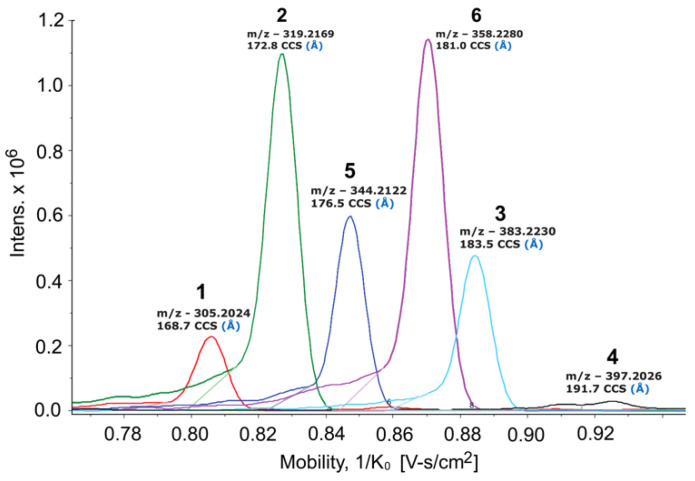
As part of our ongoing efforts to discover new natural products from marine invertebrate-associated bacteria [19], we developed a streamlined discovery platform that includes strain prioritization by metabolomics [20] and an LC/MS fractionation platform to generate screening libraries [21]. Strain WMMC1349, a marine Bacillus sp. cultivated from the sponge Cinachyrella apion, drew our attention since one of its fractions displayed activity against methicillin-resistant Staphylococcus aureus (MRSA). Further purification of the active fraction by HPLC resulted in enrichment of an active subfraction. Interestingly, subsequent analytical HPLC revealed this fraction to be a represented by a broad peak (Figure S1) despite the clear presence of a mixture of compounds as revealed by 1H NMR (Figure S1). Fortunately, Bruker timsTOF (trapped ion mobility spectrometry) MS data for this subfraction indicated a series of new molecules with m/z values of 305.2, 319.2, 344.4, 358.2, 383.2, and 397.2, respectively (Figure 1). Exhaustive attempts to separate each different m/z species (see Figures S1–S55), as initially visualized by timsTOF MS, ultimately afforded HPLC conditions amenable to clean separation and isolation of each discreet compound. The six new imidazolium-containing compounds, now differentiated from each other, were termed bacillimidazoles A–F (1–6, Figure 2), and all were assessed for in vitro activity against MRSA, B. subtilis, and E. coli; only compound 6 was found to be active (MRSA). Isotopic labelling of these metabolites using isotopically enriched culture media, and bioinformatic analysis were conducted to decipher the means by which the bacillimidazoles are biosynthesized.
Figure 1.
Bruker timsTOF MS spectrum of the bioactive wells against MRSA. Peak/compound assignments are shown above each relevant signal.
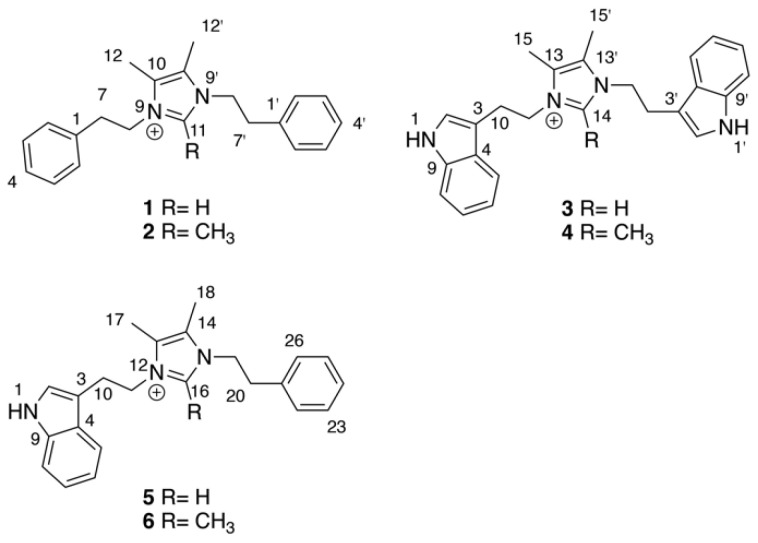
Figure 2.
Structures of bacillimidazoles A–F (1–6) with central imidazolium numberings indicated for each subgroup.
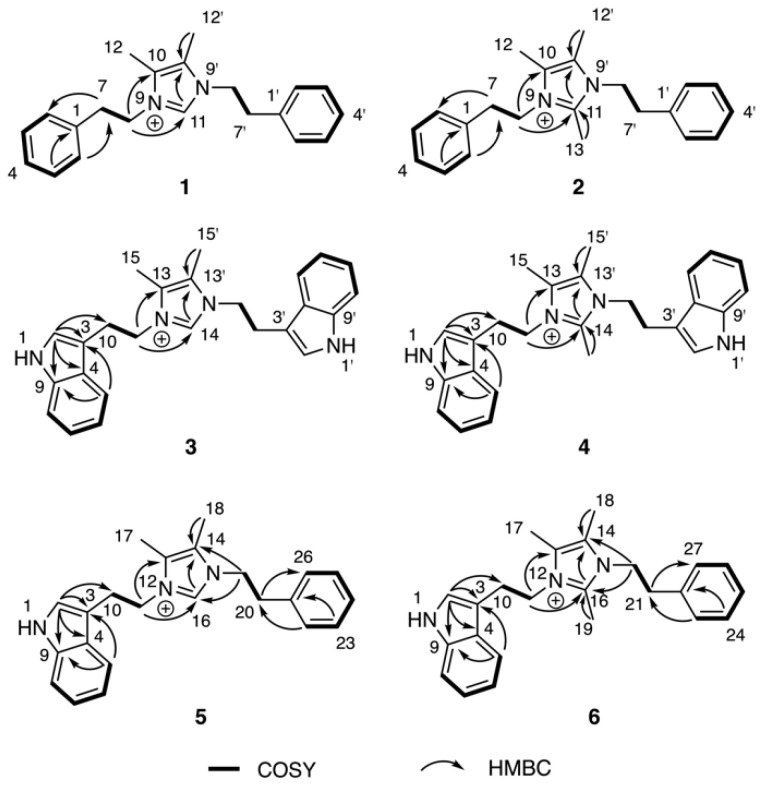
The molecular formulae of bacillimidazoles A (1) and B (2) were determined to be C21H25N2+ (m/z = 305.2029, M+, calcd 305.2012, Figure S37) and C22H27N2+ (m/z = 319.2171, M+, calcd 319.2169, Figure S38), respectively, based on HRESIMS data. In the 13C NMR spectra of 1 and 2 in Table 1, only 11 and 12 carbon signals were observed, respectively, suggesting symmetric scaffolds for both 1 and 2. Furthermore, comparisons of 1H and 13C NMR data revealed a high degree of similarity between compound 1 and lepidiline A, an imidazolium-containing alkaloid isolated from the South American plants Lepidium meyenii Walp [22]. These similarities suggested the presence of a 4,5-dimethyl imidazolium cyclic structure and two phenyl-ring containing substituents in compound 1. More highly refined datasets revealed that H2-7 (δH 3.06) and H2-8 (δH 4.35) showed COSY correlations (Figure 3) to each other. The HMBC correlations (Figure 3) were also observed from H2-7 to C-2 (δC 130.0) and from H2-8 (δH 4.35) to C-10 (δC 128.5), C-11 (δC 135.5), suggesting that –CH2CH2– groups linked the central imidazolium ring to two terminal phenyl rings, one on either side of the imidazolium. Therefore, the structure of 1 was assigned as a 1,3-difunctionalized imidazolium-containing structure. The NMR dataset of 2 was compared to that obtained for 1 and the only observable difference was an additional methyl group (δH, 2.01, H3-13; δC 9.7, CH3) substitution on C-11 (δC 144.0), which was determined by careful interpretation of the well resolved HMBC correlation from H3-13 to C-11 (Figure 3).
Table 1.
Summary of 1H and 13C NMR data for 1–4 (600 MHz for 1H (500 MHz for 4), 125 MHz for 13C, CD3OD).
| Position | 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|---|
| δC, Type | δH, (J in Hz) | δC, Type | δH, (J in Hz) | δC, Type | δH, (J in Hz) | δC, Type | δH, (J in Hz) | |
| 1, 1′ | 137.8, qC | 138.2, qC | ||||||
| 2, 2′ | 130.0, CH | 7.12, dd (8.0, 1.8) | 130.2, CH | 7.11, dd (8.0, 1.8) | 124.4, CH | 6.96, s | 124.5, CH | 6.98, s |
| 3, 3′ | 130.0, CH | 7.34, t (7.4) | 130.1, CH | 7.34, t (7.8) | 110.5, qC | 110.8, qC | ||
| 4, 4′ | 128.4, CH | 7.30, t (7.4) | 128.6, CH | 7.32, t (7.8) | 128.3, qC | 128.4, qC | ||
| 5, 5′ | 130.0, CH | 7.34, t (7.4) | 130.1, CH | 7.34, t (7.8) | 118.4, CH | 7.32, d (7.6) | 118.2, CH | 7.24, d (8.0) |
| 6, 6′ | 130.0, CH | 7.12, dd (8.0, 1.8) | 130.2, CH | 7.11, dd (8.0, 1.8) | 120.1, CH | 7.03, t (7.6) | 120.2, CH | 7.03, t (8.0) |
| 7, 7′ | 37.1, CH2 | 3.06, t (6.6) | 36.3, CH2 | 3.02, t (6.8) | 122.8, CH | 7.15, t (7.6) | 122.9, CH | 7.14, t (8.0) |
| 8, 8′ | 49.3, CH2 | 4.35, t (6.6) | 47.9, CH2 | 4.31, t (6.8) | 112.6, CH | 7.39, d (7.8) | 112.7, CH | 7.39, d (8.0) |
| 9, 9′ | 138.1, qC | 138.0, qC | ||||||
| 10, 10′ | 128.5, qC | 127.1, qC | 26.8, CH2 | 3.07, t (6.8) | 26.0, CH2 | 3.03, t (6.0) | ||
| 11, 11′ | 135.5 CH | 8.51, s | 144.0, qC | 48.7, CH2 | 4.25, t (6.8) | 47.5, CH2 | 4.19, t (6.0) | |
| 12, 12′ | 7.9, CH3 | 2.08, s | 8.2, CH3 | 2.08, s | ||||
| 13, 13′ | 9.7, CH3 | 2.01, s | 128.2, qC | 127.0, qC | ||||
| 14 | 135.4, CH | 8.12, s | 144.0, qC | |||||
| 15, 15′ | 7.9, CH3 | 2.09, s | 8.3, CH3 | 2.18, s | ||||
| 16 | 9.3, CH3 | 1.64, s | ||||||
Figure 3.
1H–1H COSY and key HMBC correlations of compounds 1–6.
The molecular formulae of bacillimidazole C (3) and bacillimidazole D (4) were identified as C25H27N4+ (m/z = 383.2228, M+, calcd 383.2230) and C26H29N4+ (m/z = 397.2385, M+, calcd 397.2387), respectively, by analyzing their HRMS data (Figures S39 and S40). Analysis of their 13C NMR data also suggested symmetrical structures for both 3 and 4. In particular, detailed 1D NMR data analyses of 3 and 4 suggested that the central portion of each molecule bore a common imidazolium functionality. Overall, five sets of unassigned aromatic protons (H-2, δH 6.96; H-5, δH 7.32; H-6, δH 7.03; H-7, δH 7.15; H-8, δH 7.39) and eight unassigned aromatic carbons (C-2, δC 124.4; C-3, δC 110.5; C-4, δC 128.3; C-5, δC 118.4; C-6, δC 120.1; C-7, δC 122.8; C-8, δC 112.6; C-9, δC 138.1) were characteristic of the 3-indoyl structural motifs in 3; the validity of this idea was verified by the observation of HMBC correlations from H-2 to C-3/C-4/C-9 and from H-5 to C-3/C-9 (Figure 3). Similar to 1 and 2, the connection between –CH2CH2– groups and the aromatic building blocks in 3, as well as the central imidazolium ring, was deduced by HMBC correlations from H-3 to C-10 and from H2-11 to C-13/C-14 (Figure 3), respectively, establishing the full structural assignment of bacillimidazole C (3). Finally, in a fashion similar to that applied to 3, the structure of 4, with its additional imidazolium-linked methyl group, was elucidated.
Approaches employed to solve the structures of 1–4 also were applied to determine the structures of 5 and 6 in Table 2. Analysis of HRMS data for bacillimidazole E (5) and bacillimidazole F (6) made clear their molecular formulae as C23H26N3+ (m/z = 344.2134, M+, calcd 344.2121) and C24H28N3+ (m/z = 358.2283, M+, calcd 358.2278), respectively. In addition, review of their 13C NMR data suggested asymmetrical structures for both 5 and 6 since 23 13C NMR signals were observed in 5 (24 signals for 6). Additionally, comparisons of NMR data for 5 to those of 1 and 3 suggested the presence of an imidazolium ring, a phenyl ring, and a 3-indole ring in 5. Furthermore, H-2 (δH 7.03), assigned to the indole ring, showed an HMBC correlation to C-10 (δH 26.5); and H-22 (δH 7.04), assigned to the phenyl ring, showed an HMBC correlation to C-20 (δC 36.7) (Figure 3). COSY correlations were observed between H2-10 (δH 3.23) and H2-11 (δH 4.36), H2-19 (δH 4.18) and H2-20 (δH 2.84). H2-11 and H2-19 both showed HMBC correlations to C-16 (δC 135.4), which was assigned to the imidazolium ring (Figure 3). Therefore, 5 was believed to contain an indole ring, an imidazolium ring, and a phenyl moiety (Figure 2). In addition, two –CH2CH2– groups were found to intervene the three different cyclic structural motifs. In applying the established correlation data, we thus elucidated structure 5 as shown in Figure 2. The structure of 6 was determined in a fashion similar to that employed for 5, and was ultimately identified as an analog of 5 bearing a methyl group on the central imidazolium ring.
Table 2.
Summary of 1H and 13C NMR data for 5 and 6 (600 MHz for 1H. (500 MHz for 6), 125 MHz for 13C, CD3OD).
| Position | 5 | 6 | ||
|---|---|---|---|---|
| δC, Type | δH, (J in Hz) | δC, Type | δH, (J in Hz) | |
| 1 | ||||
| 2 | 124.6, CH | 7.03, s | 124.6, CH | 7.02, s |
| 3 | 110.4, qC | 110.8, qC | ||
| 4 | 128.4, qC | 128.5, qC | ||
| 5 | 112.7, CH | 7.40, dd (8.2, 1.0) | 112.7, CH | 7.37, d (7.8) |
| 6 | 120.2, CH | 7.04, dt (7.6, 1.2) | 120.3, CH | 7.00, t (7.8) |
| 7 | 122.9, CH | 7.15, dt (8.0, 1.0) | 123.9, CH | 7.12, t (7.8) |
| 8 | 118.4, CH | 7.34, dd (8.0, 1.0) | 118.2, CH | 7.23, d (7.8) |
| 9 | 138.1, qC | 138.1, qC | ||
| 10 | 26.5, CH2 | 3.23, t (6.6) | 26.0, CH2 | 3.17, t (6.2) |
| 11 | 48.9, CH2 | 4.38, t (6.6) | 47.7, CH2 | 4.30, t (6.2) |
| 12 | ||||
| 13 | 128.3, qC | 127.1, qC | ||
| 14 | 128.3, qC | 127.0, qC | ||
| 15 | ||||
| 16 | 135.4, CH | 8.27, s | 144.0, qC | |
| 17 | 8.0, CH3 | 2.12, s | 8.3, CH3 | 2.15, s |
| 18 | 7.9, CH3 | 2.04, s | 8.2, CH3 | 2.07, s |
| 19 | 48.7, CH2 | 4.18, t (7.2) | 9.4, CH3 | 1.77, s |
| 20 | 36.7, CH2 | 2.84, t (7.2) | 47.6, CH2 | 4.11, t (6.8) |
| 21 | 137.8, qC | 36.3, CH2 | 2.77, t (6.8) | |
| 22 | 130.0, CH | 7.04, d (7.2) | 138.0, qC | |
| 23 | 129.9, CH | 7.29, m | 130.1, CH | 7.03, dd (7.3, 1.6) |
| 24 | 128.3, CH | 7.28, m | 130.0, CH | 7.30, m |
| 25 | 129.9, CH | 7.29, m | 128.5, CH | 7.26, m |
| 26 | 130.0, CH | 7.04, d (7.2) | 130.0, CH | 7.30, m |
| 27 | 130.1, CH | 7.03, dd (7.3, 1.6) | ||
Following their structural elucidation, bacillimidazoles A–F (1–6) were tested for antibacterial activity against MRSA, B. subtilis, and E. coli (Table S1). All compounds failed to show any significant activity against B. subtilis and E. coli, although bacillimidazole F (6) did display weak activity against MRSA with an MIC of 38.3 μM.
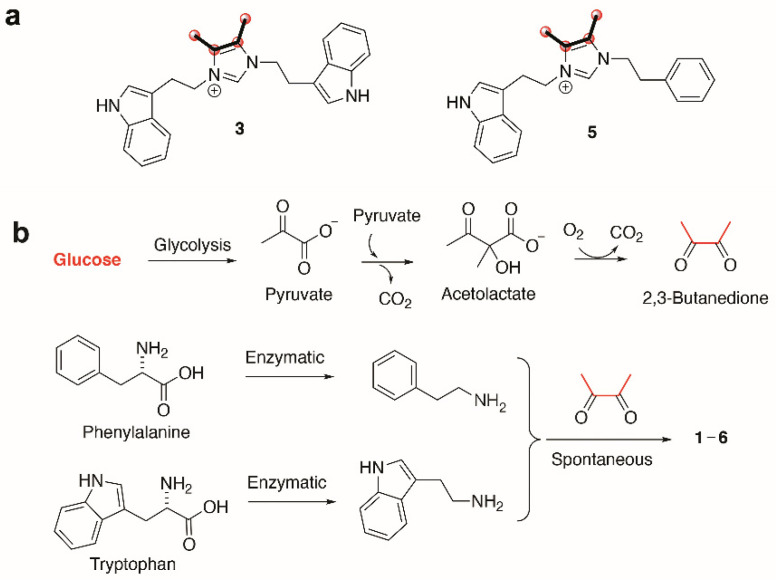
To better understand the biosynthetic mechanisms involved in generating the uncommon imidazolium structures found in the bacillimidazoles, isotopic enrichment studies were carried out. In particular, we employed 13C enriched culture media to glean vital insight into how the bacillimidazoles are constructed. By substituting two carbon sources, starch and d-glucose, with 13C6-d-glucose, we found that the four carbons composing the vicinal dimethyl olefin of bacillimidazoles C (3) and E (5) (Figures S43–S48) underwent substantial 13C enrichment relative to all other carbons (Figure 4a). On the basis of these findings, we theorized that all carbons of the basic framework originate from amino acids whereas the dimethyl olefin elements of the bacillimidazoles are derived from glucose, presumably via a glycolytic process (Figure 4b). This logic is, of course, buoyed by the resemblance of the bacillimidazole sidechains to those found in phenylalanine and tryptophan. On the basis of these findings, we investigated the possibility of a biosynthetic pathway, as shown in Figure 4. It is well known that glucose is readily converted to 2,3-butadione along the canonical glycolytic pathway to acetolactate and subsequent processing of acetolactate to the essential 2,3-butadione, which can spontaneously react with either tryptamine or phenethylamine, both of which are common bacterial metabolites formed from the aromatic amino acids tryptophan and phenylalanine. These three components—two amines and a dione—provide most of the atoms of the imidazole moiety tethered through both nitrogens to various side chains. This diimine could undergo further spontaneous reactions with either one or two carbon carboxylic acids, or derivatives, and appropriate redox agents (or enzymes) to form the central imidazolium ring. Acetyl-CoA, or an equivalent, would produce the methylated compounds and formate, or a derivative, would produce the unmethylated bacillimidazoles. There is an alternative possibility in which the relevant amino acids condense with the butadione moiety prior to decarboxylation, and that the decarboxylation(s) of such intermediates might expedite imidazolium ring formation. While it is not possible at this stage to propose a detailed stepwise biosynthesis for the bacillimidazoles, an overall path with both enzymatic and spontaneous steps is likely, and the 13C-glucose feeding studies clearly indicate the importance of glucose processing en route to imidazole assembly. That the production of bacillimidazoles by WMMC1349 is driven by secondary metabolism biosynthetic machineries and is not relegated only to primary metabolic events and/or extract workup conditions, is supported by the fact that, of six bacilli strains evaluated, only two (including WMMC1349) proved to be bacillimidazole producers (Figures S4–S6, S49–S54 and Table S2).
Figure 4.
Biosynthesis of bacillimidazoles. (a) Isotopic labeling of bacillimidazoles C (3) and E (5). Carbons highlighted with bold bonds and red spheres showed high levels of 13C incorporation. (b) Proposed biosynthetic pathway to compounds 1–6 calls for enzymatic production of tryptamine, phenethylamine and 2,3-butanedione (from glucose); all subsequent steps may proceed spontaneously.
Based on the biosynthetic insights gained from labelling experiments, we set out to identify the gene cluster responsible for the biosynthesis of 1–6. We sequenced and assembled the genome of the producer, Bacillus sp. WMMC1349 [23]. The genome sequence was analyzed with state-of-the-art biosynthetic pipelines, yet no likely biosynthetic pathway for the biosynthesis of 1–6 was identified. We therefore mined the genome for the presence of the genes involved in acetoin biosynthesis, of which 2,3-butanedione, the proposed building block of 1–6, is a precursor. We identified all of the genes responsible for acetoin biosynthesis in the genome of Bacillus sp. WMMC1349. Acetoin is biosynthesized from two molecules of pyruvate that are condensed to generate acetolactate. Acetolactate can either be enzymatically transformed into acetoin by a decarboxylase or can undergo spontaneous decarboxylation to yield the building block of 1–6, 2,3-butanedione. A diacetyl reductase subsequently converts 2,3-butanedione into acetoin. Whole genome alignments of different Bacillus spp. revealed that the region upstream of the acetoin biosynthetic genes differs significantly from other Bacillus spp., including the model strain B. subtilis 168 (insertion of a large low-density coding region in Bacillus sp. WMMC1349), while the downstream region is homologous in all analyzed genomes. To our surprise, we were not able to identify a copy of the gene family encoding butanediol-dehydrogenases which was present in all other analyzed Bacillus spp. The absence of genes involved in acetoin catabolism would be expected to increase the concentration of the precursor 2,3-butanedione. Genome mining (Section 4.5 below) of the bacillimidazole producer revealed the presence of a gene encoding an aromatic-l-amino acid decarboxylase, which is the proposed second enzyme essential for 1–6 biosynthesis en route to building blocks tryptamine and phenethylamine, respectively. No homologs of the aromatic-l-amino acid decarboxylase gene were identified in any other Bacillus genome analyzed, including the model strain Bacillus subtilis 168, indicating that the gene is facultative for the genus Bacillus. Relative to the biosynthetic machineries of most bacterial secondary metabolites, genes involved in acetoin biosynthesis and the aromatic-l-amino acid decarboxylase gene are not clustered. This observation suggests that the bacillimidazoles may form spontaneously from high-abundance primary metabolites with congruent reactivities. The biosynthesis of the bacillimidazoles is yet another example of the growing number of natural products that are produced partly by genetically coded instructions and partly by spontaneous reactivity—the joining of reactive intermediates, or products, from different pathways [24,25,26]. The initial formation of the bis-imines represents, for example, the unsurprising coupling of highly reactive primary amines with a very reactive alpha-diketone.
3. Conclusions
The isolation and structural elucidation of bacillimidazoles A–F, six imidazolium-containing heterocycles from marine Bacillus sp. WMMC1349 represent the discovery of a new class of marine-derived heterocyclic natural products. Isotopic labeling of 1–6 using 13C enriched culture media, biomimetic synthetic approaches and bioinformatic analyses were performed in order to gain insights into the biosynthetic assembly of these interesting compounds. To the best of our knowledge, there are only a few reports of natural products containing 1,3-difunctionalized imidazolium moieties, and this is the first report of naturally occurring imidazolium-containing heterocycles. We contend, based on these findings, that the discovery of 1–6 contributes to our advancing knowledge of significantly underexplored biosynthetic pathways, specifically those that intersect primary and secondary metabolic pathways. We anticipate that the lessons learned here will help to expedite efforts to more fully understand and exploit the full biosynthetic potential of Bacillus spp.
4. Materials and Methods
4.1. General Experimental Procedures
UV spectra were recorded on an Aminco/OLIS UV-Vis spectrophotometer. IR spectra were measured with a Bruker Equinox 55/S FT–IR spectrophotometer. NMR spectra were obtained in CD3OD (δH 3.34 ppm, δC 49.0 ppm) with a Bruker Avance 600 III MHz (Billerica, MA, USA) spectrometer equipped with a 1H{13C/15N/31P} cryoprobe, a Bruker Avance III 500 MHz (Billerica, MA, USA) spectrometer equipped with a 13C/15N{1H} cryoprobe, and a Bruker Avance III HD 400 MHz (Billerica, MA, USA) spectrometer. HRMS data were acquired with a Bruker MaXis™ 4G ESI-QTOF (Billerica, MA, USA) mass spectrometer. RP HPLC was performed using a Shimadzu Prominence HPLC system and a Phenomenex Gemini C18 column (250 × 30 mm). UHPLC-HRMS was acquired using a Bruker MaXis™ 4G ESI-QTOF (Billerica, MA, USA) mass spectrometer coupled with a Waters Acquity UPLC system operated by Bruker Hystar software and a C18 column (Phenomenex Kinetex 2.6 μm, 2.1 mm × 100 mm). Bruker timsTOF Pro instrument (Billerica, MA, USA) was used for the trapped ion mobility MS analysis using direct infusion with 0.003 mL/min of flow rate and ESI+ ionization source. Nebulizer gas 0.4 bar, dry gas 3.5 L/min, source temperature 220 °C, ESI voltage 4200V (+). MS spectra were collected using the following parameters: tims ramp time = 350 ms, PASEF on, scan range (m/z, 20–1000; 1/k0, 0.70–1.00 V·s/cm2.
4.2. Biological Material
Sponge specimens were collected on 27 May 2015 near the west shore of Ramrod Key (24°39′38.1″ N, 81°25′25.0″ W) in Florida. A voucher specimen is housed at the University of Wisconsin−Madison. For cultivation, a sample of sponge (1 cm3) was ground in 500 μL sterile seawater, and dilutions were made using 500 μL sterile seawater. Subsequently, 400 μL of diluted sponge sample was added to 200 μL of sterile artificial seawater and 100 μL was plated using a sterile L-shaped spreader. Diluted samples were plated on Gauze 1 media supplemented with artificial seawater. Each medium was supplemented with 50 μg/mL cycloheximide, 25 μg/mL nystatin, and 25 μg/mL nalidixic acid. Plates were incubated at 28 °C and colonies were isolated over the course of two months.
4.3. Fermentation, Extraction and Isolation
Two 10 mL seed cultures (25 × 150 mm tubes) in medium DSC (20 g soluble starch, 10 g glucose, 5 g peptone, 5 g yeast extract per liter of artificial seawater) were inoculated with strain WMMC-1349 and shaken (200 RPM, 28 °C) for seven days. Two-liter flasks (1 × 500 mL) containing ASW-A (20 g soluble starch, 10 g d-glucose, 5 g peptone, 5 g yeast extract, 5 g CaCO3 per liter of artificial seawater. For 13C enriched ASW-A media, 10 g d-glucose (U-13C6, 99%) was used instead of soluble starch and d-glucose) were inoculated with 20 mL seed culture and were incubated (200 RPM, 28 °C) for seven days. Four-liter flasks (10 × 1 L) containing medium ASW-A with Diaion HP20 (7% by weight) were inoculated with 50 mL from the 500 mL culture and shaken (200 RPM, 28 °C) for seven days. For producing artificial sea water, solutions I (415.2 g NaCl, 69.54 g Na2SO4, 11.74 g KCl, 3.40 g NaHCO3, 1.7 g KBr, 0.45 g H3BO3, 0.054 g NaF) and II (187.9 g MgCl2·6H2O, 22.72 g CaCl2·2H2O, 0.428 g SrCl2·6H2O) were made up separately using distilled water and combined to give a total volume of 20 L.
Filtered HP20 and cells were washed with H2O and extracted with acetone. The acetone extract was subjected to liquid-liquid partitioning using 30% aqueous MeOH and CHCl3 (1:1). The CHCl3-soluble partition (3.12 g) was fractionated by Sephadex LH20 column chromatography (column size 500 × 40 mm, CHCl3:MeOH = 1:1, 20 mL for each fraction). Fractions containing 1–6 (1.1 g) were subjected to RP HPLC (20%/80% to 100%/0% MeOH/H2O (with 0.1% acetic acid), 23.5 min, 20 mL/min) using a Phenomenex Gemini C18 column (250 × 30 mm). The fraction collected between 16–18 min was further fractionated by RP HPLC (22%/78% to 51%/49% MeCN/H2O (with 0.05% trifluoroacetic acid), 29.5 min, 20 mL/min) using a Phenomenex Gemini C18 column (250 × 30 mm), yielding 1 (50.2 mg, tR 28.2 min), 2 (35.8 mg, tR 27.6 min), 3 (2.2 mg, tR 27.3 min), 4 (3.1 mg, tR 28.9 min), 5 (40.5 mg, tR 28.6 min), 6 (32.3 mg, tR 29.3 min).
Bacillimidazole A (1): light yellow solid, UV-Vis (MeOH): λmax (log ε) 211 nm (3.83), 259 nm (2.65), 279 nm (2.55), 291 nm (2.55); IR (ATR): υmax 3385, 3144, 3033, 2935, 2873, 2834, 1781, 1679, 1563, 1498, 1456, 1399, 1357, 1199, 1129, 1083, 1029, 831, 801, 751, 720, 702 cm−1; HRMS M+ m/z = 305.2029 (calcd. for C21H25N2+ 305.2012). 1H NMR (600 MHz, MeOD) δH 8.51 (s, 1H), 7.34 (t, J = 7.2 Hz, 4H), 7.31 (d, J = 7.2 Hz, 2H), 7.12 (d, J = 7.0 Hz, 4H), 4.35 (t, J = 7.0 Hz, 4H), 3.06 (t, J = 6.9 Hz, 4H), 2.08 (s, 6H). 13C NMR (126 MHz, MeOD) δC 137.83, 135.53, 130.02, 129.97, 128.45, 128.44, 49.28, 37.07, 7.94.
Bacillimidazole B (2): light yellow solid, UV-Vis (MeOH): λmax (log ε) 217 nm (3.85), 274 nm (3.02), 282 nm (3.04), 290 (2.97); IR (ATR): υmax 3383, 3065, 3034, 2934, 2870, 1681, 1564, 1525, 1497, 1440, 1400, 1355, 1200, 1128, 1030, 934, 840, 801, 749, 722, 703 cm−1; HRMS M+ m/z = 319.2171 (calcd. for C22H27N2+ 319.2169). 1H NMR (600 MHz, MeOD) δH 7.36–7.33 (m, 6H), 7.11 (d, J = 6.4 Hz, 4H), 4.31 (t, J = 6.7 Hz, 4H), 3.02 (t, J = 6.7 Hz, 4H), 2.08 (s, 6H), 2.01 (s, 3H). 13C NMR (126 MHz, MeOD) δC 144.04, 138.15, 130.22, 130.08, 128.56, 127.14, 47.87, 36.33, 9.67, 8.21.
Bacillimidazole C (3): light yellow solid, UV-Vis (MeOH): λmax (log ε) 223 nm (4.25), 274 nm (3.67), 282 nm (3.69), 290 nm (3.61); IR (ATR): υmax 3357, 2946, 2835, 1679, 1564, 1449, 1432, 1341, 1203, 1185, 1137, 1024, 838, 802, 746, 722 cm−1; HRMS M+ m/z = 383.2228 (calcd. for C25H27N4+ 383.2230). 1H NMR (600 MHz, MeOD) δH 8.12 (s, 1H), 7.39 (d, J = 8.1 Hz, 2H), 7.32 (d, J = 7.9 Hz, 2H), 7.15 (t, J = 7.4 Hz, 2H), 7.03 (t, J = 7.4 Hz, 2H), 6.96 (s, 2H), 4.25 (t, J = 6.7 Hz, 4H), 3.07 (t, J = 6.7 Hz, 4H), 2.09 (s, 9H). 13C NMR (126 MHz, MeOD) δC 138.05, 135.37, 128.30, 128.19, 124.42, 122.83, 120.13, 118.43, 112.64, 110.49, 26.81, 7.94.
Bacillimidazole D (4): light yellow solid, UV-Vis (MeOH): λmax (log ε) 224 nm (3.98), 281 nm (3.44), 290 nm (3.38); IR (ATR): υmax 3360, 3292, 2925, 2854, 1729, 1648, 1561, 1456, 1411, 1342, 1256, 1235, 1181, 1105, 1073, 1025, 926, 744 cm−1; HRMS M+ m/z = 397.2385 (calcd. for C26H29N4+ 397.2387). 1H NMR (500 MHz, MeOD) δH 7.39 (d, J = 8.0 Hz, 1H), 7.24 (d, J = 7.9 Hz, 1H), 7.14 (t, J = 7.5 Hz, 1H), 7.03 (t, J = 7.5 Hz, 1H), 6.98 (s, 1H), 4.19 (t, J = 5.9 Hz, 2H), 3.03 (t, J = 5.7 Hz, 2H), 2.18 (s, 3H), 1.64 (s, 1H). 13C NMR (126 MHz, MeOD) δC 143.95, 138.00, 128.40, 126.96, 124.50, 122.88, 120.21, 118.23, 112.67, 110.76, 47.50, 25.97, 9.30, 8.29.
Bacillimidazole E (5): light yellow solid, UV-Vis (MeOH): λmax (log ε) 221 nm (3.87), 274 nm (3.17), 282 nm (3.18), 290 (3.11); IR (ATR): υmax 3376, 2991, 2950, 2836, 1677, 1564, 1497, 1456, 1398, 1356, 1341, 1201, 1134, 1078, 1025, 934, 834, 801, 747, 721, 703 cm−1; HRMS M+ m/z = 344.2134 (calcd. for C23H26N3+ 344.2121). 1H NMR (600 MHz, MeOD) δH 8.27 (s, 1H), 7.40 (d, J = 8.2 Hz, 1H), 7.34 (d, J = 7.9 Hz, 1H), 7.32–7.26 (m, 3H), 7.15 (t, J = 7.4 Hz, 1H), 7.07–7.02 (m, 4H), 4.38 (t, J = 6.5 Hz, 2H), 4.18 (t, J = 7.2 Hz, 2H), 3.23 (t, J = 6.5 Hz, 2H), 2.84 (t, J = 7.2 Hz, 2H), 2.12 (s, 3H), 2.04 (s, 3H). 13C NMR (126 MHz, MeOD) δC 138.08, 137.82, 135.44, 129.95, 129.87, 128.38, 128.34, 128.29, 128.27, 124.55, 122.85, 120.17, 118.37, 112.68, 110.42, 37.03, 26.76, 7.97, 7.89.
Bacillimidazole F (6): light yellow solid, UV-Vis (MeOH): λmax (log ε) 225 nm (3.96), 274 nm (3.55), 282 nm (3.56), 290 nm (3.51); IR (ATR): υmax 3356, 3275, 3062, 3001, 2971, 2928, 2830, 1647, 1562, 1497, 1454, 1401, 1353, 1234, 1203, 1179, 1108, 1077, 1028, 924, 745, 703 cm−1; HRMS M+ m/z = 358.2283 (calcd. for C24H28N3+ 358.2278). 1H NMR (500 MHz, MeOD) δH 7.37 (d, J = 8.0 Hz, 1H), 7.28 (m, 3H), 7.23 (d, J = 7.9 Hz, 1H), 7.11 (t, J = 7.5 Hz, 1H), 7.06 – 6.97 (m, 3H), 4.31 (t, J = 5.5 Hz, 2H), 4.11 (t, J = 6.6 Hz, 2H), 3.17 (t, J = 5.5 Hz, 2H), 2.76 (t, J = 6.6 Hz, 2H), 2.15 (s, 2H), 2.07 (s, 3H), 1.77 (s, 3H). 13C NMR (126 MHz, MeOD) δC 143.99, 138.09, 138.03, 130.07, 130.02, 128.47, 128.45, 127.06, 127.00, 124.64, 122.91, 120.28, 118.18, 112.72, 110.76, 47.74, 47.59, 36.27, 26.00, 9.43, 8.32, 8.18.
4.4. Antibacterial Testing
Bacillimidazoles A–F (1–6) were tested for antibacterial activity against E. coli (ATCC #25922), B. subtilis strain NRS-231, and Methicillin-resistant Staphylococcus aureus (MRSA) (ATCC #33591), and MICs were determined using a dilution antimicrobial susceptibility test for aerobic bacteria. Compounds 1–6 were dissolved in DMSO and serially diluted to 10 concentrations (0.25–128 μg/mL) in 96-well plates. Vancomycin was used as a positive control against B. subtilis and MRSA, and exhibited MIC values of 0.25 μg/mL. Gentamicin was used as a positive control against E. coli, and exhibited an MIC of 4 μg/mL. Bacillimidazoles, vancomycin, and gentamicin were tested in triplicate. On each plate, there were six untreated media controls. The plates were incubated at 37 °C for 18 h. The MICs were determined as the lowest concentration that inhibited visible growth of bacteria.
4.5. Sequencing and Identification of Candidate Bacillimidazole Biosynthetic Genes
16S rDNA sequencing was conducted as previously described [27]. WMMC1349 was identified as a Bacillus sp. The 16S sequence for WMMC1349 was deposited in GenBank (accession number MK892477). PacBio sequencing data were converted from BAM to FASTQ format using bedtools [28], and this fastq file was then corrected, trimmed, and assembled using Canu v1.8 [29], with an estimated genome size of 4.5 megabases (Mb). The resulting assembly was 4.677Mb over 10 contigs. It has an N50 of 4.03Mb and an L50 of 1.
Genome sequence (accession number JABJUQ000000000) was subjected to antiSMASH 5.0 analysis [30]. Results were analyzed by BLAST analysis. Acetoin and amino acid biosynthetic genes (KEGG) were identified in the bacillimidazole producer and selected model Bacillus spp. By BLAST analysis using Geneious 11.1.3 [31], and verified using additional online platforms such as Phyre2 [32]. Genomes of the producer of 1–6 and selected model Bacillus spp. were aligned using the MAUVE algorithm [33]. Promoter regions were identified using the BPROM algorithm [34].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/md20010043/s1. Detailed experimental discussions of isotopic enrichment studies and validation of bacillimidazoles as genuine natural products as well as Tables S1 & S2 summarizing all antibacterial data (Table S1), and bacillimidazole productivity of six Bacilli strains (Table S2). Also: Figure S1. Analytical HPLC analysis and 1H NMR analysis of active subfractions. Figure S2. LC-MS analysis (EIC m/z 383) of 3. Figure S3. LC-MS analysis (EIC m/z 397) of 4. Figure S4. LC-MS analysis of chemical reaction of tryptamine (10.0 mg/mL) and 2,3 butanedione (0.5 equivalent) in MeOH solution without HP-20 resin for 2 h. Figure S5. LC-MS analysis of chemical reaction of tryptamine (10.0 mg/mL) and 2,3 butanedione (0.5 equivalent) in MeOH solution with HP-20 resin for 2 h. Figure S6. LC-MS analysis of chemical reaction of tryptamine (10.0 mg/mL) and 2,3 butanedione (0.5 equivalent) in CHCl3:MeOH = 1:1 solution for 24 h. Figure S7. 1H NMR Spectrum of Bacillimidazole A (1, 600 MHz, CD3OD). Figure S8. 13C NMR Spectrum of Bacillimidazole A (1, 125 MHz, CD3OD). Figure S9. gCOSY Spectrum of Bacillimidazole A (1, 600 MHz, CD3OD). Figure S10. gHSQC Spectrum of Bacillimidazole A (1, 600 MHz, CD3OD). Figure S11. gHMBC Spectrum of Bacillimidazole A (1, 600 MHz, CD3OD). Figure S12. 1H NMR Spectrum of Bacillimidazole B (2, 600 MHz, CD3OD). Figure S13. 13C NMR Spectrum of Bacillimidazole B (2, 125 MHz, CD3OD). Figure S14. gCOSY Spectrum of Bacillimidazole B (2, 600 MHz, CD3OD). Figure S15. gHSQC Spectrum of Bacillimidazole B (2, 600 MHz, CD3OD). Figure S16. gHMBC Spectrum of Bacillimidazole B (2, 600 MHz, CD3OD). Figure S17. 1H NMR Spectrum of Bacillimidazole C (3, 600 MHz, CD3OD). Figure S18. 13C NMR Spectrum of Bacillimidazole C (3, 125 MHz, CD3OD). Figure S19. gCOSY Spectrum of Bacillimidazole C (3, 600 MHz, CD3OD). Figure S20. gHSQC Spectrum of Bacillimidazole C (3, 600 MHz, CD3OD). Figure S21. gHMBC Spectrum of Bacillimidazole C (3, 600 MHz, CD3OD). Figure S22. 1H NMR Spectrum of Bacillimidazole D (4, 500 MHz, CD3OD). Figure S23. 13C NMR Spectrum of Bacillimidazole D (4, 125 MHz, CD3OD). Figure S24. gCOSY Spectrum of Bacillimidazole D (4, 500 MHz, CD3OD). Figure S25. gHSQC Spectrum of Bacillimidazole D (4, 500 MHz, CD3OD). Figure S26. gHMBC Spectrum of Bacillimidazole D (4, 500 MHz, CD3OD). Figure S27. 1H NMR Spectrum of Bacillimidazole E (5, 600 MHz, CD3OD). Figure S28. 13C NMR Spectrum of Bacillimidazole E (5, 125 MHz, CD3OD). Figure S29. gCOSY Spectrum of Bacillimidazole E (5, 600 MHz, CD3OD). Figure S30. gHSQC Spectrum of Bacillimidazole E (5, 600 MHz, CD3OD). Figure S31. gHMBC Spectrum of Bacillimidazole E (5, 600 MHz, CD3OD). Figure S32. 1H NMR Spectrum of Bacillimidazole F (6, 500 MHz, CD3OD). Figure S33. 13C NMR Spectrum of Bacillimidazole F (6, 125 MHz, CD3OD). Figure S34. gCOSY Spectrum of Bacillimidazole F (6, 500 MHz, CD3OD). Figure S35. gHSQC Spectrum of Bacillimidazole F (6, 500 MHz, CD3OD). Figure S36. gHMBC Spectrum of Bacillimidazole F (6, 500 MHz, CD3OD). Figure S37. Positive Ion HRESIMS of Bacillimidazole A (1). Figure S38. Positive Ion HRESIMS of Bacillimidazole B (2). Figure S39. Positive Ion HRESIMS of Bacillimidazole C (3). Figure S40. Positive Ion HRESIMS of Bacillimidazole D (4). Figure S41. Positive Ion HRESIMS of Bacillimidazole E (5). Figure S42. Positive Ion HRESIMS of Bacillimidazole F (6). Figure S43. 1H NMR Spectrum of 13C Labeled Bacillimidazole C (3, 500 MHz, CD3OD). Figure S44. 13C NMR Spectrum of 13C Labeled Bacillimidazole C (3, 125 MHz, CD3OD). Figure S45. 1H NMR Spectrum of 13C Labeled Bacillimidazole E (5, 500 MHz, CD3OD). Figure S46. 13C NMR Spectrum of 13C Labeled Bacillimidazole E (5, 125 MHz, CD3OD). Figure S47. Positive Ion HRESIMS of 13C Enriched Bacillimidazole C (3). Figure S48. Positive Ion HRESIMS of 13C Enriched Bacillimidazole E (5). Figure S49. LC-MS analysis of strain Bacillus sp. WMMC325. Figure S50. LC-MS analysis of strain Bacillus sp. WMMC331. Figure S51. LC-MS analysis of strain Bacillus sp. WMMC1349. Figure S52. LC-MS analysis of strain Bacillus sp. WMMC1350. Figure S53. LC-MS analysis of strain Bacillus sp. WMMC1351. Figure S54. LC-MS analysis of strain Bacillus sp. WMMC1352. Figure S55. Culture broth of six different marine Bacillus strains. Figure S56. Pictures for sponge hosts of six different marine Bacillus strains.
Author Contributions
Conceptualization, C.R.C., J.C. and T.S.B.; methodology, J.-X.Y., Q.W., J.C. and T.S.B.; formal analysis, J.-X.Y., Q.W., E.J.N.H., M.G.C., H.H.; investigation, J.-X.Y., Q.W., E.J.N.H., M.G.C., D.R.B., G.E.A.; resources, H.H. and T.S.B.; writing—original draft preparation, J.-X.Y., Q.W., E.J.N.H. and S.R.R.; writing—review and editing, Q.W., S.R.R., J.C., M.G.C. and T.S.B.; supervision, C.R.C., J.C. and T.S.B.; project administration, C.R.C., J.C. and T.S.B.; funding acquisition, C.R.C., J.C. and T.S.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by NIH Grants U19AI109673, U19AI142720, and R01AT009874 in addition to the University of Wisconsin-Madison School of Pharmacy and the Graduate School at the University of Wisconsin. E.J.N.H. gratefully acknowledges funding by the Swiss National Science Foundation (Postdoctoral Mobility Fellowship). We would like to thank the Analytical Instrumentation Center at the School of Pharmacy, University of Wisconsin-Madison for the facilities to acquire spectroscopic data. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grant P41GM103399 (NIGMS). Additional equipment was purchased with funds from the University of Wisconsin, the NIH (RR02781, RR08438), the NSF (DMB-8415048, OIA-9977486, BIR-9214394), and the USDA.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lewis J.R. Amaryllidaceae, Sceletium, imidazole, oxazole, thiazole, peptide and miscellaneous alkaloids. Nat. Prod. Rep. 2001;18:95–128. doi: 10.1039/a909077k. [DOI] [PubMed] [Google Scholar]
- 2.Jin Z., Li Z., Huang R. Muscarine, imidazole, oxazole, thiazole, Amaryllidaceae and Sceletium alkaloids. Nat. Prod. Rep. 2002;19:454–476. doi: 10.1039/b108923b. [DOI] [PubMed] [Google Scholar]
- 3.Blunt J.W., Copp B.R., Keyzers R.A., Munro M.H.G., Prinsep M.R. Marine Natural Products. Nat. Prod. Rep. 2017;34:235–294. doi: 10.1039/C6NP00124F. [DOI] [PubMed] [Google Scholar]
- 4.Jin Z. Muscarine, imidazole, oxazole and thiazole alkaloids. Nat. Prod. Rep. 2016;33:1268–1317. doi: 10.1039/C6NP00067C. [DOI] [PubMed] [Google Scholar]
- 5.Ziar N., Montalvão S., Hodnik Z., Nawrot D.A., Žula A., Ilaš J., Kikelj D., Tammela P., Mašič L.P. Antimicrobial activity of the marine alkaloids clathrodin and oroidin, and their synthetic analogues. Mar. Drugs. 2014;12:940–963. doi: 10.3390/md12020940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassan W., Edrada R., Ebel R., Wray V., Berg A., van Soest R., Wiryowidagdo S., Proksch P. New imidazole alkaloids from the Indonesian sponge Leucetta chagosensis. J. Nat. Prod. 2004;67:817–822. doi: 10.1021/np0305223. [DOI] [PubMed] [Google Scholar]
- 7.Dyson L., Wright A.D., Young K.A., Sakoff J.A., McCluskey A. Synthesis and anticancer activity of focused compound libraries form the natural product lead, oroidin. Bioorg. Med. Chem. 2014;22:1690–1699. doi: 10.1016/j.bmc.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 8.Jin Z. Muscarine, imidazole, oxazole and thiazole alkaloids. Nat. Prod. Rep. 2013;30:869–915. doi: 10.1039/c3np70006b. [DOI] [PubMed] [Google Scholar]
- 9.Jin Z. Muscarine, imidazole, oxazole and thiazole alkaloids. Nat. Prod. Rep. 2011;28:1143–1191. doi: 10.1039/c0np00074d. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka N., Kusama T., Takahashi-Nakaguchi A., Gonoi T., Fromont J., Kobayashi J. Nagelamides X-Z, dimeric bromopyrrole alkaloids from a marine sponge Agelas sp. Org. Lett. 2013;15:3262–3265. doi: 10.1021/ol401291n. [DOI] [PubMed] [Google Scholar]
- 11.Zhang F., Wang B., Prasad P., Capon R.J., Jia Y. Asymmetric total synthesis of (+)-dragmacidin D reveals unexpected stereocomplexity. Org. Lett. 2015;17:1529–1532. doi: 10.1021/acs.orglett.5b00327. [DOI] [PubMed] [Google Scholar]
- 12.Gong K.-K., Tang X.-L., Liu Y.-S., Li P.-L., Li G.-Q. Imidazole alkaloids form the South China Sea Sponge Pericharax heteroraphis and their cytotoxi and antiviral activities. Molecules. 2016;21:150. doi: 10.3390/molecules21020150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourguet-Kondracki M.L., Martin M.T., Guyot M. A New—Carboline Alkaloid Isolated from the Marine Sponge Hyrtios erecta. Tetrahedron Lett. 1996;37:3457–3460. doi: 10.1016/0040-4039(96)00573-4. [DOI] [Google Scholar]
- 14.Pedpradab S., Edrada R., Ebel R., Wray V., Proksch P. New beta-carboline alkaloids form the Andaman Sea sponge Dragmacidon sp. J. Nat. Prod. 2004;67:2113–2116. doi: 10.1021/np0401516. [DOI] [PubMed] [Google Scholar]
- 15.Carroll A.R., Avery V.M. Leptoclinidamines A-C, indole alkaloids from the Australian ascidian Leptoclinides durus. J. Nat. Prod. 2009;72:696–699. doi: 10.1021/np800831z. [DOI] [PubMed] [Google Scholar]
- 16.Malhotra S.V., Kumar V.A. A profile of the in vitro anti-tumor activity of imidazolium-based ionic liquids. Bioorg. Med. Chem. Lett. 2010;20:581–585. doi: 10.1016/j.bmcl.2009.11.085. [DOI] [PubMed] [Google Scholar]
- 17.Liu L.-P., Zong M.-H., Linhardt R.J., Lou W.-Y., Li N., Huang C., Wu H. Mechanistic insights into the effect of imidazolium ionic liquid on liquid production by Geotrichum fermentas. Biotechnol. Biofuels. 2016;9:266. doi: 10.1186/s13068-016-0682-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright B.D., Deblock M.C., Wagers P.O., Duah E., Robishaw N.K., Shelton K.L., Southerland M.R., DeBord M.A., Kersten K.M., McDonald L.J., et al. Anti-tumor activity of lipophilic imidazolium salts on select NSCLC cell lines. J. Med. Chem. Res. 2015;24:2838–2861. doi: 10.1007/s00044-015-1330-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyche T.P., Piotrowski J.S., Hou Y., Braun D., Deshpande R., Mcllwain S., Ong I.M., Myers C.L., Guzei I.A., Westler W.M., et al. Forazoline A: Marine-derived polyketide with antifungal in vivo activity. Angew. Chem. Int. Ed. 2014;53:11583–11586. doi: 10.1002/anie.201405990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou Y., Braun D.R., Michel C.R., Klassen J.L., Adnani N., Wyche T.P., Bugni T.S. Microbial strain prioritization using metabolomics tools for the discovery of natural products. Anal. Chem. 2012;84:4277–4283. doi: 10.1021/ac202623g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang F., Barns K., Hoffmann F.M., Braun D.R., Andes D.R., Bugni T.S. Thalassosamide, a Siderophore Discovered from the Marine-Derived Bacterium Thalassospira profundimaris. J. Nat. Prod. 2017;80:2551–2555. doi: 10.1021/acs.jnatprod.7b00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui B., Zheng B.L., He K., Zheng Q.Y. Imidazole alkaloids from Lepidium meyenii. J. Nat. Prod. 2003;66:1101–1103. doi: 10.1021/np030031i. [DOI] [PubMed] [Google Scholar]
- 23.Wu Q., Throckmorton K., Maity M., Chevrette M.G., Braun D.R., Rajski S.R., Currie C.R., Thomas M.G., Bugni T.S. Bacillibactins E and F from a Marine Sponge-Associated Bacillus sp. J. Nat. Prod. 2021;84:136–141. doi: 10.1021/acs.jnatprod.0c01170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y., Potts M.B., Colosimo D., Herrera-Herrera M.L., Legako A.G., Yousufuddin M., White M.A., MacMillan J.B. Discoipyrroles A–D: Isolation, structure determination, and synthesis of potent migration inhibitors from Bacillus hunanensis. J. Am. Chem. Soc. 2013;135:13387–13392. doi: 10.1021/ja403412y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mevers E., Saurí J., Helfrich E.J.N., Henke M., Barns K.J., Bugni T.S., Andes D., Currie C.R., Clardy J. Pyronitrins A–D: Chimeric Natural Products Produced by Pseudomonas protegens. J. Am. Chem. Soc. 2019;141:17098–17101. doi: 10.1021/jacs.9b09739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Q., Li S.W., Xu H., Wang H., Hu P., Zhang H., Luo C., Chen K.X., Nay B., Guo Y.W., et al. Complex Polypropionates from a South China Sea Photosynthetic Mollusk: Isolation and Biomimetic Synthesis Highlighting Novel Rearrangements. Angew. Chem. Int. Ed. 2020;59:12105–12112. doi: 10.1002/anie.202003643. [DOI] [PubMed] [Google Scholar]
- 27.Wyche T.P., Hou Y., Braun D., Cohen H.C., Xiong M., Bugni T.S. First natural analogs of the cytotoxic thiodepsipeptide thiocoraline A from a marine Verrucosispora sp. J. Org. Chem. 2011;76:6542–6547. doi: 10.1021/jo200661n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quinlan A.R., Hall I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S.Y., Medema M.H., Weber T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic. Acids. Res. 2019;47:81–87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.For related approach see: Kerkhof L.J., Dillon K.P., Häggblom M.M., McGuinnes L.R. Profiling bacterial communities by MinION sequencing of ribosomal operons. Microbiome. 2017;5:116. doi: 10.1186/s40168-017-0336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darling A.E., Mau B., Perna N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cassiano M.H.A., Silva-Rocha R. Benchmarking Bacterial Promoter Prediction Tools: Potentialities and Limitations. mSystems. 2020;5:e00438-20. doi: 10.1128/mSystems.00439-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.