Abstract
(1) Background: This study investigates influential risk factors for predicting 30-day readmission to hospital for Campylobacter infections (CI). (2) Methods: We linked general practitioner and hospital admission records of 13,006 patients with CI in Wales (1990–2015). An approach called TF-zR (term frequency-zRelevance) technique was presented to evaluates how relevant a clinical term is to a patient in a cohort characterized by coded health records. The zR is a supervised term-weighting metric to assign weight to a term based on relative frequencies of the term across different classes. Cost-sensitive classifier with swarm optimization and weighted subset learning was integrated to identify influential clinical signals as predictors and optimal model for readmission prediction. (3) Results: From a pool of up to 17,506 variables, 33 most predictive factors were identified, including age, gender, Townsend deprivation quintiles, comorbidities, medications, and procedures. The predictive model predicted readmission with 73% sensitivity and 54% specificity. Variables associated with readmission included male gender, recurrent tonsillitis, non-healing open wounds, operation for in-gown toenails. Cystitis, paracetamol/codeine use, age (21–25), and heliclear triple pack use, were associated with a lower risk of readmission. (4) Conclusions: This study gives a profile of clustered variables that are predictive of readmission associated with campylobacteriosis.
Keywords: hospitalisation, readmission, Campylobacter infections, machine learning, text mining, feature selection, electronic health records
1. Introduction
Campylobacteriosis is the most common form of culture-positive bacterial gastroenteritis worldwide, with the species C.jejuni and C.coli, inhabiting the intestinal tracts of both humans and animals, and accounting for up to 95% of human infections [1]. The disease burden has been estimated to be over 2.4 million people per annum in the USA [2,3]. In the UK, Campylobacter is thought to cause more than 280,000 cases of food poisoning annually, and be responsible for more than 100 deaths a year at an estimated cost of £900 million [4].
Campylobacter infections are typically attributed to the handling and consumption of chicken and, less frequently, with the consumption of unpasteurized milk, red meat, sausages, contaminated water, or transmission from household pets or farm animals. Most infections are sporadic, with relatively few identifiable outbreaks, so it is difficult to trace the sources and routes of transmission. Thus, translation of exposure to infection remains poorly understood [2,5,6,7,8].
Clinical manifestations of Campylobacter entirits typically include sudden onset abdominal pain, cramping, fever and frequent diarrhoea, with bloody stools in around one in ten patients. Fatality is most common in the elderly and those with comorbid conditions [9]. Late sequelae, such as inflammatory bowel diseases [10,11,12], rheumatologic disorders (i.e., reactive arthritis) [13,14,15,16], Guillain-Barré syndrome (GBS) [17,18], and Glomerulonephritis [19], often cause long term morbidity. In Europe, the incidence of campylobacter infection has continued to increase in the last decade, and reported increases in infection rates have necessitated the establishment of measures for prevention and control through the food chain [20]. Despite its high incidence, the factors associated with chronic infection or recurrence, and hence readmission, remain poorly understood.
Complications associated with Campylobacter infection often require hospitalisation [21,22,23], and in England and Wales approximately 10% of reported cases were admitted to hospital for treatment [24]. In an Australian provincial setting, the average anunual rate of Campylobacter-associated hospital admissions was 13.6%, and the readmission rate of Campylobacter-associated hospitalizaiton was 5.53% whthin 28 days after discharge [25]. In the USA, campylobacteriosis costs an estimated $1.3 billion a year in hospitalisation and other medical costs, surpassing salmonellosis and shigellosis [2,3], with unplanned readmission adding to the clinical and financial burden [26].
Readmission rates are utilised as indicators of hospital performance and quality of care. Absolute number and rate of readmission continue to rise in the UK, increasing by 19% between 2010 and 2017. Furthermore, readmission classified as potentially preventable is rising twice this rate, estimated at over 40% over the same time-period [27]. Preventable readmissions therefore represent an increasing burden on healthcare systems and hospitals have strong incentives to predict, at the time of discharge, patients who would be at high risk of readmission. The absence of effective predictive models currently limits the effectiveness of readmission reduction strategies. To develop a reliable predictive model, one first needs to identify modifiable predictors of readmission regarding patients and care. However, this can be challenging for diseases, such as campylobacteriosis studied here, where cases of infection are not well explained by the commonly recognized risk factors [6,28,29,30] and reliable predictors of hospitalisation have not been clearly established.
In current clinical practice, the risk of patient readmission can be evaluated using the LACE index, defined by four independent variables: length of stay (L); acuity level of admission (A); comorbidity condition (C); and use of emergency rooms (E) [29]. Use of the LACE index, assuming a linear relationship among the four variables [30,31], can result in poor predictive performance [29]. In fact, there is no standard LACE threshold to classify patients as readmission versus non-readmission, and practitioner assessment is often subjective in defining such threshold. In contrast to the LACE index, some regression models have been developed to predict readmission from patient hospital records, but majority of the models [29,31,32] were not only built from a small number of variables but also were not developed to be generalizable, often relying on a small number of coded terms from primary care records.
Following a decade of rising readmission rates in the UK, in 2011 the Department of Health introduced policies focussed on reversing this trend which included financial penalties for 30-day readmission [33]. This coincided with the US Hospital Readmission Reduction Programme, which also included punitive financial measures for underperforming hospitals [34]. The estimated cost of readmission in the USA stands at approximately $17.4 billion [33], unplanned readmission has therefore become a major concern in advanced healthcare systems. Procedures for reducing readmissions, such as education, follow-up visits, and discharge ‘teams’ have been implemented in many hospitals [34], but these methods are often impractical, costly and of limited impact. Indeed, readmission rates have continued to rise in English hospitals since the introduction of these policies [35]. In light of this, there is an urgent need to identify factors that accurately predict the risk of readmission.
Risk factors for campylobacteriosis are widely recognized [5,7,36,37] but reliable predictors of hospitalisation have not been clearly established. Recently natural language processing techniques were adopted to predicit hospitalizations with structural and unstructural data [38,39]. Here, we develop a robust, validated, and cost-effective data-driven method to identify the most informative hospital readmission predictors from primary care medical records. We use a novel incorporation of machine learning techniques with electronic health records, in which clinical terms (diagnosis codes, procedure codes and medication codes) recorded in general practice were analysed using a text mining scheme, while the prediction of the re-hospitalisation was treated as a problem of document classification in text mining.
2. Materials and Methods
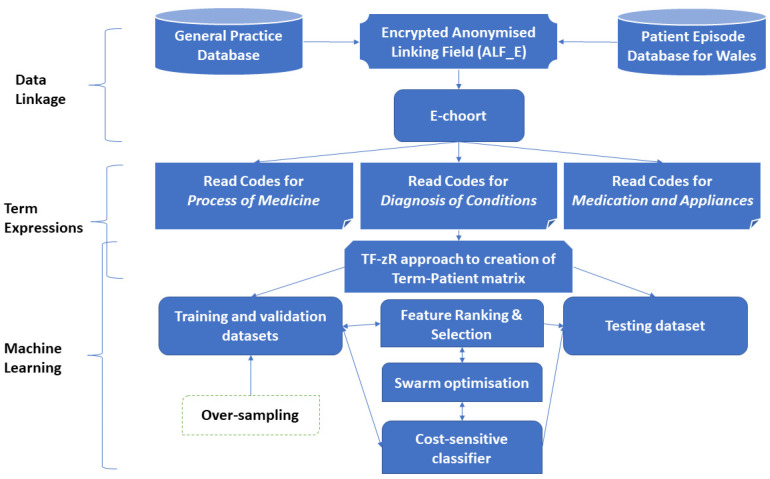
This study aims to identify key influential factors from a pool of demographic variables and clinical events recorded in primary care to predict the outcomes of campylobacteriosis patients admitted to hospital, classified as ‘readmission’ and ‘non-readmission’. Figure 1 illustrates the process of building the prediction model.
Figure 1.
The flow diagram of the process of building readmission prediction model.
2.1. Data Collection and Linkage
A GP database from the Abertawe Bro Morgannwg University (ABMU) Health Board area with Campylobacter infections between 1990 and 2015 was linked to the Patient Episode Database for Wales (PEDW) which records all episodes of inpatient and daily case activity in NHS Wales hospitals. The data linkage was conducted via the Secure Anonymised Information Linkage (SAIL) databank [40,41]. The SAIL databank brings together and links a wide range of person-based data from multiple sources relevant to health. SAIL utilises a range of measures to ensure that the data are anonymous and secure, and that they can be safely utilised for research within a robust information governance framework [41]. Each patient was given a unique Encrypted Anonymised Linking Field (ALF_E). Different databses of electronic health records hold inidviudal ALF_Es which indicate patient level data records. So, these different databases can be linked at individal level by the ALF_Es.
In this study, 12,747,826 rows of electronic health records for all patients with Campylobacter infections held in the GP database were linked with the hospital admission database by the ALF_E. The patients with Campylobacter infections, defined in terms of Read code (“A0473” for Campylobacter gastrointestinal tract infection) and in GP data and ICD 10 code (A045—Campylobacter enteritis in hospital admissions), were extracted and the date of first occurrence was selected. All admissions took place after an infection. The inclusion criteria for GP records were (i) the patient was alive at discharge and (ii) the patient was enrolled within the GP record for 12 months before the date of infection. For each patient, within 12 months after Campylobacter infection, readmission was defined as any admission taking place within 30 days following discharge from the previous admission. This readmission was regarded as a reference admission. If there was no 30-day readmission, the first admission was treated as a reference admission. In other words, for each patient, their GP records from 12 months before the infection to their reference admission were collected. If a patient was readmitted on the same day of discharge, it was counted as a single continuous admission. For patients who had multiple Campylobacter infections leading to different hospital admissions, some infections may lead to readmission, others did not. In this situation, each infection was treated separately, as they corresponded to different GP visit records. In this way, a total of 13,006 patients admitted to hospital with Campylobacter infection were obtained, 8.17% of which (1062) were readmissions. In other words, these 13,006 patients generated 12,747,826 rows of records in the GP databse.
Additional demographic variables included: deprivation status in terms of Townsend score (quintile); urban status as defined by the Office for National Statistics [42]—‘Urban > 10 k (Urban Settlements with greater than 10 k population)’, ‘Town and Fringe (located within the rural domain)’, ‘Village, Hamlet and Isolated Dwellings(located within the rural domain)’; age bands between GP event date and date of birth (0–5, 6–10, 11–15, 16–20, 21–25, 26–30, 31–35, 36–40, 41–45, 46–50, 51–55, 56–60, 60+). Thus, the initial variables were formed by gender, age groups and bands of deprivation, and medical records held electronically in general practice and in the hospital admission dataset.
The 13,006 hospital admitted patients were further randomly split into training (70% of the data), testing (15%) and validation (15%) data subsets for constructing machine learning models, selecting the optimal model and testing performance respectively.
2.2. Machine Learning Approach
This cohort study used machine learning methods to identify influential risk factors that are most predictive of readmission of Campylobacter infections from routine electronic health records. However, the linked dataset includes different health-related fields with a range of data structures, for example, age and deprivation fields are categorical values, while the majority of the factors are text terms from the GP database based on NHS Read Clinical Term system. The general practice system with 5-bytes provides around 83,000 clinical descriptive terms in hierarchical structure comprising five levels of detail, whilst each successive level offers more detail to a concept. This means, there are multiple codes for the same medication/diagnosis/procedure with a progressive level of detail. Such heterogeneous linked data presents methodological challenges for predictive analytics [43,44]. To address the challenges, we integrated machine learning and natural language processing (NLP) techniques to identify the most influential predictors associated with the readmission of Campylobacter infections from the large number of heterogeneous variables. A ‘bag of words’ (BoW) scheme [45] consisting of coded terms and other variables (words) was used to describe each patient, where the number of occurrences of each term was recorded. The prediction of readmission for each patient was thus treated as a problem of text classification. The proposed methodology is described below.
First, a Term-Patient matrix was created to represent each patient by the BoW of terms. Traditionally, the term frequency (how often a term occurs) was used to weight each term, for example, a blood pressure check may happen five times a year, a diagnosis may happen once. However, text mining studies have indicated that term frequency (TF) based classification methods often fails to effectively distinguish the individuals (patients here) [45]. Thus, in this paper, an approach called TF-zR (term frequency-zRelevance) technique was devecloped to evaluates how relevant a clinical term is to a patient in a cohort characterized by coded electronic health records (see Appendix A). The zR is a supervised term-weighting (STW) metric to assign a weight to a term based on relative frequencies of the term across different classes (i.e, readmission and non-readmission). The mechanism behind such a STW method is that the more relevant term should be the one with more concentrated frequency in one class (positive class: readmission/negative class: non-readmission) than the other class. Then the TF value and the STW metric were combined to represent each patient (see Appendix A). In this way, a quantitative digest of each patient represented by this TF-zR method in a Term-Patient matrix was obtained. This method is different from traditional unsupervised term-weighing methods which do not consider the impact of sample distributions across different classes.
For this Term-Patient matrix, each variable was then ranked using the information gain method [46] (see Appendix A) to examine its capacity of distinguishing readmission with non-readmission across all patients. Normally, the Read codes in general practice fall into the categories of “Process of Medicine” (PoM) (such as laboratory tests), “Diagnosis of Conditions” (DoC) and “Medication and Appliances” (MaA). The issue is that the PoM Read terms are frequent but carry less information, while the DoC and MaA terms, such as a diagnosis of diabetes, may occur once in a patient’s lifetime but are important and carry more information. Therefore, it is unsurprising that using a purely TF method, the PoM codes will suppress the impact of DoC and MaA, although the latter provide more meaningful clinical knowledge of patient’s health conditions in terms of diagnoses and treatments. To avoid the suppression of the PoM codes, in this study the Read codes in each category were assessed and ranked separately in terms of their capacity of distinguishing the outcomes. Then along with demographic variables, the pool of the selected codes from each category were used for constructing a classification model in the next phase.
Of the 13,006 patients admitted to hospital with Campylobacter infections, there were only 8.2% readmissions. Thus, this is an extremely imbalanced data problem. To address the problem, a cost-sensitive classification scheme [47,48] is used to provide different penalties of misclassifications of readmission and admission. Specifically, the cost of misclassifying readmission as admission is greater or more serious than misclassifying admission as readmission. Using particle swarm optimisation [49] and a weighted learning scheme, the model with the most influential predictive factors was then identified, offering the best potential of distinguishing those that were readmitted to hospital with those that were not. The read codes in the categories of DoC and MaA were given higher weighting than those in PoM. The final selected predictors were then validated with the independent unlabelled samples in a testing data subset.
The performance of the model was assessed in terms of sensitivity and specificity against 15% of the total database.
To further validate the performance of the identified clinical and demographic signals in predicting the hospitalization of Campylobacter infection, the over-sampling technique was used to adjust the class distribution of the trained data set for building machine learning models with the more balanced data set.
3. Results
This study utilised 12,747,826 health records of 13,006 patients admitted to hospital with Campylobacter infections between 1990 and 2015, while there were 1062 readmissions. So, this is a highly imbalanced data problem where the negative class has much more samples than the positive class. Table 1 shows a demographics table of Campylobacter infection admissions. Children aged 0–5 had the highest rates of hospital admissions, while patients aged between 46 and 55 had the highest rates of readmission. Children aged 6–15 had the fewest overall hospital admissions and readmissions. Due to a denser population, more people living in the urban areas had hospital admissions and readmissions than those living in town and fringe, or village, hamlet and isolated dwellings. Among patients in the 5th Townsend deprivation quintile (the most deprived), the rate of their re-admissions was 8.66%, higher than the rate of re-admissions (7.92%) among those in the 1st deprivation quintile (the most affluent). It is noted that although these statistics showed the overall impacts of demographic factors on hospital admissions, this does not mean that they are significant in predicting the readmissions as the predictors also depend on interactions between variables.
Table 1.
Demographics table of campylobacter admissions.
| Non-Readmission | Readmission | |
|---|---|---|
| Number of admitted patients | 11,944 | 1062 |
| Average age of admissions | 43.0 (SD = 22.1) | 51.8 (SD = 22.3) |
| Percentage of male admissions | 49.9% | 55.6% |
| Percentage of admissions in most affluent | 22.4% | 21.7% |
| Percentage of admission in most deprived | 17.6% | 18.7% |
| Percentage of admissions in rural domains * | 37.2% | 37.7% |
* Including “Town and Fringe (located within the rural domain)”, and “Village, Hamlet and Isolated Dwellings (located within the rural domain)”.
There were 23 categorical demographic variables generated on gender, age groups, deprivation and urbanicity. In addition, 17,483 clinical events were classified by read codes into the categories of PoM (8206 codes), DoC (3702 codes) and MaA (5575 codes). In this way, the linked dataset generated initial data with 17,506 variables. These variables were taken forward to identify the most influential predictors associated with the Campylobacter readmission.
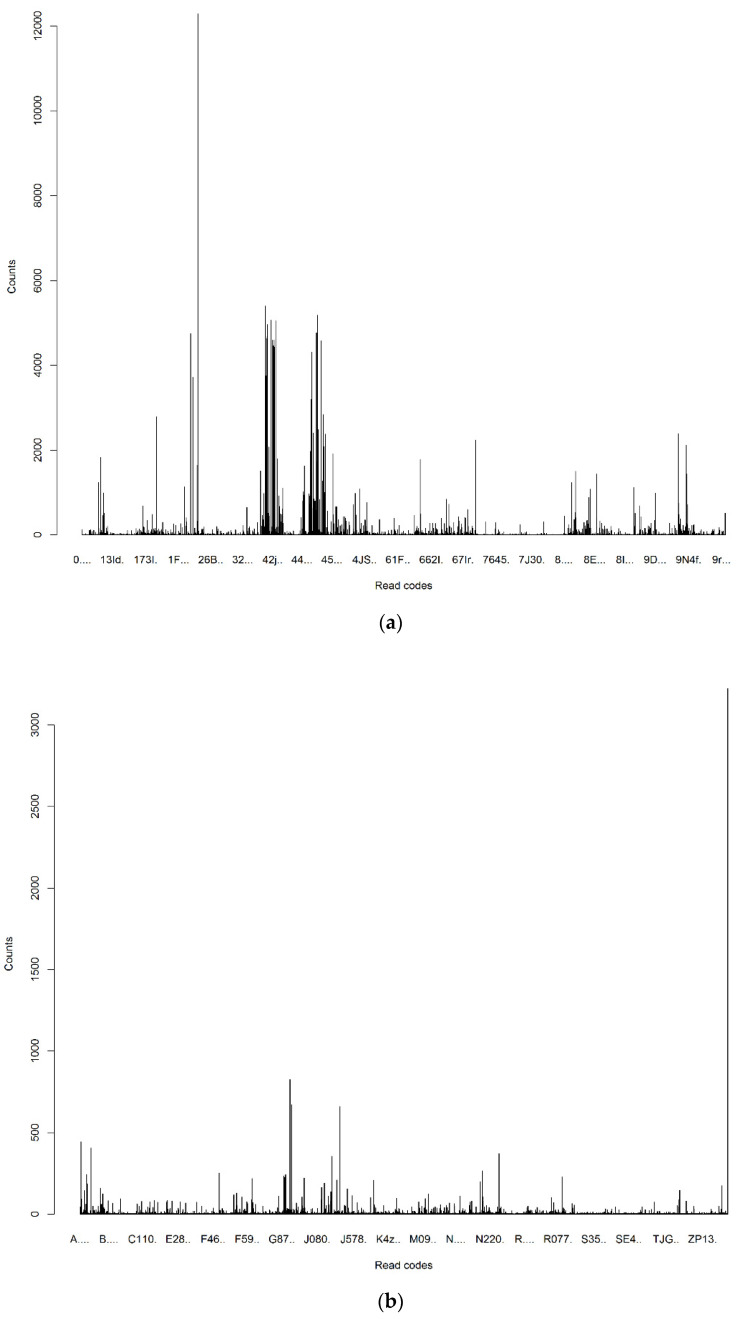
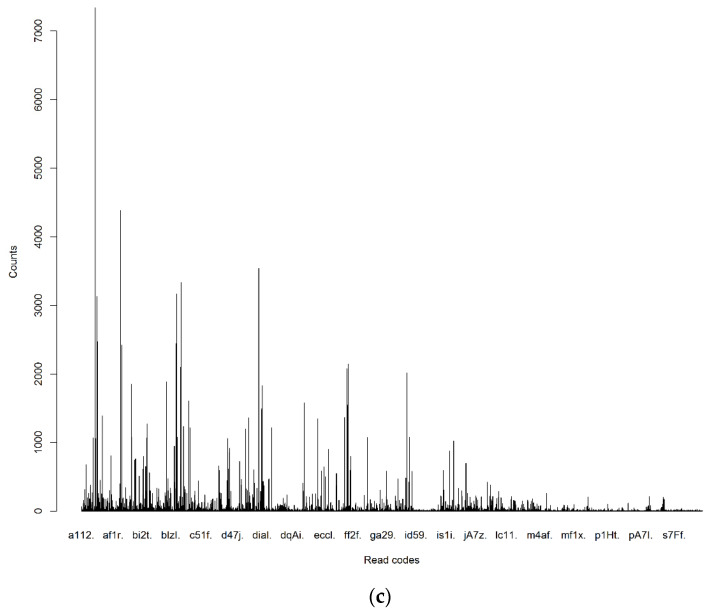
These clinical terms demonstrated a great disparity in term of TF across the different categories of PoM, DoC and MaA (Figure 2). The wide range of frequency variations exactly characterises real clinical practices, in which the PoM events often occur much more frequently than those of the DoC and MaA. It is noted that these frequency measurements did not take into account their relevance to the re-admission. Differently, the supervised term-weighting method—zR metric took into account the contributions of a term across different classes (re-admission and non-readmission) to generate the relevance measurements which show much better proportionality for different categories of read codes (Figure 3).
Figure 2.
Frequencies of read codes in different categories. (a) Frequencies of read codes in PoM; (b) frequencies of read codes in DoC; and (c) frequencies of read codes in MaA.
Figure 3.
The supervised term-weighting metric.
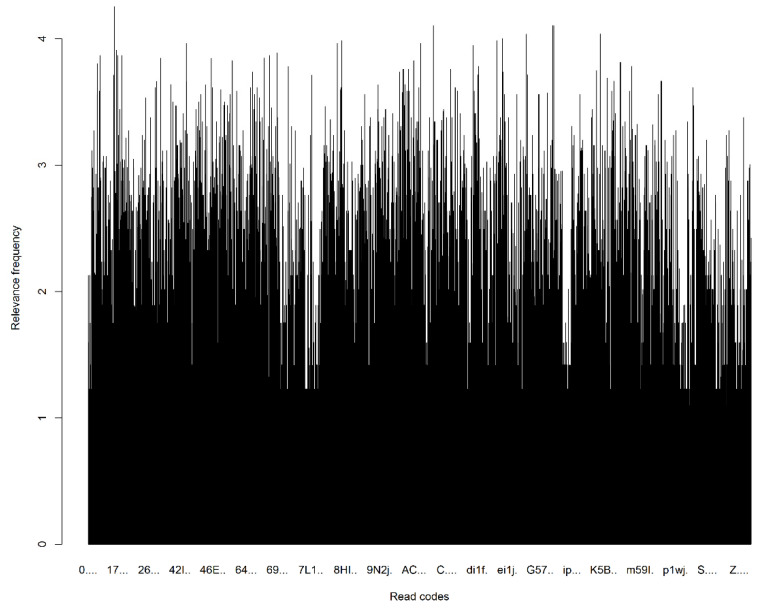
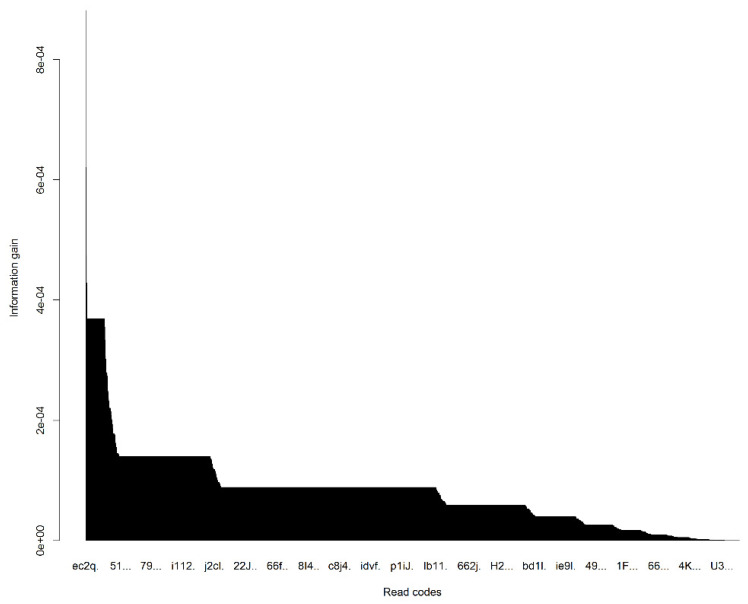
After the TF-zR method generated the Term-Patient matrix, applying information-gain to this Term-Patient matrix allowed the generation of a feature ranking metric for each variable which assessed the contribution of each variable in distinguishing between the readmission and non-readmission (Figure 4). Information-gain is normally used to determine the influential features/attributes/variables that render maximum information about a class. So in terms of information-gain, the top read codes were selected from each category of PoM, DoC, and MaA. Then together with 23 categorical demographic variables, a data space with total 623 variables was generated. From these 623 variables, the swarm optimization with weighted subset learning and cost-sensitive decision tree classifier identified the 33 optimal features that offered the best potential of predicting the hospitalisation of Campylobacter infections (Table 2). The 33 most predictive variables included an age group (ages 21~25 associated with non-readmission), gender, Townsend deprivation quintiles (bands 1 and 4), comorbidities (12 diagnostic codes), medications (11 prescription codes) and procedures (6 codes). Applying to an independent test dataset, the classifier with the 33 influential predictors performed significantly above chance to predict readmissions with sensitivity 0.73 (95% confidence interval (0.71, 0.75)), and specificity of 0.54 (95% confidence interval (0.53, 0.55)). Cystitis, paracetamol and codeine use, age (21 to 25), and heliclear triple pack, have turned up to be very efficient in classifying the outcomes of Campylobacter infections, where patients with these conditions had lower risk of readmission.
Figure 4.
The ordered information gains of variables in distinguishing between the readmissions and admissions.
Table 2.
The 33 key predictive variables identified.
| Variable Name | Description |
|---|---|
| Male | Gender |
| AGE_A21–25 | Age group: 21~25 years old |
| TOWSEND_Q1 | Townsend quintile band 1 |
| TOWSEND_Q4 | Townsend quintile band 4 |
| 67E.. | Foreign travel advice |
| 8B311 | Medication given |
| 67H.. | Lifestyle counselling |
| 9.... | Administration |
| 8H7.. | Other referral |
| 7G300 | Excision of nail bed |
| R0901 | Abdominal colic |
| K3108 | Breast infection |
| S89z. | Other open wounds NOS |
| K15.. | Cystitis |
| H170. | Allergic rhinitis due to pollens |
| H037. | Recurrent acute tonsillitis |
| A53.. | Herpes zoster |
| N05.. | Osteoarthritis and allied disorders |
| Ayu03 | Salmonella infection, unspecified |
| F501. | Infective otitis externa |
| F504. | Impacted cerumen (wax in ear) |
| N2410 | Myalgia unspecified |
| dian. | Paracetamol+codeine phosphate 500 mg/30 mg tablets |
| ka91. | Celluvisc 1% single-use eye drops |
| da7Z. | Venaxx XL 75 mg m/r capsules |
| dher. | Prochlorperazine 5 mg tablets |
| c13M. | Ventolin 200 micrograms Accuhaler |
| bs18. | Warfarin sodium 3 mg tablets |
| a6g2. | Heliclear triple pack |
| k3g1. | Fusidic acid 1% eye drops |
| e91E. | Erythromycin 125 mg/5mL sugar free suspension |
| c61z. | Beclometasone dipropionate 100 micrograms inhaler |
| da61. | Paroxetine 20 mg tablets |
To further validate the performance of the 33 predictors in this imbalanced data problem, we applied to an independent balanced dataset produced by the over-sampling technique, the 33 predictors predicted readmissions with sensitivity 0.91 (95% confidence interval (0.90, 0.913)), and specificity of 0.54 (95% confidence interval (0.52, 0.565)).
In order to demonstrate the efficiency of how the developed modelling approach tackles the issue of imbalanced classes in medical data, we further compared with logistic regression, a traditional modelling approach to readmission prediciton. Using the raw data with same training and testing datasubsets as our model, the logistic regression model with the same 33 influential predictors offered prediction of readmissions on testing datasubset with sensitivity 0.0298, and specificity 0.974. Clearly logistic regression approach cannot tackle the imblanced data issue. Then using the same oversampled data as our model, the logistic regression model with the same predictors significantly improved the prediciton performance with sensitivity 0.8196, and specificity 0.5253, but still compared unfavourably with our developed modelling approach.
4. Discussion
By integrating text mining, feature selection, and machine learning, our study provides a novel methodology for building a predictive model capable of automatically identifying influential risk factors from primary care records with good predictive performance.
Using this methodology, we identified 33 most predictive variables of age, gender, deprivation, comorbidities, medication and medical procedure. Analysis of the clinical implication of these variables revealed that most of the predictors of readmission relate to comorbidities of recurrent minor illness (e.g., recurrent tonsillitis, non- healing open wounds, ingrown toenails, impacted cerumen (wax in ear)). Males with a history of recurrent minor illnesses are at increased risk of readmission, indicating that patient profiling could help with support at discharge and more targeted use of antibiotics. Each such condition may not be directly important in the outcomes of Campylobacter infection, but combined, they give a profile of individuals that have a history of chronic minor illness and may be less well equipped to take care of themselves. These ‘at risk’ patients may require additional support at discharge to reduce readmission risk. Such support could include enhanced patient education during discharge, conducting follow-up visits or medication reconciliation [50]. These ‘at risk’ patients contrast with the profile of patients least likely to be re- admitted, typically younger females with a history of seeking treatment for bacterial infections and taking medication for illness. Cystitis has emerged in our study as the most effective variable in predicting no readmission for the campylobacter. Campylobacter infection patients with cystitis had a lower risk of readmission once they were discharged. Perhaps this signals the profile of the person with the least chance of readmission is more likely female and reports bacterial infections. The predictions identified in this study therefore provide a justification for using comorbidity as an indicator in the LACE index as assessed by Charlson comorbidity index to predict readmissions.
There are several advantages to the machine learning approach employed in this study. First, it works efficiently with a large and very high dimensional dataset for developing predictive models, which allows the predictive models to avoid the challenges of dimensionality [51]. Second, most machine learning algorithms fail to work with imbalanced datasets due to subject to a frequency bias in which more emphasis is placed on learning data observations with more occurrences. Our methodology integrates a cost-sensitive learning scheme to effectively identify the influential factors. Third, different from classic unsupervised term-weighting methods including frequency, our methodology used a supervising term weighting method to generate patient representations by considering the disparity of term distributions across data classes. This provides a foundation for identifying predictive factors with good capacity for distinguishing the outcomes of health conditions. Fourth, different from existing readmission predictive models without considering model generalisation performance during construction, our methodology centred on generalisation performance of the constructed model by adopting optimal model selection scheme and using independent data subsets for the different purposes of model constructions, hyper-parameter identification and model evaluation.
However, the proposed methodology has some limitations. It requires a high computing load to build a robust prediction model, and extensive cross-validation to evaluate the potential predictors identified. Furthermore, there are variations unexplained by this prediction model and additional information about the infections (strain, severity) and the symptoms are needed to improve the prediction performance.
This study was developed with a focus on campylobacter infection related admissions, future studies should explore the usability/fittingness of such machine learning and state-of-the-art methods of natural language processing, such as transformer models such as BERT [52], BioBERT [53], for word representations in readmission prediction.
5. Conclusions
By identifying predictors of readmission for campylobacter infections in primary care setting, we conclude that patients with a history of recurrent minor preventable illnesses may need greater support upon discharge from hospital to prevent readmission. This is important for reducing the burden on secondary care services that readmission represents and in improving care for patients. The effectiveness of this approach demonstrates the potential in machine learning methods in adopting personalised medicine to meet the goal of reducing preventable readmissions.
Appendix A
Appendix A.1. zR: Supervised Term Weighting Metric
As each patient is described by a series of read codes and additional categorical demographic variables, these codes and variables can be treated as terms as carried out in the text mining. In this way, mining electronic health records in primary care and secondary care settings corresponds to the task of text categorization which automatically classify textual documents into different predefined semantic classes. Certainly, different terms in a document (i.e., a patient record here) often make different contributions to the semantics of the document. Term weighting is an important step to assess the importance of terms in classifying unlabelled natural language documents.
In this study, we consider a term weighting scheme by which the more important term should be the one with more concentrated occurrence in one class (positive class/negative class) than the other class (negative class/positive class). To formalize this idea, we use a, b, c, and d to denote the number of different patient records, as listed below:
a = the number of patient records in the Class 1 that contains the term t.
b = the number of patient records in the Class 1 that does not contain the term t.
c = the number of patient records in the Class 0 that contains the term t.
d = the number of patient records in the in Class 0 that does not contain the term t.
where the Class 1 is the category of patients re-admitted to hospital, the Class 0 is the category of patients not re-admitted to hospital. Then we can have a contingence table of term t across the two classes of patients (see Table A1).
Table A1.
Contingence table of term t across the two classes of patients.
| Term t | |||
|---|---|---|---|
| Yes | No | Sum | |
| Class 1 | a | b | |
| Class 0 | c | d | |
| Sum | N | ||
where
| N.1 = a + c, N.0 = b + d, N1. = a + b, N0. = c + d, N = a + b + c + d. |
Then we first have the information gain of the term t defined as
where the base of this logarithmic operation (log) is 2. Information gain computes the he impurity in class elements by following the concept of entropy while aiming at decreasing the level of entropy.
We propose a supervised term weighting-based relevance metric, zRelevance (zR), to describe the situation of the term t which occurs more often in one class than in the other class.
where and are the relative frequencies (probabilities) of term t occurring across the patient records of different classes:
The weight for the term t is finally assigned as
Appendix A.2. TF-zR Approach to Creation of Term-Patient Matrix
Assuming there be m patients and n terms in the database (i.e., each patient is characterized by these n terms). Let represent the frequency of the term in the records of the patient . Then the Term-Patient (TP) matrix used to represent the relationships between clinical terms and outcomes can be generated as
where is the zR relevence metric defined above.
Author Contributions
S.B. and S.-M.Z. conceived the study. M.A.R. collected data in the cohort. S.-M.Z. conceived the data analysis methodology and conducted the experiments. S.-M.Z., S.B. and R.A.L. analyzed the results. R.A.L. advised on study design. A.H. reviewed the outcomes. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Health Data Research UK (NIWA1). This analysis was also undertaken with the support of the National Centre for Population Health and Wellbeing Research (NCPHWR) via Health and Care Research Wales (grant ref. CA02).
Institutional Review Board Statement
The data records held within the SAIL databank have been anonymised and obtained with the permission of Caldicott Guardian/Data Protection Officer; therefore, the National Research Ethics Service (NRES) has stated that no ethical review is required. Approval was obtained from the Information Governance Review Panel (IGRP) to use the SAIL System for this research question.
Informed Consent Statement
Patient consent was waived due to the electronic health records used in this study have been anonymized.
Data Availability Statement
Data are available from the SAIL (Secure Anonymised Information Linkage) Databank for researchers who meet the criteria for access to confidential data.
Conflicts of Interest
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ruiz-Palacios G.M. The Health Burden of Campylobacter Infection and the Impact of Antimicrobial Resistance: Playing Chicken. Clin. Infect. Dis. 2007;44:701–703. doi: 10.1086/509936. [DOI] [PubMed] [Google Scholar]
- 2.Eberle K.N., Kiess A.S. Phenotypic and genotypic methods for typing Campylobacter jejuni and Campylobacter coli in poultry. Poult. Sci. 2012;91:255–264. doi: 10.3382/ps.2011-01414. [DOI] [PubMed] [Google Scholar]
- 3.Campylobacter Attorney Campylobacter Costs $1.3 Billion a Year in Hospitalization and Medical Costs. (n.d.-a) [(accessed on 12 February 2017)]. Available online: http://www.campylobacterblog.com/campylobacter-information/campylobacter-costs-13-billion-a-year-in-hospitalization-and-medical-costs/
- 4.Food Standards Agency Acting on Campylobacter Together. [(accessed on 21 March 2017)]; Available online: https://www.food.gov.uk/science/microbiology/campylobacterevidenceprogramme.
- 5.Adak G.K., Cowden J.M., Nicholas S., Evans H.S. The Public Health Laboratory Service national case-control study of primary indigenous sporadic cases of campylobacter infection. Epidemiol. Infect. 1995;115:15–22. doi: 10.1017/S0950268800058076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman C.R., Hoekstra R.M., Samuel M., Marcus R., Bender J., Shiferaw B., Reddy S., Ahuja S.D., Helfrick D.L., Hardnett F., et al. Risk Factors for SporadicCampylobacterInfection in the United States: A Case-Control Study in FoodNet Sites. Clin. Infect. Dis. 2004;38((Suppl. S3)):S285–S296. doi: 10.1086/381598. [DOI] [PubMed] [Google Scholar]
- 7.Gallay A., Bousquet V., Siret V., Prouzet-Mauleon V., De Valk H., Vaillant V., Simon F., Le Strat Y., Mégraud F., Desenclos J. Risk Factors for Acquiring SporadicCampylobacterInfection in France: Results from a National Case-Control Study. J. Infect. Dis. 2008;197:1477–1484. doi: 10.1086/587644. [DOI] [PubMed] [Google Scholar]
- 8.Potter R.C., Kaneene J.B., Hall W.N. Risk Factors for Sporadic Campylobacter jejuni Infections in Rural Michigan: A Prospective Case–Control Study. Am. J. Public Health. 2003;93:2118–2123. doi: 10.2105/AJPH.93.12.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skirrow M., Blaser M. Campylobacter jejuni. In: Blaser M.J., Smith P.D., Ravdin J.I., Greenberg H.B., Guerrant R.L., editors. Infections of the Gastrointestinal Tract. Lippincott Williams and Wilkins; Philadelphia, PA, USA: 2002. p. 719. [Google Scholar]
- 10.Kaakoush N.O., Mitchell H.M., Man S.M. Role of Emerging Campylobacter Species in Inflammatory Bowel Diseases. Inflamm. Bowel Dis. 2014;20:2189–2197. doi: 10.1097/MIB.0000000000000074. [DOI] [PubMed] [Google Scholar]
- 11.Gradel K.O., Nielsen H.L., Schønheyder H.C., Ejlertsen T., Kristensen B., Nielsen H. Increased Short- and Long-Term Risk of Inflammatory Bowel Disease After Salmonella or Campylobacter Gastroenteritis. Gastroenterology. 2009;137:495–501. doi: 10.1053/j.gastro.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Jess T., Simonsen J., Nielsen N.M., Jørgensen K.T., Bager P., Ethelberg S., Frisch M. Enteric Salmonella or Campylobacter infections and the risk of inflammatory bowel disease. Gut. 2010;60:318–324. doi: 10.1136/gut.2010.223396. [DOI] [PubMed] [Google Scholar]
- 13.Locht H. Comparison of rheumatological and gastrointestinal symptoms after infection with Campylobacter jejuni/coli and enterotoxigenic Escherichia coli. Ann. Rheum. Dis. 2002;61:448–452. doi: 10.1136/ard.61.5.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hannu T., Mattila L., Rautelin H., Pelkonen P., Lahdenne P., Siitonen A., Leirisalo-Repo M. Campylobacter-triggered reactive arthritis: A population-based study. Rheumatology. 2002;41:312–318. doi: 10.1093/rheumatology/41.3.312. [DOI] [PubMed] [Google Scholar]
- 15.Fischbach L.A., Nordenstedt H., Kramer J.R., Gandhi S., Dick-Onuoha S., Lewis A., El-Serag H.B. The Association Between Barrett’s Esophagus and Helicobacter pylori Infection: A Meta-Analysis. Helicobacter. 2012;17:163–175. doi: 10.1111/j.1523-5378.2011.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falk G.W. Practical Manual of Gastroesophageal Reflux Disease. John Wiley & Sons; Hoboken, NJ, USA: 2013. Barrett’s Esophagus: Diagnosis and Surveillance; pp. 287–309. [DOI] [Google Scholar]
- 17.Poropatich K.O., Walker C.L.F., Black R. Quantifying the Association between Campylobacter Infection and Guillain-Barré Syndrome: A Systematic Review. J. Health Popul. Nutr. 2010;28:545–552. doi: 10.3329/jhpn.v28i6.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drenthen J., Yuki N., Meulstee J., Maathuis E.M., Van Doorn P.A., Visser G.H., Blok J., Jacobs B.C. Guillain-Barre syndrome subtypes related to Campylobacter infection. J. Neurol. Neurosurg. Psychiatry. 2011;82:300–305. doi: 10.1136/jnnp.2010.226639. [DOI] [PubMed] [Google Scholar]
- 19.Denneberg T., Friedberg M., Holmberg L., Mathiasen C., Nilsson K.O., Takolander R., Walder M. Combined Plasmapheresis and Hemodialysis Treatment for Severe Hemolytic-Uremic Syndrome Following Campylobacter Colitis. Acta Paediatr. 1982;71:243–245. doi: 10.1111/j.1651-2227.1982.tb09408.x. [DOI] [PubMed] [Google Scholar]
- 20.Gölz G., Rosner B., Hofreuter D., Josenhans C., Kreienbrock L., Löwenstein A., Schielke A., Stark K., Suerbaum S., Wieler L.H., et al. Relevance of Campylobacter to public health—The need for a One Health approach. Int. J. Med. Microbiol. 2014;304:817–823. doi: 10.1016/j.ijmm.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 21.Esan O.B., Perera R., McCarthy N., Violato M., Fanshawe T.R. Incidence, risk factors, and health service burden of sequelae of campylobacter and non-typhoidal salmonella infections in England, 2000–2015: A retrospective cohort study using linked electronic health records. J. Infect. 2020;81:221–230. doi: 10.1016/j.jinf.2020.05.027. [DOI] [PubMed] [Google Scholar]
- 22.Brophy S., Jones K., Rahman M.A., Zhou S.-M., John A., Atkinson M., Francis N., Lyons R.A., Dunstan F. Incidence of Campylobacter and Salmonella Infections Following First Prescription for PPI: A Cohort Study Using Routine Data. Am. J. Gastroenterol. 2013;108:1094–1100. doi: 10.1038/ajg.2013.30. [DOI] [PubMed] [Google Scholar]
- 23.Charlett A., Cowden J.M., Frost J.A., Gillespie I.A., Millward J., Neal K.R., O’Brien S.J., Painter M.J., Syed Q., Tompkins D. Ethnicity and Campylobacter infection: A population-based questionnaire survey. J. Infect. 2003;47:210–216. doi: 10.1016/S0163-4453(03)00072-0. [DOI] [PubMed] [Google Scholar]
- 24.Gillespie I.A., O’Brien S.J., Frost J.A., Adak G.K., Horby P., Swan A.V., Painter M.J., Neal K.R. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: A tool for generating hypotheses. Emerg. Infect. Dis. 2002;8:937–942. doi: 10.3201/eid0809.010817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moffatt C.R.M., Kennedy K.J., Selvey L., Kirk M.D. Campylobacter-associated hospitalisations in an Australian provincial setting. BMC Infect. Dis. 2021;21:1–10. doi: 10.1186/s12879-020-05694-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vest J.R., Gamm L.D., Oxford B.A., Gonzalez M.I., Slawson K.M. Determinants of preventable readmissions in the United States: A systematic review. Implement. Sci. 2010;5:88. doi: 10.1186/1748-5908-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris J. Emergency Readmissions: Trends in Emergency Readmissions to Hospital in England. Nuffield Trust. [(accessed on 20 September 2018)]. Available online: http://www.qualitywatch.org.uk/blog/emergency-readmissions-trends-emergency-readmissions-hospital-england#.
- 28.Scallan Walter E.J., Crim S.M., Bruce B.B., Griffin P.M. Incidence of Campylobacter-Associated Guillain-Barré Syndrome Estimated from Health Insurance Data. Foodborne Pathog. Dis. 2020;17:23–28. doi: 10.1089/fpd.2019.2652. [DOI] [PubMed] [Google Scholar]
- 29.Cotter P.E., Bhalla V.K., Wallis S.J., Biram R.W.S. Predicting readmissions: Poor performance of the LACE index in an older UK population. Age Ageing. 2012;41:784–789. doi: 10.1093/ageing/afs073. [DOI] [PubMed] [Google Scholar]
- 30.Van Walraven C., Dhalla I.A., Bell C., Etchells E., Stiell I.G., Zarnke K., Austin P.C., Forster A.J. Derivation and validation of an index to predict early death or unplanned readmission after discharge from hospital to the community. Can. Med. Assoc. J. 2010;182:551–557. doi: 10.1503/cmaj.091117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Walraven C., Wong J., Hawken S., Forster A.J. Comparing methods to calculate hospital-specific rates of early death or urgent readmission. Can. Med. Assoc. J. 2012;184:E810–E817. doi: 10.1503/cmaj.120801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Billings J., Dixon J., Mijanovich T., Wennberg D. Case finding for patients at risk of readmission to hospital: Development of algorithm to identify high risk patients. BMJ. 2006;333:327. doi: 10.1136/bmj.38870.657917.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Department of Health . Payment by Results Guidance for 2012–2013. Gateway Reference 17250. Department of Health; London, UK: 2012. [Google Scholar]
- 34.Centers for Medicare and Medicaid Services Readmissions Reduction Program (HRRP) [(accessed on 20 September 2017)]; Available online: https://www.cms.gov/Medicare/Medicare-Fee-for-Service-Payment/AcuteInpatientPPS/Readmissions-Reduction-Program.html.
- 35.What Do the Numbers Say about Emergency Readmissions to Hospital? Health Watch. [(accessed on 20 September 2018)]. Available online: https://www.healthwatch.co.uk/sites/healthwatch.co.uk/files/20171025_what_do_the_numbers_say_about_emergency_readmissions_final_0.pdf.
- 36.Eberhart-Phillips J., Walker N., Garrett N., Bell D., Sinclair D., Rainger W., Bates M. Campylobacteriosis in New Zealand: Results of a case-control study. J. Epidemiol. Community Health. 1997;51:686–691. doi: 10.1136/jech.51.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodrigues L.C., Cowden J.M., Wheeler J.G., Sethi D., Wall P.G., Cumberland P., Tompkins D.S., Hudson M.J., Roberts J.A., Roderick P.J. The study of infectious intestinal disease in England: Risk factors for cases of infectious intestinal disease with Campylobacter jejuni infection. Epidemiol. Infect. 2000;127:185–193. doi: 10.1017/S0950268801006057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lineback C.M., Garg R., Oh E., Naidech A.M., Holl J.L., Prabhakaran S. Prediction of 30-Day Readmission After Stroke Using Machine Learning and Natural Language Processing. Front. Neurol. 2021;12:649521. doi: 10.3389/fneur.2021.649521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arnaud E., Elbattah M., Gignon M., Dequen G. Deep Learning to Predict Hospitalization at Triage: Integration of Structured Data and Unstructured Text; Proceedings of the 2020 IEEE International Conference on Big Data, (Big Data); Atlanta, GA, USA. 10–13 December 2020; [DOI] [Google Scholar]
- 40.Ford D.V., Jones K.H., Verplancke J.-P., Lyons R.A., John G., Brown G., Brooks C.J., Thompson S., Bodger O., Couch T., et al. The SAIL Databank: Building a national architecture for e-health research and evaluation. BMC Health Serv. Res. 2009;9:157. doi: 10.1186/1472-6963-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyons R.A., Jones K.H., John G., Brooks C.J., Verplancke J.-P., Ford D.V., Brown G., Leake K. The SAIL databank: Linking multiple health and social care datasets. BMC Med. Inform. Decis. Mak. 2009;9:3. doi: 10.1186/1472-6947-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.ONS Rural and Urban Area Definition Metadata. [(accessed on 18 November 2016)]; Available online: https://www.ons.gov.uk.
- 43.Zhou S.-M., Fernandez-Gutierrez F., Kennedy J., Cooksey R., Atkinson M., Denaxas S., Siebert S., Dixon W., O’Neill T.W., Choy E., et al. Defining Disease Phenotypes in Primary Care Electronic Health Records by a Machine Learning Approach: A Case Study in Identifying Rheumatoid Arthritis. PLoS ONE. 2016;11:e0154515. doi: 10.1371/journal.pone.0154515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou S.-M., Lyons R.A., Bodger O.G., John A., Brunt H., Jones K., Gravenor M.B., Brophy S. Local Modelling Techniques for Assessing Micro-Level Impacts of Risk Factors in Complex Data: Understanding Health and Socioeconomic Inequalities in Childhood Educational Attainments. PLoS ONE. 2014;9:e113592. doi: 10.1371/journal.pone.0113592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feldman R., Sanger J. The text mining handbook: Advanced approaches in analyzing unstructured data. Imagine. 2007;34:410. doi: 10.1179/1465312512Z.00000000017. [DOI] [Google Scholar]
- 46.Zhou S.-M., Rahman M.A., Atkinson M., Brophy S. Mining textual data from primary healthcare records: Automatic identification of patient phenotype cohorts; Proceedings of the 2014 International Joint Conference on Neural Networks (IJCNN); Beijing, China. 6–11 July 2014; pp. 3621–3627. [DOI] [Google Scholar]
- 47.Lu H., Xu Y., Ye M., Yan K., Gao Z., Jin Q. Learning misclassification costs for imbalanced classification on gene expression data. BMC Bioinform. 2019;20:681. doi: 10.1186/s12859-019-3255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Witten I.H., Frank E., Hall M. Complementary Literature None. 3rd ed. Elsevier; New York, NY, USA: 2011. Data Mining: Practical Machine Learning Tools and Techniques. [Google Scholar]
- 49.Zhou S.-M., Lyons R.A., Bodger O., Demmler J.C., Atkinson M.D. SVM with entropy regularization and particle swarm optimization for identifying children’s health and socioeconomic determinants of education attainments using linked datasets; Proceedings of the International Joint Conference on Neural Networks; Barcelona, Spain. 18–23 July 2010; [DOI] [Google Scholar]
- 50.Koehler B.E., Richter K.M., Youngblood L., Cohen B.A., Prengler I.D., Cheng D., Masica A.L. Reduction of 30-day postdischarge hospital readmission or emergency department (ED) visit rates in high-risk elderly medical patients through delivery of a targeted care bundle. J. Hosp. Med. 2009;4:211–218. doi: 10.1002/jhm.427. [DOI] [PubMed] [Google Scholar]
- 51.Zhou S.-M., Gan J. Constructing L2-SVM-Based Fuzzy Classifiers in High-Dimensional Space With Automatic Model Selection and Fuzzy Rule Ranking. IEEE Trans. Fuzzy Syst. 2007;15:398–409. doi: 10.1109/TFUZZ.2006.882464. [DOI] [Google Scholar]
- 52.Devlin J., Chang M.W., Lee K., Toutanova K. BERT: Pre-training of deep bidirectional transformers for language understanding; Proceedings of the NAACL HLT 2019—2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; Minneapolis, MN, USA. 2–7 June 2019. [Google Scholar]
- 53.Lee J., Yoon W., Kim S., Kim D., Kim S., So C.H., Kang J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36:1234–1240. doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available from the SAIL (Secure Anonymised Information Linkage) Databank for researchers who meet the criteria for access to confidential data.