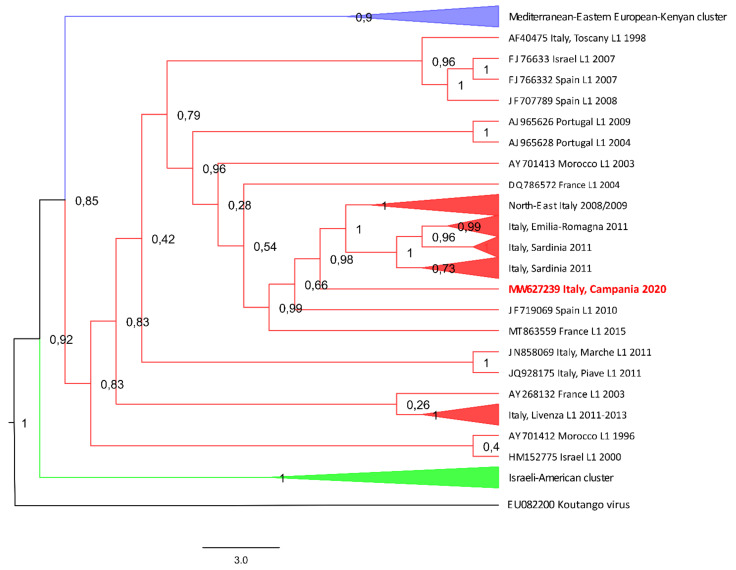
Figure 3.
Maximum likelihood phylogenetic tree of the WNV complete and partial genome sequences analyzed in this study. A detailed version of the phylogenetic tree is shown, showing all the Western Mediterranean WNV sequences. GenBank accession numbers are indicated for each strain, with country, lineage, and year of isolation. The WNV-L1 strain TE_362447_2020, obtained from the goshawk found in the Campania region in October 2020, is highlighted in red. The Koutango virus strain EU082200 has been chosen as outgroup. The tree with the highest log-likelihood is shown. The Bayesian phylogenetic analysis was performed through Bayesian Inference using a general time-reversible with gamma-distributed rate variation, a gamma category count of 4, and an invariant sites model (GTR + Γ + I). The evolutionary distances were computed using the optimal GTR + Γ + I model, with 2000 Γ-rate categories and 5000 bootstrap replications using the Shimodaira–Hasegawa (SH) test. The percentage of successful bootstrap replicate (n5000) is indicated at nodes. A Bayesian MCMC approach using BEAST with JRE v2.6.3 was employed. Ten independent MCMC runs with up to 100 million generations were performed to ensure the convergence of estimates. Tracer v.1.7.1 was used to ensure convergence during MCMC by reaching effective sample sizes greater than 100. Trees were summarized in a maximum clade-credibility tree with common ancestor heights after a 10% burn-in using TreeAnnotator v2.6.3.