Abstract
Marine sponges are prolific sources of bioactive natural products, several of which are produced by bacteria symbiotically associated with the sponge host. Bacteria-derived natural products, and the specialized bacterial symbionts that synthesize them, are not shared among phylogenetically distant sponge hosts. This is in contrast to non-symbiotic culturable bacteria in which the conservation of natural products and natural product biosynthetic gene clusters (BGCs) is well established. Here, we demonstrate the widespread conservation of a BGC encoding a cryptic ribosomally synthesized and post-translationally modified peptide (RiPP) in microbiomes of phylogenetically and geographically dispersed sponges from the Pacific and Atlantic oceans. Detection of this BGC was enabled by mining for halogenating enzymes in sponge metagenomes, which, in turn, allowed for the description of a broad-spectrum regiospecific peptidyl tryptophan-6-brominase which possessed no chlorination activity. In addition, we demonstrate the cyclodehydrative installation of azoline heterocycles in proteusin RiPPs. This is the first demonstration of halogenation and cyclodehydration for proteusin RiPPs and the enzymes catalyzing these transformations were found to competently interact with other previously described proteusin substrate peptides. Within a sponge microbiome, many different generalized bacterial taxa harbored this BGC with often more than 50 copies of the BGC detected in individual sponge metagenomes. Moreover, the BGC was found in all sponges queried that possess high diversity microbiomes but it was not detected in other marine invertebrate microbiomes. These data shed light on conservation of cryptic natural product biosynthetic potential in marine sponges that was not detected by traditional natural product-to-BGC (meta)genome mining.
Keywords: natural products, sponges, microbiome, metagenomics, RiPP, bromination, biosynthesis
Graphical Abstract
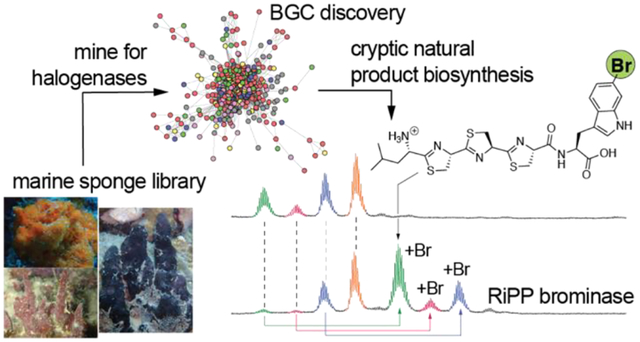
Introduction
Highly specialized small organic molecules called natural products are the universal language of symbiont-host interactions. Often, a symbiont generates a natural product that offers the host a survival and competitive advantage.1 This leads to symbiont specialization, such as by drastic genome reduction and/or by becoming a natural product superproducer by accumulating the genetic potential to generate numerous different bioactive natural products. The host, in turn, fulfills primary metabolic demands of the symbiont, which is likely why natural product producing symbionts resist laboratory cultivation. Examples of such natural product mediated symbioses are widespread in the nutrient poor benthic marine environment.2 Benthic marine invertebrates that harbor rich and diverse microbiomes, such as sponges,3–4 are engines for marine natural product discovery.5 Several sponge-derived natural products are produced by members of the bacterial microbiome that is symbiotically associated with the eukaryotic sponge host.2,6 Efforts to discover the biogenetic routes for sponge holobiont-derived natural products have been initiated by examining the chemical structures of the natural products isolated from sponges. Subsequently, a potential biosynthetic scheme to the desired natural product is postulated, with the resulting hypothetical BGCs serving as templates to mine the sponge metagenomic data. This natural product→BGC approach has connected several sponge-derived natural products to their respective BGCs, led to identification of the physiological producers from among the microbial milieu of the sponge holobiont, and illuminated microbiological and enzymological ingenuities. However, as this approach does not involve an untargeted mining of sponge microbiomes, an appreciation for the diversity and conservation of natural product BGCs present in marine sponge microbiomes has been scarce. As was recently illustrated for natural product geosmin, the BGC for which is present in nearly every streptomycete genome, conserved BGCs furnish natural products that fulfill primary ecological functions.7 While natural products in sponges are broadly postulated to serve as chemical defenses against herbivory and predation,8–9 it remains unknown whether microbiomes of different sponges share natural product BGCs, or whether certain class(es) of natural products are prioritized for conservation in sponge holobionts. Furthermore, access to cryptic natural products, the biosynthetic potential for which remains locked away in sponge microbiomes, has not been realized.
Sponges have been one of the most prolific sources of natural products and sponge-derived natural products extend across all known natural product chemical classes.5 However, it is instructive to observe that the inventory of ribosomally synthesized and post-translationally modified peptide (RiPP) natural products from marine sponges is small. The biogenetic basis of a solitary group of sponge-derived RiPPs has been established, the polytheonamides.10–12 Enzymatic reactions that furnish RiPPs are predicated upon substrate peptides that are typically divided between an N-terminal leader and a C-terminal core region.13 Amino acid residues in the core are modified by biosynthetic enzymes, and the modified core is proteolytically cleaved from the leader to furnish the mature RiPP. Genes encoding the substrate peptide and the core-modifying enzymes constitute the RiPP BGC. The polytheonamide BGC is harbored by bacterial symbionts associated with the sponge host.11–12 All other sponge-derived peptidic natural products for which BGCs have been deciphered fall under the purview of non-ribosomal peptide synthetase enzymology.14
A sub-class of RiPPs contain azole/azoline heterocycles. The azol(in)e heterocycles are derived from cysteine, serine, and threonine residues and their formation is catalyzed by ATP-dependent YcaO cyclodehydratases.15 The biosynthetic potential for azol(in)e containing RiPPs is found widespread in bacterial genomes.16 Several of these RiPPs are endowed with exquisite pharmacological potential with the azol(in)e heterocycles playing important roles in conferring bioactivity.17 Prominent examples include DNA gyrase inhibition by the archetypical linear azol(ine) peptide microcin B17,18 the ribosome-stalling activity of phazolicin,19 and the selective antibiosis activity of plantazolicin against Bacillus anthracis- the causative agent of anthrax.20 Curiously, from marine holobiont sources, the only known azol(in)e containing RiPPs are the cyanobactins that are produced by cyanobacteria symbiotically associated with tunicates.21 However, no azol(in)e containing RiPPs have been reported from sponges. Does this imply that the biosynthetic potential for the production of azol(in)e containing RiPPs is absent in sponge holobionts? Here, we demonstrate that the genetic potential for the production of azol(in)e containing RiPPs in marine sponge microbiomes is quite extensive, but the natural products themselves remain cryptic. RiPP BGCs in phylogenetically and geographically disperse marine sponges harbor a highly conserved RiPP substrate peptide and predicate the discovery of a broad-spectrum peptide tryptophanyl brominase while opening up new avenues for the combinatorial structural diversification of RiPPs.
Results
Detection of RiPP BGCs in a sponge metagenome
A recent metabolome inventory of the Floridian marine sponge Smenospongia aurea revealed the presence of brominated indoles (Table S1).22 To query the enzymatic potential for tryptophan bromination in S. aurea, we sequenced the S. aurea metagenome (Table S2). We included the RiPP microbisporicin flavin-dependent halogenase MibH23 as a query sequence to mine the S. aurea metagenome for tryptophan halogenases. MibH catalyzes the late-stage chlorination of the microbisporicin tryptophan side chain indole (Fig. S1).24–25 Several MibH homologs were detected in the S. aurea metagenome. In the neighborhood of each of these MibH homologs was a YcaO cyclodehydratase encoding gene. RiPP substrate peptides with cysteine-rich C-termini were found adjacent to the cyclodehydratases. The RiPP substrate peptides had large N-terminal leader peptides with similarity to nitrile hydratases. The presence of nitrile hydratase like leader peptides (NHLPs) has been recognized in bacterial genomes and NHLP-derived RiPPs are named proteusins.26
Surprisingly, not one, but multiple similar RiPP BGCs were identified in the S. aurea metagenome (Fig. 1, vide infra– 63 such RiPP BGCs were cataloged from S. aurea as illustrated in Fig. 2). Each BGC possessed the NHLP encoding gene in close proximity to a gene encoding the YcaO cyclodehydratase. Collectively, we have named these the sponge derived RiPP/proteusin (srp) BGCs. In addition to the NHLP, YcaO cyclodehydratase, and the halogenase, genes encoding transporters, chaperones, and proteins with catalytic functions that could not be discerned based on sequence similarity alone could also be identified in the srp BGCs. Of these, genes srpA–E and srpT1–T2, where srpE encodes the NHLP substrate, srpC encodes the YcaO cyclodehydratase, and srpT1 and srpT2 encode ATP-dependent membrane transporters were highly conserved in srp BGCs (Fig. 1). The MibH-like halogenase is encoded by srpI. While SrpI formed the basis for the discovery of the srp BGCs, srpI was not present in all srp BGCs (Fig. 1). As determined by sequence similarity using HHpred,27 srpA encodes a terpene cyclase with an N-terminal nitrile hydratase like domain while srpD encodes a prenyltransferase. Gene srpB encodes a putative bifunctional enzyme with an N-terminal metal-dependent oxidoreductase domain and a C-terminal adenyltransferase domain.
Fig. 1.
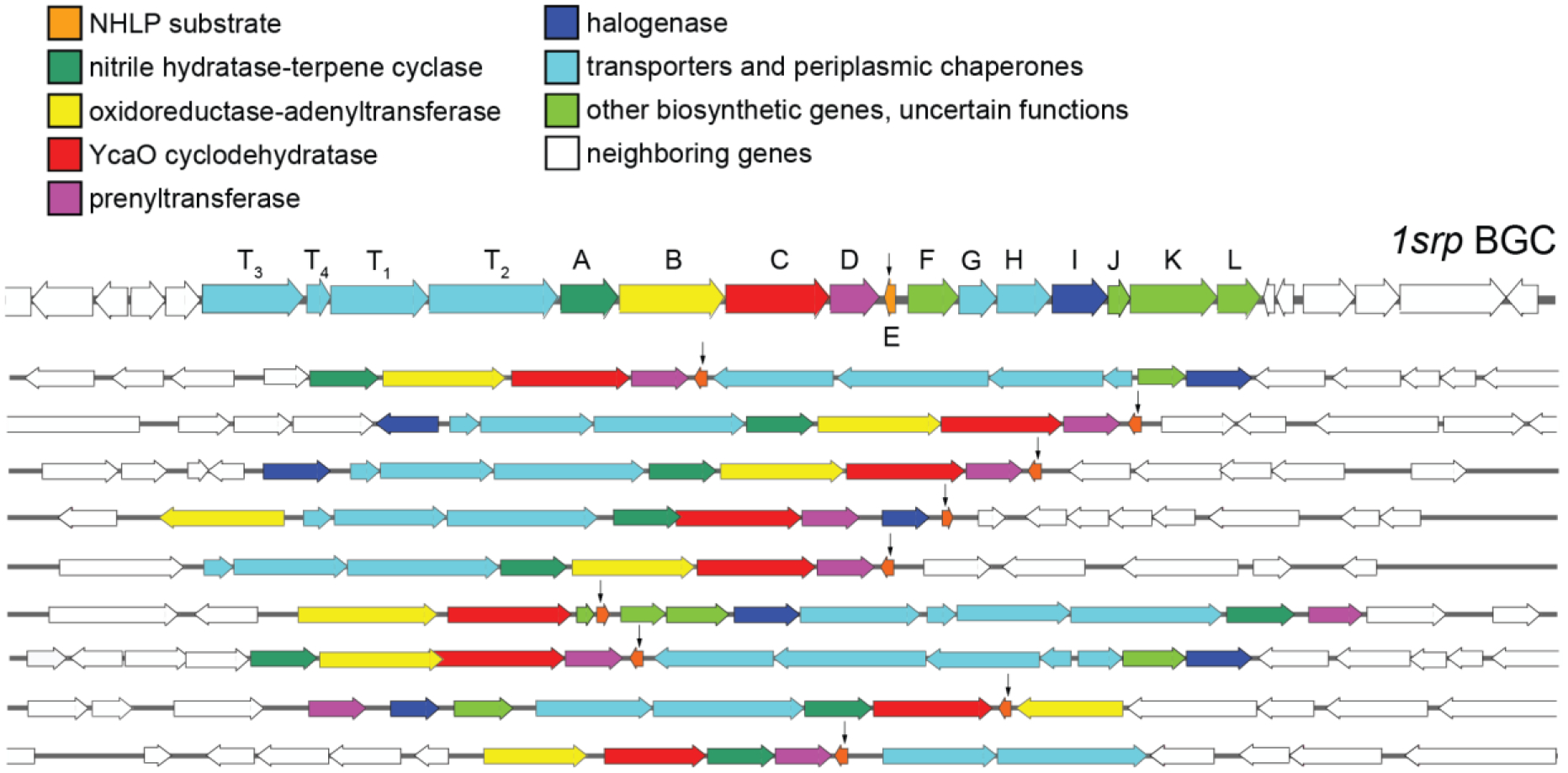
The srp BGCs detected in the S. aurea metagenome. Of the 63 srp BGCs detected in S. aurea, ten representative examples are illustrated here. The srpE genes are marked by arrows. Genes srpA–E and srpT1 and srpT2 were conserved in all srp BGCs. The halogenase is encoded by gene srpI. Gene srpT1 also possesses the peptidase that cleaves at the leader/core boundary of the NHLP SrpE substrate.
Fig. 2.
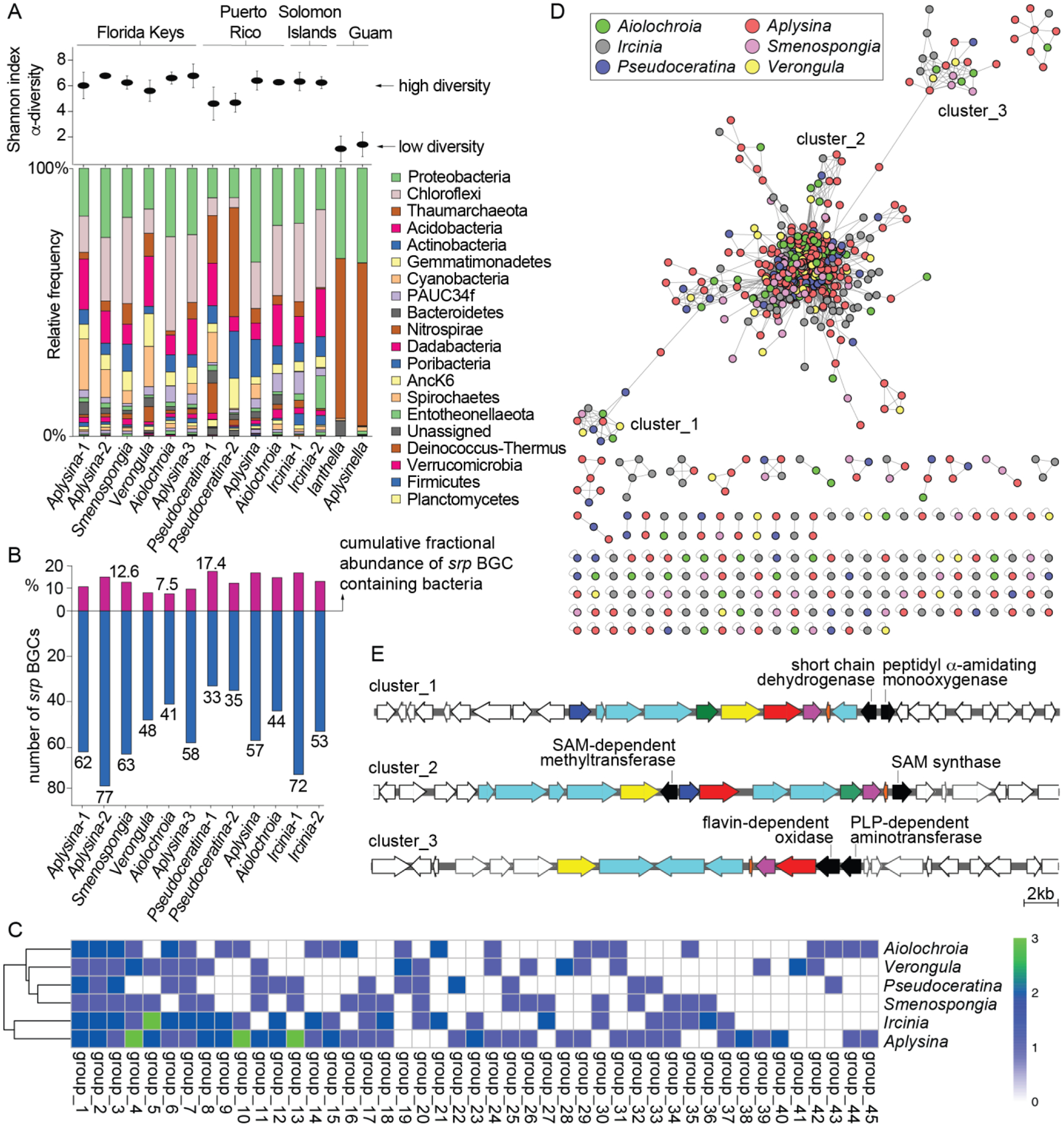
Diversity and distribution of srp BGCs. (A) (top) Sponge microbiome α-diversity as denoted by Shannon indices. (bottom) Microbiome architectures of sponge specimens used in this study. Bacterial phyla are color coded. Sponge specimens of identical genera collected from the same site are distinguished by hyphenated numerals; this nomenclature is preserved in Tables S1 and S2. (B) (top) Cumulative fractional abundance of bacteria harboring the srp BGCs. (bottom) Number of srp BGCs detected in metagenomes of sponge specimens used in this study. (C) Metagenomic bins containing srp BGCs were organized into groups according to a genome wide average nucleotide identity network (Fig. S2). Heat map showing the conservation of these groups in six different sponge genera, the values in the heat map is the number of bins that were detected in each sponge genus. The taxonomic assignments for these groups are presented in Table S2. (D) The srp BGCs were mined from sponge metagenomes using antiSMASH and parsed through BiG-SCAPE to generate a similarity network. Nodes are colored according to sponge genera. The central core corresponds to the canonical srp BGCs as illustrated in Fig. 1. (E) Representative srp BGCs from cluster_1, _2, and _3 as labeled in panel D. Putative activities of enzymes encoded by genes that were distinctly different from the canonical srp BGCs but highly conserved within the respective clusters are highlighted. Note that the srpA and srpI genes are missing in cluster_3.
Only two proteusin compound families are currently known: the polytheonamides11,28 and the landornamides.29 Neither of these two compound families bear azol(in)e heterocycles or halogenated residues (Fig. S1). Amino acid epimerases present in BGCs for both polytheonamides and landornamides were absent in the srp BGCs. As such, none of the enzymes that catalyze modifications that have been described previously for polytheonamide and landornamide proteusins were found encoded in the srp BGCs.
Of the constellation of srp BGCs detected in the S. aurea metagenome, we chose one, which we term 1srp BGC, for further characterization (Fig. 1). The 1srp BGC was present in the middle of a 117 kb metagenomic contig providing confidence that open reading frames flanking the BGC termini were not missed. The BGC boundary was determined so as to exclude open reading frames with similarity to enzyme sequences that could be rationalized to be involved in bacterial primary metabolism.
Conservation, abundance, and diversity of srp BGCs
Next, we queried if the detection of multiple srp BGCs in marine sponge metagenomes was unique to S. aurea which was collected in the Florida Keys. From the same location, we collected and sequenced the metagenomes of Aiolochroia, Aplysina, and Verongula spp. (Fig. 2A–B, Table S1). Like S. aurea, these sponge genera are ubiquitous on Floridian reefs. In this study, multiple sponge specimens of the same genus collected from the same geographical location represent morphologically distinct species. In each of these sponge specimens, we found srp BGCs to be present. These data demonstrate that the presence of srp BGCs extends across sponge hosts of different genera, families, and orders (Table S1). The abundance of srp BGCs in each metagenome was curated using antiSMASH,30 and the predicted BGCs that contain the srpE gene were selected for inventory. In the metagenome of an Aplysina specimen, at least 77 srp BGCs could be identified; the S. aurea metagenome contained 63 srp BGCs (Fig. 2B).
The presence of srp BGCs is also not constrained geographically. We found that the microbiomes of sponge specimens Aiolochroia sp., Aplysina sp., and Pseudoceratina spp. collected in Puerto Rico also possessed srp BGCs. From the Indo-Pacific, we detected srp BGCs in the microbiomes of sponge samples of Ircinia spp. collected in the Solomon Islands (Fig. 2A–B). There is limited metabolomic overlap between these sponge genera. Sponge S. aurea possesses brominated indoles in high concentration.22 However, sponges of the genera Aiolochroia, Aplysina, Verongula, and Pseudoceratina harbor bromotyrosine alkaloids,31 and Ircinia meroditerpenes.32 No RiPPs have been reported from any of these sponge genera, and none were detected in the mass spectrometry-based metabolomic data.
Characterized by the α-diversity Shannon indices, microbiomes of each of the abovementioned sponges were highly diverse (Fig. 2A). We asked if srp BGCs are restricted to sponges with highly diverse microbiomes only or do they extend to low microbial diversity sponges as well. Low microbial diversity sponge specimens belonging to genera Ianthella and Aplysinella were collected in Guam (Fig. 2A). No srpE genes were detected in these low microbial diversity sponges. This result suggests that srp BGCs may be associated only with the high microbial diversity sponges; more specimens of low microbial diversity sponges from more locations are required to further query this observation.
Despite the abundance of srp BGCs in individual sponge metagenomes, srp harboring bacteria collectively constitute a minor fraction of the respective sponge microbiomes. Illustratively, bacteria that possess the 63 srp BGCs detected in the S. aurea metagenome make up 12.6% of the microbiome fraction (Fig. 2B). The cumulative fractional abundance of srp BGC containing bacteria in sponge microbiomes ranges from 7.5% to 17.4% for samples used in this study.
At this point, our data demonstrates that the srp BGCs are not localized to a single specialized symbiont but distributed throughout the sponge microbiome. Thus, we sought to inventory which members of the microbiome harbored the srp BGCs, and if the srp BGC harboring bacteria overlapped among different sponge genera. For Aplysina, Aiolochroia, Verongula, and Pseudoceratina spp. sponges used in this study, we have reported that the microbiomes are conserved at the host genus level.31 This observation also extends to the two Ircinia spp. samples used in this study (Fig. 2A). Hence, subsequent analyses grouped sponge specimens used in this study by genus. First, all metagenomic bins in which srp BGCs were detected were grouped in a genome-wide average nucleotide identity network, which was organized according to sponge genus (Fig. S2). Clusters and individual nodes thus identified were ranked per the abundance of srp BGCs. The srp BGC abundances for the top 45 phylogenetic groups are illustrated in Fig. 2C with taxonomic assignments of these groups provided in Table S3. Bacterial groups 1–3 and 7, which are present in all six sponge genera, belong to the Latescibacterota, UBA8248, Nitrospirota, and Acidobacteriota phyla, respectively. The next most widely distributed groups, 4 and 6, belong to Proteobacteria and Gemmatimonadota phyla. Overall, Proteobacteria and Acidobacteriota were the most well represented bacterial phyla that contain srp BGCs in microbiomes of sponge specimens used in this study (Fig. S3). This finding is also supported by the %GC content of the srpE genes detected in each sponge metagenome (Fig. S4).
The inventory of over 600 srp BGCs detected here from six sponge genera was organized using the Biosynthetic Genes Similarity Clustering and Prospecting Engine (BiG-SCAPE) (Fig. 2D).33 Overall, srp BGCs clustered independently of sponge host genus. The central large network contained the canonical srp BGCs that are illustrated in Fig. 1. BGCs distant from the central core revealed the presence of additional open reading frames that were highly conserved within the respective clusters. Three such clusters are highlighted in Fig. 2D with representative srp BGCs from each of the three clusters illustrated in Fig. 2E. The srp BGCs in cluster_3 lacked the srpA and srpI genes and possessed NHLP SrpE sequences with conspicuously different C-termini (Fig. S5).
A short pentapeptide core
At present, RiPPs encoded by the srp BGCs are cryptic, meaning that we have not yet detected the natural product in situ. To gain insight into the possible natural products from srp, we focused our efforts on the 1srp BGC (Fig. 1). First, we sought to demarcate the leader/core boundary for the 1SrpE substrate peptide. A Cys-His-Asp catalytic triad containing C39 protease domain was located at the 5’-terminus of the ATP-dependent transmembrane transporter encoding gene 1srpT1 (Fig. 3A). The corresponding nucleotide sequence was optimized for expression in Escherichia coli and the recombinant protein, henceforth referred to as 1SrpT1protease, purified (Fig. S6). The optimized 83-residue peptide encoding gene 1srpE was also expressed in E. coli with an N-terminal (His)10-tag and the recombinant peptide purified. Upon in vitro incubation of 1SrpE with 1SrpT1protease, we observed a product corresponding to the 1SrpE C-terminal -LCCCW pentapeptide using liquid chromatography/mass spectrometry (LC/MS) (Fig. 3B). In addition, multiply charged ions corresponding to the inferred 78-mer leader peptide with the appended (His)10 tag were also detected with high accuracy (Fig. 3C). While even single amino acid excision natural products have been described starting from peptidyl substrates,34–35 the SrpE pentapeptide core is uncharacteristically short and is the shortest among known proteusin substrate peptides. The SrpE -LCCCW core was found to be highly conserved among sponge microbiomes independent of sponge phylogeny and geographical location with the highest variability observed at the C-terminal tryptophan residue (Fig. 3D).
Fig. 3.
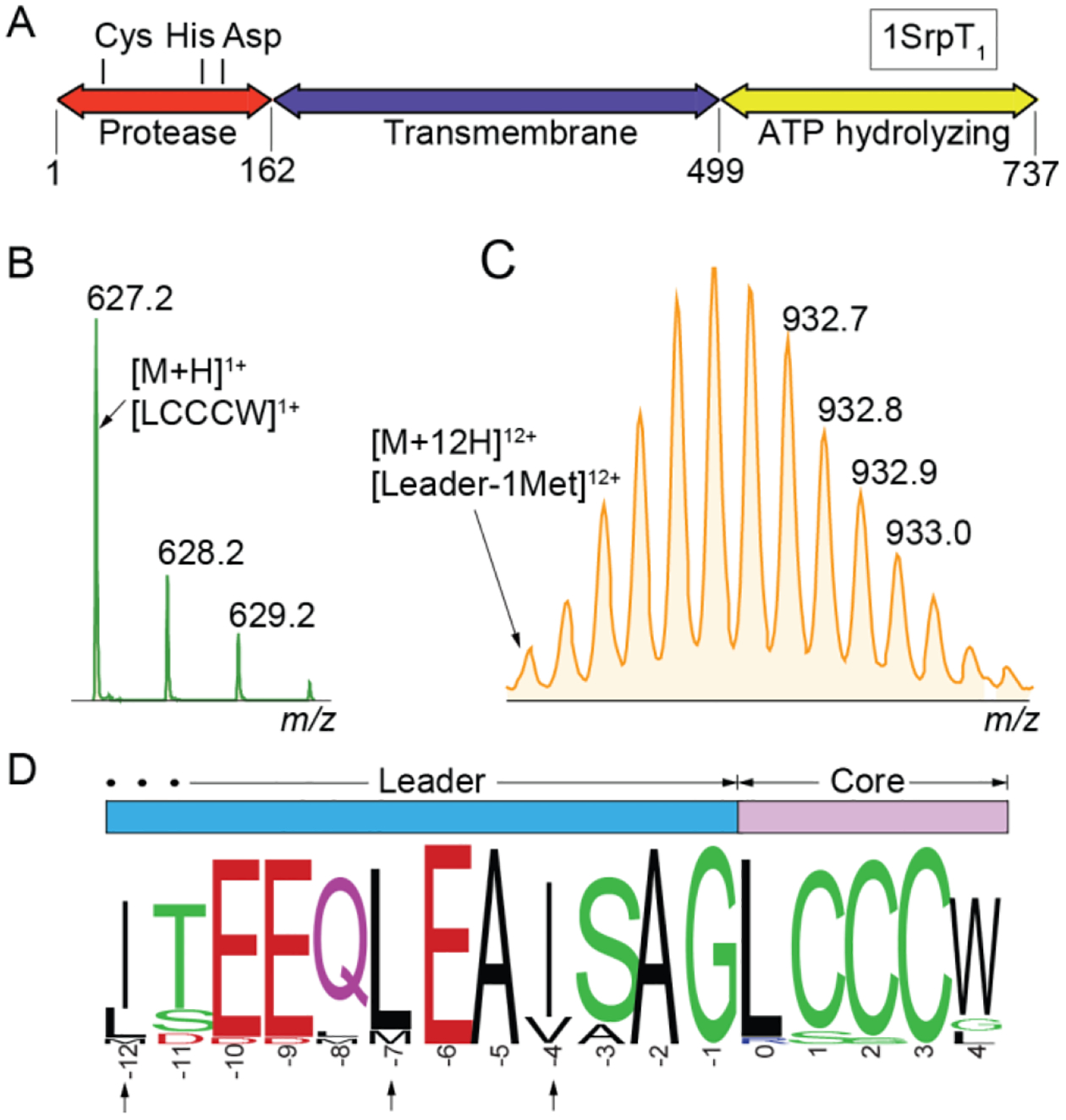
Leader/core boundary for 1SrpE substrate peptide. (A) Domain architecture for the 1SrpT1 transporter with the N-terminal 1SrpT1protease domain which possesses the Cys-His-Asp catalytic triad. LC/MS data demonstrating cleavage products observed upon treatment of 1SrpE with 1SrpT1protease. (B) Isotopic distribution for the [M+H]1+ MS1 ion corresponding to the -LCCCW core. (C) Isotopic distribution for the [M+12H]12+ MS1 ion corresponding the 1SrpE leader appended to the (His)10 tag. (D) Sequence conservation for the SrpE NHLPs. 484 SrpE sequences were used to generate the sequence logo. Residues that guide cleavage by LahT150 are marked by arrows.
Cleavage of 1SrpE was observed only when incubated with equimolar or excess 1SrpT1protease. To circumvent the low activity of 1SrpT1protease, we tested cleavage of 1SrpE by the substrate promiscuous RiPP peptidase LahT150 (Fig. S7).36 The leader peptide recognition determinants for LahT150, which are the branched aliphatic amino acid side chains at the −4, −7, and −12 positions,36 were found conserved in 1SrpE (Fig. 3D). Incubation of 1SrpE with purified LahT150 offered identical cleavage products as 1SrpT1protease but with more favorable catalyst/substrate ratio and reduced reaction times. Experiments described below utilize LahT150 in place of 1SrpT1protease.
Cyclodehydrations in a proteusin peptide
While widely represented in RiPP enzymology, ATP-dependent YcaO catalyzed cyclodehydration of cysteine, serine, and threonine residues to furnish azoline heterocycles, has not been observed for proteusin RiPPs (Fig. 4A). Intransigence of heterocyclase 1srpC expression in a soluble form in E. coli precluded in vitro experiments. Five additional SrpC homologs were tested each resulting in insoluble protein expression. However, co-expression of 1srpC with 1srpE provided an in vivo route to query the 1SrpC activity. After co-expression of 1srpE and 1srpC, purified 1SrpE was treated with the LysC protease to generate peptide fragments amenable for high resolution and high accuracy mass determination by MALDI-ToF mass spectrometry (Fig. 4B). Three products, corresponding to mono-, di-, and tridehydrated 1SrpE were observed (Fig. 4C). The monodehydrated product demonstrated two acetamide additions upon treatment with the thiol alkylating reagent iodoacetamide (Fig. S8). These data imply that each of the three cysteine residues in 1SrpE -LCCCW core could be cyclodehydrated by 1SrpC. The inferred density of azoline heterocycle installation by 1SrpC is reminiscent of plantazolicin biosynthesis in which two pairs of consecutive penta-azol(in)es are installed by a YcaO cyclodehydratase.37
Fig. 4.
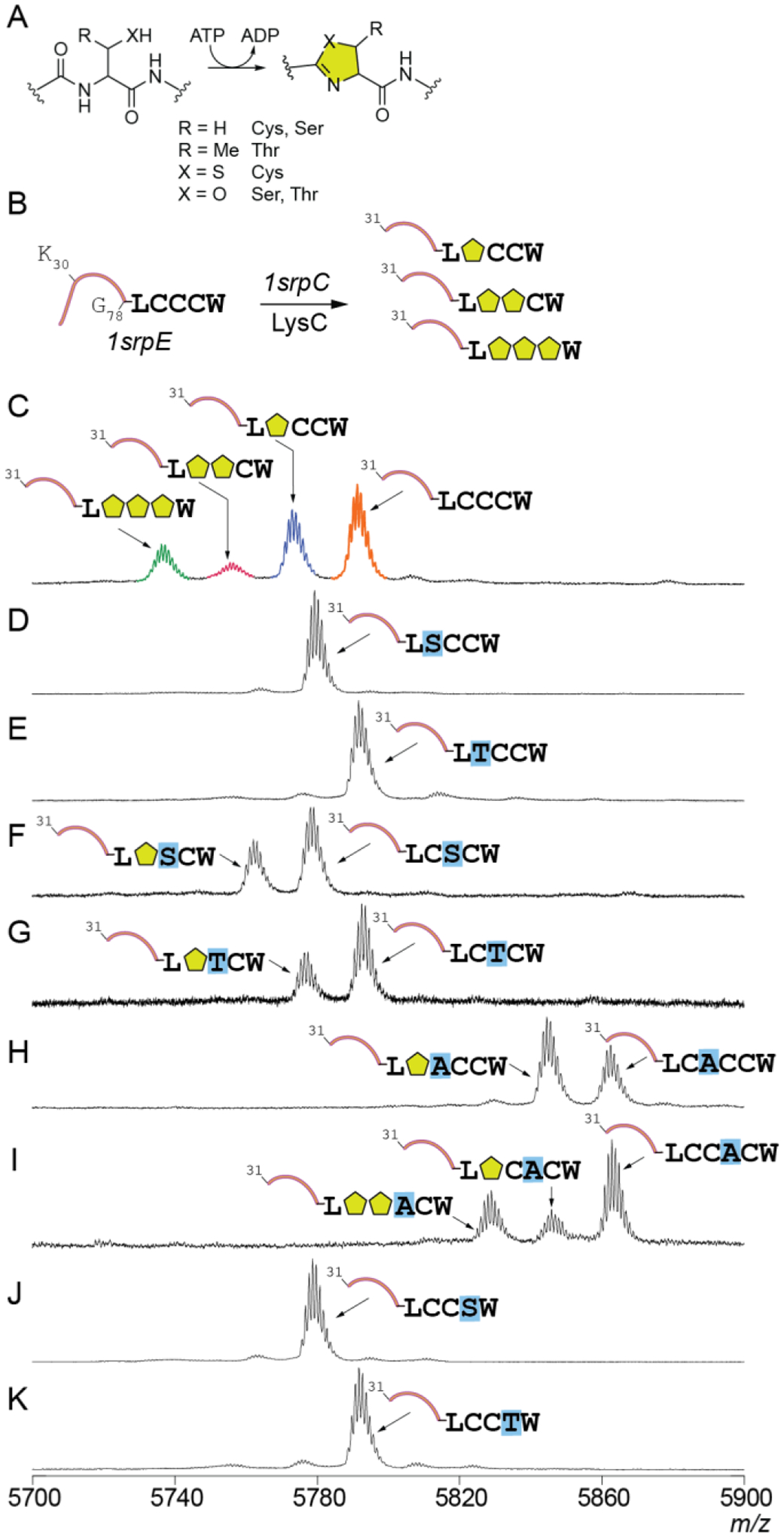
Cyclodehydration of the proteusin peptide 1SrpE. (A) Azoline formation by YcaO cyclodehydratases. Each azoline formation is accompanied by a mass decrease of 18 Da. (B) Co-expression assay procedure to characterize 1SrpC activity. The protease LysC cleaved at the C-terminus of the Lys30 residue in the 1SrpE leader. MALDI-ToF mass spectra for (C) wild type and modified (D–K) 1SrpE peptides obtained by co-expression of 1srpE and 1srpC genes in E. coli. The azoline heterocycles are denoted by yellow pentagons, mutagenized residues in the SrpE core are highlighted in blue.
Further characterization of the cyclodehydrated 1SrpE products was achieved by MS/MS fragmentation of the modified 1SrpE core furnished by LahT150-catalyzed proteolytic removal of the 1SrpE leader peptide. The monodehydrated product demonstrated MS2 fragment ions corresponding to cyclodehydration at Cys1 (1SrpE core: -LC1C2C3W) (Fig. S9). Subsequent di- and tri-cyclodehydrations were affected upon the Cys2 and Cys3 residues, respectively (Fig. S10, S11).
Numerous examples exist for RiPPs in which a single YcaO enzyme catalyzes the cyclodehydration of cysteine, serine, and threonine residues. To query whether serine and threonine residues could be modified by 1SrpC, we generated 1srpE mutants in which cysteines in the 1SrpE core were replaced by serine and threonine residues. No mutations at Cys1 (1SrpE core: -LC1C2C3W) were tolerated by 1SrpC; cyclodehydrated products were not observed in either case (Fig. 4D, 4E). Mutations at Cys2 were moderately tolerated; singly dehydrated products were observed for both the Cys2→Ser and Cys2→Thr mutants (Fig. 4F, 4G). MS/MS fragmentation established that the single heterocycle formation in the Cys2→Ser and Cys2→Thr mutants was predicated upon Cys1 (Fig. S12, S13). These data lead to the hypothesis that neither can 1SrpC modify serine and threonine residues, nor can it skip over non-modifiable residues in the 1SrpE core. To test this hypothesis, we introduced non-modifiable alanine residue after the Cys1 and Cys2 to generate the -LCACCW and -LCCACW 1SrpE core substrates. In accordance with the hypothesis, a single heterocycle at Cys1 was installed by 1SrpC for the -LCACCW core (Fig. 4H, S14). Two heterocycles at Cys1 and Cys2 could be installed by 1SrpC for the -LCCACW core (Fig. 4I). Mutations at Cys3 were not tolerated: cyclodehydrated product formation was not observed for the Cys3→Ser and Cys3→Thr mutants (Fig. 4J, 4K).
The 1SrpC specificity can be rationalized in view of the narrow sequence space explored by the SrpE cores (Fig. 3D). Contrary to inherently substrate tolerant marine RiPP biosynthetic enzymes such as those that modify expanded libraries of cyanobactin38 and prochlorosin39 substrate peptides, the SrpC YcaO cyclodehydratase is a narrow spectrum enzyme specifically tailored to modify the SrpE -LCCCW core that is highly conserved in microbiomes of diverse sponges around the world.
SrpI is a tryptophan-6-brominase
To date, a single RiPP halogenase has been described: the flavoenzyme MibH that catalyzes tryptophan-5-chlorination during biosynthesis of the lantibiotic NAI-107 (Fig. S1).24 Akin to the activity of MibH, we rationalized halogenation by SrpI to be a late-stage biosynthetic modification. Activity of 1SrpI was thus evaluated by coexpression of 1srpI with 1srpE and 1srpC. No halogenation was observed with standard media conditions (Fig. 5A). Halogenation could be realized only when 1g/L potassium bromide was added to the culture media (Fig. 5B). MS/MS fragmentation demonstrated bromination to be affected upon the tryptophanyl side chain indole (Fig. S15). Halogenases are named after the most electronegative halogen that they incorporate,40 1SrpI was thus a physiological tryptophan brominase. Mutating the catalytic lysine residue — rationalized based on characterization of other flavoenzyme halogenases — to alanine led to abolishment of activity (Fig. S16).41 Curiously, in addition to bromination of the tri-cyclodehydrated 1SrpE, we observed bromination of the mono- and di-cyclodehydrated 1SrpE (Fig. 5B). In addition to the partially cyclodehydrated 1SrpE core, brominase 1SrpI could also tolerate modifications adjacent to the tryptophan residue. The modified 1SrpE-LCCCGW core was efficiently processed by 1SrpC and 1SrpI to yield a brominated, tricyclodehydrated product (Fig. 5C). Similarly, brominated products were detected for the 1SrpE-LCCCWG (Fig. 5D) and 1SrpE-LCCCWA (Fig. S17) variants. To further explore this unexpected substrate tolerance of 1SrpI, 1srpI was coexpressed with 1srpE in the absence of 1srpC. Here, we observed the production of the brominated -LCCCW 1SrpE core without cyclodehydrations (Fig. 5E). Incubation with 1SrpT1protease furnished the brominated -LCCCW 1SrpE core (Fig. S18). Mutating the 1SrpE C-terminal tryptophan residue to tyrosine demonstrated the production of mono- and di-brominated products (Fig. 5F). Brominated product formation was not observed for 1SrpE-LCCCF.
Fig. 5.
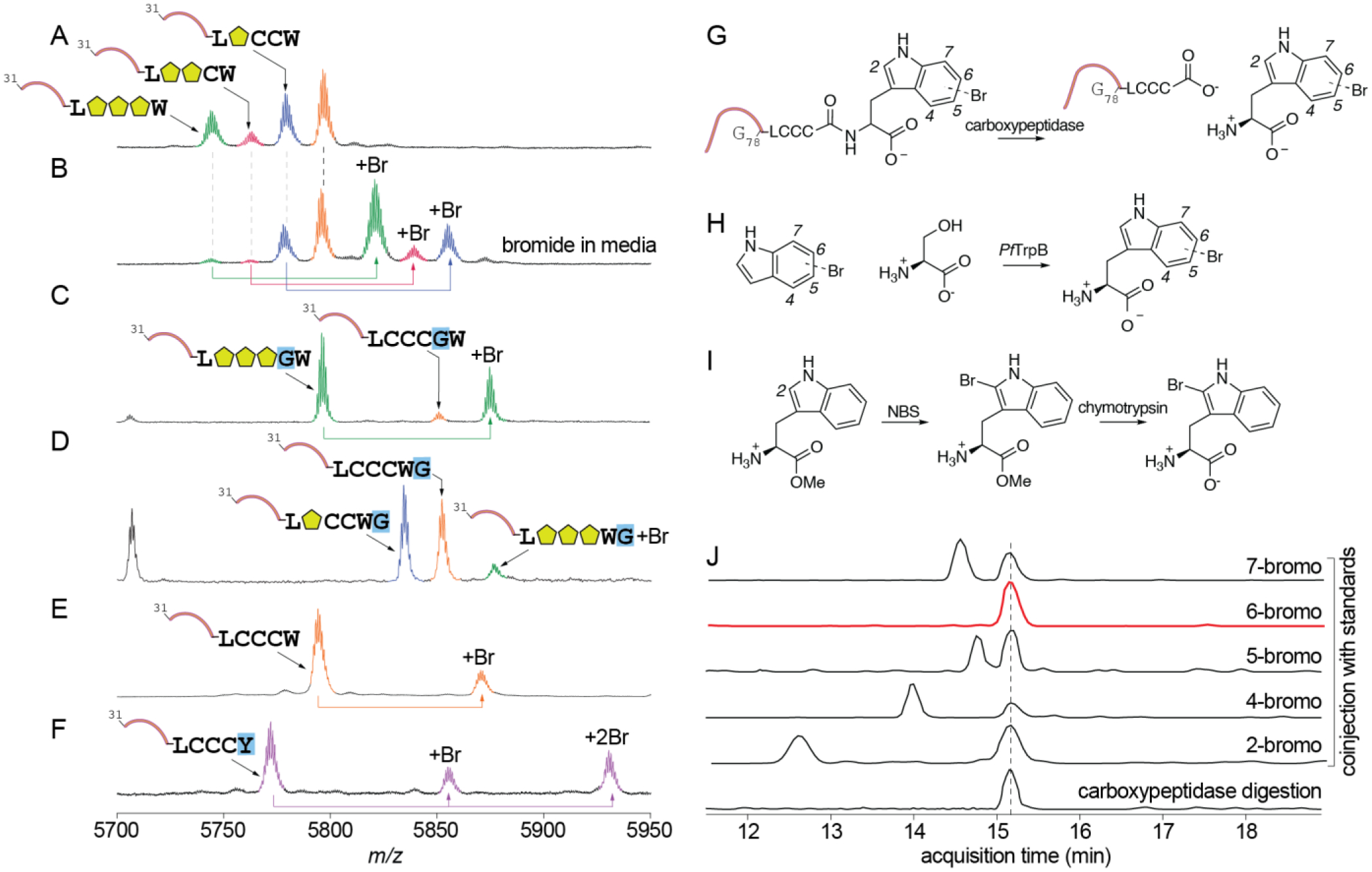
Activity of RiPP brominase 1SrpI. (A) No halogenated products were observed upon coexpression of 1sprE with 1srpC and 1srpI when culture media lacked bromide. (B) Bromination of the mono-, di-, and tri-cyclodehydrated 1SrpE was observed when culture media was supplemented with bromide. Processing of the (C) 1SrpE-LCCCGW and (D) 1SrpE-LCCCWG by 1SrpC and 1SrpE to yield cyclodehydrated-brominated products. (E) When gene 1srpC was omitted from the co-expression experiment, bromination of the unmodified 1SrpE peptide was observed. (F) Mono- and di-bromination was observed when the C-terminal tryptophan residue was modified to tyrosine. (G) To determine the regiospecificity of tryptophan halogenation by 1SrpI, gene 1srpE was co-expressed with 1srpI and the purified 1SrpE peptide product was digested with carboxypeptidase to excise the C-terminal monobrominated tryptophan residue. (H) Standards for 4-, 5-, 6-, and 7-bromotryptophan were generated by condensation of L-serine with bromoindoles using tryptophan synthase PfTrpB. (I) Scheme for synthesis of the 2-bromotryptophan standard. NBS: N-bromosuccinimide. (J) The carboxypeptidase digestion reaction was co-injected with 2-, 4-, 5-, 6-, and 7-bromotryptophan standards and the presence of monobrominated tryptophan was monitored by LCMS using extracted ion chromatograms for m/z 284.14 ± 0.1 Da. The 1SrpE-excised bromotryptophan residue from 1SrpE coeluted with the 6-bromotryptophan standard.
At this stage, the site for bromination upon the tryptophanyl indole side chain catalyzed by 1SrpI was not known. Efforts to spectroscopically characterize the brominated 1SrpE core were impeded by the low expression level of 1srpE in E. coli. To circumvent this challenge, brominated 1SrpE obtained after coexpression of 1srpE and 1srpI in E. coli was digested with carboxypeptidase to furnish the C-terminal bromotryptophan residue (Fig. 5G). Bromination by a flavin-dependent halogenase such as 1SrpI can be rationalized to be affected upon either of the 2-, 4-, 5-, 6-, and 7-positions of the indole ring. Standards for 4-, 5-, 6-, and 7-bromotryptophan were synthesized by the enzymatic ligation of serine with commercially available 4-, 5-, 6-, and 7-bromoindole by the Pyrococcus furiosus tryptophan synthase (PfTrpB, Fig. 5H).42 2-Bromotryptophan was synthesized by bromination of tryptophan methyl ester by N-bromosuccinimide followed by mild hydrolysis of the methylester by α-chymotrypsin (Fig. 5I).43 Harsher saponification conditions led to product degradation. Co-injection with the bromotryptophan standards demonstrates that 6-bromotryptophan was produced by carboxypeptidase catalyzed degradation of the 1SrpE peptide obtained by coexpression of 1srpE and 1srpI (Fig. 5J). Hence, 1SrpI is rationalized to be a peptidyl tryptophan-6-brominase.
Tolerance of other proteusin leaders by Srp enzymes
By sequence similarity, SrpC is most closely aligned with YcaO cyclodehydratases identified in marine cyanobacterial symbionts that produce macrocylic cyanobactin natural products containing azol(in)e heterocycles (Fig. S19). Cyanobactin YcaO cyclodehydratases bind their substrates using the L(S/T)EEXL sequence motif present in the substrate leader peptide.44 Introducing this recognition sequence in cyanobactin substrate peptides where it was otherwise absent rendered them competent substrates for YcaO cyclodehydratases.45 Sequence gazing identified the conservation of this YcaO recognition sequence in the proteusin substrate peptide SrpE (Fig. 6A, here, one of the cyanobactin substrate peptides TruE1 is illustrated). Curiously, even though the proteusin RiPPs landornamides and polytheonamides do not contain genes encoding YcaO cyclodehydratases in their BGCs and azol(in)e heterocycles are not present in the mature natural products, the respective substrate peptides, OspA and PoyA also contain the YcaO cyclodehydratase recognition sequence (Fig. 6A).11,29 The leader/core boundaries for TruE1, OspA, and PoyA have been described previously.11,29,46 The position of the YcaO recognition sequence relative to the leader/core boundary was identical among the four peptides (Fig. 6A).
Fig. 6.
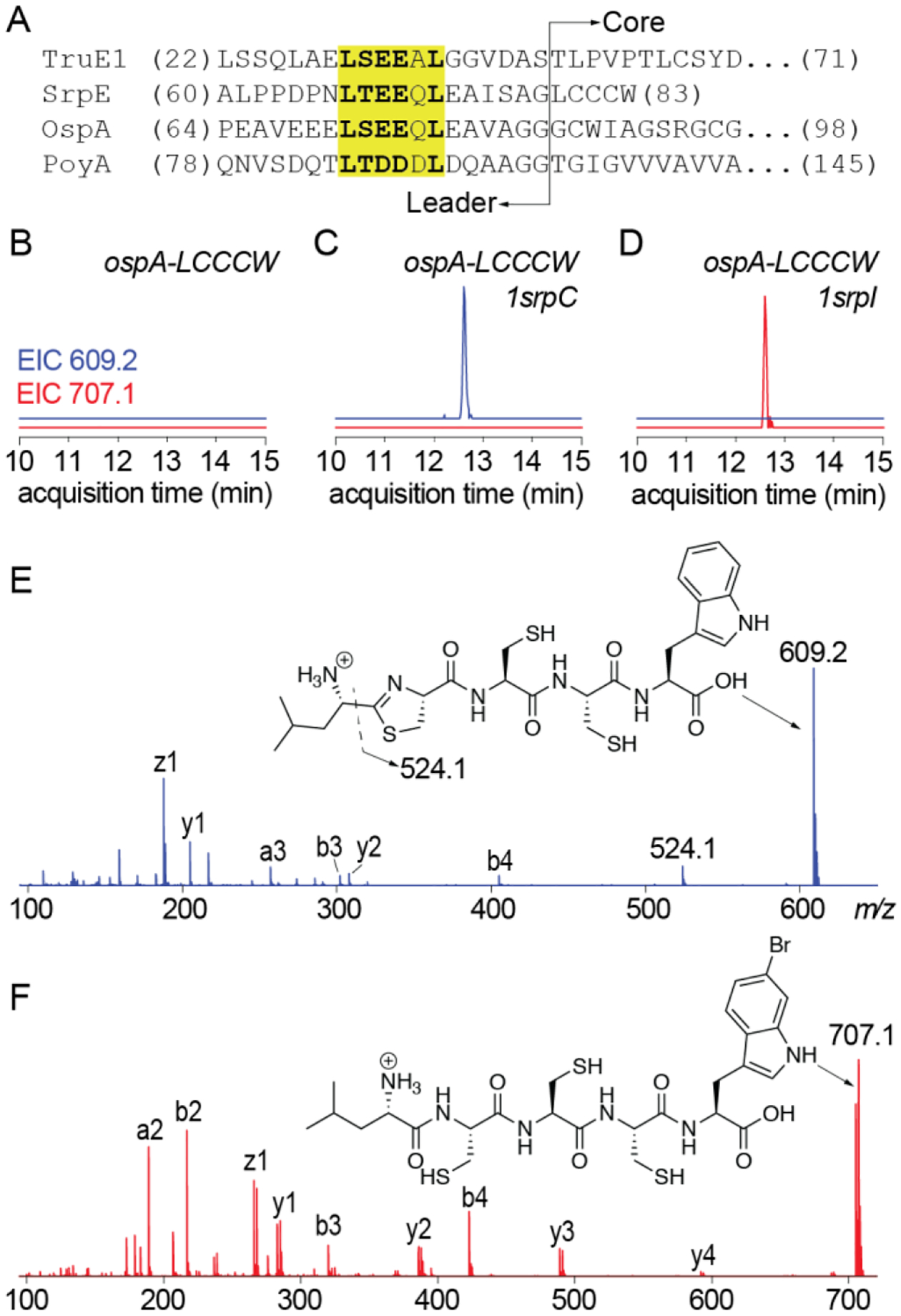
Conservation of the YcaO recognition sequences in RiPP substrates. (A) Sequence alignment of TruE1, SrpE, OspA, and PoyA substrate peptides. The YcaO cyclodehydratase recognition sequence is highlighted in yellow and the leader/core boundary is marked. Extracted ion chromatograms (EICs) for m/z 609.2 (in blue) and m/z 707.1 (in red) corresponding to the cyclodehydrated and brominated -LCCCW core obtained when the gene encoding the OspA-LCCCW chimeric substrate was expressed (B) without 1srpC or 1srpI, (C) with 1srpC, and (D) with 1srpI. Structural annotation of the (E) monocyclodehydrated product obtained after LahT150 digestion of modified OspA-LCCCW peptide by 1SrpC, and (F) the brominated OspA-LCCCW peptide by 1SrpI.
In light of this observation, we asked if the leader peptide sequences from TruE1, OspA, and PoyA could support the cyclodehydration activity of 1SrpC. Given the narrow tolerance of 1SrpC for modifications of the 1SrpE core, the physiological 1SrpE core was preserved. We generated clones encoding the TruE1-LCCCW, OspA-LCCCW, and PoyA-LCCCW chimeric peptides. Given the broad substrate scope of the brominase 1SrpI, we also queried if the activity of 1SrpI could be observed for these chimeric substrates. The three chimeric peptide encoding genes were co-expressed with 1srpC and 1srpI and the resultant peptides purified, and after digestion with LahT150, analyzed by LC/MS. For the OspA-LCCCW chimera, we observed modification by both 1SrpC and 1SrpI (Fig. 6B–D). Modifications were not observed for the TruE1-LCCCW and PoyA-LCCCW chimeras. Extracted ion chromatograms, corresponding to the cyclodehydrated -LCCCW product, m/z 609.2, demonstrated product formation when ospA-LCCCW was co-expressed with 1srpC (Fig. 6B,C). Annotation of the MS2 fragmentation spectra demonstrated that the single cyclodehydration observed was localized to Cys1 (Fig. 6E). No further cyclodehydrations could be observed. Similarly, extracted ion chromatograms, corresponding to the brominated -LCCCW product, m/z 707.1, demonstrated product formation when ospA-LCCCW was co-expressed with 1srpI (Fig. 6B, 6D). Annotation of the MS2 spectra identified the C-terminal tryptophan residue to be the site of bromination (Fig. 6F). Upon coexpression of 1srpC and 1srpI with ospA-LCCCW, bromination of unmodified and singly cyclodehydrated core peptide was observed (Fig. S20).
Discussion
First clues that bacterial symbionts could produce natural products that had been isolated from sponges came from physical separation of bacteria from sponge tissue wherein the separated bacterial fractions were found enriched in the natural products.47 Prominent examples include the colocalization of cyanobacterial filaments from Dysideidae sponges with polybrominated phenols and polychlorinated peptides and that of various polyketide and peptidic natural products isolated from sponge Theonella swinhoei with a filamentous bacterial symbiont (reviewed in refs. 2, 6). These microbe-molecule colocalization experiments benefited from the natural product producing symbiont being highly abundant in the sponge microbiome. In the post genomic age, rationalized biosynthetic schemes guided the mining of sponge metagenomes, and the colocalization experiments were subsequently validated.48–49 Within this context, work presented here provides a glimpse into the cryptic biosynthetic potential that is present in sponge microbiome members that are of lesser abundance and traditionally not viewed as natural product producing taxa.
We find the srp BGC to be distributed in high diversity sponge microbiomes without constraints of sponge host phylogeny or geographical location. While they were universal in high microbial diversity sponges in our collection, we could not detect srp BGCs in multiple other marine invertebrate metagenomes that we have sequenced. Data presented in this study likely underrepresent the distribution of srp BGCs in sponge microbiomes with a greater spread of srp BGCs to be realized as more sponge microbiomes are queried. Deeper metagenomic sequencing can also likely uncover even more srp BGCs within a sponge microbiome. Illustratively, over 200 NHLP encoding srpE genes are detected in the publicly available metagenomic data for the marine sponge Ircinia ramosa (collected in Australia).50
The presence of numerous highly similar srp BGCs within a single sponge metagenome is without precedent. At present, we do not know if some srp BGCs are plasmid encoded or not. Biosynthesis of natural products by bacterial symbionts is thought to provide the sponge host with chemical defense against predation and a competitive growth advantage.9,51 Does then the conservation of srp BGCs in marine sponge microbiomes point towards a conserved ecological role of the as yet cryptic RiPP, or represent the vestigial remnants of an ancestral BGC that is being shuttled among microbial symbionts? Within sponges, different symbionts share genetic potential to interact with the host via exchange of primary metabolites.50,52 Whether this redundancy extends to secondary metabolites with specialized ecological roles remains a tantalizing prospect.
Cyclodehydration for the installation of azol(in)e heterocycles and aromatic side chain halogenation are both activities that were absent from previously known proteusin RiPPs. Our data demonstrates that SrpC and SrpI can use their cognate core ligated to other leader peptides (Fig. 6). However, leader peptide tolerance was not universal as the TruE and PoyA leaders did not support modification. Among the three non-cognate leader peptides tested here, sequence alignment identifies OspA to be of closest similarity to SrpE (Fig. S21). Unlike the RiPP chlorinase MibH, which requires prior modifications to be installed on the RiPP core region for halogenation of the tryptophan indole, SrpI can brominate the unmodified peptide. A winged helix-turn-helix RiPP precursor peptide recognition element that is found in several RiPP biosynthetic enzymes cannot be located at either the N- or the C- termini of SrpI.53 Enzymatic bromination, without contaminating chlorination in vivo, can be thought of as an enabling platform for the cross-coupling of tryptophanyl peptides with boronic acids, as has been achieved for other classes of indolic natural products.54–55 Matching the regiospecificity of SrpI, tryptophan-6-bromination is widely detected in conotoxins,56 predicating further exploration of SrpI as a general purpose peptide modification biocatalyst.
Sequence motifs that direct the C39 protease to cleave at the leader/core boundary and the activity of the YcaO cyclodehydratase were found conserved in SrpE.36,45 Both motifs are placed towards the C-terminus of the SrpE leader. Proteusin RiPPs possess unusually long leader peptide sequences. At the N-terminus of the proteusin substrate OspA, a recognition motif was identified which directs the activity of a radical SAM epimerase which converts two L-amino acid residues in the OspA core to D-amino acids.57 While radical SAM epimerases have not been identified in our current inventory of srp BGCs, the epimerase recognition sequence in the OspA leader is partially conserved in the SrpE leader (Fig. S21). The srp BGCs contain many other genes encoding biosynthetic enzymes that are not present in other proteusin BGCs, activities and leader peptide dependencies for which remain to be determined. Overall, in addition to beginning to construct the widely distributed cryptic srp BGC encoded natural product(s), we demonstrate that the Srp biosynthetic enzymes can interact with other substrate peptides which now sets the stage for combinatorial structural diversification of proteusin RiPPs.
Conclusion
Here, we describe the discovery of a RiPP BGC that is conserved and exceptionally widely distributed in marine sponge microbiomes. Encoded within this BGC was found an obligate peptidyl tryptophan brominase the activity of which did not depend on prior modifications on the RiPP core peptide. Sequence motifs that guide biosynthetic modifications are shared within different RiPP compound classes which provides avenues for combinatorial diversification of the RiPP chemical space. This study establishes marine sponges as sources of as yet unknown cryptic natural products, in addition to the already impressive catalog of bioactive natural products that have been isolated from these benthic marine invertebrates.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (ES02660 to V.A.; GM122521 to E.W.S.). The authors thank V. J. Paul, J. S. Biggs, C. J. Freeman, and H. R. Thapa for collection of sponge specimens. Sponge specimens from Solomon Islands were acquired via export permit rp/2017/003. Assistance from the Ministry of Environment, Climate Change, Disaster Management, and Meteorology (Solomon Islands) and Solomon Islands National University is gratefully acknowledged. This work was supported by Georgia Institute of Technology’s Systems Mass Spectrometry Core and the Molecular Evolution Core facilities. The authors thank S. K. Nair for insightful discussions.
Footnotes
Supporting Information
Materials and methods used in this study are described in the Supporting Information. These include methods describing metagenomic sequencing, in vivo and in vitro biochemical assays, and analytical data.
Data Deposition
The bacterial 16S rRNA, sponge 28S rRNA, and sponge ITS-2 amplicon sequences are deposited to GenBank with BioProject ID PRJNA694437. Also included are the metagenomic reads and annotated srp BGCs. Untargeted LC/MS data for sponge extracts has been deposited to MassIVE database with ID MSV000086758.
References
- 1.Crawford JM; Clardy J, Bacterial symbionts and natural products. Chem. Commun 2011, 47 (27), 7559–7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morita M; Schmidt EW, Parallel lives of symbionts and hosts: chemical mutualism in marine animals. Nat. Prod. Rep 2018, 35 (4), 357–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas T; Moitinho-Silva L; Lurgi M; Bjork JR; Easson C; Astudillo-Garcia C; Olson JB; Erwin PM; Lopez-Legentil S; Luter H; Chaves-Fonnegra A; Costa R; Schupp PJ; Steindler L; Erpenbeck D; Gilbert J; Knight R; Ackermann G; Victor Lopez J; Taylor MW; Thacker RW; Montoya JM; Hentschel U; Webster NS, Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun 2016, 7, 11870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pita L; Rix L; Slaby BM; Franke A; Hentschel U, The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome 2018, 6 (1), 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll AR; Copp BR; Davis RA; Keyzers RA; Prinsep MR, Marine natural products. Nat. Prod. Rep 2020, 37 (2), 175–223. [DOI] [PubMed] [Google Scholar]
- 6.Paul VJ; Freeman CJ; Agarwal V, Chemical ecology of marine sponges: new opportunities through “-omics”. Integr. Comp. Biol 2019, 59 (4), 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becher PG; Verschut V; Bibb MJ; Bush MJ; Molnár BP; Barane E; Al-Bassam MM; Chandra G; Song L; Challis GL; Buttner MJ; Flärdh K, Developmentally regulated volatiles geosmin and 2-methylisoborneol attract a soil arthropod to Streptomyces bacteria promoting spore dispersal. Nat. Microbiol 2020, 5 (6), 821–829. [DOI] [PubMed] [Google Scholar]
- 8.Pawlik JR, The chemical ecology of sponges on Caribbean reefs: natural products shape natural systems. BioScience 2011, 61 (11), 888–898. [Google Scholar]
- 9.Puglisi MP; Sneed JM; Sharp KH; Ritson-Williams R; Paul VJ, Marine chemical ecology in benthic environments. Nat. Prod. Rep 2014, 31 (11), 1510–1553. [DOI] [PubMed] [Google Scholar]
- 10.Hamada T; Matsunaga S; Yano G; Fusetani N, Polytheonamides A and B, highly cytotoxic, linear polypeptides with unprecedented structural features, from the marine sponge, Theonella swinhoei. J. Am. Chem. Soc 2005, 127 (1), 110–118. [DOI] [PubMed] [Google Scholar]
- 11.Freeman MF; Gurgui C; Helf MJ; Morinaka BI; Uria AR; Oldham NJ; Sahl H-G; Matsunaga S; Piel J, Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science 2012, 338 (6105), 387–390. [DOI] [PubMed] [Google Scholar]
- 12.Rust M; Helfrich EJN; Freeman MF; Nanudorn P; Field CM; Rückert C; Kündig T; Page MJ; Webb VL; Kalinowski J; Sunagawa S; Piel J, A multiproducer microbiome generates chemical diversity in the marine sponge Mycale hentscheli. Proc. Natl. Acad. Sci. U. S. A 2020, 117 (17), 9508–9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montalbán-López M; Scott TA; Ramesh S; Rahman IR; van Heel AJ; Viel JH; Bandarian V; Dittmann E; Genilloud O; Goto Y; Grande Burgos MJ; Hill C; Kim S; Koehnke J; Latham JA; Link AJ; Martínez B; Nair SK; Nicolet Y; Rebuffat S; Sahl H-G; Sareen D; Schmidt EW; Schmitt L; Severinov K; Süssmuth RD; Truman AW; Wang H; Weng J-K; van Wezel GP; Zhang Q; Zhong J; Piel J; Mitchell DA; Kuipers OP; van der Donk WA, New developments in RiPP discovery, enzymology and engineering. Nat. Prod. Rep 2021, 38 (1), 130–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gogineni V; Hamann MT, Marine natural product peptides with therapeutic potential: Chemistry, biosynthesis, and pharmacology. Biochim. Biophys. Acta 2018, 1862 (1), 81–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burkhart BJ; Schwalen CJ; Mann G; Naismith JH; Mitchell DA, YcaO-Dependent Posttranslational Amide Activation: biosynthesis, structure, and function. Chem. Rev 2017, 117 (8), 5389–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kloosterman AM; Shelton KE; van Wezel GP; Medema MH; Mitchell DA, RRE-Finder: a genome-mining tool for class-independent RiPP discovery. mSystems 2020, 5 (5), e00267–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao L; Do T; Link AJ, Mechanisms of action of ribosomally synthesized and posttranslationally modified peptides (RiPPs). J. Ind. Microbiol. Biotechnol 2021, 0, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collin F; Maxwell A, The microbial toxin microcin B17: prospects for the development of new antibacterial agents. J. Mol. Biol 2019, 431 (18), 3400–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Travin DY; Watson ZL; Metelev M; Ward FR; Osterman IA; Khven IM; Khabibullina NF; Serebryakova M; Mergaert P; Polikanov YS; Cate JHD; Severinov K, Structure of ribosome-bound azole-modified peptide phazolicin rationalizes its species-specific mode of bacterial translation inhibition. Nat. Commun 2019, 10 (1), 4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalyon B; Helaly SE; Scholz R; Nachtigall J; Vater J; Borriss R; Süssmuth RD, Plantazolicin A and B: structure elucidation of ribosomally synthesized thiazole/oxazole peptides from Bacillus amyloliquefaciens FZB42. Org. Lett 2011, 13 (12), 2996–2999. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt EW; Nelson JT; Rasko DA; Sudek S; Eisen JA; Haygood MG; Ravel J, Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. U. S. A 2005, 102 (20), 7315–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cantrell TP; Freeman CJ; Paul VJ; Agarwal V; Garg N, Mass spectrometry-based integration and expansion of the chemical diversity harbored within a marine sponge. J. Am. Soc. Mass Spectrom 2019, 30, 1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foulston LC; Bibb MJ, Microbisporicin gene cluster reveals unusual features of lantibiotic biosynthesis in actinomycetes. Proc. Natl. Acad. Sci. U. S. A 2010, 107 (30), 13461–13466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ortega MA; Cogan DP; Mukherjee S; Garg N; Li B; Thibodeaux GN; Maffioli SI; Donadio S; Sosio M; Escano J; Smith L; Nair SK; van der Donk WA, Two flavoenzymes catalyze the post-translational generation of 5-chlorotryptophan and 2-aminovinyl-cysteine during NAI-107 biosynthesis. ACS Chem. Biol 2017, 12 (2), 548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruz JCS; Iorio M; Monciardini P; Simone M; Brunati C; Gaspari E; Maffioli SI; Wellington E; Sosio M; Donadio S, Brominated variant of the lantibiotic NAI-107 with enhanced antibacterial potency. J. Nat. Prod 2015, 78 (11), 2642–2647. [DOI] [PubMed] [Google Scholar]
- 26.Haft DH; Basu MK; Mitchell DA, Expansion of ribosomally produced natural products: a nitrile hydratase- and Nif11-related precursor family. BMC Biology 2010, 8 (1), 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabler F; Nam S-Z; Till S; Mirdita M; Steinegger M; Söding J; Lupas AN; Alva V, Protein sequence analysis using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinformatics 2020, 72 (1), e108. [DOI] [PubMed] [Google Scholar]
- 28.Bhushan A; Egli PJ; Peters EE; Freeman MF; Piel J, Genome mining- and synthetic biology-enabled production of hypermodified peptides. Nat. Chem 2019, 11 (10), 931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bösch NM; Borsa M; Greczmiel U; Morinaka BI; Gugger M; Oxenius A; Vagstad AL; Piel J, Landornamides: antiviral ornithine-containing ribosomal peptides discovered through genome mining. Angew. Chem., Int. Ed 2020, 59 (29), 11763–11768. [DOI] [PubMed] [Google Scholar]
- 30.Blin K; Shaw S; Steinke K; Villebro R; Ziemert N; Lee SY; Medema MH; Weber T, antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 2019, 47 (W1), W81–W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohanty I; Tapadar S; Moore SG; Biggs JS; Freeman CJ; Gaul DA; Garg N; Agarwal V, Presence of bromotyrosine alkaloids in marine sponges is independent of metabolomic and microbiome architectures. mSystems 2021, 6 (2), e01387–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menna M; Imperatore C; Aniello F; Aiello A, Meroterpenes from marine invertebrates: structures, occurrence, and ecological implications. Mar. Drugs 2013, 11 (5), 1602–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Navarro-Muñoz JC; Selem-Mojica N; Mullowney MW; Kautsar SA; Tryon JH; Parkinson EI; De Los Santos ELC; Yeong M; Cruz-Morales P; Abubucker S; Roeters A; Lokhorst W; Fernandez-Guerra A; Cappelini LTD; Goering AW; Thomson RJ; Metcalf WW; Kelleher NL; Barona-Gomez F; Medema MH, A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol 2020, 16 (1), 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ting CP; Funk MA; Halaby SL; Zhang Z; Gonen T; van der Donk WA, Use of a scaffold peptide in the biosynthesis of amino acid–derived natural products. Science 2019, 365 (6450), 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roush RF; Nolan EM; Lohr F; Walsh CT, Maturation of an Escherichia coli ribosomal peptide antibiotic by ATP-consuming N-P bond formation in microcin C7. J. Am. Chem. Soc 2008, 130 (11), 3603–3609. [DOI] [PubMed] [Google Scholar]
- 36.Bobeica SC; Dong S-H; Huo L; Mazo N; McLaughlin MI; Jiménez-Osés G; Nair SK; van der Donk WA, Insights into AMS/PCAT transporters from biochemical and structural characterization of a double Glycine motif protease. eLife 2019, 8, e42305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deane CD; Burkhart BJ; Blair PM; Tietz JI; Lin A; Mitchell DA, In vitro biosynthesis and substrate tolerance of the plantazolicin family of natural products. ACS Chem. Biol 2016, 11 (8), 2232–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donia MS; Hathaway BJ; Sudek S; Haygood MG; Rosovitz MJ; Ravel J; Schmidt EW, Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat. Chem. Biol 2006, 2 (12), 729–735. [DOI] [PubMed] [Google Scholar]
- 39.Cubillos-Ruiz A; Berta-Thompson JW; Becker JW; van der Donk WA; Chisholm SW, Evolutionary radiation of lanthipeptides in marine cyanobacteria. Proc. Natl. Acad. Sci. U. S. A 2017, 114 (27), E5424–E5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blasiak LC; Drennan CL, Structural perspective on enzymatic halogenation. Acc. Chem. Res 2009, 42 (1), 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agarwal V; Miles ZD; Winter JM; Eustaquio AS; El Gamal AA; Moore BS, Enzymatic halogenation and dehalogenation reactions: pervasive and mechanistically diverse. Chem. Rev 2017, 117 (8), 5619–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romney DK; Murciano-Calles J; Wehrmüller JE; Arnold FH, Unlocking reactivity of TrpB: a general biocatalytic platform for synthesis of tryptophan analogues. J. Am. Chem. Soc 2017, 139 (31), 10769–10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhandari DM; Fedoseyenko D; Begley TP, Mechanistic studies on tryptophan lyase (NosL): identification of cyanide as a reaction product. J. Am. Chem. Soc 2018, 140 (2), 542–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koehnke J; Mann G; Bent AF; Ludewig H; Shirran S; Botting C; Lebl T; Houssen WE; Jaspars M; Naismith JH, Structural analysis of leader peptide binding enables leader-free cyanobactin processing. Nat. Chem. Biol 2015, 11 (8), 558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sardar D; Pierce E; McIntosh JA; Schmidt EW, Recognition sequences and substrate evolution in cyanobactin biosynthesis. ACS Synth. Biol 2015, 4 (2), 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donia MS; Ravel J; Schmidt EW, A global assembly line for cyanobactins. Nat. Chem. Biol 2008, 4 (6), 341–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unson MD; Holland ND; Faulkner DJ, A brominated secondary metabolite synthesized by the cyanobacterial symbiont of a marine sponge and accumulation of the crystalline metabolite in the sponge tissue. Mar. Biol 1994, 119 (1), 1–11. [Google Scholar]
- 48.Wilson MC; Mori T; Rückert C; Uria AR; Helf MJ; Takada K; Gernert C; Steffens UAE; Heycke N; Schmitt S; Rinke C; Helfrich EJN; Brachmann AO; Gurgui C; Wakimoto T; Kracht M; Crüsemann M; Hentschel U; Abe I; Matsunaga S; Kalinowski J; Takeyama H; Piel J, An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 2014, 506, 58–62. [DOI] [PubMed] [Google Scholar]
- 49.Agarwal V; Blanton JM; Podell S; Taton A; Schorn MA; Busch J; Lin Z; Schmidt EW; Jensen PR; Paul VJ; Biggs JS; Golden JW; Allen EE; Moore BS, Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nat. Chem. Biol 2017, 13 (5), 537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Engelberts JP; Robbins SJ; de Goeij JM; Aranda M; Bell SC; Webster NS, Characterization of a sponge microbiome using an integrative genome-centric approach. ISME J 2020, 14 (5), 1100–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loh T-L; Pawlik JR, Chemical defenses and resource trade-offs structure sponge communities on Caribbean coral reefs. Proc. Natl. Acad. Sci. U. S. A 2014, 111 (11), 4151–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Podell S; Blanton JM; Oliver A; Schorn MA; Agarwal V; Biggs JS; Moore BS; Allen EE, A genomic view of trophic and metabolic diversity in clade-specific Lamellodysidea sponge microbiomes. Microbiome 2020, 8 (1), 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burkhart BJ; Hudson GA; Dunbar KL; Mitchell DA, A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat. Chem. Biol 2015, 11 (8), 564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma SV; Tong X; Pubill-Ulldemolins C; Cartmell C; Bogosyan EJA; Rackham EJ; Marelli E; Hamed RB; Goss RJM, Living GenoChemetics by hyphenating synthetic biology and synthetic chemistry in vivo. Nat. Commun 2017, 8 (1), 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Runguphan W; O’Connor SE, Diversification of monoterpene indole alkaloid analogs through cross-coupling. Org. Lett 2013, 15 (11), 2850–2853. [DOI] [PubMed] [Google Scholar]
- 56.Craig AG; Jimenez EC; Dykert J; Nielsen DB; Gulyas J; Abogadie FC; Porter J; Rivier JE; Cruz LJ; Olivera BM; McIntosh JM, A novel post-translational modification involving bromination of tryptophan: identification of the residue, L-6-bromotryptophan in peptide from Conus imperialis and Conus radiatus venome. J. Biol. Chem 1997, 272 (8), 4689–4698. [DOI] [PubMed] [Google Scholar]
- 57.Fuchs SW; Lackner G; Morinaka BI; Morishita Y; Asai T; Riniker S; Piel J, A Lanthipeptide-like N-terminal leader region guides peptide epimerization by radical SAM epimerases: implications for RiPP evolution. Angew. Chem., Int. Ed 2016, 55 (40), 12330–12333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


