Abstract
Objectives
Accumulating evidences show that the regulatory network of m6A modification is essential for mammalian spermatogenesis. However, as an m6A reader, the roles of YTHDF2 remain enigmatic due to the lack of a proper model. Here, we employed the germ cell conditional knockout mouse model and explored the function of YTHDF2 in spermatogenesis.
Materials and methods
Ythdf2 germ cell conditional knockout mice were obtained by crossing Ythdf2‐floxed mice with Vasa‐Cre and Stra8‐Cre mice. Haematoxylin and eosin (HE) staining, immunofluorescent staining and Western blotting were used for phenotyping. CASA, IVF and ICSI were applied for sperm function analysis. RNA‐seq, YTHDF2‐RIP‐seq and quantitative real‐time PCR were used to explore transcriptome changes and molecular mechanism analysis.
Results
Our results showed that YTHDF2 was highly expressed in spermatogenic cells. The germ cell conditional knockout males were sterile, and their sperm displayed malformation, impaired motility, and lost fertilization ability. During differentiated spermatogonia transiting to pachytene spermatocyte, most m6A‐modified YTHDF2 targets that were degraded in control germ cells persisted in pachytene spermatocytes of Ythdf2‐vKO mice. These delayed mRNAs were mainly enriched in pathways related to the regulation of transcription, and disturbed the transcriptome of round spermatid and elongated spermatid subsequently.
Conclusion
Our data demonstrate that YTHDF2 facilitates the timely turnover of phase‐specific transcripts to ensure the proper progression of spermatogenesis, which highlights a critical role of YTHDF2 in spermatogenesis.
Keywords: m6A, male sterility, mRNA clearance, spermatogenesis, Ythdf2
YTHDF2 accelerates the degradation of m6A‐modified RNA during the early stages of spermatogenesis. The timely clearance of phase‐specific transcripts mediated by YTHDF2 ensures the formation of functional sperm.
1. INTRODUCTION
N6‐methyladenosine (m6A), the most abundant chemical modification in mRNA, plays crucial roles in multiple biological processes. 1 , 2 The mRNA m6A modification is dynamic and reversible. m6A is mainly installed by METTL3 and METTL14 core complex, together with several other key components, including WTAP, VIRMA and ZC3H13, 3 , 4 , 5 , 6 and this modification can also be removed by the ‘eraser’ proteins ALKBH5 and FTO. 7 , 8 Importantly, m6A exerts its regulatory functions primarily through various ‘readers’, including YTH domain‐containing proteins (YTHDF1, YTHDF2, YTHDF3, YTHDC1 and YTHDC2), eIF3, HnRNP and IGF2BP2. 9 , 10 , 11 , 12 In different biological contexts, the reader proteins specifically decipher the m6A‐decorated RNAs by recruiting distinct effectors, which are directly involved in regulating RNA stability, translation, splicing and subsequently determining the fates of m6A‐containing transcripts. 9 , 13 , 14 , 15 , 16
Accumulating evidences showed the regulatory network of m6A modification is essential for mammalian spermatogenesis. For instance, germ cell–specific inactivation of METTL3 or METTL14 induces progressive loss of spermatogonial stem cells (SSC). 17 , 18 Depletion of ALKBH5 results in abnormal spermatogenesis, due to aberrant metabolism of m6A‐marked mRNAs. 7 , 19 Previous reports demonstrated that different ‘reader’ proteins are also critical for spermatogenesis by recognizing m6A marks on specific mRNAs and mediating various processes of RNA metabolism. Among the m6A reader proteins, YTHDC1 was uncovered to be essential for SSC survival, 20 and YTHDC2, proved to be an m6A ‘reader’ by our previous research, 21 targets meiosis‐related genes and its loss induces defects in meiosis. 21 , 22 , 23 , 24 Besides YTHDC1 and YTHDC2, the abundant expression of YTHDF1, YTHDF2 and YTHDF3 can also be detected in testis, implying that these genes may be functional during spermatogenesis. However, ablation of Ythdf1 or Ythdf3 does not cause noticeable defects in spermatogenesis. 25 As to Ythdf2, its exact roles are controversial in different studies. An initial study claimed that knockout of Ythdf2 does not affect the fertility of male mice, 26 but a more recent study showed that deletion of Ythdf2 leads to male hypofertility. 25
To clarify the biological roles and underlying mechanism of YTHDF2 during spermatogenesis, in this study, we generated a conditional knockout mouse model with a deletion of the critical domain of YTHDF2, and revealed that YTHDF2 is indispensable for spermatogenesis and deletion of Ythdf2 in male germ cells resulted in sterility phenotype. Mechanistically, YTHDF2 facilitated the timely turnover of phase‐specific transcripts to ensure the proper progression of spermatogenesis.
2. MATERIALS AND METHODS
2.1. Ythdf2 cKO mouse generation
Ythdf2‐floxed mice were generated before by our laboratory. 27 , 28 We generated germ cell–specific knockout mice by crossing Ythdf2‐floxed mice with Vasa‐Cre or Stra8‐Cre mice, 18 , 29 respectively. All mice used in this study were kept at C57BL/6 genetic background, and housed under specific pathogen‐free (SPF) conditions.
2.2. Histological and immunofluorescent staining analysis
For haematoxylin and eosin (H&E) staining, testes were fixed in modified Davidson's fluid (MDF), dehydrated with increasing concentration of ethanol, embedded in paraffin and cut into 5‐μm‐thick sections. Then, the sections were deparaffinized, rehydrated and stained with haematoxylin and eosin.
For immunofluorescent staining, the sections were deparaffinized, rehydrated and boiled in sodium citrate buffer (Maxim, MVS‐0066) for 10 min. After washing three times by 0.1% Triton ×‐100 in PBS, the sections were blocked with 5% BSA and then incubated with primary antibody overnight at 4°C. Primary antibodies used in this study were as follows: anit‐YTHDF2 (Abcam, ab220163), anti‐SYCP3 (Abcam, ab15093), and anti‐γH2AX (Abcam, ab26350). On the next day, the slides were washed three times in PBST (0.1% Tween‐20 in PBS) and incubated with the secondary antibody and Hoechst at room temperature for 2 h. Finally, the slides were washed in PBST and mounted with 50% glycerol.
For spermatocyte chromosome spreads, seminiferous tubules were placed in hypotonic extraction buffer (30 mM Tris‐HCl pH 8.5, 50 mM sucrose, 17 mM citric acid, 5 mM EDTA, 2.5 mM DTT and 1 mM PMSF) for 45 min at room temperature. Then, the tissues were minced in 100 mM sucrose. 10 μl of cell suspension was pipetted onto slides and fixed by fixative (1% PFA, 0.15% Triton ×‐100, 10 mM sodium borate) for 3 h. After washing four times by TBS (150 mM NaCl, 20 mM Tris‐HCl, pH 7.6), samples were blocked with blocking buffer (1% normal donkey serum, 0.3% BSA and 0.05% Triton ×‐100) for 1 h at room temperature, and incubated with primary antibodies overnight at 37°C. On the next day, after washing 3 times by TBS, slides were blocked with blocking buffer for 5 h, and finally incubated with secondary antibodies for 1.5 h at 37°C. After washing three times by TBS, the slides were mounted with 50% glycerol.
2.3. TUNEL assay
TUNEL assay was performed with TUNEL BrightRed Apoptosis Detection Kit (Vazyme, A113‐01) according to the manufacturer's instructions. Briefly, the paraffin sections were deparaffinized, rehydrated and incubated with PBS containing Proteinase K at room temperature for 20 min. After washing two times by PBS, the sections were incubated with equilibration buffer at room temperature for 30 min, and TdT buffer at 37°C for 1 h. Then, the sections were washed three times by PBS and incubated with DAPI at room temperature for 5 min. The slides finally were washed by PBS and mounted with 50% glycerol.
2.4. Computer‐assisted sperm analysis
Sperm was collected from cauda epididymides and incubated with HTF medium (Irvine Scientific, 90125) for 5 min at 37°C. 10 μl of sperm suspension was placed in slide chamber and analysed by IVOS II system (Hamilton Thorne) with default parameters.
2.5. In vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI)
For IVF, sperm was harvested from cauda epididymides of control and vKO mice aged 10–20 weeks and incubated in HTF medium for 1 h. Cumulus–oocyte complexes (COCs) were collected from oviduct ampullae of wild‐type mice and incubated in HTF drops. Then, the sperm was added to HTF drops containing COCs and incubated at 37°C for 5 h. Finally, the presumptive zygotes were washed several times until the cumulus cells and excess sperm were removed. Then, the zygotes were transferred into KSOM medium (Millipore, MR‐020P‐D) and cultured in a humidified atmosphere at 37°C with 5% CO2.
For ICSI, sperm was isolated from cauda epididymides of control and vKO mice aged 10–20 weeks and incubated in HTF medium for 1 h. COCs were isolated from oviduct ampullae of wild‐type mice, followed by removing the cumulus cells in medium containing 0.5 mg/ml hyaluronidase (Sigma, H3506) at 37°C. Then, the dispersed sperm was injected into the MII oocyte by a microinjector. The 1‐cell development rate is calculated by dividing the number of zygotes by the number of injected MII oocytes. The 2‐cell or blastocyst rate is defined as the ratio of 2‐cell or blastocyst to the number of zygotes.
2.6. Western blot analysis
Total protein lysates were extracted with RIPA lysis buffer. Then, the protein samples were loaded and separated in a SDS‐PAGE gel, transferred onto the PVDF membranes (Millipore, IPVH00010). The membranes were blocked with 5% skimmed milk for 2 h, and then incubated with primary antibodies overnight at 4°C. On the next day, the membranes were washed four times using TBST and incubated with secondary antibody at room temperature for 2 h. Primary antibodies were as follows: anti‐GAPDH (Santa Cruz, sc‐32233) and anti‐YTHDF2 (Abcam, ab220163).
2.7. Isolation of spermatogenic cells
The STA‐PUT method was used to isolate pachytene spermatocyte, round spermatid and elongated spermatid as described previously. 30 Briefly, mouse testes were decapsulated and digested with collagenase IV (1 mg/ml) until the seminiferous tubules were dispersed. The dispersed seminiferous tubules were collected by centrifugation at 500 g. The pellet was further digested with 0.25% trypsin and filtered through a 40‐μm Nylon Cell Strainer. The cell suspension was bottom‐loaded into a cell separation apparatus, followed by a 2%–4% bovine serum albumin (BSA) gradient. Then, the cell fractions were harvested after about 2.5 h of sedimentation. Magnetic‐activated cell sorting (MACS) was used to isolate the undifferentiated and differentiating spermatogonia. Undifferentiated spermatogonia were purified using CD90.2 Positive Selection Kit II (STEM CELL, #19851) from P6.5 testes. Differentiating spermatogonia were purified using CD117 (cKIT) Positive Selection Kit (STEM CELL, #18757) from P10.5 testes.
2.8. RNA extraction and qRT‐PCR
Total RNA was extracted using RNAiso Plus (TaKaRa, #9109) according to the manufacturer's instructions. cDNA synthesis was carried out using the PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, RR047B). Real‐time RT–PCR analysis was performed using AceQ qPCR SYBR Green Master Mix (Vazyme, Q141‐03). Primer sequences are listed in Table S1.
2.9. RNA‐seq and data analysis
For bulk RNA‐seq libraries, total RNA was extracted with TRIzol from purified germ cells. mRNA libraries were prepared with VAHTS mRNA‐seq V3 Library Prep Kit for Illumina (Vazyme, NR611) according to the manufacturer's instructions, and sequenced on Illumina NovaSeq platform.
Quality control, removal of overrepresented sequences and sequencing adapters were performed with FastQC and Trim Galore, respectively. Filtered reads were aligned to GRCm38 using HISAT2 (version 2.2.1) with default parameters. Then, the gene expression level (read counts) was quantified by featureCounts (version 1.6.0). Differential expression testing was performed with R package DESeq2 (version 1.38.0). Genes with false discovery rate (FDR)–adjusted p‐value <0.05 and fold change >1.5 were marked as differentially expressed genes (DEGs). Gene ontology (GO) enrichment analysis was performed using DAVID. DEGs are listed in Table S2.
2.10. YTHDF2‐RIP‐seq and data analysis
Whole testes or purified germ cells were lysed with lysis buffer (150 mM KCl, 20 mM Tris‐HCl, pH 7.5, 2 mM EDTA, 0.5% NP‐40, 0.5 mM DTT, 1:100 protease inhibitor cocktail and 1:100 SUPERase•In RNase Inhibitor) for 30 min with gentle rotary shaking at 4°C, and then centrifuged at 16,000 g for 20 min. The supernatant was pre‐cleared with Dynabeads protein A/G beads for 1 h at 4°C. Meanwhile, 100 μl of protein A/G beads was coated with YTHDF2 antibody (ProteinTech, 24744–1‐AP) for 1 h at 4°C. 1/10 of pre‐cleared samples were saved as input, and the rest samples were incubated with the pre‐coated beads for 4 h at 4 °C. After washing 6 times with NT2 buffer (200 mM NaCl, 2 mM EDTA, 0.05% NP‐40, 50 mM Tris‐HCl pH 7.5, 0.5 mM DTT, 1:1 000 protease inhibitor and 1:1 000 SUPERase•In RNase Inhibitor), the RNAs bound to the Dynabeads were extracted with TRIzol. The recovered and input RNAs were used to generate RNA‐seq libraries following the Smart‐seq2 protocol as described previously. 31
Quality control of sequencing data was performed by FastQC and Trim Galore. FPKM was calculated by using RSEM with default parameters. The RIP target was defined as Input FPKM >1 and IP FPKM / Input FPKM >1.2. The intersection of RIP targets between two replicates was identified as the final target genes of YTHDF2. YTHDF2‐targeted genes are listed in Table S3.
The intersection of the following four data sets was identified as delayed‐decay RNAs upon YTHDF2 depletion: (1) the significant down‐regulated genes (adjusted p values <0.05 and fold change >1.5) between two development stages of wild‐type germ cells (such as SG to P period or P to RS period); (2) the m6A‐modified genes in wild‐type germ cells of previous stage (these data were acquired from the published data 18 ); (3) the YTHDF2‐targeted genes; and (4) the genes with an up‐regulated tendency between two stages in Ythdf2‐null mice (fold change >1.2).
3. RESULTS
3.1. YTHDF2 in germ cells is essential for male fertility
Considering that Ythdf2 is highly and dynamically expressed in male germ cells 29 (Figure S1A), we postulated that YTHDF2 likely plays functional roles during spermatogenesis. Immunofluorescent staining showed that YTHDF2 was mainly located in the cytoplasm of germ cells and its protein level was highest in spermatocytes, followed by spermatogonia, and lowest in haploid germ cells (Figure 1A). To determine the function of YTHDF2 in spermatogenesis, we generated Ythdf2‐KO first mouse by flanking the exon 4 with loxP sites, which encodes the critical YTH domain of YTHDF2 (Figure 1B and Figure S1B). By mating with FLPeR mice, sequentially with Vasa‐Cre transgenic mouse (Cre recombinase is initially expressed in primordial germ cells), Ythdf2 germ cell–specific knockout mice were obtained (hereafter referred to as Ythdf2‐vKO) (Figure 1B). Western blot and immunofluorescence analyses confirmed the germ cell–specific depletion of YTHDF2 in Ythdf2‐vKO testes (Figure 1C,D). In addition, the onset of high expression of YTHDF2 in P18 testis also indicated that it was mostly enriched in spermatocytes (Figure 1C). We then sought to investigate the fertility of Ythdf2‐vKO male mice by mating them with wild‐type females. Although copulation plugs could be observed normally, no offspring of Ythdf2‐vKO male mice was born (Figure 1E), suggesting that knockout of Ythdf2 in germ cells caused loss of male fertility.
FIGURE 1.
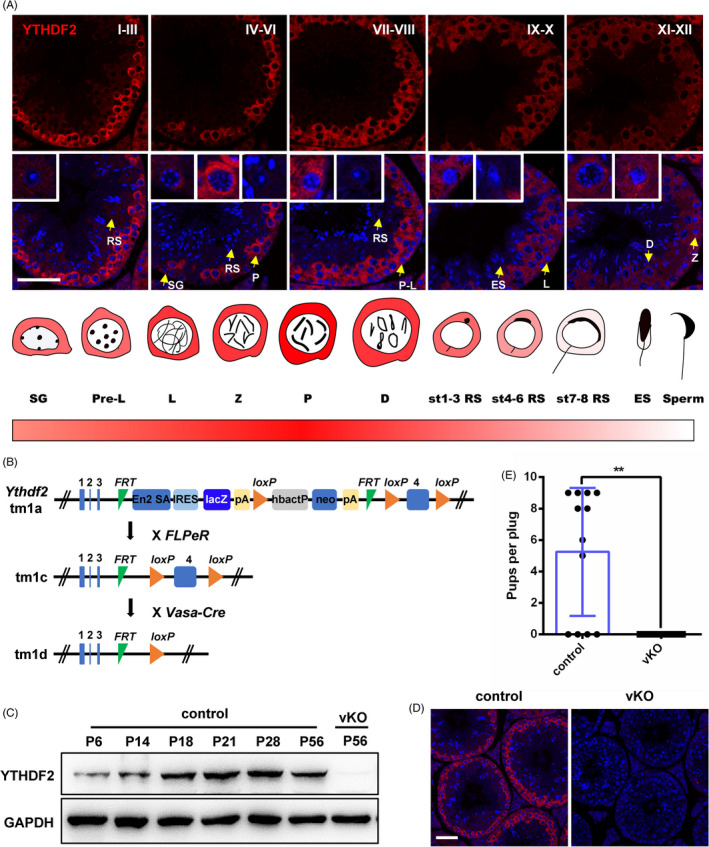
YTHDF2 is essential for male mouse fertility. (A) Immunofluorescent staining of YTHDF2 in adult testis. Scale bar, 50 μm. I‐XII indicates the cycle of the seminiferous epithelium. SG, spermatogonia; Pre‐L, pre‐leptotene spermatocyte; L, leptotene spermatocyte; Z, zygotene spermatocyte; P, pachytene spermatocyte; D, diplotene spermatocyte; RS, round spermatid; ES, elongated spermatid; st, stage. (B) Schematic diagram for the Ythdf2 targeting strategy. (C) Western blot analysis of YTHDF2 in control and Ythdf2‐vKO testes at different ages. P, postnatal day. (D) Immunofluorescent staining of YTHDF2 in control and Ythdf2‐vKO testes. Scale bar, 50 μm. (E) Litter size per plug from Ythdf2‐vKO and control mice. Data are presented as means ± SD (n ≥ 9 for each group). Significance was calculated with unpaired two‐tailed Student's t‐test (** p < 0.01)
3.2. Ythdf2 deficiency leads to sperm defects and loss of fertilization capacity
To examine the male infertility of Ythdf2‐vKO mice in detail, we first analysed the gross morphology of testes. No noticeable difference was observed in testis size and weight between Ythdf2‐vKO and control mice (Figure 2A,B). Then, we performed computer‐assisted sperm analysis (CASA) and found that the count of epididymal sperm significantly reduced in Ythdf2‐vKO mice (Figure 2C). CASA result showed that the motility and progressive motility of Ythdf2‐vKO sperm were markedly impaired (Figure 2D,E). Histological analysis of sperm collected from cauda epididymides indicated that more than 40% of sperms were deformed with various structural abnormalities (Figure 2F,G and Figure S2A). TUNEL assay revealed that the apoptotic cells increased significantly in Ythdf2‐vKO testes compared with the control (Figure 2H,I), and the apoptosis signal was mainly detected in elongated spermatids, suggesting that these sperm cells may be retained and phagocytosed by Sertoli cells (Figure 2H).
FIGURE 2.
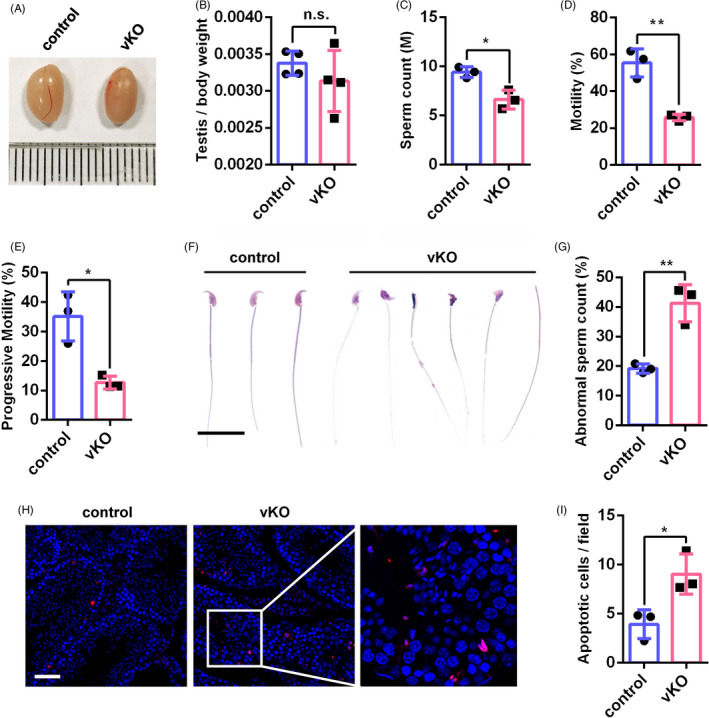
Ythdf2 deletion results in sperm defects. (A) Gross morphology of representative testes from adult control and age‐matched Ythdf2‐vKO. (B) The testis/body weight ratio in adult control and Ythdf2‐vKO mice. (C) The sperm count in cauda epididymides of adult control and age‐matched Ythdf2‐vKO mice. M, million. (D) and (E) CASA assay of motility and progressive motility of sperm from adult control and age‐matched Ythdf2‐vKO mice. (F) Haematoxylin and eosin‐stained sperm collected from control and Ythdf2‐vKO cauda epididymides. Scale bar, 20 μm. (G) The percentage of abnormal sperm in adult control and Ythdf2‐vKO mice. (H) TUNEL assay of adult control and age‐matched Ythdf2‐vKO testes. Scale bar, 50 μm. (I) Quantification of apoptotic cells in adult control and age‐matched Ythdf2‐vKO testes. Data are presented as means ± SD (B, n = 4 for each group; C‐E, G and I, n = 3 for each group). Significance was calculated with unpaired two‐tailed Student's t‐test (n.s., not significant, * p < 0.05, ** p < 0.01)
To investigate whether the Ythdf2‐vKO sperm is functionally normal in fertilization, in vitro fertilization (IVF) was carried out and these sperm failed to fertilize the wild‐type oocytes (Figure S2B). Interestingly, intracytoplasmic sperm injection (ICSI) of Ythdf2‐vKO sperm showed that fertilized eggs were able to develop to blastocytes normally, in spite of lower fertilization rates of Ythdf2‐null sperm than that of wild‐type sperm (Figure S2C, D). These results suggested that Ythdf2 deficiency impaired the natural fertilization ability of sperm.
Since YTHDF2 is highly expressed in spermatogonia and spermatocytes, we reasoned that Ythdf2‐vKO mice might display abnormalities in the early stage of spermatogenesis. Unexpectedly, Ythdf2‐vKO mice showed normal spermatogenesis cycle (Figure S3A). Then, we performed chromosome spreads of spermatocytes and immunofluorescent staining with γH2AX (DNA double strand break marker) and SYCP3 (an axial element protein of synaptonemal complex) to examine the meiosis process. The results indicated that the expression and localization of γH2AX and SYCP3 appeared normal at the four stages (leptotene, zygotene, pachytene and diplotene) of Ythdf2‐vKO spermatocytes (Figure S3B), and there was no significant difference in the ratio of the four kinds of spermatocytes compared with the control mice (Figure S3C), suggesting that the absence of YTHDF2 has no obvious effect towards meiosis.
A previous study reported that YTHDF2 regulates the migration and proliferation of GC‐1 spermatogonial cell line. 32 Then, we asked whether YTHDF2 functions in spermatogonia in vivo. Immunofluorescent staining indicated that there is no noticeable difference in the number of PLZF‐positive spermatogonia between Ythdf2‐vKO and control mice (Figure S4A, B), implying that YTHDF2 is unlikely to be essential for spermatogonia. To further narrow down the exact stage when YTHDF2 regulates spermatogenesis, we crossed Ythdf2‐floxed mice with Stra8‐Cre mice, which induces recombination starting from type A1 spermatogonia, and obtained a Ythdf2‐null model before meiosis initiation (Ythdf2‐sKO). As expected, immunofluorescent staining result demonstrated that YTHDF2 was still readily detected in spermatogonia, while it disappeared thereafter in Ythdf2‐sKO testes (Figure S5A). Similar to Ythdf2‐vKO mice, although the overall morphology of Ythdf2‐sKO testis was comparable with the control (Figure S5B, C), the sperm count, motility and progressive motility in Ythdf2‐sKO were all substantially decreased (Figure S5D‐F), along with the increased number of deformed sperm and loss of fertilizing capacity (Figure S5G‐I). The TUNEL result also revealed that the apoptotic sperm in the testes of Ythdf2‐sKO mice increased significantly (Figure S5J).
Taken together, these results demonstrate that Ythdf2 deficiency apparently does not affect the early stage of spermatogenesis, but leads to sperm defects and loss of fertilization capacity.
3.3. YTHDF2 depletion in germ cells disturbs transcriptome during spermatogenesis
To explore the underlying mechanism of sperm defects upon YTHDF2 depletion, we systematically illuminated genome‐wide gene expression changes induced by Ythdf2 deletion in germ cells. Firstly, we isolated undifferentiated spermatogonia, differentiated spermatogonia, pachytene spermatocytes, round spermatids and elongated spermatids. Then, we performed RNA sequencing of the purified germ cells and gene expression analysis. The result demonstrated that the transcriptomic profiles of various stages of germ cells from control mice were comparable to the previous report, 33 indicating a high purity of the isolated germ cells (Figure S6A). Principal component analysis (PCA) of gene expression showed the transcriptome diverged along with spermatogenesis between control and Ythdf2‐vKO mice (Figure 3A). Only a few number of genes are altered in undifferentiated and differentiated spermatogonia of Ythdf2‐vKO compared with the control mice (Figure 3B and Figure S6B, C), consistent with our above observation that Ythdf2 deficiency does not influence the self‐renewal and differentiation of spermatogonia (Figure S4A, B). After pachytene stage, the number of differentially expressed genes (DEGs) between Ythdf2‐vKO and control germ cells steadily increased (Figure 3B and Figure S6D‐F). For instance, in differentiated spermatogonia, loss of YTHDF2 does not cause considerable transcriptomic changes, while in the elongated spermatid, there were as many as 1056 genes up‐regulated and 2309 genes down‐regulated (Figure 3B). Gene ontology (GO) analysis showed that the down‐regulated DEGs in round spermatid were significantly enriched in sperm‐egg fusion, spermatid development and acrosome assembly (Figure 3C); in elongated spermatid, the down‐regulated DEGs were mainly associated with sperm motility, cilium assembly, binding of sperm to zona pellucida and chromosome condensation (Figure 3D). Many DEGs in elongated spermatid have been reported to be closely related to spermiogenesis. For example, Smcp, Sord, Atp1a4, Cdc14a and Pgk2 are all important for sperm motility 34 , 35 , 36 , 37 , 38 ; Akap4 and Spem1 deletion results in sperm deformity 39 , 40 ; and Pmis2 null sperm fails to bind to the zona pellucida 41 (Figure 3E). Together, the transcriptome of Ythdf2‐vKO germ cells changed mainly thereafter pachytene stage, and the DEGs were coincident with the sperm malformation, impaired motility and loss of fertilization capacity in Ythdf2‐vKO mice.
FIGURE 3.
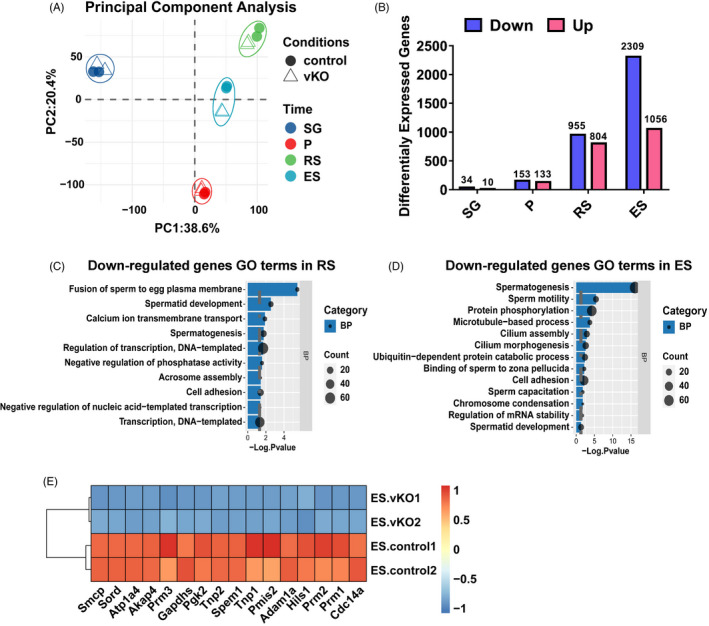
YTHDF2 depletion disrupts transcriptome in late spermatogenesis. (A) PCA of transcriptome from various stages of purified germ cells. (B) The number of dysregulated genes in various stages of purified germ cells. (C) GO analysis of the down‐regulated genes in round spermatids of Ythdf2‐vKO mice. (D) GO analysis of the down‐regulated genes in elongated spermatids of Ythdf2‐vKO mice. (E) Expression of DEGs in elongated spermatids of control and Ythdf2‐vKO mice
3.4. YTHDF2 is required for m6A‐modified mRNA clearance
Previous studies have shown that YTHDF2 preferentially recognizes m6A‐modified mRNA and mediates their rapid degradation in many biological contexts. 27 , 42 , 43 , 44 , 45 Based on the expression pattern of YTHDF2 and the phenotype of Ythdf2‐vKO mice, we focused on gene expression changes in stages from differentiated spermatogonia‐to‐pachytene spermatocyte transition, and pachytene spermatocyte‐to‐round spermatid transition. Transcriptomic analysis showed that there were 4645 genes down‐regulated during the transition from differentiated spermatogonia to pachytene spermatocyte (Figure 4A), and there were 7041 genes down‐regulated during the transition from pachytene spermatocyte to round spermatid (Figure S8B). To explore the biological mechanism of YTHDF2 in testis, we carried out YTHDF2‐RIP‐seq to identify the direct targets of YTHDF2. The correlation of two replicates of YTHDF2‐targeted transcripts from adult whole testes and pachytene spermatocytes was high positive and moderate positive, respectively (Figure S7A, E). The YTHDF2 enrichment genes were comparable between two replicates (Figure S7B‐D, F‐H), indicating that the targets of YTHDF2 we obtained were reliable. The RIP‐seq of whole testes identified 4273 YTHDF2‐targeted transcripts, among which 2509 targets were m6A‐modified (Figure 4A). RIP‐seq of pachytene spermatocytes identified 3012 YTHDF2‐targeted transcripts, of which 2063 targets were also marked with m6A (Figure S8B).
FIGURE 4.
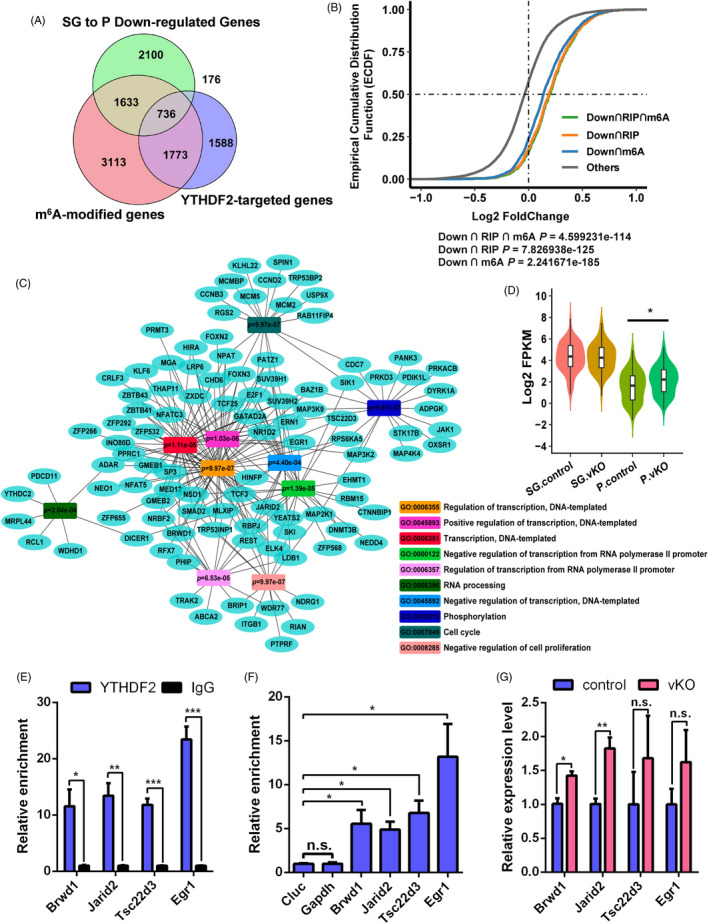
YTHDF2 promotes the degradation of its target mRNA. (A) Venn diagram showing that overlaps among genes down‐regulated from control differentiated spermatogonia to pachytene spermatocyte, m6A‐modified genes and YTHDF2‐targeted genes. The m6A‐seq data were acquired from a published data 18 . YTHDF2‐targeted genes were identified in adult whole testes. (B) Cumulative distributions of relative stability change between control and Ythdf2‐vKO pachytene spermatocytes. (C) GO analysis of genes with delayed degradation from differentiated spermatogonia to pachytene spermatocyte. (D) The RNA expression level of transcription‐associated genes at different stages of germ cells. Significance was calculated with two‐tailed Mann‐Whitney test (*p < 0.05). (E) YTHDF2‐targeted candidates verified by YTHDF2‐RIP‐qPCR in control and Ythdf2‐vKO testes. IgG was used as negative control. (F) m6A‐enriched candidates verified by m6A‐RIP‐qPCR in control and Ythdf2‐vKO testes. Non‐m6A‐modified gene Cluc was used as a negative control. (G) qPCR validation of delayed genes in control and Ythdf2‐vKO pachytene spermatocytes. Data of (E‐G) are presented as means ± SEM (n = 3 for each group). Significance was calculated with unpaired two‐tailed Student's t‐test (n.s., not significant, *p < 0.05, **p < 0.01, ***p < 0.001)
We next sought to compare the degradation rates of YTHDF2‐targeted mRNA during spermatogenesis. During differentiated spermatogonia transiting to pachytene spermatocyte, 736 m6A‐modified YTHDF2 targets were degraded in control germ cells (Figure 4A), while in Ythdf2‐vKO germ cells, almost all of them persisted in pachytene spermatocytes, with 233 genes fold change >1.2 (Figure 4B and Figure S8A). Remarkably, GO analysis showed that these delayed mRNAs were mainly enriched in pathways related to the regulation of transcription (Figure 4C), and the transcription‐associated genes showed a higher level in Ythdf2‐vKO than that in control mice (Figure 4D). Among the aberrant degradation of transcription‐associated genes, such as Brwd1, Jarid2, Egr1 and Tsc22d3, all of these genes were m6A‐modified and YTHDF2‐targeted, and they could be independently validated by real‐time qPCR (Figure 4E‐G). Interestingly, BRWD1 contains a Bromodomain and is closely related to chromatin remodelling 46 ; JARID2 is a transcription repressor that recruits Polycomb repressive complex 2 (PRC2) to regulate gene expression. 47 The delayed degradation of these transcription‐associated genes may cause abnormal transcription in the subsequent stages via a transcriptional cascade effect. 48 Indeed, from pachytene spermatocyte to elongated spermatid, we observed a 10.8‐fold increase (286 to 3365) in the number of DEGs (Figure 3B).
In the transition from pachytene spermatocytes to round spermatid, 1481 m6A‐modified YTHDF2 targets were degraded (Figure S8B), and 389 of them showed delayed decay in Ythdf2‐vKO round spermatid (Figure S8C, D). GO analysis showed that most of these delayed mRNAs were engaged in cell cycle and transcription (Figure S8E). Although YTHDF2 is more likely involved in this process based on its expression pattern and the phenotype of Ythdf2‐vKO mice, we cannot rule out the possibility that these alterations were caused indirectly, because a large number of transcription‐related factors have been abnormally expressed in the previous stage. Considering that YTHDF2 is still low expressed in round spermatid, we intended to explore whether the DEGs in round spermatid are directly regulated by YTHDF2 via m6A modification. Only 8.27% (79/955) down‐regulated genes and 4.48% (36/804) up‐regulated genes were both YTHDF2‐targeted and m6A‐modified (Figure S9A, B), and these genes had no obvious relationship with round spermatid development, suggesting YTHDF2 may have no direct effect on round spermatid development.
In summary, the improper accumulation of mRNA induced by YTHDF2 depletion may be the cause of abnormalities in the later spermatogenesis stages, ultimately leading to male sterility.
4. DISCUSSION
m6A is the most prevalent modification of eukaryotic mRNA. 49 It has been reported to be involved in almost every aspect of RNA metabolism, including mRNA splicing, transport, stability and translation. 9 , 13 , 50 An increasing body of studies have demonstrated that various biological processes are tightly regulated by m6A modification, such as haematopoiesis, neurodevelopment and spermatogenesis. 18 , 19 , 51 , 52 Spermatogenesis is a complex and multi‐stage biological process involving self‐renewal and differentiation of spermatogonia, meiosis and spermiogenesis. 53 , 54 During spermatogenesis, the germ cells undergo dramatic changes in transcriptomic, epigenomic, proteomic and metabolic levels, with the biogenesis and removal of various biomolecules in distinct stages. 55 Single‐cell RNA‐seq has shown that spermatogenic cells of different stages have their own unique transcriptomic characteristics, which are sophisticatedly regulated. 29 YTHDF2, an m6A binding protein, is reported to be critical for mediating mRNA decay. 14 , 45 By deleting exon 2 of Ythdf2, an initial study found these conventional knockout males on a mixed genetic background possess normal reproductive capability, 26 while in a later study, the authors used CRISRP/Cas9 to delete exon 4 of Ythdf2, and found that the surviving KO males on a mixed genetic background are subfertile. 25 These findings indicate that different targeting strategies and genetic background might lead to distinct outcomes. In this study, we generated the germ cell–specific knockout mice on a C57BL/6 genetic background to dissect the exact role of YTHDF2 during spermatogenesis, and uncovered that the global transcriptional dysregulation may be the major cause that contributes to sperm deformity and infertility upon Ythdf2 deletion. Mechanistically, we demonstrated that the proper and timely mRNA clearance mediated by YTHDF2 is essential for spermatogenesis.
As m6A ‘reader’ proteins, YTH family members have been shown to be crucial for mRNA processing, stability and translation. Interestingly, their expressions are quite dynamic in germ cells. During spermatogenesis, Ythdc1 deletion results in progressive loss of spermatogonia. 20 Ythdc2 KO mice are infertile and exhibit meiotic arrest due to inefficient translation and degradation of YTHDC2‐targeted mRNA. 21 , 22 In our study, we showed that Ythdf2 deletion has no effect on the self‐renew and differentiation of spermatogonia, but it is indispensable in later developmental stages of spermatogenesis. Our results showed that YTHDF2 is mainly expressed in spermatogonia and spermatocytes, and its expression is decreased rapidly in the later stages. We collected different stages of germ cells to perform RNA‐seq analysis, which showed that the dysregulated genes are significantly increased at spermatid. Given YTHDF2 expression is very low in spermatid, most likely, the transcriptomic disorder might be a secondary effect. Similarly, Tang et al. reported that ALKBH5 depletion results in abnormal spermatogenesis by affecting transcript changes at pachytene stage and round spermatid stage. 19 Mice with inactivated both Mettl3 and Mettl14 in advanced germ cells with Stra8‐Cre show normal meiosis, decreased sperm count and destroyed sperm motility, 18 which is very similar to the phenotype of our Ythdf2‐vKO and Ythdf2‐sKO mice, indicating that YTHDF2 is in charge of regulating the m6A‐decorated transcripts in late spermatogenesis.
An analysis of the m6A methylome of spermatogenic cells at different developmental stages uncovered that there are a lot of identical transcripts marked by m6A at several different stages. 18 Our analysis of YTHDF2‐RIP‐seq at different developmental stages indicated that some of the above‐mentioned transcripts are also YTHDF2 targets at different stages, such as Brwd1 and Jarid2. They are all expressed at both spermatogonia and pachytene spermatocyte, meanwhile, they are also targeted by YTHDF2 at both stages. It raises an open question concerning how YTHDF2 is switched on at the right time and place. An attractive hypothesis is that YTHDF2 may accelerate mRNA clearance only when it becomes redundant, which deserves further study.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHOR CONTRIBUTIONS
BS, HC and XJG conceived the experiments and designed the experiments. MQ performed all experiments with the help of BS, YG, YZ, XYG, XC and FW. HS and JJ performed all computational analyses. HM and XJG critically revised the manuscript. MQ, BS and HC wrote the manuscript. All authors contributed to the study and approved the submitted version.
Supporting information
Supplementary Material
Table S2
Table S3
ACKNOWLEDGMENTS
We would like to thank Dr. Ming‐Han Tong at the CAS Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, for kindly gifting the Stra8‐Cre mice. This work was supported by the National Key Research and Development Program of China (2016YFC1000600 to BS) and the National Natural Science Foundation of China (31970796 to BS).
Qi M, Sun H, Guo Y, et al. m6A reader protein YTHDF2 regulates spermatogenesis by timely clearance of phase‐specific transcripts. Cell Prolif.2022;55:e13164. doi: 10.1111/cpr.13164
Meijie Qi, Haifeng Sun and Yueshuai Guo contributed equally to this work.
Contributor Information
Xuejiang Guo, Email: guo_xuejiang@njmu.edu.cn.
Hao Chen, Email: chenh7@sustech.edu.cn.
Bin Shen, Email: binshen@njmu.edu.cn.
DATA AVAILABILITY STATEMENT
The high‐throughput sequencing data have been deposited to the NCBI Sequence Read Archive (SRA) database (accession ID, PRJNA761327).
REFERENCES
- 1. Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187‐1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heck AM, Wilusz CJ. Small changes, big implications: the impact of m(6)A RNA methylation on gene expression in pluripotency and development. Biochim Biophys Acta Gene Regul Mech. 2019;1862(9):194402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu J, Yue Y, Han D, et al. A METTL3‐METTL14 complex mediates mammalian nuclear RNA N6‐adenosine methylation. Nat Chem Biol. 2014;10(2):93‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ping XL, Sun BF, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6‐methyladenosine methyltransferase. Cell Res. 2014;24(2):177‐189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wen J, Lv R, Ma H, et al. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self‐renewal. Mol Cell. 2018;69(6):1028‐1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yue Y, Liu J, Cui X, et al. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jia G, Fu Y, Zhao X, et al. N6‐methyladenosine in nuclear RNA is a major substrate of the obesity‐associated FTO. Nat Chem Biol. 2011;7(12):885‐887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shi H, Wei J, He C. Where, when, and how: context‐dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74(4):640‐650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alarcón CR, Goodarzi H, Lee H, et al. HNRNPA2B1 is a mediator of m(6)A‐dependent nuclear RNA processing events. Cell. 2015;162(6):1299‐1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang H, Weng H, Sun W, et al. Recognition of RNA N(6)‐methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meyer KD, Patil DP, Zhou J, et al. 5’ UTR m(6)A promotes Cap‐independent translation. Cell. 2015;163(4):999‐1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roignant JY, Soller M. m(6)A in mRNA: an ancient mechanism for fine‐tuning gene expression. Trends Genet. 2017;33(6):380‐390. [DOI] [PubMed] [Google Scholar]
- 14. Du H, Zhao Y, He J, et al. YTHDF2 destabilizes m(6)A‐containing RNA through direct recruitment of the CCR4‐NOT deadenylase complex. Nat Commun. 2016;7:12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xiao W, Adhikari S, Dahal U, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61(4):507‐519. [DOI] [PubMed] [Google Scholar]
- 16. Wang X, Zhao BS, Roundtree IA, et al. N(6)‐methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388‐1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu K, Yang Y, Feng GH, et al. Mettl3‐mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017;27(9):1100‐1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin Z, Hsu PJ, Xing X, et al. Mettl3‐/Mettl14‐mediated mRNA N(6)‐methyladenosine modulates murine spermatogenesis. Cell Res. 2017;27(10):1216‐1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tang C, Klukovich R, Peng H, et al. ALKBH5‐dependent m6A demethylation controls splicing and stability of long 3'‐UTR mRNAs in male germ cells. Proc Natl Acad Sci USA. 2018;115(2):E325‐E333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kasowitz SD, Ma J, Anderson SJ, et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018;14(5):e1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsu PJ, Zhu Y, Ma H, et al. Ythdc2 is an N(6)‐methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27(9):1115‐1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wojtas MN, Pandey RR, Mendel M, et al. Regulation of m(6)A transcripts by the 3'–>5’ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol Cell. 2017;68(2):374‐387. [DOI] [PubMed] [Google Scholar]
- 23. Jain D, Puno MR, Meydan C, et al. ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. Elife. 2018;7:e30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bailey AS, Batista PJ, Gold RS, et al. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. Elife. 2017;6:e26116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lasman L, Krupalnik V, Viukov S, et al. Context‐dependent functional compensation between Ythdf m(6)A reader proteins. Genes Dev. 2020;34(19–20):1373‐1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ivanova I, Much C, Di Giacomo M, et al. The RNA m(6)A reader YTHDF2 is essential for the post‐transcriptional regulation of the maternal transcriptome and oocyte competence. Mol Cell. 2017;67(6):1059‐1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Z, Qian P, Shao W, et al. Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 2018;28(9):904‐917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang H, Zuo H, Liu J, et al. Loss of YTHDF2‐mediated m(6)A‐dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res. 2018;28(10):1035‐1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen Y, Zheng Y, Gao Y, et al. Single‐cell RNA‐seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 2018;28(9):879‐896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gan H, Wen L, Liao S, et al. Dynamics of 5‐hydroxymethylcytosine during mouse spermatogenesis. Nat Commun. 2013;4:1995. [DOI] [PubMed] [Google Scholar]
- 31. Picelli S, Faridani OR, Bjorklund AK, et al. Full‐length RNA‐seq from single cells using Smart‐seq2. Nat Protoc. 2014;9(1):171‐181. [DOI] [PubMed] [Google Scholar]
- 32. Huang T, Liu Z, Zheng Y, et al. YTHDF2 promotes spermagonial adhesion through modulating MMPs decay via m(6)A/mRNA pathway. Cell Death Dis. 2020;11(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Green CD, Ma Q, Manske GL, et al. A comprehensive roadmap of murine spermatogenesis defined by single‐cell RNA‐seq. Dev Cell. 2018;46(5):651‐667 e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nayernia K, Adham IM, Burkhardt‐Gottges E, et al. Asthenozoospermia in mice with targeted deletion of the sperm mitochondrion‐associated cysteine‐rich protein (Smcp) gene. Mol Cell Biol. 2002;22(9):3046‐3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cao W, Aghajanian HK, Haig‐Ladewig LA, et al. Sorbitol can fuel mouse sperm motility and protein tyrosine phosphorylation via sorbitol dehydrogenase. Biol Reprod. 2009;80(1):124‐133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McDermott JP, Sanchez G, Chennathukuzhi V, et al. Green fluorescence protein driven by the Na, K‐ATPase alpha4 isoform promoter is expressed only in male germ cells of mouse testis. J Assist Reprod Genet. 2012;29(12):1313‐1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wen Z, Zhu H, Zhang A, et al. Cdc14a has a role in spermatogenesis, sperm maturation and male fertility. Exp Cell Res. 2020;395(1):112178. [DOI] [PubMed] [Google Scholar]
- 38. Danshina PV, Geyer CB, Dai Q, et al. Phosphoglycerate kinase 2 (PGK2) is essential for sperm function and male fertility in mice. Biol Reprod. 2010;82(1):136‐145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fang X, Huang LL, Xu J, et al. Proteomics and single‐cell RNA analysis of Akap4‐knockout mice model confirm indispensable role of Akap4 in spermatogenesis. Dev Biol. 2019;454(2):118‐127. [DOI] [PubMed] [Google Scholar]
- 40. Zheng H, Stratton CJ, Morozumi K, et al. Lack of Spem1 causes aberrant cytoplasm removal, sperm deformation, and male infertility. Proc Natl Acad Sci USA. 2007;104(16):6852‐6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yamaguchi R, Fujihara Y, Ikawa M, et al. Mice expressing aberrant sperm‐specific protein PMIS2 produce normal‐looking but fertilization‐incompetent spermatozoa. Mol Biol Cell. 2012;23(14):2671‐2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang X, Lu Z, Gomez A, et al. N6‐methyladenosine‐dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117‐120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen M, Wei L, Law CT, et al. RNA N6‐methyladenosine methyltransferase‐like 3 promotes liver cancer progression through YTHDF2‐dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254‐2270. [DOI] [PubMed] [Google Scholar]
- 44. Zhang C, Huang S, Zhuang H, et al. YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene. 2020;39(23):4507‐4518. [DOI] [PubMed] [Google Scholar]
- 45. Li M, Zhao X, Wang W, et al. Ythdf2‐mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 2018;19(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Philipps DL, Wigglesworth K, Hartford SA, et al. The dual bromodomain and WD repeat‐containing mouse protein BRWD1 is required for normal spermiogenesis and the oocyte‐embryo transition. Dev Biol. 2008;317(1):72‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Celik H, Koh WK, Kramer AC, et al. JARID2 functions as a tumor suppressor in myeloid neoplasms by repressing self‐renewal in hematopoietic progenitor cells. Cancer Cell. 2018;34(5):741‐756 e748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: initial rates, not steady state, determine network kinetics. Proc Natl Acad Sci USA. 2003;100(16):9371‐9376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149(7):1635‐1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Geula S, Moshitch‐Moshkovitz S, Dominissini D, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347(6225):1002‐1006. [DOI] [PubMed] [Google Scholar]
- 51. Paris J, Morgan M, Campos J, et al. Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25(1):137‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Widagdo J, Anggono V. The m6A‐epitranscriptomic signature in neurobiology: from neurodevelopment to brain plasticity. J Neurochem. 2018;147(2):137‐152. [DOI] [PubMed] [Google Scholar]
- 53. Griswold MD. Spermatogenesis: the commitment to meiosis. Physiol Rev. 2016;96(1):1‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Clermont Y. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiol Rev. 1972;52(1):198‐236. [DOI] [PubMed] [Google Scholar]
- 55. Gan H, Cai T, Lin X, et al. Integrative proteomic and transcriptomic analyses reveal multiple post‐transcriptional regulatory mechanisms of mouse spermatogenesis. Mol Cell Proteomics. 2013;12(5):1144‐1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Table S2
Table S3
Data Availability Statement
The high‐throughput sequencing data have been deposited to the NCBI Sequence Read Archive (SRA) database (accession ID, PRJNA761327).


