Abstract
Current clinical phenomenological diagnosis in psychiatry neither captures biologically homologous disease entities nor allows for individualized treatment prescriptions based on neurobiology. In this report, we studied two large samples of cases with schizophrenia, schizoaffective, and bipolar I disorder with psychosis, presentations with clinical features of hallucinations, delusions, thought disorder, affective, or negative symptoms. A biomarker approach to subtyping psychosis cases (called psychosis Biotypes) captured neurobiological homology that was missed by conventional clinical diagnoses. Two samples (called “B-SNIP1” with 711 psychosis and 274 healthy persons, and the “replication sample” with 717 psychosis and 198 healthy persons) showed that 44 individual biomarkers, drawn from general cognition (BACS), motor inhibitory (stop signal), saccadic system (pro- and anti-saccades), and auditory EEG/ERP (paired-stimuli and oddball) tasks of psychosis-relevant brain functions were replicable (r’s from .96–.99) and temporally stable (r’s from .76–.95). Using numerical taxonomy (k-means clustering) with nine groups of integrated biomarker characteristics (called bio-factors) yielded three Biotypes that were virtually identical between the two samples and showed highly similar case assignments to subgroups based on cross-validations (88.5%–89%). Biotypes-1 and -2 shared poor cognition. Biotype-1 was further characterized by low neural response magnitudes, while Biotype-2 was further characterized by overactive neural responses and poor sensory motor inhibition. Biotype-3 was nearly normal on all bio-factors. Construct validation of Biotype EEG/ERP neurophysiology using measures of intrinsic neural activity and auditory steady state stimulation highlighted the robustness of these outcomes. Psychosis Biotypes may yield meaningful neurobiological targets for treatments and etiological investigations.
Keywords: psychosis, classification, Biotypes, biomarkers, EEG, cognition, saccades
Conventional systems for psychosis diagnosis1–3 are primarily experiential; they do not incorporate biomarkers for differentiating individual cases by subtype. Hyman4 stated “…a laboratory-based system will be required [for] additional substantial improvements” in psychiatric diagnoses. The Bipolar-Schizophrenia Network for Intermediate Phenotypes (B-SNIP1) consortium sought to identify biomarker features distinguishing the three leading psychosis diagnoses, schizophrenia (SZ), schizoaffective disorder (SAD), and bipolar I disorder with psychosis (BDP). Two key elements were required: (1) large samples across multiple diagnoses were needed to capture heterogeneity within and between diagnoses and support the required computations. Small samples could fail to demarcate neurobiologies and/or to adequately capture hard-to-classify, nonprototypical cases; (2) a broad range of biomarkers5 encompassing the neuro-cognitive6–10 and physiological11–13 correlates of these heterogeneous syndromes.
Based on experience within the Psychiatric Genetics Consortium, Sullivan et al14 (p. 25) stated the range of genetic findings using conventional clinical diagnoses “strongly suggest that our diagnostic categories do not define pathophysiological entities.” Identification of promising neurobiological entities within idiopathic psychosis could support such etiological investigations and advance treatment developments. Previous investigators have proposed neurobiologically distinct subgroups of psychoses using biomarkers.15–17 B-SNIP1 also developed a neurobiological transdiagnostic model using numerical taxonomy of biomarker data yielding three “psychosis Biotypes”.18 B-SNIP’s strengths included large transdiagnostic samples estimating the relative proportions of psychosis cases from different classes, a more diverse biomarker panel than previously available, use of first-degree relatives, measures not used in numerical taxonomy for concurrent and construct validation,19 and unique parsing of biomarker variance in psychosis only.
Ioannidis20 questioned the replicability of many scientific findings, notably in psychology and psychiatry.21 Replication supports confidence in outcomes,4,22,23 so replicating the B-SNIP1 psychosis Biotypes was crucial given their implications for psychosis subtyping, development of laboratory-assisted diagnosis in psychiatry, and future treatment developments via patient stratification. We present such a replication and cross-validation, plus construct validation of critical biomarker features, in a new sample of similarly large size. The remarkable similarities between B-SNIP-1 and replication samples illustrate the promise of biomarker-defined psychosis Biotypes for capturing actionable neurobiological knowledge for treatment targeting.24
Methods
The National Institute of Mental Health (NIMH), through the National Data Archive (NDA), is responsible for storage of and managing access to all data used in this manuscript. Instructions for requesting access are provided at the beginning of the Supplementary Methods. Data collection strategies were the same for B-SNIP125–29 and replication samples.30–33 Data analyses for our initial Biotypes paper18 were updated for electroencephalography (EEG) and event-related potentials (ERPs), with data for B-SNIP1 and replication projects quantified using the same procedures. This meant re-quantifying all B-SNIP1 subjects using updated procedures. Differences from B-SNIP1 quantification procedures and outcomes are underlined in the main text and Supplementary Methods.
Subjects
Subject recruitment, interviews, and laboratory data collection were completed at B-SNIP consortium sites (full details on recruitment and clinical and demographic characteristics for B-SNIP1 are available in Tamminga et al34; those same procedures were followed for the replication sample). The Institutional Review Board at every participating institution approved the projects; all subjects provided informed consent prior to participation after they obtained a complete study description. Clinically stable participants were administered the Structured Clinical Interview for Diagnostic and Statistical Manual of Mental Disorders IV (DSM-IV-TR35). Persons meeting a diagnosis for SZ, SAD, or BDP were rated on the Birchwood Social Functioning,36 Global Assessment of Functioning, Montgomery-Asberg Depression Rating,37 Positive and Negative Syndrome,38 and Young Mania Rating39 scales. Healthy persons were free of lifetime psychosis syndromes, recurrent mood syndromes, and a history of psychosis or bipolar disorders in their first-degree relatives.40 All participants were rated on the Hollingshead Two-Factor Socioeconomic Rating Scale. B-SNIP1 had 711 psychosis and 274 healthy participants18; the replication sample had 717 psychosis cases and 198 healthy persons. See Supplementary tables 1–2 for demographics and clinical characteristics, Supplementary tables 3 for clinical characteristics group comparisons, and Supplementary tables 4 and 5 for medication comparisons. As shown in those tables, the two samples were remarkably similar on demographic characteristics.
Biomarker Panel for Biotypes
Biomarkers were selected given known deviations in psychosis at the time of B-SNIP1 initiation,41 and included laboratory tests indexing neurocognitive, perceptual, and physiological systems of relevance to psychosis. The included measures are traditional “endophenotypes” 42: (1) Brief Assessment of Cognition in Schizophrenia (BACS43,44), assesses global neuropsychological functioning (psychosis cases have impaired cognition25,30); (2) prosaccades measure speed of visual orienting (psychosis cases show variably slowed or speeded responses45,46); (3) anti-saccades assess inhibitory control under perceptual conflict because the visual stimulus and required response location are incompatible47 (psychosis cases have increased error rates18,26,31); (4) the stop-signal test (SST48) measures adequacy of adapting speeded motor responses to a visual stimulus under conditions requiring more or less inhibitory control (psychosis cases have poor adaptive response times and increased errors18,27,30); and auditory brain responses (electroencephalography (EEG) and event-related brain potentials (ERP)) to (5) repetitive stimuli (paired-stimuli paradigm49,50) and (vi) targets randomly interspersed with nontarget (or standard) stimuli (oddball paradigm51,52). The EEG/ERP paradigms assess the brain physiology of preparation for, and recovery from, sensory activations, neural responses to stimulus salience and relevance, context updating in working memory, and nonspecific (or intrinsic) brain activity, all of which are deviant in psychosis.11,18,28,29,32,33 These paradigms have substantial evidence of utility,25–33,43–45,50–64 and their intermediate level of neurobiological targeting allow for links both downward (more molecular) and upward (more clinical/observational) in the causal chain, an approach supporting discovery of genotype-clinical phenotype associations for etiologically complex diseases.65,66
For each paradigm, individual manuscripts from B-SNIP125–29 and replication samples30–33 detail the data collection and quantification methodologies. The same procedures for data collection and updated quantifications were used for B-SNIP1 and the replication sample (as briefly described below and further detailed in Supplementary Methods).
BACS
The BACS has six subtests covering four cognitive domains (Verbal Memory, Processing Speed, Reasoning and Problem Solving, Working Memory).
Pro- and Antisaccade Tasks
For prosaccades, three fixation conditions (gap, synchronous, and overlap) were administered (32 trials per condition). Participants fixated a central cross and moved their eyes quickly and accurately to a peripheral cue once it appeared. For antisaccades, an “overlap” condition was used because it is most sensitive to psychosis.67 Participants fixated a central cross and when the peripheral cue appeared they were to move their eyes quickly and accurately to the mirror image location of the cue (opposite direction, same distance from central fixation; 80 total trials). Each saccade was scored for (1) direction (to evaluate correct or error response) and (2) onset latency (time from cue illumination to saccade initiation).
SST
Subjects sat before a computer monitor displaying a white central fixation cross. A green circle (Go cue) appeared to the left or right. On 40% of trials, a red Stop Signal was presented at central fixation.27,30 Participants were instructed to respond quickly and accurately unless they encountered the Stop Signal. On failed Stop trials, a red “X” appeared over the Stop Signal to provide performance feedback; these trials were counted as errors. A baseline task of 50 consecutive Go-only trials was administered to assess baseline reaction time. Strategic slowing (difference between response latencies on baseline Go trials and Go trials during Stop Signal performance) and proportion of Stop Signal errors were used in Biotype construction.18
Auditory Paired Stimuli Task
Subjects passively listened through headphones to broadband auditory click pairs with 500 msec interclick interval (B-SNIP1: 150 pairs, replication sample: 120 pairs) occurring every 9.5 s on average (9–10 s inter-pair interval).
Auditory Oddball Task
Subjects listened through headphones to 567 standard (1000 Hz) and 100 target (1500 Hz) tones presented in pseudorandom order (1300 ms intertrial interval) and pressed a button when a target was detected (to maintain vigilance).
EEG Recording and Data Reduction
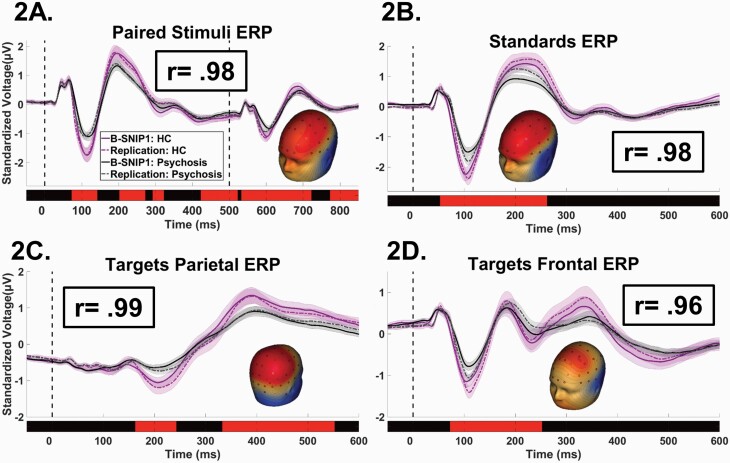
EEG was recorded from 64 Ag/AgCl sensors with nose reference and forehead ground. Data from good trials were averaged to create 64-sensor event-related potentials (ERPs). In order to maximize use of available spatial, temporal, and oscillatory information in the evoked response, a frequency-wise PCA of evoked power28,29 was conducted across all subjects to empirically define frequency bands for analysis, resulting in low, beta, and gamma ranges. A spatial PCA28,29,68,69 was also conducted on the broadband grand-averaged ERP waveforms (used for conventional ERP analyses) and then on each frequency band PCA outcome. PCA weights were multiplied by the 64-sensor data for each ERP and power waveform and summed across sensors, yielding “virtual sensors” that were used for data analysis. This resulted in four sets of waveforms that were analyzed instead of 64 separate sensors, efficiently summarizing the spatial distributions, minimizing the number of statistical comparisons, and maximizing the signal/noise ratio of the ERP data.68 Modifications from B-SNIP1 (see Supplementary Methods) simplified and improved frequency domain scoring and ensured standardized data quality control between projects.32,33 Time bins selected for Biotypes analyses were based on the same significant group effects from B-SNIP118 that were indistinguishable in the replication sample32,33 (see below). Figure 1 (voltage) and Supplementary figure 1 (frequency) show time courses and spatial topographies for each waveform (ERP-voltage, low, beta, and gamma).
Fig. 1.
Event related potential waveforms by stimulus type and project. Standardized and grand averaged voltage (y-axis) by time (msec; x-axis) for ERP waveforms (“virtual sensors”). Time 0 on the x-axis indicates the time of initial stimulus delivery. For each plot, waveforms are shown for the healthy (shades of purple) and psychosis cases (shades of gray), with the solid lines indicating B-SNIP1 and the dotted lines indicating replication sample. Confidence interval clouds (99%tile) are shown for each line. Red bars above the x-axis show time ranges of significant differences between healthy and psychosis groups. Head inserts show the surface topography of the individual virtual sensors. Boxed r-values are correlations between the B-SNIP1 and replication sample waveforms. (A). Paired-stimuli paradigm—dotted lines indicate the time of S1 (first stimulus) and S2 (second stimulus); oddball task waveforms—(B) standard stimuli; (C) parietal cortex response (P3b) to target stimuli; (D) frontal cortex response (P3a) to target stimuli.
Data Integration for Bio-Factor Creation
We evaluated whether individual biomarker variables assessed shared and replicable aspects of brain functioning (e.g., whether pro- and anti-saccade response latencies both index the same speed of visual orienting construct, whether the N100 ERP during the paired-stimuli task assesses a highly similar neural response as the N100 ERP during the oddball task). Reducing variable redundancy via methods like PCA enhances group comparisons, reduces the number of such comparisons, and increases the accuracy of numerical taxonomy methods like k-means clustering.70 PCA reduces data dimensionality (maximizing signal/noise) by replacing a group of variables with linear combinations of those variables, thus creating statistically efficient domains for subsequent analyses. PCA was conducted within paradigm sets (BACS—see Supplementary Methods, saccades, SST, EEG/ERP). The outcomes of these data integrations and their consistency between B-SNIP1 and replication samples are shown in the Results section. These integrated biomarker composites were called “bio-factors.”
Longitudinal Stability Analysis
Data were collected using identical procedures at three time points spaced six months apart (baseline, 6-month, and 12-month). Ninety-four replication sample participants (72 psychosis and 22 healthy) had data at the three time points. Intraclass correlations (ICCs71) were used on bio-factor data across the three time points to determine longitudinal stability.
Construct Validation Measures
These measures were not used in Biotype construction. They were included here to evaluate specific hypotheses that arose from the Biotypes outcomes to probe specific critical features differentiating subgroups.
Intrinsic EEG Activity (IEA)
Data came from the 9–10 s interpair interval of the paired-stimuli task.72 Epochs consisted of EEG from 500 ms after the second click of each trial to 500 ms before the first click of the next trial. No stimuli were presented during this period and there was no task other than waiting for the next stimuli pair, so these data capture nonspecific (intrinsic) brain activity. EEG data were pre-processed following methods described above and in Thomas et al.72 Data were transformed into the frequency domain, with frequency bands empirically determined using PCA,72 resulting in four primary bands (97% variance explained): delta/theta, alpha, beta, and gamma (Supplementary figure 2).
Auditory Steady State Response
Participants from Parker et al73 who overlapped with the replication sample were used in auditory steady state analyses (n = 437). In steady state paradigms, stimuli are modulated at a known frequency for an extended period (40-hz for 1500 ms here; 40-hz is optimal for probing auditory cortex74). There is a known input (40-hz stimulation) and an expected output (40-hz oscillations in the EEG). Subjects listened through headphones to 50 trials of stimuli amplitude modulated at 40 Hz for 1500 ms.
ERPs to the steady state stimuli were calculated for each sensor and subject. There were specific hypotheses about magnitudes of ERP responses so data integration with PCA was not necessary. Following the exact methods of Parker et al,73 sensors with peak auditory response (“F1,” “Fz,” “F2,” “FC3,” “FC1,” “FCz,” “FC2,” “FC4,” “C1,” “Cz,” “C2”) were averaged to define the ERPs. Steady state stimulation allows for quantification of two separate responses: (1) the onset response, like any other ERP, occurs to the initial stimuli onset. The time-period from 90 to 110 ms defined the N100, and from 180 to 220 ms defined the P200—average voltage in these time ranges was used to quantify strength of neural responses; and (2) oscillatory EEG activity to continual 40-Hz stimulation allows for quantification of neural activity in relation to sustained stimulation. Single-trial voltage data for each subject were converted to the time-frequency domain following previously published methods.73,75 Neural power at 40 Hz was averaged over the steady state period.
Data Analyses
Group effects were tested using analysis of variance in SPSS, and post-hoc comparisons using Tukey’s method (HSD or Tukey–Kramer where appropriate). For statistical significance in omnibus tests, the Holm–Bonferroni procedure76 was used to maintain the family-wise alpha at .05.
Numerical taxonomy to construct psychosis Biotypes was obtained using k-means clustering in SPSS (see Supplementary Methods). Only psychosis cases were used at this stage because bio-factors were created using variables that differentiated psychosis and healthy persons, so the problem was now meaningfully parsing bio-factor variance within psychosis. The number of clusters given the data were determined using the gap statistic77 and two-step pre-clustering procedure78,79 in SPSS, as in our previous Biotypes paper.18 For both the B-SNIP1 and replication samples, gap statistic and Two-Step outcomes are shown in Supplementary figure 3 and Supplementary table 6, respectively. In all cases, the most parsimonious solution was three clusters given the bio-factor data.
Canonical discriminant analyses in SPSS were used to efficiently capture group differences (DSM diagnoses or Biotypes; see Supplementary Methods). Group membership was the classification variable, and the biomarker data were the predictors. This analysis eased visualization of group differentiations, allowed a simple metric for comparing groups on multiple measures simultaneously, and allowed calculation of optimal effect size separations between subgroups.
Results
Replication of Individual Biomarkers and Bio-factor Structures
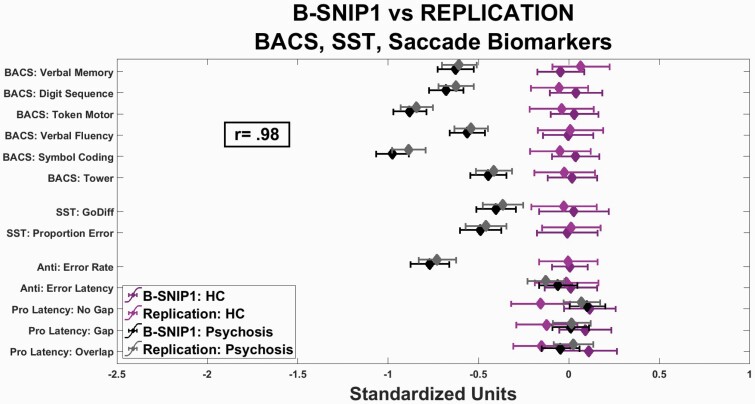
The first step identified variables that differentiated psychosis and healthy participants; there were 44 such variables: 6 BACS, 5 saccade, 2 SST, and 31 EEG/ER (figures 1 and 2, and Supplementary figure 1). The same patterns were observed with or without adjustments for demographic variables. The biomarker patterns between the two projects were consistent. The ICCs demonstrated remarkable similarity between the pattern of means for the BACS, saccade, and SST variables (figure 2: ICC = 0.98) and the EEG/ERP variables (figure 1 and Supplementary figure 1: ICCs = 0.92 to 0.99).
Fig. 2.
Cognition and saccade variables by project. Standardized scores (x-axis) for individual variables (y-axis) from the BACS, SST, and anti (Anti)- and pro (Pro)-saccade tasks, comparing healthy persons (shades of purple) and psychosis cases (shades of gray). Darker symbols and lines show means and standard deviations for B-SNIP1, and lighter symbols and lines show means and standard deviations for the replication sample. The correlation of mean performances between B-SNIP1 and the replication sample across all measures was .98.
We also investigated associations with medication usage among the psychosis cases. Of the 44 biomarkers by 18 medications regressions (792 total), only one (0.1%) showed 3%–4%, five (0.6%) showed 2%–3%, and 27 (3.4%) showed 1%–2% of uniquely shared variance (see Supplementary table 7 for complete listing of medication associations). All other associations (95.9%) accounted for less than 1% of uniquely shared variance.
PCA analyses consistently identified nine components, for both B-SNIP1 and replication samples, that accounted for the variance of the 44 biomarker variables (one BACS, two saccade, 1 SST, and 5 EEG/ERP; see Supplementary figure 4 for scree plots). We called these integrated linear components “bio-factors.” The value of such bio-factors for numerical taxonomy is supported to the extent they can be successfully replicated. Supplementary figure 5 shows the pattern matrices for all nine bio-factors, along with their similarities between the B-SNIP1 and replication samples. The ICCs between the two projects, across bio-factors, show remarkable similarities (ICCs from 0.89 to 1.0), indicating robustness of this data reduction step. These bio-factors were used in the following analyses.
Temporal Stability of Bio-Factors
In addition to showing the similarity of group differences and bio-factor patterns between B-SNIP1 and replication samples, the temporal stability of bio-factors, in the absence of any effort to intervene on them, is important for demonstrating enduring biological features of psychosis for which specific and effective treatments could be developed.80 Repeated testing identifies more trait- versus state-like biomarkers. High ICCs indicate stability in spite of other changes (e.g., in symptoms or medications). The nine bio-factors all showed high temporal stabilities (ICCs: BACS = 0.95, antisaccade = 0.86, SST = 0.83, latency = 0.76; N100 = 0.92, P300 = 0.89; P200 = 0.87; paired-stimuli S2 = 0.78; ongoing high frequency = 0.87; see on-line Methods). Thus, bio-factors are capturing stable neurobiological features, in the absence of treatments targeting those specific neurobiological deviations.
DSM Diagnoses are Similar on Bio-factors Between B-SNIP1 and Replication Samples
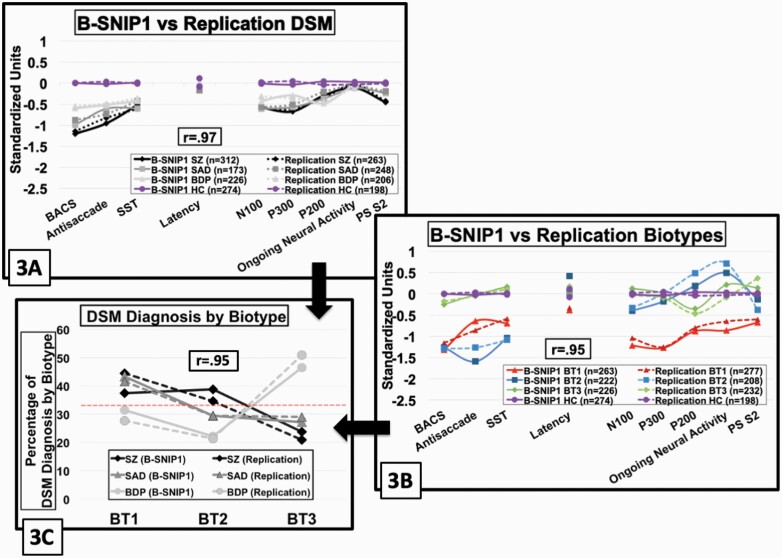
Biomarkers are not included in DSM criteria, but such information could possibly aid differential psychosis diagnosis.81 To evaluate this possibility, we compared SZ, SAD, and BDP on the nine bio-factors (figure 3A). First, bio-factor patterns between B-SNIP1 and replication samples for DSM diagnoses were highly similar (ICC = 0.97). Second, there were group effects on DSM analyses for all bio-factors but saccade latency and ongoing EEG high-frequency activity (Holm–Bonferroni adjusted P-values—significant effect F’s = 15.2 to 154.8, P’s < .001; nonsignificant effect F’s = 0.8 to 2.7, P’s > .047; figure 4A). Only the BACS significantly differentiated all four groups [SZ < SAD < BDP < healthy], capturing a severity continuum. The following bio-factors showed a similar severity continuum as the BACS: N100/P300 ERPs [(SZ = SAD) < BDP < healthy], antisaccade [SZ < (SAD = BDP) < healthy], and paired-stimuli S2 ERP [SZ < SAD < (BDP = healthy)]. The only bio-factor to indicate modestly greater deviation for BDP was the P200 ERP [BDP < (SZ = SAD) < healthy] (figure 4A; BDP effect size from SZ/SAD = 0.18). The SST is a psychosis biomarker within DSM diagnoses [(SZ = SAD = BDP) < healthy], with modest effect size (0.50 of psychosis from healthy).
Fig. 3.
Bio-factor patterns by DSM psychosis diagnosis and psychosis biotypes between B-SNIP1 and replication samples. Bio-factor means by standardized scores (y-axis) are displayed by group and project, with color-coding and symbol differentiations. Solid lines indicate B-SNIP1 and dotted lines indicate replication sample. Boxed r-values are the correlations between the B-SNIP1 and replication samples. Lines link conceptually related variables (cognition variables on the left and ERP variables on the right). The legend shows the number of observations by group. Purple lines and symbols show the healthy data, with their y-axis values adjusted so the average value of B-SNIP1 and the replication sample is zero; the healthy values are the same in plots (A) and (B). The psychosis groups are displayed as a function of their difference from the healthy means. In relation to the healthy subjects, values below zero indicate deficient values and those above zero indicate exuberant values. (A) DSM diagnoses; (B) psychosis biotypes; (C) the proportion of cases within each DSM diagnosis that had each biotype.
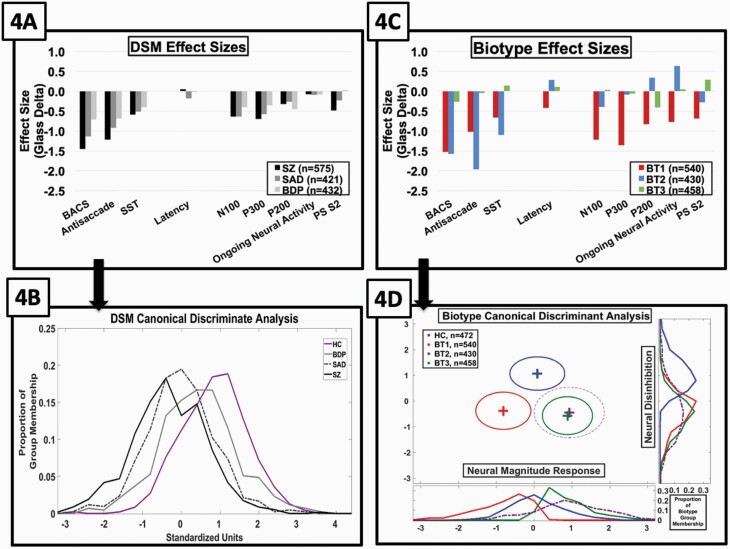
Fig. 4.
Effect size separations from healthy by DSM psychosis diagnoses and psychosis Biotypes subgroups. B-SNIP1 and replication samples were combined for these analyses given their high degree of similarity. Glass effect sizes (y-axis) by bio-factor (x-axis) are shown from the healthy for DSM diagnoses (A) and psychosis biotypes (C). In both plots, the healthy sample means fall at the zero line on the y-axis. The outcome of canonical discriminant analyses, using all bio-factors to create functions that optimally separate groups are shown in (B) and (D). (B) There was one significant function that differentiated the DSM diagnosis psychosis groups. Plots show proportion of cases within each group (y-axis) as a function of their standardized discriminant function scores (x-axis). (D) There were two discriminant functions that differentiated the psychosis Biotype groups. The first function on the x-axis captured “Neural Response Magnitude,” and the second function on the y-axis captured “Neural Disinhibition.” Frequency polygons show the proportion of cases by group at the bottom (Neural Response Magnitude) and right (Neural Disinhibition) of the central plot that shows the centroids and standard deviation ellipses by group.
BACS was the only individual bio-factor that differentiated all three DSM psychosis subgroups. It may be possible to improve group separations by using all bio-factors simultaneously. Canonical discriminant analysis using the bio-factors to optimize DSM group separations yielded one significant function (Wilks’ lambda = 0.88, Χ2 = 101.7, P < .001). The most substantial contributions to this function were BACS (structure matrix r = .70; lower is worse BACS), paired stimuli S2 ERP (r = .55; lower is smaller amplitude), antisaccade (r = −.47; higher is more errors), and P300 ERP (r = .46; lower is smaller amplitude). Cases were ordered on a severity continuum with SZ < SAD < BDP (within psychosis comparison, F(2,1425) = 78.3, P < .001; figure 4B), and all three groups were less than healthy (all groups comparison, F(3,1896) = 152.7, P < .001; SZ < SAD < BDP < healthy). This function modestly increased the separation between the extreme psychosis groups (SZ to BDP—from 0.74 to 0.89 standard deviation units).
Psychosis Biotypes are Neurobiologically Distinct, and Outcomes are Indistinguishable Between B-SNIP1 and Replication Samples
Bio-factor analyses were only modestly effective for differentiating conventional psychosis diagnoses, consistent with the conclusion of Sullivan et al.14 A possible alternative for psychosis was to examine a biomarker-based classification18 to assist identification of neurobiologically specific subgroups.4 This approach relied on bio-factor variance within psychosis independent of clinical features.
Psychosis Biotypes formation and replication of bio-factor patterns
The k-means solutions were obtained separately for B-SNIP1 and the replication samples, and the algorithm achieved cluster stability within 22 iterations for the former and 16 iterations for the latter. The k-means outcomes resulted in numbers of observations per cluster (Biotypes) as described in Supplementary table 1 and figure 3B.
All bio-factors showed between-Biotype differentiations (Holm-Bonferroni adjusted significance, F’s = 28.1 to 306.5, P’s < .001; figure 4C), as might have been expected because numerical taxonomy used these measures to create maximally homogeneous and distinct subgroups. Of note, however, the bio-factor patterns between B-SNIP1 and replication samples for psychosis Biotypes were highly similar (ICC = 0.95), indicating the differentiating patterns are robust. In addition, bio-factor patterns within each Biotype were the same regardless of DSM diagnosis (see Supplementary figure 9). Biotypes and conventional diagnoses are also not redundant because although Biotype-1 was mostly SZ (40.7%) and SAD (42.3%), Biotype-2 was mostly SZ (36.9%) and less BDP (21.8%), and Biotype-3 was largely BDP (48.6%), all DSM diagnoses were represented in all Biotypes (figure 3C shows all percentages).
Specific patterns of bio-factor deviations characterized psychosis Biotypes (figure 4C). Biotype-1 and Biotype-2 show marked deficit on general cognitive ability as measured by BACS [(BT1 = BT2) < BT3]. Biotype-1’s defining feature is deficient neural activation revealed by substantially low N100 [BT1 < BT2 < BT3] and P300 [BT1 < (BT3 = BT2)] ERP magnitudes indicating difficulty detecting stimulus salience, modestly low P200 ERP magnitude [BT1 < BT3 < BT2] indicating compromised ability to properly invest in salient stimuli,82–84 low ongoing EEG neural activity [BT1 < BT3 < BT2], low amplitude responses to repeated auditory stimuli as indexed by the PS S2 ERP [BT1 < BT2 < BT3], and slowed response latencies to saccade stimuli [BT1 < (BT3 = BT2)]. Biotype-2’s defining features, however, are greater deviation on cognitive tasks that require inhibitory control in sensorimotor performance as indexed by antisaccade [BT2 < BT1 < BT3] and SST [BT2 < BT1 < BT3], and excessive ongoing EEG neural activity not clearly locked to stimulus registration and the related accentuation of P200 ERP.72 While Biotype-3 has modest deviations on general cognition and P200 ERP, and modestly larger responses to repeated auditory stimuli (PS S2 ERP), they are similar to healthy subjects on bio-factors.
Canonical discriminant analysis using the bio-factors to optimize psychosis Biotype group separations yielded two significant functions (Function 1: Wilks’ lambda = 0.22, Χ2 = 1201.3, P < .001; Function 2: Wilks’ lambda = 0.48, Χ2 =5 59.7, P < .001). The first function had the most substantial contributions from P300 ERP (structure matrix r = .59; lower is smaller amplitude), N100 ERP (r = .52; lower is smaller amplitude), ongoing neural activity (r = .40; lower is less activity), BACS (r = .38; lower is worse BACS) and paired-stimuli S2 ERP (r = .34; lower is smaller amplitude) bio-factors. This function captured the deficient neural activation of Biotype-1 ([BT1 < BT2 < (BT3 = healthy)]; see figure 4D). The second function had the most substantial contributions from ongoing neural activity (r = .52; higher is more activity), antisaccade performance (r = .52; higher is more errors), P200 ERP (r = .45; higher is larger amplitude) and SST performance (r = −.41; lower is worse performance). This function captured the neural overactivity and concomitant inhibitory failures typifying Biotype-2 ([(BT3 = healthy = BT1) < BT2]; see figure 4D). These two functions increased the effect size separation between psychosis groups in Cartesian space (BT1 to BT2 = 2.07; BT1 to BT3 = 2.43; BT2 to BT3 = 2.64; BT3 and healthy did not significantly differ).
Cross-Validation of Psychosis Biotype classifications
In addition to independently replicating the bio-factor patterns for B-SNIP1 and replication samples (figure 4C), we cross-validated group membership assignments in two ways (see Supplementary table 8). First, we applied the B-SNIP1 k-means solution to the replication sample data and compared the classification similarity to the independently obtained replication sample group memberships (89.0% similarity). Second, we applied the replication sample k-means solution to the B-SNIP1 data and compared the classification similarity to the independently obtain B-SNIP1 sample group memberships (88.5% similarity).
Construct Validating Physiological Indicators of Psychosis Biotypes
The above information demonstrates (1) relevant biomarkers are replicable, (2) data integration for forming bio-factors is robust, (3) bio-factors are temporally stable, (4) bio-factor patterns by group are replicable, and (5) psychosis Biotypes from numerical taxonomy are robust. In addition to demonstrating such internal consistency, it is important to show that psychosis Biotypes capture discriminations on pertinent measures not involved in Biotype formulation.
The most important neurophysiological features of psychosis Biotypes were low neural response to salient stimuli (characteristic of Biotype-1 and directly indexed by the N100 and P300 ERPs) and neural overactivity (characteristic of Biotype-2 and estimated by P200 ERP and ongoing neural activity72). Neurophysiological theories of psychoses have leaned on nonspecific (or intrinsic) activity as an important translational biomarker,11,12,85 but intrinsic activity fails to consistently differentiate conventional clinical psychosis diagnoses.18,72,86,87 Intrinsic activity was only estimated because we did not, a priori, include a measure of this construct in biomarker quantification.
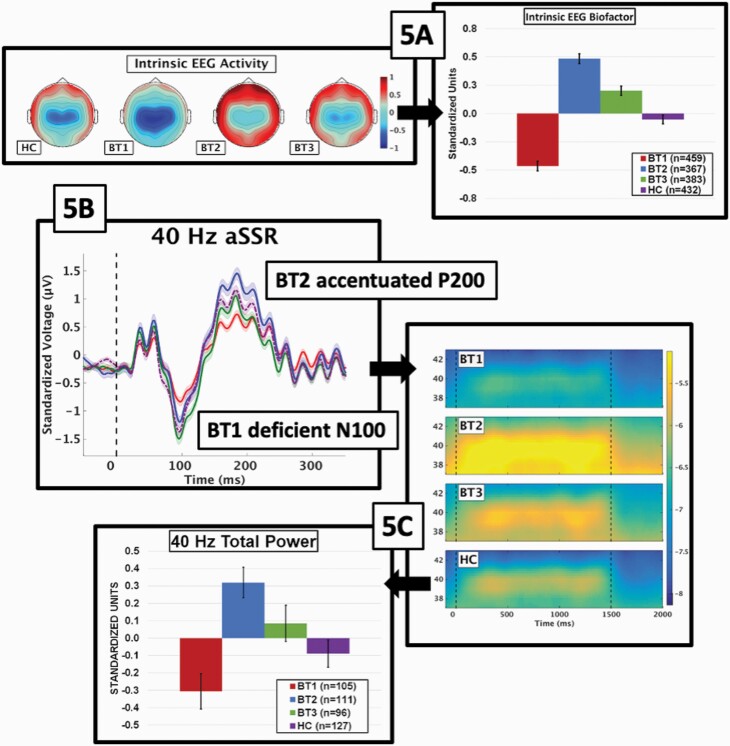
We probed the prominence of intrinsic EEG activity (IEA) for differentiating Biotypes in two ways. First, we used IEA from EEG recording during which participants had no stimulus processing requirements.72 All empirically derived frequency bands (Supplementary figure 2) significantly differentiated Biotypes (F’s from 31.5 to 81.4, all P’s < .001), but did not differentiate DSM diagnoses (F’s from 0.9 to 1.6, all P’s > .195). An additional PCA using these four frequency bands was used to create an IEA bio-factor (figure 5A). The pattern of group differentiation on the IEA bio-factor [BT1 < healthy < BT3 < BT2], indicated that Biotype-1 had low and Biotype-2 had high IEA, fortifying the outcome of numerical taxonomy shown in figures 3B and 4C.
Fig. 5.
Validation of biotype neurophysiological features. (A) Top-down topographies on the left show strength of neural response for the intrinsic EEG activity (IEA) bio-factor. The scale of neural power from FFT is to the right of the topographies, with deeper red indicating stronger neural response and deeper blue indicating weaker neural response. The bar chart shows the means and standard errors for the IEA bio-factor by group. Parts (B) and (C) show different aspects of neural response from 40-Hz auditory steady-state stimulation. (B) The auditory ERP from 50 ms before to 350 ms after stimulation onset. The location and effects for the N100 (BT1 deficient) and P200 (BT2 accentuated) are shown. The “divots” in the P200 ERP response are the beginning of the neural oscillations to the 40-Hz stimuli. (C) Following the ERP, auditory cortical neurons oscillate at 40hz throughout stimulation. The time-frequency plot to the right of the ERP shows single trial power by group centered on 40-Hz. The single trial power scale is shown to the right, with brighter yellow indicating stronger response and deeper blue indicating weaker response. The associated bar chart shows the means and standard errors by group for strength of the 40-Hz response.
The auditory steady state73 response provided our second validating measure. ERPs at the beginning of steady-state stimulation (figure 5B) replicated the N100 (low amplitude in Biotype-1; F(3,435) = 8.6, P < .001; [(BT3 = healthy) < (healthy = BT2) < BT1]) and P200 (higher amplitude in Biotype-2; F(3,435) = 10.2, P < .001; [(BT1 = BT3) < (BT3 = healthy) < BT2]) effects. Coincident with the P200, “divots” in the ERP show initiation of the oscillating 40-hz response. The steady-state response at 40-Hz is shown in figure 5C. The strength of this response replicated the effects seen for both ongoing neural activity (figures 3B and 4C) and IEA (figure 5A), with Biotype-2 showing high and Biotype-1 showing low activity in response to prolonged stimulation of auditory cortex (F(3,435) = 8.2, P < .001; [BT1 < (healthy = BT3) < BT2]). Multiple other external validators published in other papers, from MRI to social functioning, showed the possible advantages of psychosis Biotypes for capturing neurobiologically distinctive psychosis subgroups.18,24,86–89
Discussion
Andreasen90 encouraged identification of the psychosis biotype via comprehensive laboratory evaluation to facilitate the quest for pathophysiology and etiology. The clinical phenomenotype untethered from neurobiology may be ill equipped to support this mission.4,91–94 Using two large datasets, we demonstrate repeatability of biomarkers and bio-factors. We show high correspondence between the two samples on bio-factor patterns for DSM diagnoses, which showed modest neurobiological distinction. Alternatively, B-SNIP psychosis Biotypes were neurobiologically distinctive with remarkably similar features across samples. This is a promising demonstration, replication, cross-validation, and construct validation within psychosis that supports the possibility of transitioning psychosis subtyping to a laboratory discipline.
This outcome confirms that neurobiologically buttressed psychosis subtypes are derivable and robust. Their identification required two variations from typical psychosis research. First, biomarker testing covered a range of brain deviations5 to capture heterogeneity in psychosis at an intermediate level of neurobiological targeting.42 Using multiple tests that indexed the same deviation also enhanced signal/noise and maximized the ability to capture meaningful psychosis-relevant variance in a stable, repeatable fashion. Second, we recruited large and diverse samples across the bipolar-schizophrenia spectrum to support the requisite computations and capture variability in neurobiology across idiopathic psychoses. These were not epidemiological samples, but cases came from academic and community mental health centers, small towns with large universities, large cities, inner cities, rural regions, affluent and less affluent areas. These cases ranged from four standard deviations below to two standard deviations above the healthy mean on multiple biomarkers. The breadth and severity of clinical features also highlights the diversity of these samples (see Supplementary table 2).
Neurobiological stratification could facilitate the search for specific etiology and improved treatment targeting.81 Developing precision therapeutics based on clinical features alone is difficult because multiple causes can yield the same clinical features.95 Elsewhere in medicine, biomarker data re-shuffle cases with similar clinical presentations into distinct pathologies with distinct treatments; idiopathic psychosis may be similar. Of interest in this regard, we have shown that clinical features distinguishing B-SNIP psychosis Biotypes are different from those distinguishing DSM psychosis diagnoses.24,96
Psychosis Biotypes and conventional diagnoses, therefore, are neither neurobiologically nor clinically redundant. Adding neurobiological information to treatment targeting efforts provides an opportunity to match interventions to pathophysiology. In this regard, B-SNIP biomarkers and Biotypes may advantage clinical care. It is unlikely Biotype-1 and Biotype-2 cases will benefit from the same treatments given their different physiologies; and treatments appropriate for those Biotypes would most likely be less effective for Biotype-3. We explore such possibilities in a recent paper on the promise of neurobiologically informed treatments for psychosis.24
Biomarker-targeted treatments are uncommon in all of psychiatry – the field awaits robust characterization of biomarkers that index therapeutic changes in relevant cerebral systems. Psychiatry lacks known measures like white blood cell count for leukemia. It is worth considering, however, whether discovery of disease and treatment efficacy markers for psychosis can be facilitated through case stratification via neurobiology. We present one possible approach to this problem using specific measures of psychosis-relevant brain functioning. The utility of B-SNIP psychosis Biotypes for practical application in the clinic will motivate our continuing work.
Supplementary Material
Acknowledgments
United Stated Public Health Service, National Institute of Health grants MH103366, MH096900, MH103368, MH077851, MH096913, MH078113, MH096942, MH077945, MH096957.
References
- 1. Biedermann F, Fleischhacker WW. Psychotic disorders in DSM-5 and ICD-11. CNS Spectr. 2016;21(4):349–354. [DOI] [PubMed] [Google Scholar]
- 2. American Psychiatric Association., American Psychiatric Association. DSM-5 Task Force. Diagnostic and Statistical Manual of Mental Disorders: DSM-5. 5th ed. American Psychiatric Association; 2013:xliv; 947 p. [Google Scholar]
- 3. First MB, Reed GM, Hyman SE, Saxena S. The development of the ICD-11 Clinical Descriptions and Diagnostic Guidelines for Mental and Behavioural Disorders. World Psychiatry. 2015;14(1):82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hyman SE. The diagnosis of mental disorders: the problem of reification. Annu Rev Clin Psychol. 2010;6:155–179. [DOI] [PubMed] [Google Scholar]
- 5. Price ND, Magis AT, Earls JC, et al. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat Biotechnol. 2017;35(8):747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McTeague LM, Goodkind MS, Etkin A. Transdiagnostic impairment of cognitive control in mental illness. J Psychiatr Res. 2016;83:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McTeague LM, Huemer J, Carreon DM, Jiang Y, Eickhoff SB, Etkin A. Identification of common neural circuit disruptions in cognitive control across psychiatric disorders. Am J Psychiatry. 2017;174(7):676–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Knöchel C, Reuter J, Reinke B, et al. Cortical thinning in bipolar disorder and schizophrenia. Schizophr Res. 2016;172(1-3):78–85. [DOI] [PubMed] [Google Scholar]
- 9. Weinberg D, Lenroot R, Jacomb I, et al. Cognitive subtypes of schizophrenia characterized by differential brain volumetric reductions and cognitive decline. JAMA Psychiatry. 2016;73(12):1251–1259. [DOI] [PubMed] [Google Scholar]
- 10. Patel Y, Parker N, Shin J, et al. Virtual histology of cortical thickness and shared neurobiology in 6 psychiatric disorders. JAMA Psychiatry. 2021;78(1):47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rolls ET, Loh M, Deco G, Winterer G. Computational models of schizophrenia and dopamine modulation in the prefrontal cortex. Nat Rev Neurosci. 2008;9(9):696–709. [DOI] [PubMed] [Google Scholar]
- 12. Spencer KM. Time to be spontaneous: a renaissance of intrinsic brain activity in psychosis research? Biol Psychiatry. 2014;76(6):434–435. [DOI] [PubMed] [Google Scholar]
- 13. Javitt DC, Sweet RA. Auditory dysfunction in schizophrenia: integrating clinical and basic features. Nat Rev Neurosci. 2015;16(9):535–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sullivan PF, Agrawal A, Bulik CM, et al. ; Psychiatric Genomics Consortium . Psychiatric genomics: an update and an agenda. Am J Psychiatry. 2018;175(1):15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hall MH, Smoller JW, Cook NR, et al. Patterns of deficits in brain function in bipolar disorder and schizophrenia: a cluster analytic study. Psychiatry Res. 2012;200(2-3):272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sponheim SR, Iacono WG, Thuras PD, Beiser M. Using biological indices to classify schizophrenia and other psychotic patients. Schizophr Res. 2001;50(3):139–150. [DOI] [PubMed] [Google Scholar]
- 17. John ER, Prichep LS, Alper KR, et al. Quantitative electrophysiological characteristics and subtyping of schizophrenia. Biol Psychiatry. 1994;36(12):801–826. [DOI] [PubMed] [Google Scholar]
- 18. Clementz BA, Sweeney JA, Hamm JP, et al. Identification of distinct psychosis biotypes using brain-based biomarkers. Am J Psychiatry. 2016;173(4):373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cronbach LJ, Meehl PE. Construct validity in psychological tests. Psychol Bull. 1955;52(4):281–302. [DOI] [PubMed] [Google Scholar]
- 20. Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2(8):e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pashler H, Wagenmakers EJ. Editors’ Introduction to the Special Section on Replicability in Psychological Science: A Crisis of Confidence? Perspect Psychol Sci. 2012;7(6):528–530. [DOI] [PubMed] [Google Scholar]
- 22. Piper SK, Grittner U, Rex A, et al. Exact replication: foundation of science or game of chance? PLoS Biol. 2019;17(4):e3000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bzdok D, Varoquaux G, Steyerberg EW. Prediction, not association, paves the road to precision medicine. JAMA Psychiatry. 2021;78(2):127–128. [DOI] [PubMed] [Google Scholar]
- 24. Clementz BA, Trotti RL, Pearlson GD, et al. Testing psychosis phenotypes from bipolar-schizophrenia network for intermediate phenotypes for clinical application: biotype characteristics and targets. Biol Psychiatry Cogn Neurosci Neuroimaging. 2020;5(8):808–818. [DOI] [PubMed] [Google Scholar]
- 25. Hill SK, Reilly JL, Keefe RS, et al. Neuropsychological impairments in schizophrenia and psychotic bipolar disorder: findings from the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) study. Am J Psychiatry. 2013;170(11):1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reilly JL, Frankovich K, Hill S, et al. Elevated antisaccade error rate as an intermediate phenotype for psychosis across diagnostic categories. Schizophr Bull. 2014;40(5):1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ethridge LE, Soilleux M, Nakonezny PA, et al. Behavioral response inhibition in psychotic disorders: diagnostic specificity, familiality and relation to generalized cognitive deficit. Schizophr Res. 2014;159(2-3):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ethridge LE, Hamm JP, Pearlson GD, et al. Event-related potential and time-frequency endophenotypes for schizophrenia and psychotic bipolar disorder. Biol Psychiatry. 2015;77(2):127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hamm JP, Ethridge LE, Boutros NN, et al. Diagnostic specificity and familiality of early versus late evoked potentials to auditory paired stimuli across the schizophrenia-bipolar psychosis spectrum. Psychophysiology. 2014;51(4):348–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gotra MY, Hill SK, Gershon ES, et al. Distinguishing patterns of impairment on inhibitory control and general cognitive ability among bipolar with and without psychosis, schizophrenia, and schizoaffective disorder. Schizophr Res. 2020;223:148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang L, Jackson B, Rodrigue A, et al. Antisaccade error rate and gap effects in psychosis syndromes from B-SNIP2. Psychol Med. 2021;in press [DOI] [PubMed] [Google Scholar]
- 32. Parker DA, Trotti RL, McDowell JE, et al. Auditory paired-stimuli responses across the psychosis and bipolar spectrum and their relationship to clinical features. Biomarkers Neuropsychiatry. 2020;3:100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parker D, Trotti R, McDowell J, et al. Auditory oddball responses across the schizophrenia-bipolar spectrum and their relationship to cognitive and clinical features. Am J Psychiatry. 2021;in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tamminga CA, Ivleva EI, Keshavan MS, et al. Clinical phenotypes of psychosis in the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP). Am J Psychiatry. 2013;170(11):1263–1274. [DOI] [PubMed] [Google Scholar]
- 35. American Psychiatric Association. Diagnostic Criteria from DSM-IV-TR. Washington, DC: American Psychiatric Association; 2000: xii: 370 p. [Google Scholar]
- 36. Birchwood M, Smith J, Cochrane R, Wetton S, Copestake S. The Social Functioning Scale. The development and validation of a new scale of social adjustment for use in family intervention programmes with schizophrenic patients. Br J Psychiatry. 1990;157:853–859. [DOI] [PubMed] [Google Scholar]
- 37. Montgomery SA, Asberg M. A new depression scale designed to be sensitive to change. Br J Psychiatry. 1979;134:382–389. [DOI] [PubMed] [Google Scholar]
- 38. Lançon C, Auquier P, Nayt G, Reine G. Stability of the five-factor structure of the Positive and Negative Syndrome Scale (PANSS). Schizophr Res. 2000;42(3):231–239. [DOI] [PubMed] [Google Scholar]
- 39. Young RC, Biggs JT, Ziegler VE, Meyer DA. A rating scale for mania: reliability, validity and sensitivity. Br J Psychiatry. 1978;133:429–435. [DOI] [PubMed] [Google Scholar]
- 40. Andreasen NC, Rice J, Endicott J, Reich T, Coryell W. The family history approach to diagnosis. How useful is it? Arch Gen Psychiatry. 1986;43(5):421–429. [DOI] [PubMed] [Google Scholar]
- 41. Tamminga CA, Pearlson GD, Stan AD, et al. Strategies for advancing disease definition using biomarkers and genetics: the bipolar and schizophrenia network for intermediate phenotypes. Biol Psychiatry Cogn Neurosci Neuroimaging. 2017;2(1):20–27. [DOI] [PubMed] [Google Scholar]
- 42. Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160(4):636–645. [DOI] [PubMed] [Google Scholar]
- 43. Keefe RS, Harvey PD, Goldberg TE, et al. Norms and standardization of the Brief Assessment of Cognition in Schizophrenia (BACS). Schizophr Res. 2008;102(1− 3):108–115. [DOI] [PubMed] [Google Scholar]
- 44. Keefe RS, Goldberg TE, Harvey PD, Gold JM, Poe MP, Coughenour L. The brief assessment of cognition in schizophrenia: reliability, sensitivity, and comparison with a standard neurocognitive battery. Schizophr Res. 2004;68(2− 3):283–297. [DOI] [PubMed] [Google Scholar]
- 45. McDowell JE, Clementz BA. Behavioral and brain imaging studies of saccadic performance in schizophrenia. Biol Psychol. 2001;57(1− 3):5–22. [DOI] [PubMed] [Google Scholar]
- 46. Reilly JL, Harris MS, Khine TT, Keshavan MS, Sweeney JA. Reduced attentional engagement contributes to deficits in prefrontal inhibitory control in schizophrenia. Biol Psychiatry. 2008;63(8):776–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hallett PE, Adams BD. The predictability of saccadic latency in a novel voluntary oculomotor task. Vision Res. 1980;20(4):329–339. [DOI] [PubMed] [Google Scholar]
- 48. Lipszyc J, Schachar R. Inhibitory control and psychopathology: a meta-analysis of studies using the stop signal task. J Int Neuropsychol Soc. 2010;16(6):1064–1076. [DOI] [PubMed] [Google Scholar]
- 49. Adler LE, Pachtman E, Franks RD, Pecevich M, Waldo MC, Freedman R. Neurophysiological evidence for a defect in neuronal mechanisms involved in sensory gating in schizophrenia. Biol Psychiatry. 1982;17(6):639–654. [PubMed] [Google Scholar]
- 50. Freedman R, Adler LE, Gerhardt GA, et al. Neurobiological studies of sensory gating in schizophrenia. Schizophr Bull. 1987;13(4):669–678. [DOI] [PubMed] [Google Scholar]
- 51. Linden DE. The p300: where in the brain is it produced and what does it tell us? Neuroscientist. 2005;11(6):563–576. [DOI] [PubMed] [Google Scholar]
- 52. Polich J. Updating P300: an integrative theory of P3a and P3b. Clin Neurophysiol. 2007;118(10):2128–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Turetsky BI, Greenwood TA, Olincy A, et al. Abnormal auditory N100 amplitude: a heritable endophenotype in first-degree relatives of schizophrenia probands. Biol Psychiatry. 2008;64(12):1051–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bramon E, Rabe-Hesketh S, Sham P, Murray RM, Frangou S. Meta-analysis of the P300 and P50 waveforms in schizophrenia. Schizophr Res. 2004;70(2-3):315–329. [DOI] [PubMed] [Google Scholar]
- 55. Hall MH, Schulze K, Rijsdijk F, et al. Heritability and reliability of P300, P50 and duration mismatch negativity. Behav Genet. 2006;36(6):845–857. [DOI] [PubMed] [Google Scholar]
- 56. Patterson JV, Hetrick WP, Boutros NN, et al. P50 sensory gating ratios in schizophrenics and controls: a review and data analysis. Psychiatry Res. 2008;158(2):226–247. [DOI] [PubMed] [Google Scholar]
- 57. Johannesen JK, O’Donnell BF, Shekhar A, McGrew JH, Hetrick WP. Diagnostic specificity of neurophysiological endophenotypes in schizophrenia and bipolar disorder. Schizophr Bull. 2013;39(6):1219–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cheng CH, Chan PS, Liu CY, Hsu SC. Auditory sensory gating in patients with bipolar disorders: a meta-analysis. J Affect Disord. 2016;203:199–203. [DOI] [PubMed] [Google Scholar]
- 59. Thomas ML, Green MF, Hellemann G, et al. Modeling deficits from early auditory information processing to psychosocial functioning in schizophrenia. JAMA Psychiatry. 2017;74(1):37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Turetsky BI, Bilker WB, Siegel SJ, Kohler CG, Gur RE. Profile of auditory information-processing deficits in schizophrenia. Psychiatry Res. 2009;165(1− 2):27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Turetsky BI, Dress EM, Braff DL, et al. The utility of P300 as a schizophrenia endophenotype and predictive biomarker: clinical and socio-demographic modulators in COGS-2. Schizophr Res. 2015;163(1− 3):53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Perlman G, Foti D, Jackson F, Kotov R, Constantino E, Hajcak G. Clinical significance of auditory target P300 subcomponents in psychosis: differential diagnosis, symptom profiles, and course. Schizophr Res. 2015;165(2− 3):145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wada M, Kurose S, Miyazaki T, et al. The P300 event-related potential in bipolar disorder: a systematic review and meta-analysis. J Affect Disord. 2019;256:234–249. [DOI] [PubMed] [Google Scholar]
- 64. Lundin NB, Bartolomeo LA, O’Donnell BF, Hetrick WP. Reduced electroencephalogram responses to standard and target auditory stimuli in bipolar disorder and the impact of psychotic features: analysis of event-related potentials, spectral power, and inter-trial coherence. Bipolar Disord. 2018;20(1):49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ingelsson E, Knowles JW. Leveraging human genetics to understand the relation of LDL cholesterol with type 2 diabetes. Clin Chem. 2017;63(7):1187–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rubio-Perez C, Guney E, Aguilar D, et al. Genetic and functional characterization of disease associations explains comorbidity. Sci Rep. 2017;7(1):6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. McDowell JE, Clementz BA. The effect of fixation condition manipulations on antisaccade performance in schizophrenia: studies of diagnostic specificity. Exp Brain Res. 1997;115(2):333–344. [DOI] [PubMed] [Google Scholar]
- 68. Carroll CA, Kieffaber PD, Vohs JL, O’Donnell BF, Shekhar A, Hetrick WP. Contributions of spectral frequency analyses to the study of P50 ERP amplitude and suppression in bipolar disorder with or without a history of psychosis. Bipolar Disord. 2008;10(7):776–787. [DOI] [PubMed] [Google Scholar]
- 69. Dien J, Khoe W, Mangun GR. Evaluation of PCA and ICA of simulated ERPs: promax vs. infomax rotations. Hum Brain Mapp. 2007;28(8):742–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ding C, He X.. K-means clustering via principal component analysis. presented at: Proceedings of the twenty-first international conference on Machine learning; 2004; Banff, Alberta, Canada. [Google Scholar]
- 71. Hedeker DR, Gibbons RD. Longitudinal data analysis. Wiley Series in Probability and Statistics. Hoboken, NJ: Wiley-Interscience; 2006;xx: 337 p. [Google Scholar]
- 72. Thomas O, Parker D, Trotti R, et al. Intrinsic neural activity differences in psychosis biotypes: findings from the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) consortium. Biomarkers Neuropsychiatry. 2019;1:100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Parker DA, Hamm JP, McDowell JE, et al. Auditory steady-state EEG response across the schizo-bipolar spectrum. Schizophr Res. 2019;209:218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Picton TW, John MS, Dimitrijevic A, Purcell D. Human auditory steady-state responses. Int J Audiol. 2003;42(4):177–219. [DOI] [PubMed] [Google Scholar]
- 75. Hamm JP, Gilmore CS, Clementz BA. Augmented gamma band auditory steady-state responses: support for NMDA hypofunction in schizophrenia. Schizophr Res. 2012;138(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6(2):65–70. [Google Scholar]
- 77. Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. J Roy Stat Soc B-Stat Methodol. 2001;63:411–423. [Google Scholar]
- 78. Norusis MJ. SPSS 15.0 Advanced Statistical Procedures Companion. Upper Saddle River, NJ: Prentice Hall; 2007:xiv: 418 p. [Google Scholar]
- 79. Tkaczynski A. Segmentation Using Two-Step Cluster Analysis. In: Dietrich T, Rundle-Thiele S, Kubacki K, eds. Segmentation in Social Marketing: Process, Methods and Application. Singapore: Springer; 2017;109–125. [Google Scholar]
- 80. Lema YY, Gamo NJ, Yang K, Ishizuka K. Trait and state biomarkers for psychiatric disorders: Importance of infrastructure to bridge the gap between basic and clinical research and industry. Psychiatry Clin Neurosci. 2018;72(7):482–489. [DOI] [PubMed] [Google Scholar]
- 81. Council NR. Toward precision medicine: building a knowledge network for biomedical research and a new taxonomy of disease. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Upper Saddle River, NJ: The National Academies Press; 2011. The National Academies Collection: Reports funded by National Institutes of Health. [PubMed] [Google Scholar]
- 82. Crowley KE, Colrain IM. A review of the evidence for P2 being an independent component process: age, sleep and modality. Clin Neurophysiol. 2004;115(4):732–744. [DOI] [PubMed] [Google Scholar]
- 83. Allison BZ, Polich J. Workload assessment of computer gaming using a single-stimulus event-related potential paradigm. Biol Psychol. 2008;77(3):277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Horat SK, Herrmann FR, Favre G, et al. Assessment of mental workload: a new electrophysiological method based on intra-block averaging of ERP amplitudes. Neuropsychologia. 2016;82:11–17. [DOI] [PubMed] [Google Scholar]
- 85. Clementz BA. Time for change in psychosis research. In: Tamminga C, Ivleva, E., Reininghaus, U, van Os, J., eds. Psychotic Disorders: Comprehensive Conceptualization and Treatments. New York, NY: Oxford University Press; 2020. [Google Scholar]
- 86. Hudgens-Haney ME, Ethridge LE, McDowell JE, et al. Psychosis subgroups differ in intrinsic neural activity but not task-specific processing. Schizophr Res. 2018;195:222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hudgens-Haney ME, Ethridge LE, Knight JB, et al. Intrinsic neural activity differences among psychotic illnesses. Psychophysiology. 2017;54(8):1223–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ivleva EI, Clementz BA, Dutcher AM, et al. Brain structure biomarkers in the psychosis biotypes: findings from the bipolar-schizophrenia network for intermediate phenotypes. Biol Psychiatry. 2017;82(1):26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Guimond S, et al. A diagnosis and biotype comparison across the psychosis spectrum: investigating volume and shape amygdala-hippocampal differences from the B-SNIP Study. Schizophr Bull. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Andreasen NC. The diagnosis of schizophrenia. Schizophr Bull. 1987;13(1):9–22. [DOI] [PubMed] [Google Scholar]
- 91. Wexler BE. Beyond the Kraepelinean dichotomy. Biol Psychiatry. 1992;31(6):539–541. [DOI] [PubMed] [Google Scholar]
- 92. Hyman SE. Revolution stalled. Sci Transl Med. 2012;4(155):155cm11. [DOI] [PubMed] [Google Scholar]
- 93. Casey BJ, Craddock N, Cuthbert BN, Hyman SE, Lee FS, Ressler KJ. DSM-5 and RDoC: progress in psychiatry research? Nat Rev Neurosci. 2013;14(11):810–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. McHugh PR. Psychiatry at stalemate. Cerebrum. 2009. https://www.dana.org/article/updating-the-diagnostic-and-statistical-manual-of-mental-disorders/ [Google Scholar]
- 95. Fischer BA, Carpenter WT Jr. Will the Kraepelinian dichotomy survive DSM-V? Neuropsychopharmacology. 2009;34(9):2081–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Reininghaus U, Böhnke JR, Chavez-Baldini U, et al. Transdiagnostic dimensions of psychosis in the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP). World Psychiatry. 2019;18(1):67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.