Abstract
Astrocytes are the most abundant cell type in the human brain and are important regulators of several critical cellular functions, including synaptic transmission. Although astrocytes are known to play a central role in the etiology and pathophysiology of schizophrenia, little is known about their potential involvement in clinical response to the antipsychotic clozapine. Moreover, astrocytes display a remarkable degree of morphological diversity, but the potential contribution of astrocytic subtypes to disease biology and drug response has received little attention. Here, we used state-of-the-art human induced pluripotent stem cell (hiPSC) technology to derive a morphological subtype of astrocytes from healthy individuals and individuals with schizophrenia, including responders and nonresponders to clozapine. Using functional assays and transcriptional profiling, we identified a distinct gene expression signature highly specific to schizophrenia as shown by disease association analysis of more than 10 000 diseases. We further found reduced levels of both glutamate and the NMDA receptor coagonist d-serine in subtype astrocytes derived from schizophrenia patients, and that exposure to clozapine only rescued this deficiency in cells from clozapine responders, providing further evidence that d-serine in particular, and NMDA receptor-mediated glutamatergic neurotransmission in general, could play an important role in disease pathophysiology and clozapine action. Our study represents a first attempt to explore the potential contribution of astrocyte diversity to schizophrenia pathophysiology using a human cellular model. Our findings suggest that specialized subtypes of astrocytes could be important modulators of disease pathophysiology and clinical drug response, and warrant further investigations.
Keywords: hiPSC, astrocyte diversity, transcription, glutamate, d-serine
Introduction
Astrocytes are ubiquitous cells in the central nervous system (CNS). Previously viewed as a passive and homogenous cell population whose main function was to provide structural support for neurons, these glial cells have recently emerged as an integral constituent of the CNS, where they actively participate in a variety of biological processes which are vital for proper brain functioning, including synaptogenesis and modulation of NMDA receptor-mediated glutamatergic neurotransmission.1 Paralleling the increasingly popular glutamate hypothesis of schizophrenia (SCZ),2 mounting evidence from postmortem, brain imaging, and genetic studies suggests that astrocytes play a critical role in the pathophysiology of SCZ.3,4 However, the precise molecular mechanisms underlying this astrocytic contribution to SCZ biology remain poorly understood, partly due to the inaccessibility of human CNS tissue and the lack of appropriate animal and cellular models. Moreover, astrocytes display a remarkable degree of morphological, regional, and cellular diversity,5 and although at least five distinct populations have been described in the human brain,6,7 very little is known about their molecular characterization. Most functional and molecular studies published in recent years have employed rodent models,5,8,9 which may not fully recapitulate human astrocyte heterogeneity as substantial species differences exist.7 Additionally, different astrocytic subtypes may mediate distinct pathophysiological processes in uniquely human disorders, but this emerging area of astrocyte biology research remains unexplored.9 Recent method developments in human induced pluripotent stem cells (hiPSCs) biology have provided a feasible platform for the elucidation of cellular and molecular anomalies in SCZ.10 Here, we study an hiPSC-derived morphological subtype of astrocytes generated from healthy individuals and individuals with SCZ, including patients responsive and nonresponsive to the antipsychotic clozapine (CLZ), and explore the transcriptional architectures underlying SCZ and CLZ response.
Methods
Overview of Experimental Design
A schematic of the overall design of the study can be found in supplementary figure 1. In total, morphological subtype astrocytes were generated from two healthy control (HC) and six schizophrenia (SCZ) hiPSC lines. To compare the transcriptional profiles of subtype astrocytes to classical astrocytes, these two cell types were derived from one of the HC lines and differentiated for 180 days, using three replicates of a single clone from each group of astrocytes for RNA-sequencing (RNA-seq) analysis. In addition, subtype astrocytes were generated from a human embryonic stem cell (hESC) line (CCTL14)11 to compare its differentiation propensity to that of hiPSCs using immunocytochemistry (ICC). The subtypes astrocytes derived from HC and SCZ hiPSC lines for RNA-seq analysis were differentiated for 200 days.
Recruitment of SCZ Patients and Determination of CLZ Response
Patients were recruited through psychiatric units in Brno, Czech Republic. The main inclusion criteria were: confirmed SCZ diagnosis, prior or current treatment with CLZ, male sex, age between 20 and 60 years, and signed informed consent. Patients with another psychiatric disorder, somatic disorder affecting brain function, or IQ < 70 were excluded. Supplementary table 1 summarizes the demographic, clinical, and CLZ response characteristics of all patient donors. Sources of data for diagnosis and determination of CLZ response status were medical records and an interview with a structure analogous to the Comprehensive Assessment of Symptoms and History (CASH) structured interview.12 In addition, two board-certified psychiatrists verified the diagnosis and assessed symptoms at the time of the recruitment using the Mini-International Neuropsychiatric Interview (MINI) structured interview.13 CLZ response status was determined on the basis of an initial evaluation of the medication used at the time of recruitment and a subsequent analysis of the medical records. CLZ responders (CLZ-R) had to (1) be treated with only CLZ at the initial assessment, and (2) show either full or partial but sufficient effect of CLZ on positive and negative symptoms. Patients were considered CLZ nonresponders (CLZ-NR) if (1) they were treated with any other antipsychotic alone or in combination with CLZ at the initial assessment, and (2) showed either no effect of CLZ or its combination with other medications, or experienced relapse of the symptoms during the CLZ treatment necessitating acute treatment with other drugs. A detailed description of the clinical procedures can be found in the online supplementary information.
Generation of hiPSC Lines and Derivation of Subtype Astrocytes
The hiPSC lines were generated from human dermal fibroblasts obtained from the recruited participants using the CytoTune-iPS 2.0 Sendai Reprogramming Kit (Invitrogen). To generate subtype astrocytes, hESCs were plated onto Matrigel-coated plates at 1000 cells/cm2 in the chemically defined conditional medium, whereas hiPSC lines were plated onto Matrigel-coated plates in clumps by manual cut-outs or aided with UltraPure EDTA (Invitrogen, 15575020) in Essential 8 Medium (Gibco, A1517001) according to our previous studies.14,15 For RNA-seq analysis and d-serine and glutamate measurements, subtype astrocytes were enriched at 90–120 days of differentiation by manually cutting out the cell colonies and growing them in separate Matrigel-coated plates until 180 or 200 days of differentiation.
Functional Assays
The subtype astrocytes derived from one of the HC hiPSC lines were prepared for calcium imaging as described previously.14,15 Glutamate concentrations were measured using an ELISA assay (Abcam, ab83389). To measure d-serine, cell samples were treated with and without 1 μM CLZ after 166 days of differentiation for 2 weeks, fixed immediately on day 180, and stained with d-serine antibody (Abcam, ab6472) the next day. ICC was performed as described previously.14,15 The stained cells were examined using a confocal microscope (Zeiss LSM 700).
RNA Sequencing and Data Processing
Total RNA was extracted from the astrocyte samples using the RNeasy Plus Mini Kit (Qiagen). Sequencing libraries were prepared with the TruSeq Stranded mRNA kit (Illumina) and sequenced on an Illumina HiSeq 4000 platform at an average depth of 50 million reads per sample using a read length of 150 bp and an insert size of 350 bp. Raw sequencing reads were quality assessed with FastQC (Babraham Institute) and further processed with Trimmomatic V0.32.16 Hisat217 was then used to first build a transcriptome index based on Ensembl annotations, and then to map the trimmed reads to the human GRCh38 reference genome. To quantify gene expression levels, mapped reads were summarized at the gene level using featureCounts18 guided by Ensembl annotations.
Statistical Analyses
All differential expression (DE) analyses were performed using DESeq2.19 A gene was considered significant if the FDR was <0.1. Over-representation tests of GO terms (biological processes) were conducted with clusterProfiler,20 while disease association analysis was performed with the R package disgenet2r.21 In both cases, FDR <0.05 was used as the threshold to determine significant enrichment. A detailed description of the materials and methods used in this study can be found in the online supplementary information.
Results
Derivation and Molecular Characterization of Subtype Astrocytes
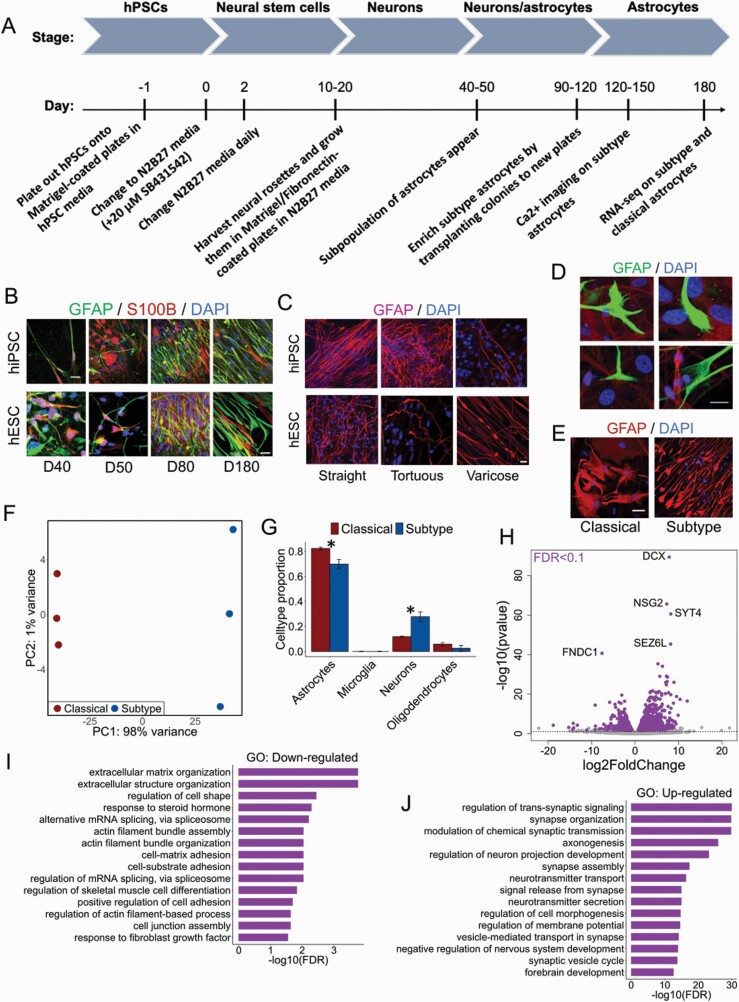
To study the transcriptional profiles of hiPSC-derived subtype astrocytes and their alterations in SCZ and CLZ response, we generated astrocytes from a control hiPSC line and a human embryonic stem cell (hESC) line using a modified version of our previously published neural differentiation protocol14 (figure 1A, supplementary figure 1). After ~45 days of differentiation, a morphological subpopulation of GFAP and S100B-positive cells started to appear and were subsequently enriched by physical translocation (figure 1A). This astrocytic subpopulation was characterized by a distinctive morphology which was elongated and unbranched (figure 1B), exhibiting structural similarities to the primate-specific interlaminar astrocytes residing in the upper cortical layers.22 To our knowledge, this morphological subpopulation of astrocytes has not been assessed previously in vitro. The astrocytes exhibited structural diversity along their single process, which displayed a predominantly oblong morphology, but could also be tortuous and to a lesser extent varicose (figure 1C). The tail-ends were also morphologically diverse, typically fanning out in brush-like structures of various shapes (figure 1D). As astrocytes are known to be involved in the modulation of NMDA receptor-mediated glutamatergic neurotransmission, both by responding to, and the potential release of, neurotransmitters,23 we used calcium imaging and immunocytochemistry (ICC) to examine the presence of glutamate-evoked calcium transients and to assess protein expression of d-serine in the derived astrocytes, respectively. We found that exposure to glutamate elicited pulsatile and synchronous calcium transients (supplementary movie 1), and that d-serine was expressed and distributed throughout the cell, including the soma, process, and tail-ends (supplementary figure 2), suggesting that these subtype astrocytes may respond to glutamatergic signaling in the brain.
Fig. 1.
Derivation and characterization of astrocytic subtype population. (A) Outline of the neural differentiation protocol used to derive subtype astrocytes from a hESC and a hiPSC cell line (hPSCs). RNA-seq for comparison between classical and subtype astrocytes was performed on 180 days old hiPSCs only. (B) Differentiation of hESC and hiPSC-derived subtype astrocytes from day 40 to 180. Scale bar: 50 μm. (C) Morphological diversity of derived subtype astrocytes at day 100 of differentiation. Scale bar: 20 μm. (D) Diverse tail-ends of subtype astrocytes. Scale bar: 10 μm. All experiments in B-D were performed three times. (E) Representative images of GFAP-positive classical and subtype astrocytes derived from control hiPSC lines and used for RNA-seq analysis. Scale bar: 20 μm. (F) Principal component analysis (PCA) of three replicates of hiPSC-derived classical and subtype astrocytes based on the top 500 genes with largest expression variance across samples. (G) Computational deconvolution of four major cell types in the human brain. Data is shown as mean ± SD. *Two-sample t-test: P < .05. (H) Volcano plot showing genes with differential expression (DE) in subtype astrocytes compared to classical astrocytes. The dotted line represents FDR <0.1. The five top genes with strongest association are labeled. (I) GO over-representation test of downregulated DE genes. The top 15 significant (FDR < 0.05) GO terms (biological processes) are displayed. (J) GO over-representation test of upregulated DE genes. The top 15 significantly enriched GO terms are shown.
Transcriptional Profiling of Subtype Astrocytes
As structural heterogeneity implies variability in the underlying patterns of gene expression, RNA sequencing (RNA-seq) was used to compare the transcriptional profiles of the derived subtype astrocytes with those of “classical” astrocytes displaying a typical branched morphology (figure 1E). Both subtype and classical astrocytes were derived from the same hiPSC line (three replicates each), and global gene expression was assessed after 180 days of differentiation (figure 1A). Principal component analysis (PCA) showed a complete separation on cell type across the first two components, explaining 98% of the variation in gene expression (figure 1F). Computational deconvolution of four major brain cell types suggested that some of the observed variation may be attributed to cell type composition, as two cell types (astrocytes and neurons) were found to be present in significantly different abundances across the two astrocytic groups (figure 1G, supplementary table 2). Consequently, the effects of cell type composition were corrected for in downstream analyses. Differential expression (DE) analysis identified 1720 (11%) genes with genome-wide significance (FDR < 0.1), of which 1050 were upregulated and 670 were downregulated in subtype astrocytes compared to classical astrocytes (figure 1H, supplementary table 3). The upregulated DE genes were enriched for several biological processes (GO terms) related to synaptic transmission (figure 1I and J, supplementary table 4), suggesting a potential role of the derived astrocytes in synapse-related processes. As no specific markers for different subpopulations of human astrocytes have yet been validated,5 we compared the transcriptional profiles of the derived astrocytes with the profiles of primary brain cells isolated from human cortical samples24 to assess their similarities. Hierarchical clustering and correlation analysis showed that our derived astrocytes both clustered together and correlated strongly with human adult astrocytes, suggesting that they bear transcriptional resemblance to the astrocytes found in vivo (supplementary figure 3).
Subtype Astrocytes from SCZ Patients Display Disease-Specific Signatures
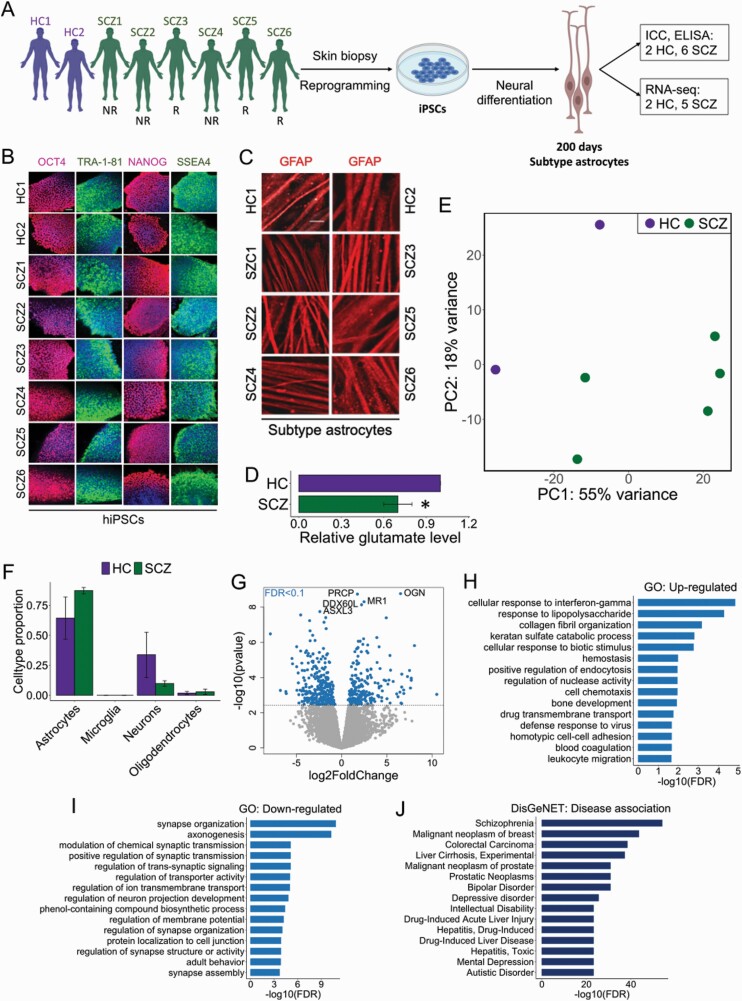
Specific subsets of human astrocytes may contribute to neuropsychiatric disorders, but this emerging area of research is in its infancy and little is known about the underlying molecular mechanisms.9 To investigate the transcriptional landscape of subtype astrocytes in the context of SCZ, the same neural differentiation protocol was employed to derive subtype astrocytes from two healthy controls (HC) and six SCZ patients, including three CLZ responders (CLZ-R), and three nonresponders (CLZ-NR) (figure 2A, supplementary figure 1). These astrocytes were differentiated for 20 additional days (200 days in total) in order to increase the purity of the cell cultures. All samples stained positive for the pluripotency markers OCT4, TRA-1–81, NANOG, and SSEA4 at the iPSC stage (figure 2B), as well as the astrocytic marker GFAP at the mature astrocyte stage (figure 2C). ELISA measurements of intracellular glutamate concentrations revealed significantly reduced glutamate levels in SCZ astrocytes compared to HCs (figure 2D). A corresponding decrease in mRNA levels of the major glutamate transporter genes SLC1A2 and SLC1A3 was not found (supplementary figure 4), suggesting that the reduction in intracellular glutamate was likely driven by dysfunctional de novo synthesis25 rather than decreased glutamate uptake from the extracellular space. Clustering of samples by PCA showed a clear separation on diagnosis (figure 2E), and although the effects of cell type composition may explain some of this variation, no significant difference in cell type abundances was found (figure 2F, supplementary table 5). In total, 504 genes were differentially expressed (FDR < 0.1) across the two diagnostic groups, 211 of these were upregulated and 293 were downregulated in SCZ astrocytes (Fig. 2G, Suppl. Table 6). The upregulated genes were enriched for several immune-related biological processes, which is in line with known inflammatory disturbances in SCZ26 and the important role of astrocytes in regulating neuroinflammation.27 The downregulated genes were enriched for multiple GO terms related to synaptic function, indicating a disrupted regulation of synaptic neurotransmission or altered astroglial sensitivity to synaptic signals (figure 2H and I, supplementary table 7). We performed additional DE and pathway analyses on three SCZ and three HC astrocytes from our previous study28 which used a nearly identical differentiation protocol to generate classical astrocytes from hiPSC cells. We identified fewer DE genes compared to the present study, and the genes were mostly enriched for ribosomal and mitochondrial functions (supplementary figure 5), which indicates that our findings are related to astrocytic diversity rather than representing general SCZ mechanisms present in all astrocytes. Importantly, disease association analysis showed SCZ as the most enriched disease term among a curated set of 10,370 diseases (figure 2J, supplementary table 8, supplementary figure 6), strongly supporting the SCZ-specific nature of the transcriptional signature identified.
Fig. 2.
Potential role of subtype astrocytes in SCZ. (A) Schematic illustration of the reprogramming and differentiation process of subtype astrocytes derived from HC and SCZ patients. Clozapine responders (R) and nonresponders (NR) are indicated. (B) All HC and SCZ hiPSCs expressed typical pluripotency markers. (C) All HC and SCZ hiPSCs stained positively for GFAP. (D) Intracellular glutamate levels (fold change) in SCZ subtype astrocytes relative to HC astrocytes. Measurements were performed in triplicates. Data is shown as mean ± SD. *Mann–Whitney U test: P < .05. (E) PCA analysis of two HC and five SCZ subtype astrocyte samples for which RNA-seq data was available. The PCA analysis was based on the top 500 genes with largest expression variance. PC: principal component. (F) Computational deconvolution of human brain cell types in HC and SCZ samples. Data shown as mean ± SD. (G) Volcano plot showing significant DE genes (FDR < 0.1, dotted line). Top 5 genes with lowest P-values are labeled. (H) GO analysis of upregulated DE genes. Top 15 enriched (FDR < 0.05) GO terms (biological processes) are shown. (I) GO analysis of downregulated DE genes. Top 15 enriched (FDR < 0.05) GO terms are shown. (J) Top 15 disease terms of the CURATED DisGeNET database (10 370 diseases) associated with the identified SCZ-related DE genes.
CLZ Rescues Deficient D-Serine Levels in Subtype Astrocytes Derived from Responders
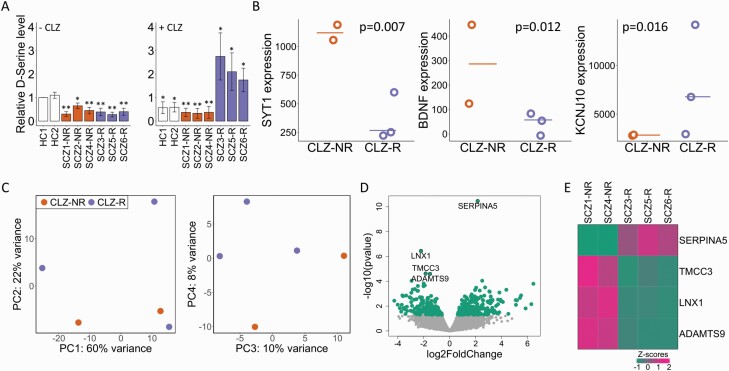
Clozapine (CLZ) is the primary antipsychotic used in treatment-resistant SCZ, and 60%–70% of patients treated with CLZ show a response.29 Although CLZ is primarily a serotonin/dopamine modulator, it has been proposed that it could also induce its therapeutic effects through glutamatergic regulation.30 However, studies have shown conflicting evidence as to the precise role played by the NMDA receptor coagonist d-serine.31 Using ICC, we found a significant and CLZ response-independent decrease in d-serine levels in SCZ astrocytes relative to HCs. Intriguingly, exposing the cell cultures to CLZ rescued the d-serine deficiency in CLZ responders but not in nonresponders (figure 3A, supplementary figure 7), indicating that d-serine could be an important mediator of CLZ response in the brain. To further elucidate the molecular mechanisms of CLZ response in human astrocytes, we conducted both a targeted (72 genes of interest related to glutamate and d-serine-metabolism and transport) and a global DE analysis. The targeted analysis identified 3 genes (SYT1, BDNF, and KCNJ10) with differential expression across CLZ-R and CLZ-NR groups (figure 3B, supplementary table 9), with the strongest association (P = .007) seen for SYT1, considered the master switch responsible for neurotransmitter release.32 In the global analysis, four genes (SERPINA5, LNX1, TMCC3, and ADAMTS9) were significantly associated (FDR < 0.1) with CLZ response (figure 3C and D, supplementary table 10). Interestingly, SERPINA5 encodes a member of the serine protease inhibitor family of proteins, or serpins, which has been implicated in synaptic plasticity and memory formation.33 Heatmap analysis showed no overlap in expression levels of the four genes between the two CLZ response groups, indicating a strong correlation with response status (figure 3E).
Fig. 3.
Potential role of subtype astrocytes in CLZ response. (A) Fold change of d-serine levels (measured as the number of d-serine-positive puncta) relative to HC1 before CLZ treatment. Left: Before CLZ treatment, Right: after CLZ treatment. The experiments were performed three times. Data shown as mean ± SD. Mann–Whitney U test: *P < .05 and **P < 0.01. (B) Expression levels of genes related to glutamate and d-serine metabolism and transport with nominally significant (P < .05) differences in CLZ-R vs. CLZ-NR samples. (C) PCA plots showing partial and complete clustering on CLZ response status across PCs 1–2 (left) and PCs 3–4 (right), respectively. The plots are based on the top 500 genes with largest expression variation across SCZ samples. (D) Global DE analysis between CLZ-R and CLZ-NR samples. Significant DE genes (FDR < 0.1) are labeled. (E) Heatmap of the four DE genes with differential expression in CLZ responders (R) compared to nonresponders (NR). Expression levels are shown as standardized z scores.
Discussion
In the present study, we derived a morphological subpopulation of hiPSC astrocytes and examined its gene expression profile with respect to SCZ pathology and CLZ response, with a particular focus on genes and cellular processes related to glutamatergic processes. An important finding was a significant decrease in intracellular glutamate concentrations in SCZ astrocytes which was likely driven by deficient glutamate synthesis rather than reduced uptake capacity. This is in line with our previous study showing no differences in glutamate uptake between hiPSC-derived SCZ and HC astrocytes.28 Several studies have reported excessive extracellular glutamate concentrations in SCZ patients, and this is thought to be a direct consequence of NMDA receptor hypofunction and reduced glutamate clearance by astrocytes.34 Our results suggest that de novo biosynthesis of glutamate from glucose, which primarily takes place in astrocytes,34 could act as a feedback mechanism by which increased levels of synaptic glutamate are counterbalanced, and thus represent an important component of glutamatergic regulation.34
Another important finding was a significant reduction in d-serine levels in SCZ astrocytes and a subsequent CLZ-R-selective rescue induced by CLZ treatment. d-Serine is released by both astrocytes and neurons and binds to the glycine site of NMDA receptors to facilitate receptor activation by glutamate. Decreased levels of d-serine have been reported in the cerebrospinal fluid (CSF) and blood of SCZ patients, and accumulating clinical evidence suggests that treatment with d-serine, especially in high doses, significantly improves positive, negative, and cognitive symptoms in SCZ.35,36 Remarkably, CLZ response in our study was defined based on improvement in positive and negative symptoms, and CLZ treatment led to full recovery of d-serine in CLZ-R astrocytes only, providing further support for the positive effects of d-serine on these symptom groups. CLZ treatment has also been found to improve glial d-serine release in the rat cortex and in cultured astrocytes,37 but whether d-serine-related processes may act as mediators of clinical response has not been investigated. Our results suggest that the therapeutic properties of CLZ may by be mediated by its ability to maintain a high d-serine pool and a consequent improvement in release capacity, indicating that the regulation of d-serine biogenesis and metabolism may be exploited as a potential target for the development of novel therapeutic agents.
The systematic investigation of astrocyte diversity is a nascent field of astrocyte biology research which was partly motivated by the early detection of morphological heterogeneity between astrocytes.5,9 Researchers have since identified additional forms of diversity, including cellular, regional, and functional, and are beginning to decipher their relevance in health and disease.8 Most of these studies, however, have been carried out using rodent models, which may prove inadequate to recapitulate astrocyte diversity in humans as important species differences exist.1,7 The main strength and novelty of the present study was the use of a human iPSC-derived cell model to explore the transcriptional mechanisms of astrocyte diversity in the context of SCZ and CLZ response. We identified distinct patterns of gene expression that differentiated this subpopulation from other astrocytes with a classical morphology, and found that subtype astrocytes from SCZ patients displayed a disease-specific expression signature, which indicates a potential role in SCZ pathology and may facilitate a more targeted approach towards pharmaceutical interventions.
Despite its strengths, however, our study also has limitations which need to be addressed in subsequent work. First, the small sample size necessitates caution when interpreting the results as the reduced statistical power may be insufficient to disentangle the biological effects of interest from donor-specific variation. Second, additional functional analyses are needed to demonstrate that the cellular identity of the derived subtype astrocytes corresponds to known subsets in the human brain.
In conclusion, our study represents a first attempt to investigate the potential role of astrocyte diversity in SCZ using human iPSCs derived from healthy individuals and SCZ patients. Our findings warrant additional investigations, in particular in-depth functional characterization of astrocyte populations, in order to further clarify the precise contribution of astrocytic heterogeneity to SCZ biology.
Supplementary Material
Acknowledgments
This work was supported by grants from the Czech Health Research Council (15-31063A, Y.-M.S. and T.K.), the South-Eastern Norway Regional Health Authority (#2018094, S.D.), the Research Council of Norway (#223273, S.D.), and the European Structural and Investment Funds (CETOCOEN PLUS project: CZ.02.1.01/0.0/0.0/15_003/0000469, E.B.). We thank Mr. Lars Hansson for preparing samples for RNA sequencing. The sequencing service was provided by the Norwegian Sequencing Centre (www.sequencing.uio.no), a national technology platform hosted by the University of Oslo and supported by the Functional Genomics and Infrastructure programs of the Research Council of Norway and the South-Eastern Regional Health Authorities.
Author Contributions
Conceptualized and designed the study: I.A., Y-M.S. Obtained human skin biopsies: T.K. Carried out stem cell experiments: Y-M.S., H.H., M.G. Performed RNA-seq analyses: I.A., E.B. Wrote the manuscript: I.A. Supervision and funding acquisition: Y.-M.S., S.D., O.A.A. All authors critically read and approved the final version of the manuscript.
Conflicts of Interest
O.A.A. is a consultant to HealthLytix and has received speaker’s honorarium from Lundbeck and Synovion. The other authors declare no conflict of interest.
References
- 1. Vasile F, Dossi E, Rouach N. Human astrocytes: structure and functions in the healthy brain. Brain Struct Funct. 2017;222(5):2017–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Uno Y, Coyle JT. Glutamate hypothesis in schizophrenia. Psychiatry Clin Neurosci. 2019;73(5):204–215. [DOI] [PubMed] [Google Scholar]
- 3. Kim R, Healey KL, Sepulveda-Orengo MT, Reissner KJ. Astroglial correlates of neuropsychiatric disease: from astrocytopathy to astrogliosis. Prog Neuropsychopharmacol Biol Psychiatry. 2018;87(Pt A):126–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tarasov VV, Svistunov AA, Chubarev VN, et al. . Alterations of astrocytes in the context of schizophrenic dementia. Front Pharmacol. 2019;10:1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khakh BS, Deneen B. The emerging nature of astrocyte diversity. Annu Rev Neurosci. 2019;42:187–207. [DOI] [PubMed] [Google Scholar]
- 6. Oberheim NA, Goldman SA, Nedergaard M. Heterogeneity of astrocytic form and function. Methods Mol Biol. 2012;814:23–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oberheim NA, Takano T, Han X, et al. . Uniquely hominid features of adult human astrocytes. J Neurosci. 2009;29(10):3276–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khakh BS, Sofroniew MV. Diversity of astrocyte functions and phenotypes in neural circuits. Nat Neurosci. 2015;18(7):942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Westergard T, Rothstein JD. Astrocyte diversity: current insights and future directions. Neurochem Res. 2020;45(6):1298–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Balan S, Toyoshima M, Yoshikawa T. Contribution of induced pluripotent stem cell technologies to the understanding of cellular phenotypes in schizophrenia. Neurobiol Dis. 2019;131:104162. [DOI] [PubMed] [Google Scholar]
- 11. Adewumi O, Aflatoonian B, Ahrlund-Richter L, et al. ; International Stem Cell Initiative . Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25(7):803–816. [DOI] [PubMed] [Google Scholar]
- 12. Andreasen NC, Flaum M, Arndt S. The Comprehensive Assessment of Symptoms and History (CASH). An instrument for assessing diagnosis and psychopathology. Arch Gen Psychiatry. 1992;49(8):615–623. [DOI] [PubMed] [Google Scholar]
- 13. Sheehan DV, Lecrubier Y, Sheehan KH, et al. . The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry. 1998;59(Suppl 20):22–33;quiz 34. [PubMed] [Google Scholar]
- 14. Grabiec M, Hříbková H, Vařecha M, et al. . Stage-specific roles of FGF2 signaling in human neural development. Stem Cell Res. 2016;17(2):330–341. [DOI] [PubMed] [Google Scholar]
- 15. Hříbková H, Grabiec M, Klemová D, Slaninová I, Sun YM. Calcium signaling mediates five types of cell morphological changes to form neural rosettes. J Cell Sci. 2018;131(3):jcs206896. [DOI] [PubMed] [Google Scholar]
- 16. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. [DOI] [PubMed] [Google Scholar]
- 19. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, et al. . The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48(D1):D845–D855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Colombo JA, Reisin HD. Interlaminar astroglia of the cerebral cortex: a marker of the primate brain. Brain Res. 2004;1006(1):126–131. [DOI] [PubMed] [Google Scholar]
- 23. Perea G, Navarrete M, Araque A. Tripartite synapses: astrocytes process and control synaptic information. Trends Neurosci. 2009;32(8):421–431. [DOI] [PubMed] [Google Scholar]
- 24. Zhang Y, Sloan SA, Clarke LE, et al. . Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89(1):37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schousboe A, Scafidi S, Bak LK, Waagepetersen HS, McKenna MC. Glutamate metabolism in the brain focusing on astrocytes. Adv Neurobiol. 2014;11:13–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Müller N. Inflammation in schizophrenia: pathogenetic aspects and therapeutic considerations. Schizophr Bull. 2018;44(5):973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Colombo E, Farina C. Astrocytes: key regulators of neuroinflammation. Trends Immunol. 2016;37(9):608–620. [DOI] [PubMed] [Google Scholar]
- 28. Akkouh IA, Ueland T, Hansson L, et al. . Decreased IL-1β-induced CCL20 response in human iPSC-astrocytes in schizophrenia: potential attenuating effects on recruitment of regulatory T cells. Brain Behav Immun. 2020;87:634–644. [DOI] [PubMed] [Google Scholar]
- 29. Lally J, Gaughran F, Timms P, Curran SR. Treatment-resistant schizophrenia: current insights on the pharmacogenomics of antipsychotics. Pharmgenomics Pers Med. 2016;9:117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goldstein ME, Anderson VM, Pillai A, Kydd RR, Russell BR. Glutamatergic neurometabolites in clozapine-responsive and -resistant schizophrenia. Int J Neuropsychopharmacol. 2015;18(6):pyu117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Samanaite R, Gillespie A, Sendt KV, McQueen G, MacCabe JH, Egerton A. Biological predictors of clozapine response: a systematic review. Front Psychiatry. 2018;9:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rizo J, Rosenmund C. Synaptic vesicle fusion. Nat Struct Mol Biol. 2008;15(7):665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Almonte AG, Sweatt JD. Serine proteases, serine protease inhibitors, and protease-activated receptors: roles in synaptic function and behavior. Brain Res. 2011;1407:107–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mei YY, Wu DC, Zhou N. Astrocytic regulation of glutamate transmission in schizophrenia. Front Psychiatry. 2018;9:544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kantrowitz JT, Malhotra AK, Cornblatt B, et al. . High dose d-serine in the treatment of schizophrenia. Schizophr Res. 2010;121(1-3):125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. MacKay MB, Kravtsenyuk M, Thomas R, Mitchell ND, Dursun SM, Baker GB. d-serine: potential therapeutic agent and/or biomarker in schizophrenia and depression? Front Psychiatry. 2019;10:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tanahashi S, Yamamura S, Nakagawa M, Motomura E, Okada M. Clozapine, but not haloperidol, enhances glial d-serine and L-glutamate release in rat frontal cortex and primary cultured astrocytes. Br J Pharmacol. 2012;165(5):1543–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.