Abstract
Comparative evaluation of the semiautomated COBAS AMPLICOR hepatitis B virus (HBV) MONITOR Test (COBAS-HBV) and manual AMPLICOR HBV MONITOR Test (AMPLICOR-HBV) on 208 serum samples revealed no significant difference in the sensitivities of the two assays. Twenty samples tested HBV DNA negative and 183 samples tested HBV DNA positive by both assays. Three samples tested positive by COBAS-HBV only and two samples tested positive by AMPLICOR-HBV only. HBV DNA concentrations determined by the two assays were significantly related (n = 183, r = 0.97, P < 0.0001), which indicates that COBAS-HBV could replace AMPLICOR-HBV. The major inconvenience of COBAS-HBV is the required performance of appropriate predilutions of high-titer samples in order to extend the narrow dynamic range of the assay.
Hepatitis B virus (HBV) causes a number of different diseases, ranging from clinically inapparent infection to severe, fulminant hepatitis, cirrhosis, hepatocellular carcinoma, and death (8). Although a number of tests are employed to follow the course of HBV-associated diseases and to predict the long-term outcome of the infection, quantitative measurement of HBV DNA in serum samples appears to be the most reliable method for monitoring chronically infected patients (2, 8, 10). Quantitative determination of HBV DNA is important for monitoring HBV replication activity and disease progression, as well as for assessing responses to antiviral treatment of patients with chronic hepatitis B (8, 10). The maternal HBV DNA load during the perinatal period is the most important determinant of infection outcome in infants and has been identified as a stronger predictor of persistent infection in infants than maternal HBeAg status (3). Furthermore, the detection of HBV DNA is useful in resolving diagnostic uncertainties following serological testing for markers of HBV infection that are caused by HBV genetic variations (21).
Several assays for the quantitative measurement of HBV DNA have been developed and used in research and diagnostic virology laboratories (1, 5, 6, 8, 11, 15–18, 20). Unfortunately, these assays generate highly divergent results due to the lack of standardization and the differences in dynamic ranges (7, 18, 19, 22). The first standardized commercially available method based on quantitative PCR, the AMPLICOR HBV MONITOR test (AMPLICOR-HBV), was introduced a few years ago by Roche Molecular Systems (Branchburg, N.J.) (6, 12). The test is based on the coamplification of HBV template and an internal quantitation standard (QS) and on a subsequent enzyme-linked immunosorbent assay detection of captured amplicons. The HBV DNA is then calculated by comparing HBV/QS ratios of optical density values to a standard curve set up for each run. Although AMPLICOR-HBV performed well in both research and clinical settings (4, 6, 12–14, 18, 19), this manual test is rather laborious, requires much hands-on time, and carries a risk of technical errors. To overcome these problems, the manufacturer recently adopted a test for automated processing by a COBAS analyzer (16). In comparison to other COBAS-based quantitative tests (e.g., for HCV and human immunodeficiency virus) in the COBAS AMPLICOR HBV MONITOR Test (COBAS HBV) the amount of HBV DNA in each specimen is calculated from the ratio of the total HBV absorbance to the total QS absorbance and the total input number of QS molecules, using a simple algorithm rather than using a standard curve like that used in the manual version of the test. However, adaptation for automated processing significantly changed the dynamic range of the test: the span narrowed from 4 × 102 to 4 × 107 HBV DNA copies/ml (AMPLICOR-HBV) to 2 × 102 to 2 × 105 HBV DNA copies/ml (COBAS-HBV).
Since the analytical performances of the premarketing version of COBAS-HBV, such as linearity, reproducibility, and precision, have been recently evaluated on panels of reference samples (16), the present study describes the results of a comparative evaluation of COBAS-HBV and AMPLICOR-HBV on clinical samples under the routine conditions of a diagnostic virology laboratory. A total of 208 serum samples obtained from 172 HBsAg-positive Slovenian patients (34 HBeAg-positive and 138 HBeAg-negative carriers) at various clinical stages of chronic infection was included in the study. All serum samples were stored in aliquots at −70°C and thawed only once, prior to testing. Quantification of HBV DNA using the AMPLICOR-HBV was performed from August to December 1999 as a part of routine assessment of our HBsAg-positive patients. According to our laboratory strategy, based on 3 years of experience with AMPLICOR-HBV, all HBeAg-negative samples were initially tested undiluted and all the HBeAg-positive samples were tested after a predilution of 1:1,000 in negative human serum. The same 208 samples were tested by the premarketing version of COBAS-HBV in February 2000. In order to extend the narrow dynamic range of COBAS-HBV, serum samples were prediluted in negative human serum according to the following criteria: samples quantified previously by AMPLICOR-HBV that contained between 2 × 103 and 2 × 107 HBV DNA copies/ml were diluted 1:50, samples containing between 2 × 105 and 2 × 109 HBV DNA copies/ml were diluted 1:5,000, and samples containing between 2 × 107 and 2 × 1011 HBV DNA copies/ml were diluted 1:500,000.
Determination of HBV DNA by the semiautomated COBAS-HBV and manual AMPLICOR-HBV with 208 HBsAg-positive serum samples revealed no significant difference in the sensitivities of the two tests (Table 1). Thus, 183 samples tested HBV DNA positive and 20 samples tested HBV DNA negative by both tests. Discordant results were obtained with five samples: HBV DNA was detected in three samples only by the semiautomated test and in two samples only by the manual test (Table 1). Three patients with discordant results (one healthy carrier and two patients undergoing interferon-α therapy; patients 1, 2, and 4, respectively; Table 1) became HBV DNA negative by AMPLICOR HBV, as well as by COBAS-HBV, over the following 3 to 8 months. However, for another patient (the healthy carrier, patient 5; Table 1), both tests detected low titers of HBV DNA (COBAS-HBV, 1, 560 copies/ml; AMPLICOR-HBV, 1,920 copies/ml) in a follow-up sample obtained 4 months after initial testing. Finally, one patient (patient 3; Table 1) died due to non-liver-related disease 10 weeks after the initial testing.
TABLE 1.
Comparison of the results obtained by COBAS-HBV and AMPLICOR-HBV with 208 serum samples
| AMPLICOR-HBV result | COBAS-HBV result
|
Total | |
|---|---|---|---|
| Positive (>200 HBV DNA copies/ml) | Negative (<200 HBV DNA copies/ml) | ||
| Positive (>400 HBV DNA copies/ml) | 183 | 2a | 185 |
| Negative (<400 HBV DNA copies/ml) | 3b | 20 | 23 |
| Total | 186 | 22 | 208 |
These two samples were quantified by AMPLICOR-HBV at 520 HBV DNA copies/ml (patient 1) and 1,200 HBV DNA copies/ml (patient 2).
These three samples were quantified by COBAS-HBV at 383 (patient 3), 646 (patient 4), and 1,820 (patient 5) HBV DNA copies/ml.
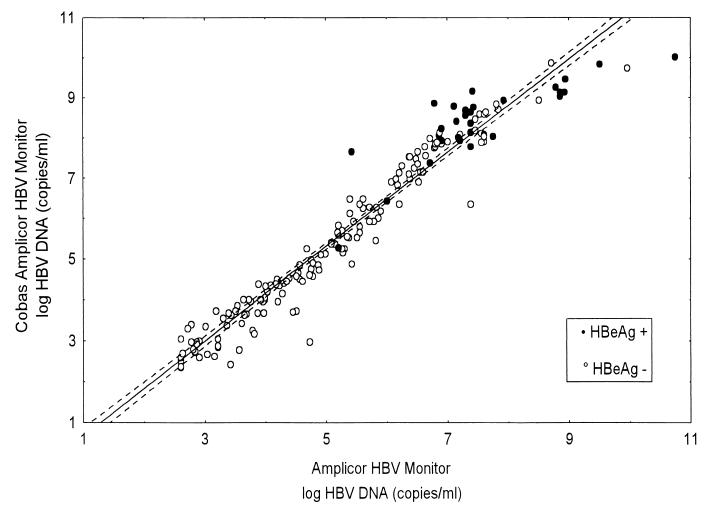
To determine the relationship between the HBV DNA results obtained by the two tests using the same samples, linear regression analysis (Pearson correlation) was performed. Statistical analysis was carried out using the Statistica 6.0 program (StatSoft Inc., Tulsa, Okla.). HBV DNA concentrations determined by the two tests in a given sample were significantly related (n = 183, r = 0.97, P < 0.0001) . Figure 1 shows a plot of log10 AMPLICOR-HBV results versus log10 COBAS-HBV results with a fitted regression line described by the following equation: y = −0.4930 + 1.1618 × x. The median HBV DNA levels as measured by COBAS-HBV and AMPLICOR-HBV for the HBeAg-positive samples were 108.3 and 107.3 copies/ml, respectively, and those for the HBeAg-negative samples were 104.8 and 104.7 copies/ml, respectively.
FIG. 1.
Correlation between COBAS-HBV- and AMPLICOR-HBV-determined HBV DNA concentrations in the analysis of HBV DNA in 183 serum samples.
The results of our study show that HBV DNA concentrations determined by the two tests are highly related and indicate that the semiautomated COBAS-HBV could replace the manual microwell plate-based AMPLICOR-HBV. Similar conclusions were also made by Noborg et al., who recently comparatively evaluated both tests with 153 samples originating from Sweden (16). However, in our opinion, determining which HBV DNA viral load test is more suitable for routine clinical virology laboratory is not simple. In comparison to manual AMPLICOR-HBV, the use of semiautomated COBAS-HBV certainly simplifies the amplification and detection of PCR products and calculation of results and reduces the hands-on time and risk of technical or computational errors. However, the major drawback of COBAS-HBV, from a laboratory perspective, is a narrowed dynamic range of this test, since the upper limit of detection was reduced from 4 × 107 (AMPLICOR-HBV) to 2 × 105 (COBAS-HBV) copies/ml. Thus, for a significant number of samples, HBV DNA quantification by COBAS-HBV was possible only after an appropriate predilution of a particular sample. Since the manufacturer doesn't recommend any effective strategy concerning sample predilution, each laboratory has to empirically determine the most appropriate predilution for a particular clinical sample. In order to minimize the costs linked to repeated testing of samples, we recommend that the laboratory not quantify HBV DNA by COBAS-HBV until the HBeAg status of a particular sample has been determined. If a sample is recognized as HBeAg positive and the laboratory has already used AMPLICOR-HBV, we suggest prediluting a sample, which should be quantified by COBAS-HBV by using a dilution factor that represents a median log10 HBV-DNA level determined by AMPLICOR-HBV among HBeAg-positive samples in a particular laboratory substracted by 2.5 log10. If we had used this strategy in our study, it would have allowed us to quantify 32 (94.1%) out of 34 HBeAg-positive serum samples by COBAS-HBV without additional testing. For laboratories that plan to start HBV DNA quantification by directly using COBAS-HBV or for those who have used quantification methods other than AMPLICOR-HBV, we recommend, as recently suggested by Noborg et al. (16), 1:105 as the most suitable predilution for HBeAg-positive samples. If we had used this strategy in our study, it would have allowed us to quantify 30 (88.2%) out of 34 HBeAg-positive serum samples by COBAS-HBV without additional testing.
However, HBeAg-positive samples represent only a minority of the samples sent to a clinical virology laboratory for the quantification of HBV DNA (ranging approximately from 15 to 25% of the total samples). Therefore, we tried to find, just as we did for HBeAg-positive samples, the most appropriate dilution strategy for HBeAg-negative samples. Although we checked different strategies, no single dilution allowed us to quantify more than 46% of the HBeAg-negative samples by using COBAS-HBV without additional testing. Finally, we found out that the largest number of HBeAg-negative samples could have been quantified using COBAS-HBV without additional testing when initially tested undiluted. If we had used this “undiluted samples” strategy in our study, we would have quantified 59% of HBeAg-negative serum samples without additional testing using COBAS-HBV, and 41% of samples would have been tested again. If we had continued the quantification of these samples with a 1:103 predilution, there still would have remained 11 samples that would have been categorized as containing more than 2 × 108 HBV DNA copies/ml; we would have needed to dilute further to determine the exact quantity of HBV DNA. On the contrary, if we had applied this undiluted samples strategy using AMPLICOR-HBV, we would have quantified 94% of our HBeAg-negative samples without additional testing. All the remaining HBeAg-negative samples would have been quantified using AMPLICOR-HBV with a single additional 1:105 dilution.
The proposed strategies for the most cost-effective testing of both HBeAg-positive and HBeAg-negative samples using COBAS-HBV should be, however, verified in a prospective manner, which is the purpose of our present work.
In conclusion, we noted that the new semiautomated COBAS-HBV has many advantages over AMPLICOR-HBV, but based on the preliminary European list prices as of July 2000, we calculated that the per-test cost for COBAS-HBV is at least 20% higher than that of AMPLICOR-HBV, mainly because of repeated testing, which appears to be necessary to determine exactly the HBV DNA load in a significant number of HBeAg-negative samples. However, we allow the possibility that COBAS-HBV would perform better in laboratories which have different serum sample patterns (e.g., if testing mostly patients with chronic hepatitis B on antiviral therapy) or demonstrate a significant savings in labor, hands-on time, disposables, and biohazardous waste when using a semiautomated test instead of a manual test for HBV DNA quantification.
REFERENCES
- 1.Barlet V, Cohard M, Thelu M A, Chaix M J, Baccard C, Zarski J P, Seigneurin J M. Quantification detection of hepatitis B virus DNA in serum using chemiluminescence: comparison with radioactive solution hybridization assay. J Virol Methods. 1994;49:141–152. doi: 10.1016/0166-0934(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 2.Berger A, Braner J, Doerr H W, Weber B. Quantification of viral load: clinical relevance for human immunodeficiency virus, hepatitis B virus and khepatitis C virus infection. Intervirology. 1998;41:24–34. doi: 10.1159/000024912. [DOI] [PubMed] [Google Scholar]
- 3.Burk R D, Hwang L Y, Ho G Y F, Shafritz D A, Beasley R P. Outcome of perinatal hepatitis B virus exposure is dependent on maternal virus load. J Infect Dis. 1994;170:1418–1423. doi: 10.1093/infdis/170.6.1418. [DOI] [PubMed] [Google Scholar]
- 4.Cacciola I, Pollicino T, Squadrito G, Cerenzia G, Villari D, de Franchis R, Santantonio T, Brancatelli S, Colucci G, Raimondo G. Quantification of intrahepatic hepatitis B virus (HBV) DNA in patients with chronic HBV infection. Hepatology. 2000;31:507–512. doi: 10.1002/hep.510310235. [DOI] [PubMed] [Google Scholar]
- 5.Chen T, Luk J M, Cheung S T, Yu W C, Fan S T. Evaluation of quantitative PCR and branched-chain DNA assay for detection of hepatitis B virus DNA in sera from hepatocellular carcinoma and liver transplant patients. J Clin Microbiol. 2000;38:1977–1980. doi: 10.1128/jcm.38.5.1977-1980.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerken G, Gomes J, Lampertico P, Colombo M, Rothaar T, Trippler M, Colucci G. Clinical evaluation and applications of the Amplicor HBV Monitor™ test, a quantitative HBV DNA PCR assay. J Virol Methods. 1998;74:155–165. doi: 10.1016/s0166-0934(98)00081-0. [DOI] [PubMed] [Google Scholar]
- 7.Heermann K-H, Gerlich W H, Chudy M, Schaefer S, Thomssen R the EUROHEP Pathobiology Group. Quantitative detection of hepatitis B virus DNA in two international reference plasma preparations. J Clin Microbiol. 1999;37:68–73. doi: 10.1128/jcm.37.1.68-73.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendricks D A, Stowe B J, Hoo B S, Kolberg J L, Irvine B D, Neuwald P D, Urdea M S, Perrillo R P. Quantitation of HBV DNA in human serum using a branched DNA (bDNA) signal amplification assay. Am J Clin Pathol. 1995;104:537–546. doi: 10.1093/ajcp/104.5.537. [DOI] [PubMed] [Google Scholar]
- 9.Ho S K N, Chan T M, Cheng I K P, Lai K N. Comparison of the second-generation Digene Hybrid-Capture assay with the branched-DNA assay for measurement of hepatitis B virus DNA in serum. J Clin Microbiol. 1999;37:2461–2465. doi: 10.1128/jcm.37.8.2461-2465.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodinka R L. Laboratory diagnosis of viral hepatitis. In: Specter S, editor. Viral hepatitis: diagnosis, therapy, and prevention. Totowa, N.J: Humana Press; 1999. pp. 193–249. [Google Scholar]
- 11.Kaneko S, Miller R H, Feinstone S M, Unoura M, Kobayashi K, Hattori N, Purcell R H. Detection of serum hepatitis B virus DNA in patients with chronic hepatitis using the polymerase chain reaction assay. Proc Natl Acad Sci USA. 1989;86:312–316. doi: 10.1073/pnas.86.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kessler H H, Pierer K, Dragon E, Lackner H, Santner B, Stünzner D, Stelzl E, Waitzl B, Marth E. Evaluation of a new assay for HBV DNA quantitation in patients with chronic hepatitis B. Clin Diagn Virol. 1998;9:37–43. doi: 10.1016/s0928-0197(97)10008-3. [DOI] [PubMed] [Google Scholar]
- 13.Kessler H H, Preininger S, Stelzel E, Daghofer E, Santner B, Marth E, Lackner H, Stauber R E. Identification of different states of hepatitis B virus infection with a quantitative PCR assay. Clin Diagn Lab Immunol. 2000;7:298–300. doi: 10.1128/cdli.7.2.298-300.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagata I, Colucci G, Gregorio G V, Cheeseman P, Williams R, Mieli-Vergani G, Vergani D. The role of HBV DNA quantitative PCR in monitoring the response to interferon treatment in chronic hepatitis B virus infection. J Hepatol. 1999;30:965–969. doi: 10.1016/s0168-8278(99)80247-5. [DOI] [PubMed] [Google Scholar]
- 15.Niesters H G M, Krajden M, Cork L, de Medina M, Hill M, Fries E, Osterhaus A D M E. A multicenter study evaluation of the Digene Hybrid Capture II signal amplification technique for detection of hepatitis B virus DNA in serum samples and testing of EUROHEP standards. J Clin Microbiol. 2000;38:2150–2155. doi: 10.1128/jcm.38.6.2150-2155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noborg U, Gusdal G A, Pisa E K, Hedrum A, Lindh M. Automated quantitative analysis of hepatitis B virus DNA by using the Cobas Amplicor HBV Monitor test. J Clin Microbiol. 1999;37:2793–2797. doi: 10.1128/jcm.37.9.2793-2797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pawlotsky J M, Bastie A, Lonjon I, Rémiré J, Darthuy F, Soussy C J, Dhumeaux D. What technique should be used for routine detection and quantification of HBV DNA in clinical samples? J Virol Methods. 1997;65:245–253. doi: 10.1016/s0166-0934(97)02196-4. [DOI] [PubMed] [Google Scholar]
- 18.Pawlotsky J M, Bastie A, Hézode C, Lonjon I, Darthuy F, Rémiré J, Dhumeaux D. Routine detection and quantification of hepatitis B virus DNA in clinical laboratories: performance of three commercial assays. J Virol Methods. 2000;85:11–21. doi: 10.1016/s0166-0934(99)00149-4. [DOI] [PubMed] [Google Scholar]
- 19.Quint W G V, Heijtink R A, Schirm J, Gerlich W H, Niesters H G M. Reliability of methods for hepatitis B virus DNA detection. J Clin Microbiol. 1995;33:225–228. doi: 10.1128/jcm.33.1.225-228.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ranki M, Schätzl H M, Zachoval R, Uusi-Oukari M, Lehtovaara P. Quantification of hepatitis B virus DNA over a wide range from serum for studying viral replicative activity in response to treatment and in recurrent infection. Hepatology. 1995;21:1492–1499. [PubMed] [Google Scholar]
- 21.Van Deursen F J, Hino K, Wyatt D, Molyneaux P, Yates P, Wallace L A, Dow B C, Carman W F. Use of PCR in resolving diagnostic difficulties potentially caused by genetic variation of hepatitis B virus. J Clin Pathol. 1998;51:149–153. doi: 10.1136/jcp.51.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaaijer H L, ter Borg F, Cuypers H T M, Hermus M C A H, Lelie P N. Comparison of methods for detection of hepatitis B virus DNA. J Clin Microbiol. 1994;32:2088–2091. doi: 10.1128/jcm.32.9.2088-2091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]