Abstract
Campylobacter jejuni recovered from patients with Guillain-Barré syndrome (GBS) in different geographical locations and bearing different heat-labile and heat-stable antigens were found to have identical amino acid sequences in their flagellar flaA short variable region, suggesting that it may be a potentially useful marker for GBS association.
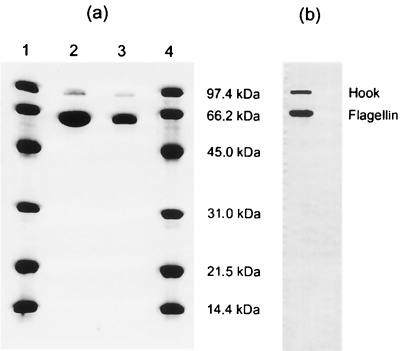
Gastroenteritis caused by Campylobacter jejuni is one of the common antecedent infections associated with Guillain-Barré Syndrome (GBS), an acute demyelinating polyneuropathy (9). In both the United States and Japan, many GBS-associated C. jejuni strains were found to belong to the Penner heat-stable (HS) serotype O19 (3). We have recently identified the heat-labile (HL) antigens of two O19 GBS strains and the O19 serostrain as HL77 and HL84. Also HL77, and HL84 serostrains were found to have the HS O-antigen 19 (11). This serological data supports the clonal nature of serotype O19 strains (2, 6). Further studies in our laboratory using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) immunoblot of whole-cell lysates with absorbed anti-HL77 and anti-HL84 serotyping antisera revealed that the HL77 and HL84 serodeterminants reside on the flagellar components of these O19 strains (unpublished data). Flagellar preparations, containing the flagellar hook proteins and the flagellin subunits, were prepared from the HL77 and HL84 type strains (Fig. 1a), and both the flagellar hook protein and the flagellin were shown to react with the anti-HL77 (data not shown) and anti-HL84 (Fig. 1b) typing antisera by SDS-PAGE immunoblot (10).
FIG. 1.
(a) SDS-PAGE of flagellar preparations from HL type strains 77 (lane 2) and 84 (lane 3). Lanes 1 and 4 show the migration of molecular weight markers. (b) SDS-PAGE immunoblot of flagellar preparation from HL type strain 84 reacting with absorbed anti-HL84.
DNA sequencing and alignment of the structural flagellum gene flaA was performed in an attempt to understand the role of the flagella in contributing to the serospecificity and pathogenesis of GBS. The flaA gene was amplified by PCR (7) from 1 μl of DNA template prepared by boiling in 0.4 ml of distilled water a loopful of bacteria from an overnight culture grown on sheep blood agar plate incubated at 37°C under microaerophilic conditions. The PCR forward (FLA4F) and reverse (FLA1728R) primer sequences were 5′-GGATTTCGTATTAACACAAATGGTGC and 5′-CTGTAGTAATCTTAAAACATTTTG, respectively (5, 7). Both strands of DNA containing flaA were sequenced using an automated system (ABI/Applied Biosystems, Inc., Foster City, Calif.) and primers including FLA4F, FLA1728R, 5′-GTGCAACCCAGTCTTCAAA, 5′-AAACCTGAACCTGCCGAGAA, 5′-AGGTTTCTCAGCAGGCAGTGG, and 5′-TGATTTGAGCTTGAACCGATTTG. The sequence data was compiled (Seqman 4.0; DNASTAR, Inc., Madison, Wis.), compared, and aligned with published data in GenBank (1; http://www.ncbi.nlm.nih.gov). The flaA gene sequences of five serotype O19 strains with HL77 and HL84 antigens used in our previous study were determined and compared, including two isolates from a pair of Japanese siblings suffering from GBS (OH4382 and OH4384), the O19 serostrain, and the HL77 and HL84 serostrains (11). The flaA genes of the two Japanese C. jejuni O19 GBS strains (OH4382 and OH 4384) are identical to the flaA gene of the O19 serostrain but have a one-base difference from the two identical flaA sequences of the HL77 and HL84 serostrains (Fig. 2). However, the mutation at coordinate 327 in the two HL serostrains is silent and does not result in any amino acid changes. The similarity of flaA sequences therefore confirms that the strains have identical HL antigens and supports the finding that HL77 and HL84 specificities reside on the flagella.
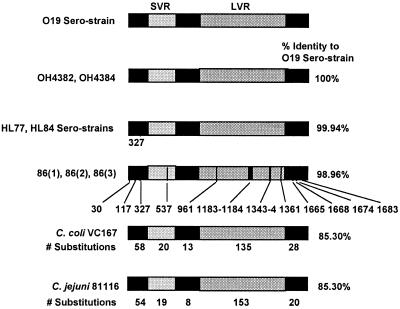
FIG. 2.
A diagrammatic representation of the flaA gene structures of different Campylobacter strains showing locations of sequence homology and variance. The SVR is located between coordinates 450 and 600 of flaA of L17 (AF050192), and the LVR is located between coordinates 700 to 1450 of flaA of C. jejuni L17. The difference in DNA sequence from the O19 serostrain was indicated with a vertical bar, except for C. coli VC167 and C. jejuni 81116, for which the number of changes in each region of flaA is indicated below the bar diagrams. Strains OH4382 and OH4384 are Japanese GBS strains; CH86(1), -(2), and -(3) are Chilean GBS strains.
The flaA gene of three C. jejuni isolates from multiple stool specimens of a GBS patient in Chile was also sequenced and found to be identical. These strains were typed as non-O19 and non-HL77 and HL84 and hence were antigenically different from the Japanese OH4382 and OH4384 isolates. Comparison of flaA sequences of the Chilean GBS isolates showed 98.96% homology to the flaA of the Japanese GBS isolates (Fig. 2 and 3). The differences between the Chilean and Japanese GBS strains were found at coordinates 30 (A to G), 117 (C to T), 327 (C to T), 537 (C to T), 961 (A to G), 1343 to 1344 (AA to CC), 1361 (C to T), 1665 (T to C), 1668 (T to A), 1674 (A to T), and 1683 (A to G) and at a six-nucleotide (TTCAAG) insertion at coordinates 1183 and 1184 (Fig. 2). The nucleotide differences that were at the 5′ end and the short variable region (SVR; Fig. 2) of flaA did not result in changes in the amino acids. However, the nucleotide differences at coordinates 961, 1343 to 1344, and 1361 that were in the long variable region (LVR; Fig. 2) resulted in amino acid conversions from threonine (Thr) to alanine (Ala), glutamic acid (Glu) to alanine, and alanine to valine (Val), respectively. Also, the Chilean isolate have a 6-bp insertion in the LVR that codes for valine-glutamine after amino acid 394.
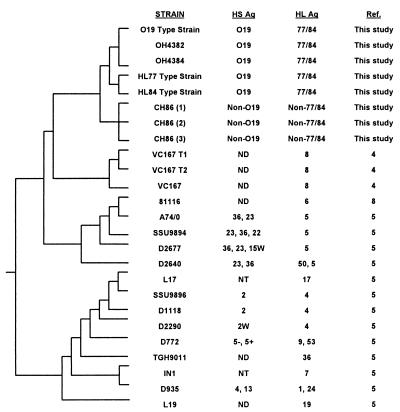
FIG. 3.
Dendrogram of aligned flaA sequences to show the relationships of different Campylobacter strains. The difference between O19 serostrains with HL77 and HL84 serostrains, Chilean GBS strains, C. coli VC167, C. jejuni 81116, and C. jejuni L17 were 1, 18, 254, 254, and 389 bp, respectively. NT, not typeable; ND, not determined. Strains OH4382 and OH4384 are Japanese GBS strains; strains CH86(1), -(2), and -(3) are Chilean GBS strains.
When the entire flaA sequences were compared with sequences in GenBank (1) and a dendrogram was generated to study the relationship of GBS-related and non-GBS-associated Campylobacter strains (Fig. 3), it was apparent that the GBS-related isolates were clustered together. Strains (C. coli VC167 and C. jejuni 81116) that were most closely related to the GBS isolates have about 15% sequence differences in their flaA genes, with 19 to 20 bp differences in their SVR and 135 to 153 bp differences in their LVR (Fig. 2 and 3). The genetic differences in the SVR of C. coli VC167 and C. jejuni 81116 also specify for very different amino acids in this part of their flagella. Sequence analysis of the C. jejuni SVR has also been proposed to be a useful epidemiology tool for strain discrimination (5). The alignment of all the 42 SVR sequences in GenBank showed that only the GBS strain C. jejuni D445 O19:HL77 (5; GenBank accession no. AF015106) is 100% homologous to the O19 GBS strains in the present study. The SVR of C. jejuni D445 is identical to two other GBS strains of the same serotype (5). It is thus surprising and unusual to find that strains of C. jejuni isolated from very different geographic locations (America, Japan, and Chile) and having very different antigenic formulas (O19:HL77 or HL84 or non-O19 and non-HL77 or HL84) would have identical amino acid sequences at their flagellar SVR. The common denominator in these strains is their association with GBS. Therefore, we hypothesize that the SVR of Campylobacter flagella may serve as an unique marker for their association with GBS. It is also interesting to note that the genetic and hence amino acid differences in the flagellar LVR of the Chilean and Japanese GBS isolates may specify for their different HL specificities. Indeed, further studies are required to confirm whether a certain region, such as the SVR in the flagellar antigen of Campylobacter, may be a useful marker for GBS association, while other regions may be associated with serospecificity, and to show how the flagellar may contribute to the induction and/or pathogenesis of GBS.
Nucleotide sequence accession numbers.
The GenBank accession numbers assigned to the C. jejuni flaA gene sequences determined in this study are as follows: O19 serostrain, AF290496; HL77 serostrain, AF290497; HL84 serostrain, AF290498; OH4382, AF290499; OH4384, AF290500; 86–1, AF290501; 86–2, AF290502; and 86-3, AF290503.
Acknowledgments
This work was supported in part by the Canadian Bacterial Diseases Network.
We thank the DNA Core Facility, National Microbiology Laboratory, for the preparation of the primers and the DNA sequencing work and Ed Taboada for his technical assistance.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Miller W, Lipman D J. Gapped BLAST and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujimoto S B, Allos M, Misawa N, Patton C M, Blaser M J. Restriction fragment length polymorphism analysis and random amplified polymorphic DNA analysis of Campylobacter jejuni strains isolated from patients with Guillain-Barré syndrome. J Infect Dis. 1997;176:1105–1108. doi: 10.1086/516522. [DOI] [PubMed] [Google Scholar]
- 3.Kuroki S, Saida T, Nukina M, Haruta T, Yoshioka M, Kobayashi Y, Naklanishi H. Campylobacter jejuni strains isolated from patients with Guillain-Barré syndrome belong mostly to Penner serogroup O19 and contain β-N-acetylglucosamine residues. Ann Neurol. 1993;33:234–247. doi: 10.1002/ana.410330304. [DOI] [PubMed] [Google Scholar]
- 4.Logan S M, Trust T J, Guerry P. Evidence for posttranslational modification and gene duplication of Campylobacter flagellin. J Bacteriol. 1989;169:3031–3038. doi: 10.1128/jb.171.6.3031-3038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meinersmann R J, Holsel L O, Fields P I, Hiett K L. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J Clin Microbiol. 1997;35:2801–2814. doi: 10.1128/jcm.35.11.2810-2814.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Misawa N, Allos B M, Blaser M J. Differentiation of Campylobacter jejuni serotype O19 strains from non-O19 strains by PCR. J Clin Microbiol. 1998;36:3567–3573. doi: 10.1128/jcm.36.12.3567-3573.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nachamkin I, Bohachick K, Patton C M. Flagellin gene typing of Campylobacter jejuni by restriction fragment length polymorphism analysis. J Clin Microbiol. 1993;31:1531–1536. doi: 10.1128/jcm.31.6.1531-1536.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nuijten P J, van Asten F J, Gaastra W, vander Zeijst B A. Structural and functional analysis of two Campylobacter jejuni flagellin genes. J Biol Chem. 1990;265:17798–17804. [PubMed] [Google Scholar]
- 9.Rees J H, Soudain S E, Gregson N A, Hughes R A. Campylobacter jejuni infection and Guillain-Barré Syndrome. N Engl J Med. 1995;333:1374–1379. doi: 10.1056/NEJM199511233332102. [DOI] [PubMed] [Google Scholar]
- 10.Tsang R S W, Luk J M C, Woodward D L, Johnson W M. Immunochemical characterization of haemagglutinating antigen of Arcobacter spp. FEMS Microbiol Lett. 1996;136:209–213. doi: 10.1111/j.1574-6968.1996.tb08051.x. [DOI] [PubMed] [Google Scholar]
- 11.Tsang R S W, Frosk P, Johnson W M. Heat labile serotyping of two Campylobacter jejuni strains isolated from patients with Guillain-Barré syndrome and belonging to serotype O19 (Penner) J Clin Microbiol. 2000;38:2021–2022. doi: 10.1128/jcm.38.5.2021-2022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]