Dear Editor,
Omicron (B.1.1.529), is a heavily mutated and highly contagious SARS-CoV-2 variant, which is currently causing large outbreaks in many countries. Protection provided by current vaccines is substantially reduced against Omicron.1,2 Moreover, many immunocompromised individuals cannot effectively be protected by vaccines.3 Hence, antiviral therapies will be essential to protect the most vulnerable individuals from severe COVID-19.
Different antibody therapies have been approved as COVID-19 therapies.4 Moreover, a range of antiviral small-molecule drugs are under investigation or already approved for the treatment of COVID-19. Remdesivir, an intravenous inhibitor of the viral RNA-dependent RNA polymerase (nsp12), was the first antiviral drug to be approved for the treatment of COVID-19.4 Molnupiravir and PF-07321332 are oral antiviral drugs that are hoped to be able to overcome the issues associated with an intravenous agent.4 Molnupiravir, a derivative of the broad-spectrum antiviral drug ribavirin, is metabolized into the active compound EIDD-1931, which is incorporated into the complementary RNA strand that is used as a template for the synthesis of viral genomic RNA during replication of the SARS-CoV-2 RNA genome. EIDD-1931 incorporation into the template strand causes excessive mutations in newly synthesized viral genomes, which affect their functionality in a process called ‘error catastrophe’ or ‘lethal mutagenesis’.5 Molnupiravir is approved in the UK and treatment of vulnerable SARS-CoV-2-infected individuals early after diagnosis has started.
The combination of PF-07321332 (nirmatrelvir) and ritonavir (which reduces PF-07321332 metabolism), also known as paxlovid, has been reported to reduce hospitalization of SARS-CoV-2-infected individuals in clinical trials.4 Other antiviral drug candidates for SARS-CoV-2 include the protease inhibitors, camostat, nafamostat, and aprotinin, which inhibit cleavage and activation of the viral spike (S) protein by host cell proteases and, in turn, SARS-CoV-2 entry into host cells.6
Reduced activity against the Omicron variant has been reported for antibody therapies.2 However, the effects of antiviral drugs against the Omicron remain to be investigated. Here, we tested the effects of EIDD-1931, ribavirin, remdesivir, favipravir (an additional RNA-dependent RNA polymerase inhibitor that displayed anti-SARS-CoV-2 activity in phase III clinical trials),7 PF-07321332, nafamostat, camostat, and aprotinin on the replication of two SARS-CoV-2 Omicron (B.1.1.529) isolates (Omicron 1, Omicron 2, see Supplementary information, Data S1) and one Delta (B.1.167.2) isolate (see Supplementary information, Data S1)8 in Caco-2 and Calu-3 cells as previously described.9
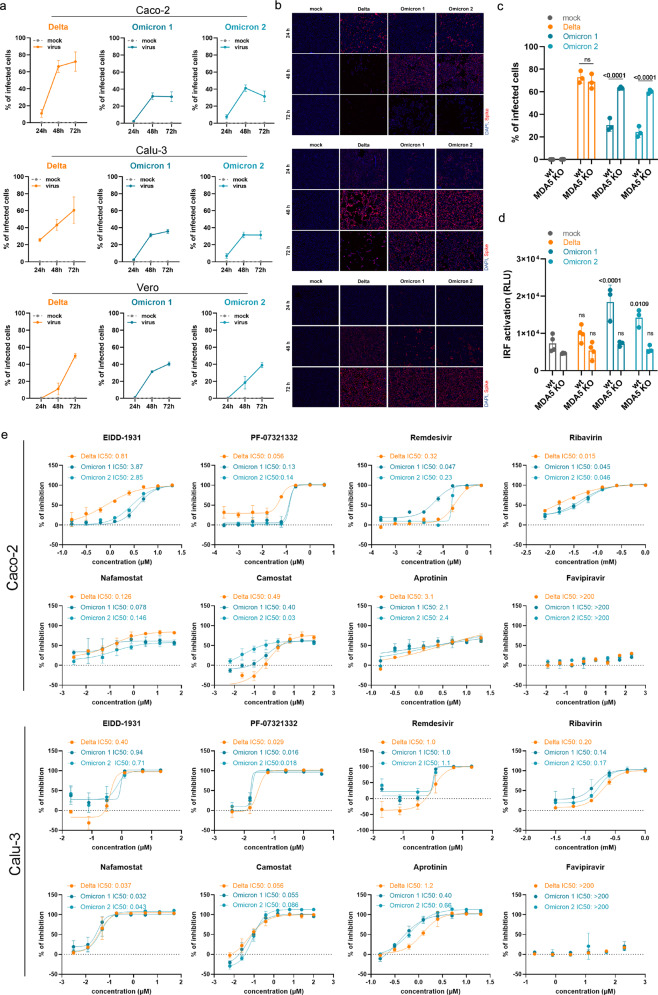
The Omicron isolates infected fewer cells in Calu-3 and Caco-2 cell cultures when compared with the Delta isolate (Fig. 1a, b), which is in agreement with previous findings in Calu-3 cells10 and in the hamster upper respiratory tract.11
Fig. 1. Interferon antagonism and antiviral therapy against novel SARS-CoV-2 variant Omicron.
a Caco-2 and Calu-3 cells were infected with SARS-CoV-2 variant Delta (GenBank ID: MZ315141), Omicron 1 (GenBank ID: OL800702) and Omicron 2 (GenBank ID: OL800703) at an MOI of 0.01. The number of infected cells at different time points post infection was determined by immunofluorescence staining of the SARS-CoV-2 S protein. Graphs represent means ± SD of 12 biological replicates. b Representative immunofluorescence images of a are shown (4× magnification). c Virus infection rates in A549-ACE2/TMPRSS2 MDA5-WT (wt) and A549-ACE2/TMPRSS2 MDA5 KO (MDA5 KO) cells 72 h post infection as determined by immunofluorescence staining of the S protein. Graph represents data of four biological replicates. d Induction of IRF transcriptional activity 24 h post infection in a promotor reporter assay. Graph displays means ± SD of four biological replicates. e Dose-dependent effects of selected antiviral compounds on SARS-CoV-2 Omicron and Delta variant isolates. Compounds were added to confluent monolayers and cells were subsequently infected with viral variants at MOI of 0.01. The inhibition rate was evaluated 24 h (Caco-2) and 48 h (Calu-3) post infection by staining of the S protein. Graphs depict means ± SD of three biological replicates. P-values were calculated using two-way ANOVA (c, d). ns, not significant.
However, all three isolates displayed comparable infection patterns in Vero cells (Fig. 1a, b). In contrast to Caco-2 and Calu-3 cells, Vero cells have a defective interferon response and represent an established model for studying virus replication in an interferon-deficient host cell background.12 Hence, the differences in Omicron virus replication in interferon-competent (Caco-2, Calu-3) and interferon-deficient (Vero) cells suggest that Omicron viruses may be less effective in antagonizing cellular interferon signaling than Delta viruses.
In agreement, the Delta isolate displayed superior infection patterns in A549 cells transduced with ACE2 (cellular receptor for the SARS-CoV-2 S protein) and TMPRSS2 (cleaves and activates S), but not in the same cell model with defective interferon signaling due to MDA5 knockout13 (Fig. 1c). Moreover, the Omicron isolates, but not the Delta isolate, activated interferon signaling as indicated by activation of the interferon response factor (IRF) promotor in A549 cells, which was prevented by MDA5 knockout (Fig. 1d). Taken together, these data show that Omicron viruses are less effective than Delta viruses in antagonizing the interferon response in human cells, which may contribute to the lower pathogenicity of the Omicron variant observed in patients.14 Notably, SARS-CoV-2 proteins known to inhibit the host cell interferon response including S, NSP3, NSP6, NSP14, nucleocapsid (N), and membrane (M) are mutated in the Omicron variant.15
Antiviral testing indicated a similar sensitivity of Omicron and Delta isolates to EIDD-1931, PF-07321332, remdesivir, favipravir, ribavirin, nafamostat, camostat, and aprotinin and, hence, to a range of drugs representing different mechanisms of action (Fig. 1e). This shows that the mutations in the Omicron variant do not cause substantial changes in the drug sensitivity profiles of the viruses.
For drugs targeting the RNA-dependent RNA polymerase and the replication of the viral genome, this may not come as too much of a surprise. Across the replicase-transcriptase complex (nsp7, nsp8, nsp9, nsp10, nsp12, nsp14), only two missense mutations were present in the investigated Omicron isolates, both of which are part of the set of mutations that define the Omicron variant. The RNA-dependent RNA polymerase nsp12 contains a single change, P323L, which was also present in the Alpha, Beta, and Gamma variants. P323L is far removed from the RNA binding site (Supplementary information, Fig. S1), and would not be expected to impact on RNA replication based on a structural analysis.
One further variant-defining mutation was present in the exonuclease (nsp14), resulting in an I42V change, which is present near the interface site with nsp10. This is a conservative substitution of two small hydrophobic side chains. Structural analysis shows the I42 side chain contacting V40 and N41, which directly contact nsp10 (Supplementary information, Fig. S2). However, this is a minor change that seems unlikely to have a significant impact on the interaction with nsp10 or on antiviral drug activity.
In contrast to our study, which did not detect differences between the sensitivity of Omicron and Delta isolates to TMPRSS2 inhibitors, one previous study found that an Omicron isolate was less sensitive to camostat than a Delta isolate.10 Given that this study compared two isolates in one cell line, it is possible that genomic differences between these isolates, which are independent of those defining the Delta and Omicron variants, were responsible for the observed differences. Notably, we detected in Caco-2 cells a 16.3-fold difference between the camostat IC50 for our Delta isolate (0.49 µM) and that for the Omicron 2 isolate (0.03 µM) (Fig. 1e). However, the Omicron 1 isolate displayed a camostat IC50 (0.40 µM) very close to that obtained for the Delta isolate, and we did not observe a similar difference in Calu-3 cells (Fig. 1e).
Moreover, Omicron mutations are only detected in close vicinity to one of the S cleavage sites. H655Y, N679K, and P681H are close to the 685 furin cleavage site. Among these mutations, only N679K is specific to Omicron (numbering of residues based on the reference virus protein sequence). There is no structure for this region of S, because it is a disordered, flexible region. N679K (and P681H) increases the positive charge, but there is no obvious indication that these mutations might affect S cleavage.
In conclusion, our comparison of Omicron and Delta isolates in different cellular models shows that Omicron viruses remain sensitive to a broad range of anti-SARS-CoV-2 drugs and drug candidates with a broad range of mechanisms of action. Moreover, Omicron viruses are less effective at antagonizing the host cell interferon response, which may explain why they cause less severe disease.14
Supplementary information
Acknowledgements
This work was supported by the Frankfurter Stiftung für krebskranke Kinder. The authors thank Lena Stegmann, Kerstin Euler and Sebastian Grothe for their technical assistance.
Author contributions
D.B., M.M., and J.C. conceived and designed the study. D.B., M.W., M.N.W., and J.C. performed the experiments. All authors analyzed the data. M.M. wrote the manuscript. D.B., M.N.W., M.M., and J.C. revised the manuscript. All authors have read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Contributor Information
Mark N. Wass, Email: M.N.Wass@kent.ac.uk
Martin Michaelis, Email: M.Michaelis@kent.ac.uk.
Jindrich Cinatl, Jr, Email: Cinatl@em.uni-frankfurt.de.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-022-00619-9.
References
- 1.Collie, S., Champion, J., Moultrie, H., Bekker, L. G. & Gray, G. N. Engl. J. Med.10.1056/NEJMc2119270 (2021).
- 2.Planas, P. et al. Nature.10.1038/d41586-021-03827-2 (2021).
- 3.Embi PJ, et al. MMWR Morb. Mortal. Wkly Rep. 2021;70:1553–1559. doi: 10.15585/mmwr.mm7044e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.García-Lledó A, et al. Rev. Esp. Quimioter. 2021 doi: 10.37201/req/158.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kabinger F, et al. Nat. Struct. Mol. Biol. 2021;28:740–746. doi: 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaur U, et al. Curr. Drug Targets. 2021;22:192–201. doi: 10.2174/1389450121666200924113243. [DOI] [PubMed] [Google Scholar]
- 7.Ruzhentsova TA, et al. Am. J. Transl. Res. 2021;13:12575–12587. [PMC free article] [PubMed] [Google Scholar]
- 8.Wilhelm, A. et al. medRxiv.10.1101/2021.12.07.21267432 (2021).
- 9.Bojkova D, et al. Cells. 2020;9:2377. doi: 10.3390/cells9112377. [DOI] [Google Scholar]
- 10.Zhao, H. et al. Emerg. Microbes Infect. 11, 277–283 (2022). [DOI] [PMC free article] [PubMed]
- 11.Abdelnabi, R. et al. bioRxiv10.1101/2021.12.24.474086 (2021).
- 12.García-Sastre A, et al. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 13.Yin X, et al. Cell Rep. 2021;34:108628. doi: 10.1016/j.celrep.2020.108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maslo, C. et al. JAMA. 10.1001/jama.2021.24868 (2021).
- 15.Xu, D., Biswal, M., Neal, A. & Hai, R. Curr. Res. Virol. Sci.10.1016/j.crviro.2021.100013 (2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.