Abstract
Omicron, the most recent SARS-CoV-2 variant of concern (VOC), harbours multiple mutations in the spike protein that were not observed in previous VOCs. Initial studies suggest Omicron to substantially reduce the neutralizing capability of antibodies induced from vaccines and previous infection. However, its effect on T cell responses remains to be determined. Here, we assess the effect of Omicron mutations on known T cell epitopes and report data suggesting T cell responses to remain broadly robust against this new variant.
Keywords: SARS-CoV-2, COVID-19, Omicron, variants, T cell epitopes, mutations, insertions, deletions, peptide-HLA binding
1. Main Text
Omicron (B.1.1.529), detected in Botswana, South Africa, and Hong Kong in November 2021, has emerged as a new SARS-CoV-2 variant of concern (VOC). Based on SARS-CoV-2 genome data available on GISAID (https://www.gisaid.org (accessed on 25 December 2021)), Omicron has been identified in at least 85 countries, raising concerns about its potentially high transmissibility. The most worrying aspect of Omicron is the abundance of mutations in the spike (S) protein, with some shared with previous VOCs and some new. Preliminary studies have reported a drastic reduction in the neutralization efficacy of infection- and vaccine-elicited antibodies and sera against Omicron [1,2,3,4,5], indicating a strong capability of Omicron to evade humoral immune responses. However, the extent to which Omicron is capable to evade cellular immune responses, the other arm of the adaptive immune system mediated by T cells, is not yet clear.
T cell responses are a key armament against viral infections which, in addition to assisting B cell activation for generating antibodies, help in providing protection from disease by eliminating virus-infected cells. SARS-CoV-2 T cell responses induced by either natural infection or vaccines have been linked to rapid viral clearance and reduced disease severity [6,7,8], even when the neutralizing antibody response is reduced [9] or absent [10]. Thus, if SARS-CoV-2 T cell responses hold up, they are likely to assist in limiting disease severity in infections caused by Omicron that seemingly escapes neutralizing antibodies [1,2,5]. This antibody escape is reported to be facilitated by Omicron-defining mutations present in the N-terminal domain of S, as well as in all four classes of neutralizing antibody epitopes located in the receptor binding domain [3,4]. If mutations in Omicron result in T cell escape, it could also limit the protection provided by T cells.
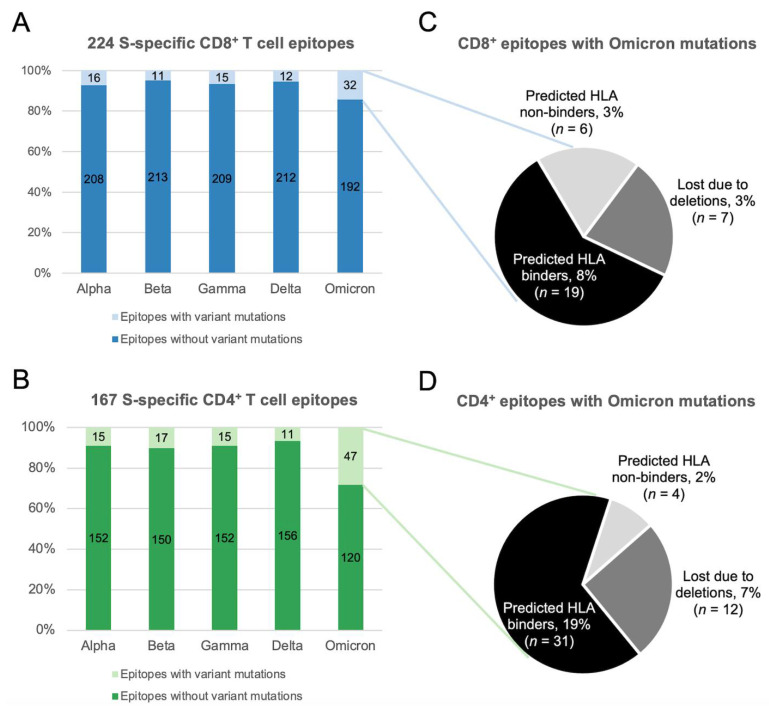
Here we provide a preliminary investigation into the robustness of T cell responses against Omicron by leveraging information of T cell epitopes known to be targeted in COVID-19 infected and/or vaccinated individuals. We first focused on epitopes derived from the S protein. This is of primary interest in the context of T cell responses elicited by COVID-19 vaccines, with many such vaccines employing S-specific antigens. T cell responses against S have also been shown to be immunodominant upon natural infection [11]. We considered all S-specific 224 CD8+ and 167 CD4+ SARS-CoV-2 T cell epitopes available at IEDB [12] (accessed on 9 December 2021) and screened them for Omicron-defining mutations (obtained from https://covariants.org (accessed on 9 December 2021)). This revealed that 14% of CD8+ and 28% of CD4+ T cell epitopes comprise at least one position harbouring an Omicron mutation (Figure 1A,B), indicating that a large majority of both CD8+ and CD4+ T cell epitopes (86% and 72%, respectively) remain unaffected by Omicron.
Figure 1.
Percentage of S-specific SARS-CoV-2 T cell epitopes with and without mutations present in the five VOCs: (A) CD8+ T cell epitopes. (B) CD4+ T cell epitopes. Only epitopes of canonical lengths (CD8+: 8–12 residues and CD4+:15 residues) were included in the analysis. VOC-defining mutations (that also include deletions) were obtained from https://covariants.org (accessed on 9 December 2021). Predicted effect of Omicron mutations on peptide-HLA binding of SARS-CoV-2 (C) CD8+ and (D) CD4+ T cell epitopes. NetMHCpan-4.1 and NetMHCpanII-4.0 were employed for predicting peptide-HLA binding using the default parameters [13].
The number of epitopes encompassing Omicron mutations (32 CD8+ and 47 CD4+ epitopes) are nonetheless notably higher than for other VOCs, especially in the case of CD4+ T cell epitopes (Figure 1A,B). To further assess the capacity of Omicron mutations to evade responses against these epitopes, we employed widely-used computational tools [13] to predict the impact of Omicron mutations on binding of these S-specific epitopes to their cognate human leukocyte antigen (HLA) alleles. Such HLA-epitope binding is a necessary requirement for T cell recognition. Inspection of these mutations revealed that the multiple deletions in the S protein of Omicron lead to loss of seven CD8+ and 12 CD4+ epitopes, while only six CD8+ and four CD4+ epitopes (listed in Tables S1 and S2, respectively) are predicted to become non-binders (Figure 1C,D). In the case of CD8+ epitopes, half (3/6) of the epitopes predicted to abrogate HLA binding are exclusive to Omicron, while the remaining were also observed in previous VOCs (Table S1). Since the majority of the CD8+ and CD4+ epitopes harbouring Omicron mutations are predicted to retain HLA binding (Figure 1C,D), this lowers the possibility of T cell escape. A similar trend regarding the preservation of peptide-HLA binding is also observed for other VOCs (Table S3).
Taken collectively, the presented data would suggest that SARS-CoV-2 T cell immunity acquired by S-focused COVID-19 vaccines or previous infection would remain broadly robust against Omicron, as was the case for other VOCs [14,15].
As an interesting by-product, our analysis revealed that a new peptide EPEDLPQGF, gained for the first time due to the unique three amino acid insertion ‘EPE’ following position 214 in the S protein of Omicron, is predicted to bind strongly with two common HLA alleles (HLA-B*35:01 and B*53:01) and weakly with four others (HLA-A*26:01, B*07:02, B*44:02, and B*51:01). This peptide may constitute a unique Omicron-specific T cell epitope, if confirmed to be targeted.
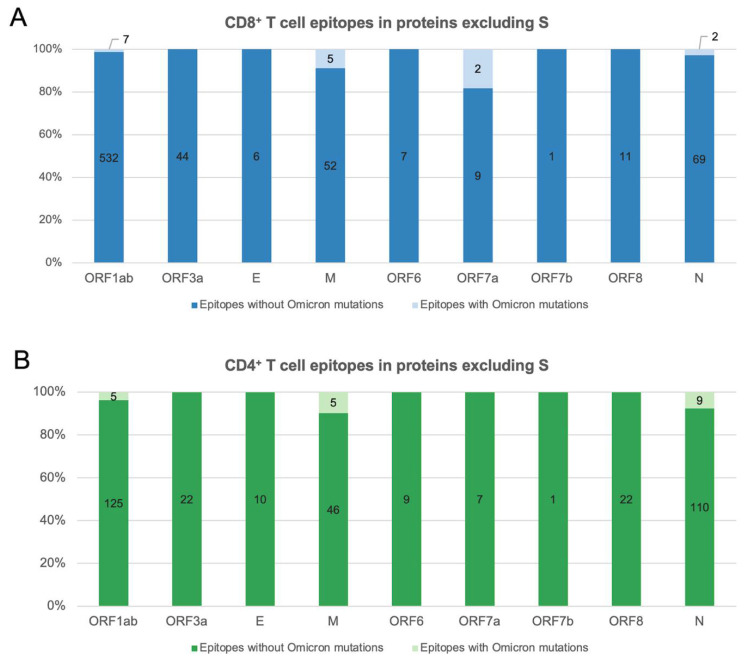
We next considered SARS-CoV-2 T cell epitopes derived from other proteins, besides S. This is of interest in the context of T cell responses induced by prior natural infection or inactivated COVID-19 vaccines, which have been found to target epitopes derived from multiple SARS-CoV-2 proteins [16,17]. Investigating all 745 CD8+ and 373 CD4+ SARS-CoV-2 T cell epitopes derived from proteins besides S (available at IEDB [12]; accessed on 9 December 2021) revealed that an overwhelming majority of them (98% and 95%, respectively) remained unaffected by mutations observed in Omicron (Figure 2). Among those affected, deletions accounted for the putative loss of five epitopes, with four (1 CD8+ and 3 CD4+) being from the immunodominant nucleocapsid (N) protein.
Figure 2.
Percentage of SARS-CoV-2 T cell epitopes derived from proteins besides S with and without Omicron-defining mutations: (A) CD8+ T cell epitopes. (B) CD4+ T cell epitopes. Only epitopes of canonical lengths (CD8+: 8–12 residues and CD4+:15 residues) were included in the analysis. Omicron-defining mutations (that also include deletions) were obtained from https://covariants.org (accessed on 9 December 2021).
It is the case that some T cell epitopes are preferentially targeted in the population. Escape from responses against the most commonly targeted epitopes should be carefully scrutinized, since it could potentially affect a large fraction of the population. To investigate this, we assessed whether Omicron mutations affect immunoprevalent CD8+ T cell epitopes that have been reported to be commonly targeted by convalescent individuals from different geographical regions [18]. As was the case for other VOCs, none of the Omicron mutations were present in any of the 20 immunoprevalent epitopes. Coupling these results with the fact that multiple T cell epitopes are targeted within an individual [11], suggests that the acquired T cell immunity across proteins would be expected to remain largely intact against Omicron.
There are multiple limitations of this study. First, the study is based on a set of experimentally-determined SARS-CoV-2 T cell epitopes that have been reported so far. While this set includes a large number of epitopes, it may not be exhaustive. Second, assessing the robustness of T cell responses against a SARS-CoV-2 variant using the fraction of epitopes encompassing mutations present in the variant is a first order analysis. While it can assist to provide preliminary estimates, further targeted experiments are required to confirm the robustness of T cell responses against Omicron, and also to test the capacity of specific epitope mutations to confer T cell escape. Some initial experimental studies along these lines have already started to appear while this paper was under review, which have arrived at consistent conclusions [19,20,21].
Overall, given that most of the experimental T cell epitopes known to be targeted in vaccinated and/or previously infected individuals (collectively, accounting for ~60% of the global population as of 25 December 2021 [22]) are unaffected by Omicron mutations, our preliminary analysis suggests that the effectiveness of pre-existing T cell immunity will remain intact. T cell responses alone, however, do not block infection and therefore do not prevent transmission. Thus, while the number of infections may rise considerably as a consequence of Omicron’s ability to evade antibodies [23,24], robust T cell immunity provides hope that, similar to other VOCs [25], the level of protection against severe disease would remain high. Emerging data is suggestive of a reduced risk of hospitalisation and death from Omicron [26,27]; however, the degree to which pre-existing T cell immunity is contributing to this still needs to be clearly established.
Acknowledgments
This analysis was made possible by the open sharing of immunological data of experimentally determined SARS-CoV-2 epitopes by research groups from around the world through the IEDB database. We gratefully acknowledge the contributions of all the researchers, scientists and technical staff involved. We would also like to thank the service of South African research teams involved in the early detection of Omicron, without which the world would not have known about it.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v14010079/s1, Table S1: List of CD8+ T cell epitopes with Omicron mutations that are predicted to become non-binders. NetMHCpan-4.1 was employed for predicting peptide-HLA binding using the default parameters; Table S2: List of CD4+ T cell epitopes with Omicron mutations that are predicted to become non-binders. NetMHCpanII-4.0 was employed for predicting peptide-HLA binding using the default parameters; Table S3: Summary of SARS-CoV-2 T cell epitopes affected by mutations in other VOCs. NetMHCpan-4.1 and NetMHCpanII-4.0 were employed for predicting peptide-HLA binding using the default parameters.
Author Contributions
Conceptualization: S.F.A., A.A.Q. and M.R.M.; methodology: S.F.A., A.A.Q. and M.R.M.; software: S.F.A.; validation: S.F.A. and A.A.Q.; formal analysis: S.F.A. and A.A.Q.; investigation: S.F.A., A.A.Q. and M.R.M.; resources: A.A.Q. and M.R.M.; data curation: S.F.A.; writing—original draft preparation: S.F.A., A.A.Q. and M.R.M.; writing—review and editing: S.F.A., A.A.Q. and M.R.M.; visualization: A.A.Q.; supervision: A.A.Q. and M.R.M.; project administration: A.A.Q. and M.R.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the General Research Fund of the Hong Kong Research Grants Council (RGC) [Grant no. 16213121].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors have filed for patent protection for various aspects of SARS-CoV-2 T cell epitope and vaccine design work.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wilhelm A., Widera M., Grikscheit K., Toptan T., Schenk B., Pallas C., Metzler M., Kohmer N., Hoehl S., Helfritz F.A., et al. Reduced neutralization of SARS-CoV-2 Omicron variant by vaccine sera and monoclonal antibodies. medRxiv. 2021 doi: 10.1101/2021.12.07.21267432. [DOI] [Google Scholar]
- 2.Roessler A., Riepler L., Bante D., von Laer D., Kimpel J. SARS-CoV-2 B.1.1.529 variant (Omicron) evades neutralization by sera from vaccinated and convalescent individuals. medRxiv. 2021 doi: 10.1101/2021.12.08.21267491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.-H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2021 doi: 10.1038/d41586-021-03827-2. [DOI] [PubMed] [Google Scholar]
- 4.Liu L., Iketani S., Guo Y., Chan J.F.-W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2021 doi: 10.1038/d41586-021-03826-3. [DOI] [PubMed] [Google Scholar]
- 5.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2021 doi: 10.1038/d41586-021-03796-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalimuddin S., Tham C.Y.L., Qui M., de Alwis R., Sim J.X.Y., Lim J.M.E., Tan H.-C., Syenina A., Zhang S.L., Le Bert N., et al. Early T cell and binding antibody responses are associated with COVID-19 RNA vaccine efficacy onset. Med. 2021;2:682–688.e4. doi: 10.1016/j.medj.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertoletti A., Tan A.T., Le Bert N. The T-cell response to SARS-CoV-2: Kinetic and quantitative aspects and the case for their protective role. Oxford Open Immunol. 2021;2:iqab006. doi: 10.1093/oxfimm/iqab006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geers D., Shamier M.C., Bogers S., den Hartog G., Gommers L., Nieuwkoop N.N., Schmitz K.S., Rijsbergen L.C., van Osch J.A.T., Dijkhuizen E., et al. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci. Immunol. 2021;6:eabj1750. doi: 10.1126/sciimmunol.abj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steiner S., Schwarz T., Corman V.M., Sotzny F., Bauer S., Drosten C., Volk H.-D., Scheibenbogen C., Hanitsch L.G. Reactive T cells in convalescent COVID-19 patients with negative SARS-CoV-2 antibody serology. Front. Immunol. 2021;12:687449. doi: 10.3389/fimmu.2021.687449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanojevic M., Geiger A., Ostermeier B., Sohai D., Lazarski C., Lang H., Jensen-Wachspress M., Webber K., Burbelo P., Cohen J., et al. Spike-directed vaccination elicits robust spike-specific T-cell response, including to mutant strains. Cytotherapy. 2021;24:10–15. doi: 10.1016/j.jcyt.2021.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarke A., Sidney J., Methot N., Yu E.D., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., Wang E., et al. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep. Med. 2021;2:100355. doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng Y., Li Y., Yang R., Tan W. SARS-CoV-2-specific T cell immunity to structural proteins in inactivated COVID-19 vaccine recipients. Cell. Mol. Immunol. 2021;18:2040–2041. doi: 10.1038/s41423-021-00730-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quadeer A.A., Ahmed S.F., McKay M.R. Landscape of epitopes targeted by T cells in 852 individuals recovered from COVID-19: Meta-analysis, immunoprevalence, and web platform. Cell Rep. Med. 2021;2:100312. doi: 10.1016/j.xcrm.2021.100312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keeton R., Tincho M.B., Ngomti A., Baguma R., Benede N., Suzuki A., Khan K., Cele S., Bernstein M., Karim F., et al. SARS-CoV-2 spike T cell responses induced upon vaccination or infection remain robust against Omicron. medRxiv. 2021 doi: 10.1101/2021.12.26.21268380. [DOI] [Google Scholar]
- 20.Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., et al. SARS-CoV-2 vaccination induces immunological memory able to cross-recognize variants from Alpha to Omicron. bioRxiv. 2021 doi: 10.1101/2021.12.28.474333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou R., To K.K.-W., Peng Q., Chan J.M.-C., Huang H., Yang D., Lam B.H.-S., Chuang V.W.-M., Cai J.-P., Liu N., et al. Vaccine-breakthrough infection by the SARS-CoV-2 Omicron variant elicits broadly cross-reactive immune responses. bioRxiv. 2021 doi: 10.1101/2021.12.27.474218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie H., Mathieu E., Rodés-Guirao L., Appel C., Giattino C., Ortiz-Ospina E., Hasell J., Macdonald B., Beltekian D., Roser M. Coronavirus Pandemic (COVID-19) 2020. [(accessed on 25 December 2021)]. Our World in Data. Available online: https://ourworldindata.org/coronavirus.
- 23.Kuhlmann C., Mayer C.K., Claassen M., Maponga T.G., Sutherland A.D., Suliman T., Shaw M., Preiser W. Breakthrough infections with SARS-CoV-2 Omicron variant despite booster dose of mRNA vaccine. SSRN Electron. J. 2021 doi: 10.2139/ssrn.3981711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pulliam J.R.C., van Schalkwyk C., Govender N., von Gottberg A., Cohen C., Groome M.J., Dushoff J., Mlisana K., Moultrie H. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv. 2021 doi: 10.1101/2021.11.11.21266068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.UK Health Security Agency COVID-19 Vaccine Surveillance Report—Week 49. [(accessed on 11 December 2021)]; Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1039677/Vaccine_surveillance_report_-_week_49.pdf.
- 26.Wolter N., Jassat W., Walaza S., Welch R., Moultrie H., Groome M., Amoako D.G., Everatt J., Bhiman J.N., Scheepers C., et al. Early assessment of the clinical severity of the SARS-CoV-2 Omicron variant in South Africa. medRxiv. 2021 doi: 10.1101/2021.12.21.21268116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Public Health England SARS-CoV-2 Variants of Concern and Variants under Investigation in England: Technical Briefing 33. [(accessed on 25 December 2021)]; Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1043680/technical-briefing-33.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.