Abstract
A quantitative real-time PCR assay was developed to measure human cytomegalovirus (HCMV) DNA load in peripheral blood leukocytes (PBLs). The HCMV DNA load in PBLs was normalized by means of the quantification of a cellular gene (albumin). The results of the real-time PCR assay correlated with those of the HCMV pp65-antigenemia assay (P < 0.0001).
Human cytomegalovirus (HCMV) infection is characterized by a primary infection leading to a lifelong persistence of the viral genome. Periodically, the virus reactivates from latency and recovers its ability to multiply. HCMV is a major cause of morbidity and mortality in bone marrow or solid-organ transplant recipients and in AIDS patients. Early diagnosis of HCMV infection in high-risk patients is essential in order to start preemptive treatments (3, 13, 15). The detection of the pp65 antigen in leukocytes is a sensitive method widely used for the early diagnosis of HCMV infection, but it is labor-intensive, requires immediate processing, and relies on a subjective interpretation of the slides (1, 7). Qualitative PCR detection of HCMV DNA in leukocytes or plasma is considered the most sensitive method, but it lacks specificity for the diagnosis of HCMV disease (2, 6, 16). Quantification of HCMV DNA has been proposed to be more specifically associated with the disease (1, 18). Real-time PCR based on the TaqMan technology (4, 5) provides an accurate means to quantify viral DNA with the major advantage of avoiding post-PCR handling that can be the source of DNA carryover. Recent studies report the utility of real-time PCR for the quantification of HCMV DNA (9, 11, 12, 14, 17). In these studies, the primers used for PCR were located in the major immediate-early gene (12, 14, 17), the US17 gene (9), or the glycoprotein B gene (17). As suggested by Yun et al., the sensitivity of quantitative PCR may be dependent on the viral target gene (17); however, the most appropriate region for amplification has not been established, as the sequence diversity of clinical isolates remains to be characterized. In the present study, a real-time PCR assay was developed to quantify HCMV DNA in peripheral blood leukocytes (PBLs) using a target sequence located in the UL83 gene which codes for the lower matrix protein detected in the pp65 antigenemia test. The HCMV DNA load in PBLs was normalized by means of the quantification of a cellular gene (albumin) and the results were compared to those of the pp65 antigenemia assay.
DNA extractions were performed in all experiments using the QIAamp blood kit (QIAGEN S. A., Courtabœuf, France) according to the manufacturer's recommendations, except that DNA was eluted in 200 μl of distilled water. To amplify HCMV DNA, primers and probe were defined in the UL83 region as follows: pp549s (direct primer), 5′-GTCAGCGTTCGTGTTTCCCA-3′; pp812as (reverse primer), 5′-GGGACACAACACCGTAAAGC-3′; and pp770s (fluorogenic probe), 5′FAM-CCCGCAACCCGCAACCCTTCATG-3′TAMRA. No cross-reactivity was observed when the specificity of the primers and probe was tested for other human herpes viruses (herpes simplex virus types 1 and 2, varicella-zoster virus, human herpes virus 6, and Epstein-Barr virus) and human fibroblast DNA (data not shown). The real-time PCRs were carried out using the TaqMan PCR core reagent kit (PE Applied Biosystems, Courtabœuf, France). Ten microliters of DNA was added to a PCR mixture containing 1× PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 6-carboxy-x-rhodamine), 5 mM MgCl2, 0.2 mM dATP, dCTP, and dGTP, 0.4 mM dUTP, 0.2 mM concentrations of each primer, 0.1 mM fluorogenic probe, 1.25 U of AmpliTaq Gold, and 0.5 U of AmpErase. The PCR conditions consisted of 1 cycle of 2 min at 50°C and 1 cycle of 10 min at 95°C followed by 45 cycles of 15 s at 95°C and 1 min at 65°C. The reaction, data acquisition, and analysis were performed using the ABI PRISM 7700 sequence detection system (PE Applied Biosystems). The number of target copies in the reaction was deduced from the threshold cycle (CT) values corresponding to the fractional cycle number at which the released fluorescence exceeded 20 times the standard deviation of the mean baseline emission. A plasmid containing one copy of the UL83 target sequence (pKS-pp65K7) was used as a standard for HCMV DNA quantification. To construct the pKS-pp65K7 standard, DNA from the HCMV AD169 strain was amplified with primers pp549s and pp812as and cloned into the pBluescript II KS vector (Stratagene, Amsterdam, The Netherlands). The concentration of purified pKS-pp65K7 plasmid DNA was determined with a spectrophotometer at 260 nm and the corresponding copy number was calculated.
To evaluate HCMV DNA quantification on clinical samples, 46 PBL samples that were positive in the pp65 antigenemia assay were collected from solid-organ transplant recipients or AIDS patients. The pp65 antigenemia assay was carried out on fresh PBL samples using the CINAkit (Argene Biosoft, Varilhes, France) following the manufacturer's instructions. DNA was extracted from 2 × 106 PBLs isolated from EDTA-treated blood samples. For each sample, 2 aliquots of DNA, corresponding to 105 cells, were used to quantify HCMV DNA using the standard curve constructed with pKS-pp65K7. Two other aliquots were used to quantify the human albumin gene using primers and probe described by Laurendeau et al. (8) with some modifications: the forward primer was 5′-GCTGTCATCTCTTGTGGGCTGT-3′, the reverse primer was 5′-AAACTCATGGGAGCTGCTGGTT-3′, and the probe was 5′FAM-CCTGTCATGCCCACACAAATCTCTCC-3′ TAMRA. The conditions used to amplify the albumin gene were identical to those used for HCMV DNA, allowing both reactions to be carried out simultaneously in the same run. The standard curve for quantification of the albumin gene was constructed with 10-fold serial dilutions ranging from 2 × 105 to 20 copies of human genomic DNA (Roche Molecular Biochemicals, Meylan, France) tested in triplicate. The normalized value of the HCMV DNA load was expressed as the number of HCMV copies per 2 × 105 PBLs calculated as the ratio (HCMV DNA mean copy number/[albumin DNA mean copy number/2]) × 2 × 105. This allowed sample-to-sample variations, which can lead to a misinterpretation of the HCMV DNA copy number, to be normalized in terms of cell count, yield of DNA extraction, and PCR efficiency.
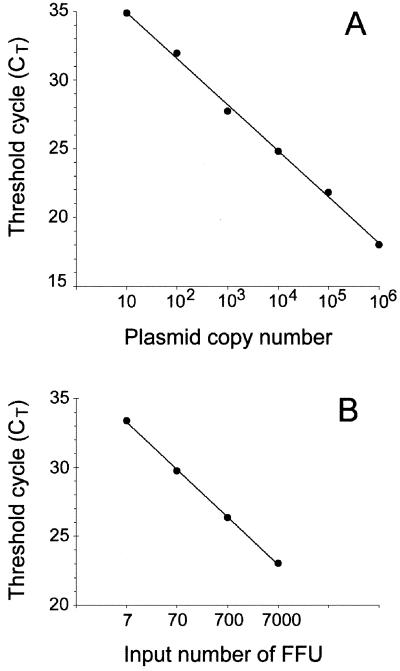
The sensitivity and the intra-assay reproducibility of the real-time PCR assay were evaluated by using plasmid pKS-pp65K7 (containing one copy of the UL83 target sequence) as a standard. One copy of the plasmid could be detected in one in five assays, while 10 plasmid DNA copies were detected with 100% sensitivity. Thus, the lowest limit for quantification was considered to be 10 copies of the target sequence. The intra-assay variation of the CT values was evaluated using five replicates of plasmid dilutions containing 106, 105, 104, 103, 102, and 10 copies. The coefficients of variation (CV) were 1.4, 1.9, 0.6, 1.3, 1.1, and 2.5%, respectively. Plasmid pKS-pp65K7 was used to construct the standard curve for HCMV DNA quantification. Tenfold serial dilutions ranging from 106 to 10 copies of plasmid were tested in triplicate and the CT values were plotted against the copy number. The linear correlation between the CT and the logarithm of the DNA copy number was repeatedly greater than 0.995 (Fig. 1A).
FIG. 1.
Standard curves for HCMV DNA quantification. (A) Plasmid pKS-pp65K7 containing one copy of the UL83 target sequence was used to construct the standard curve for HCMV DNA quantification. Tenfold serial dilutions ranging from 106 to 10 copies of plasmid were tested in triplicate and the mean CT values were plotted against the copy number. The correlation coefficient was 0.997 and the slope was −3.36. The amplification efficiency, calculated as [10(−1/slope) −1] × 100, was 98%. (B) Serial dilutions of HCMV strain AD169 (obtained from a cell culture supernatant with a titer of 7 × 106 FFU/ml in a shell vial assay) were tested by real-time PCR and the mean CT values were plotted against the number of FFU input in the reaction mixture. The correlation coefficient was 0.999. The slope was −3.46, corresponding to 95% amplification efficiency.
To determine whether the results of HCMV DNA quantification by real-time PCR were consistent with those obtained by virus titration in a shell vial assay, serial dilutions of a viral stock of AD169 with a titer of 7 × 106 infectious foci (FFU)/ml were quantified with the TaqMan assay (Fig. 1B). One FFU corresponded to about 20 HCMV DNA copies. Moreover, as shown in Fig. 1, the DNA amplification efficiencies obtained with AD169 and pKS-pp65K7 were similar (95 and 98%, respectively). This result gave support to the use of plasmid pKS-pp65K7 as a relevant standard for HCMV DNA quantification. The interassay reproducibility of the TaqMan assay was evaluated using 3 serial dilutions of the AD169 stock (103, 102, and 10 FFU). For each dilution, four independent DNA extractions were performed and HCMV DNA was quantified in four independent TaqMan runs. The CV values of the HCMV DNA copy numbers were 17, 12, and 21%, respectively, for the 3 dilutions tested.
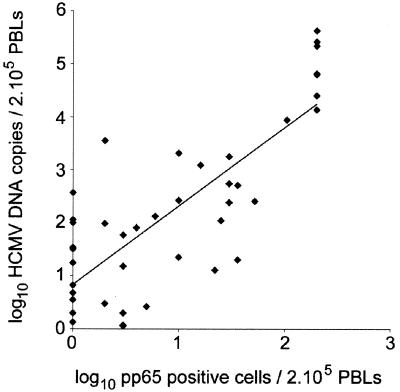
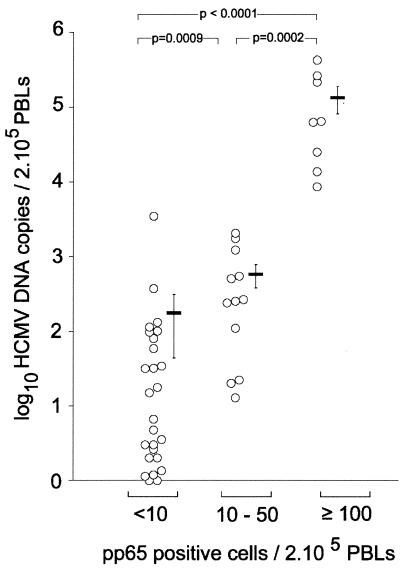
To evaluate the HCMV DNA TaqMan assay on clinical samples, we tested 46 PBL samples that were positive in the pp65 antigenemia assay. HCMV DNA quantification in PBL samples was performed in parallel with the quantification of the albumin gene in order to determine the amount of cellular DNA input in each reaction. The mean value of the albumin gene copy number for the 46 PBL samples tested was 6.2 × 104 copies (ranging from 7.2 × 103 to 2 × 105 copies). This corresponded to a mean DNA extraction yield of 31%, compared to the theoretical value of 2 × 105 albumin copies expected for the amount of cellular DNA input in the reaction mixture (equivalent to 105 cells). Forty-four out of 46 PBL samples were positive for HCMV DNA with the TaqMan assay. For the 44 HCMV DNA-positive samples, the normalized values of the HCMV DNA load ranged from 1 to 4.26 × 105 copies per 2 × 105 cells. As shown in Fig. 2, a statistically significant correlation was observed between the HCMV DNA copy number and the pp65-positive cell number (Spearman rank test; r = 0.657, P < 0.0001). As the level of HCMV antigenemia is usually considered to correlate with HCMV disease (1), samples from patients were classified into three groups according to the results of the pp65 antigenemia assay. Group 1 (n = 26) corresponded to samples with low HCMV antigenemia values (<10 positive cells/2 × 105 PBLs), group 2 (n = 12) corresponded to moderate values (10 to 50 positive cells/2 × 105 PBLs), and group 3 (n = 8) corresponded to high values (≥100 positive cells/2 × 105 PBLs). As shown in Fig. 3, the high pp65-positive samples from group 3 had significantly higher HCMV DNA copy numbers than the moderate pp65-positive samples from group 2 (Mann-Whitney U test; P = 0.0002) and the low pp65-positive samples from group 1 (P < 0.0001). The HCMV DNA load values were also significantly different between groups 1 and 2 (P = 0.0009). The mean HCMV DNA copy number per 2 × 105 PBLs was 1.8 × 102 (range, 0 to 3.5 × 103) for group 1 samples, 5.8 × 102 (range, 20 to 2 × 103) for group 2, and 1.3 × 105 (range, 8.7 × 103 to 4.6 × 106) for group 3.
FIG. 2.
Correlation between the HCMV DNA copy number and the pp65-positive cell number in PBL. HCMV DNA load was quantified by real-time PCR in 46 PBL samples that were positive in the pp65 antigenemia assay. The copy numbers of HCMV DNA were normalized by means of the quantification of the albumin gene and plotted against the pp65-positive cell numbers obtained by the antigenemia assay (pp65 values greater than 200 positive cells per 2 × 105 PBLs were set to 200). The two samples negative for HCMV DNA were not figured on this logarithmic graph. The correlation between the HCMV DNA copy number and the pp65-positive cell number was examined by the nonparametric Spearman rank test and was found to be significant, with a correlation coefficient of r = 0.657 (P < 0.0001).
FIG. 3.
Comparison of HCMV DNA load with pp65 antigenemia levels in PBL samples. The HCMV DNA load was quantified by real-time PCR in 46 PBL samples and compared to the pp65 antigenemia level. High pp65-positive samples (≥100 positive cells/2 × 105 PBLs; n = 8) had significantly higher HCMV DNA copy numbers than the moderate pp65-positive samples (10 to 50 positive cells; n = 12) and the low pp65-positive samples (<10 positive cells; n = 26). p indicates the significance of the nonparametric Mann-Whitney U test. Bars show the mean HCMV DNA copy number and standard error of the mean for each group.
The development of real-time PCR technology is a promising improvement for the quantification of HCMV DNA in clinical samples and will be useful for the follow-up of patients with a high risk of developing HCMV disease. The real-time PCR assay developed in this study allowed the quantification of HCMV DNA over a large dynamic range (10 to 106 copies), as previously reported for other real-time PCR assays (9, 11, 12, 14, 17). The assay was reproducible as indicated by the intra-assay CV values obtained with the plasmid standard (0.6 to 2.5%) and by the interassay CV values obtained with the HCMV AD169 strain (12 to 21%). The fact that the evaluation of interassay reproducibility took into account the DNA extraction step could explain why the CV values were higher than those (<10%) reported by Nitsche et al. with a quantitative HCMV DNA TaqMan-based assay (12). Because only one PBL sample was available for each patient, replicates of DNA extraction and quantification could not be performed in order to determine the CV values for clinical specimens. For such specimens the DNA extraction step could have a major effect on the assay's reproducibility. Indeed, the quantification of the albumin gene in PBL samples showed a wide range of sample-to-sample variation in the DNA extraction yield, emphasizing the need to normalize the HCMV load values for cell samples as proposed by others (10). This was achieved by referring the HCMV copy number not to the initial cell count but to the actual amount of cellular DNA estimated by quantification of the albumin gene.
The HCMV DNA copy number in PBL samples determined with real-time PCR correlated to the pp65-positive cell number, in agreement with the results of other TaqMan-based assays (9, 14). However, some discrepancies were observed. First, two samples with low antigenemia pp65 values (2 and 4 positive cells/2 × 105 PBLs) were negative for HCMV DNA. For both samples, the albumin gene quantification indicated a poor DNA extraction yield (≤10%). Such cases of low DNA yield should lead to cautious interpretation of negative HCMV DNA results. It was not possible to retest these two samples due to insufficient material. Secondly, although 21 out of 26 samples from group 1 (pp65 antigenemia was <10 positive cells/2 × 105 PBLs) had an HCMV copy number per 2 × 105 PBLs of less than 100, 4 samples from this group ranged from 101 to 372 copies, and 1 sample had 3.5 × 103 copies. Others have reported a similar difference between antigenemia and quantitative PCR results (10, 16), but the clinical significance of a high HCMV DNA load associated with a low antigenemia remains to be established.
As for other quantitative PCR techniques, real-time PCR applied to the quantification of HCMV DNA needs to be standardized in order to ameliorate the reproducibility and to ensure the most accurate follow-up of the patients. The region of the viral genome which is the most appropriate target for HCMV quantitative PCR remains to be determined, and a comparative evaluation of TaqMan assays based on different HCMV target genes will be useful for this purpose. Moreover, the standardization of the technique requires the amplification of a control (such as a cellular gene) to monitor the efficiency of the reaction. As the TaqMan technology allows multiplex PCR to be performed by using two fluorogenic probes labeled with different dyes, the dual quantification of HCMV DNA and the albumin gene in a single-tube format will be the next step in the development of our assay.
Acknowledgments
This work was supported in part by an ANRS (AC 11, groupe CMV) grant and by the MESRT grant Programme de Recherches Fondamentales en Microbiologie, Maladies infectieuses et Parasitologie.
We thank Ann Beaumont for careful reading and editing of the manuscript.
REFERENCES
- 1.Boeckh M, Boivin G. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin Microbiol Rev. 1998;11:533–554. doi: 10.1128/cmr.11.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drouet E, Colimon R, Michelson S, Fourcade N, Niveleau A, Ducerf C, Boibieux A, Chevallier M, Denoyel G. Monitoring levels of human cytomegalovirus DNA in blood after liver transplantation. J Clin Microbiol. 1995;33:389–394. doi: 10.1128/jcm.33.2.389-394.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Einsele H, Ehninger G, Hebart H, Wittkowski K M, Schuler U, Jahn G, Mackes P, Herter M, Klingebiel T, Löffler J, Wagner S, Müller C A. Polymerase chain reaction monitoring reduces the incidence of cytomegalovirus disease and the duration and side effects of antiviral therapy after bone marrow transplantation. Blood. 1995;86:2815–2820. [PubMed] [Google Scholar]
- 4.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 5.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanj S S, Sharara A I, Clavien P A, Hamilton J D. Cytomegalovirus infection following liver transplantation: review of the literature. Clin Infect Dis. 1996;22:537–549. doi: 10.1093/clinids/22.3.537. [DOI] [PubMed] [Google Scholar]
- 7.Landry M L, Ferguson D, Cohen S, Huber K, Wetherill P. Effect of delayed specimen processing on cytomegalovirus antigenemia test results. J Clin Microbiol. 1995;33:257–259. doi: 10.1128/jcm.33.1.257-259.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laurendeau I, Bahuau M, Vodovar N, Larramendy C, Olivi M, Bieche I, Vidaud M, Vidaud D. TaqMan PCR-based gene dosage assay for predictive testing in individuals from a cancer family with INK4 locus haploinsufficiency. Clin Chem. 1999;45:982–986. [PubMed] [Google Scholar]
- 9.Machida U, Kami M, Fukui T, Kazuyama Y, Kinoshita M, Tanaka Y, Kanda Y, Ogawa S, Honda H, Chiba S, Mitani K, Muto Y, Osumi K, Kimura S, Hirai H. Real-time automated PCR for early diagnosis and monitoring of cytomegalovirus infection after bone marrow transplantation. J Clin Microbiol. 2000;38:2536–2542. doi: 10.1128/jcm.38.7.2536-2542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nazzari C, Gaeta A, Lazzarini M, Castelli T D, Mancini C. Multiplex polymerase chain reaction for the evaluation of cytomegalovirus DNA load in organ transplant recipients. J Med Virol. 2000;61:251–258. [PubMed] [Google Scholar]
- 11.Nitsche A, Steuer N, Schmidt C A, Landt O, Ellerbrok H, Pauli G, Siegert W. Detection of human cytomegalovirus DNA by real-time quantitative PCR. J Clin Microbiol. 2000;38:2734–2737. doi: 10.1128/jcm.38.7.2734-2737.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nitsche A, Steuer N, Schmidt C A, Landt O, Siegert W. Different real-time PCR formats compared for the quantitative detection of human cytomegalovirus DNA. Clin Chem. 1999;45:1932–1937. [PubMed] [Google Scholar]
- 13.Roberts T C, Brennan D C, Buller R S, Gaudreault-Keener M, Schnitzler M A, Sternhell K E, Garlock K A, Singer G G, Storch G A. Quantitative polymerase chain reaction to predict occurrence of symptomatic cytomegalovirus infection and assess response to ganciclovir therapy in renal transplant recipients. J Infect Dis. 1998;178:626–635. doi: 10.1086/515383. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka N, Kimura H, Iida K, Saito Y, Tsuge I, Yoshimi A, Matsuyama T, Morishima T. Quantitative analysis of cytomegalovirus load using a real-time PCR assay. J Med Virol. 2000;60:455–462. doi: 10.1002/(sici)1096-9071(200004)60:4<455::aid-jmv14>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 15.van der Meer J T, Drew W L, Bowden R A, Galasso G J, Griffiths P D, Jabs D A, Katlama C, Spector S A, Whitley R J. Summary of the international consensus symposium on advances in the diagnosis, treatment and prophylaxis and cytomegalovirus infection. Antiviral Res. 1996;32:119–140. doi: 10.1016/s0166-3542(96)01006-6. [DOI] [PubMed] [Google Scholar]
- 16.Weber B, Nestler U, Ernst W, Rabenau H, Braner J, Birkenbach A, Scheuermann E H, Schoeppe W, Doerr H W. Low correlation of human cytomegalovirus DNA amplification by polymerase chain reaction with cytomegalovirus disease in organ transplant recipients. J Med Virol. 1994;43:187–193. doi: 10.1002/jmv.1890430217. [DOI] [PubMed] [Google Scholar]
- 17.Yun Z, Lewensohn-Fuchs I, Ljungman P, Vahlne A. Real-time monitoring of cytomegalovirus infections after stem cell transplantation using the TaqMan polymerase chain reaction assays. Transplantation. 2000;69:1733–1736. doi: 10.1097/00007890-200004270-00037. [DOI] [PubMed] [Google Scholar]
- 18.Zipeto D, Baldanti F, Zella D, Furione M, Cavicchini A, Milanesi G, Gerna G. Quantification of human cytomegalovirus DNA in peripheral blood polymorphonuclear leukocytes of immunocompromised patients by the polymerase chain reaction. J Virol Methods. 1993;44:45–55. doi: 10.1016/0166-0934(93)90006-d. [DOI] [PubMed] [Google Scholar]