Abstract
The family Paramyxoviridae consists of a group of large, enveloped, negative-sense, single-stranded RNA viruses and contains many important human and animal pathogens. Molecular and biochemical characterization over the past decade has revealed an extraordinary breadth of biological diversity among this family of viruses. Like all enveloped viruses, paramyxoviruses must fuse their membrane with that of a receptive host cell as a prerequisite for viral entry and infection. Unlike most other enveloped viruses, the vast majority of paramyxoviruses contain two distinct membrane-anchored glycoproteins to mediate the attachment, membrane fusion and particle entry stages of host cell infection. The attachment glycoprotein is required for virion attachment and the fusion glycoprotein is directly involved in facilitating the merger of the viral and host cell membranes. Here we detail important functional, biochemical and structural features of the attachment and fusion glycoproteins from a variety of family members. Specifically, the three different classes of attachment glycoproteins are discussed, including receptor binding preference, their overall structure and fusion promotion activities. Recently solved atomic structures of certain attachment glycoproteins are summarized, and how they relate to both receptor binding and fusion mechanisms are described. For the fusion glycoprotein, specific structural domains and their proposed role in mediating membrane merger are illustrated, highlighting the important features of protease cleavage and associated tropism and virulence. The crystal structure solutions of both an uncleaved and a cleavage-activated metastable F are also described with emphasis on how small conformational changes can provide the necessary energy to mediate membrane fusion. Finally, the different proposed fusion models are reviewed, featuring recent experimental findings that speculate how the attachment and fusion glycoproteins work in concert to mediate virus entry.
Keywords: paramyxovirus, entry, attachment glycoprotein, fusion glycoprotein, membrane fusion, receptor, ephrin-B2, CD46, SLAM, nectin, neuraminidase, sialic acid, heptad
Introduction to Paramyxoviruses
The family Paramyxoviridae is an interesting group of large, enveloped, negative-sense, single-stranded RNA viruses that includes many important human and animal viruses such as measles virus (MeV), mumps virus, respiratory syncytial virus (RSV) and the human parainfluenza viruses (hPIV) in addition to animal viral agents such as and Sendai virus (SeV), parainfluenza virus type 5 (PIV5) and canine distemper virus (CDV).1 Several paramyxoviruses, including Newcastle disease virus (NDV) and rinderpest, pose major economic threats due to their possible impact on poultry and livestock industries.2, 3 In addition, more recently discovered members of the paramyxovirus family, namely Hendra virus (HeV) and Nipah virus (NiV), have been shown to possess a broad host range with the ability to infect and cause disease in a number of animal species as well as humans.4, 5
Paramyxoviruses were originally classified as “myxoviruses” in the family Orthomyxoviridae due to the shared properties of hemagglutination and neuraminidase activity of the envelope glycoproteins of some members. However, paramyxoviruses differ from orthomyxoviruses in a number of critical aspects including genome organization, protein expression and replication strategies and more closely resemble other families in the order Mononegavirales, including Rhabdoviridae, Filoviridae and Bornaviridae.6 The family Paramyxoviridae is divided into two subfamilies, Paramyxovirinae and Pneumovirinae. The classification is based on the organization of the genome, the molecular properties and biological activities of the encoded proteins, and morphological criteria.1 Existing and proposed genera with examples of family members and unclassified viruses are summarized in Table 1. In 2002 two new genera were added to the Paramyxovirinae subfamily such that there are now 5 genera including Respirovirus, Rubulavirus, Morbillivirus, Henipavirus and Avulavirus.7 NDV and other avian paramyxoviruses were removed from the genus Rubulavirus and reclassified in a new taxon, Avulavirus, due to differences in genome organization8, RNA editing profiles and phylogenetic comparisons.9, 10 The formation of the genus Henipavirus, which presently includes HeV and NiV, was justified in large part because of genome size, unique genome termini, limited homology with other family members and the different biological activities of various encoded proteins.5, 11 The Pneumovirinae includes the genera Pneumovirus and Metapneumovirus, and members include RSV and human metapneumovirus (hMPV), respectively.
Table 1.
Existing and proposed genera in the family Paramyxoviridae
| Subfamily | Genus | Species |
|---|---|---|
| Paramyxovirinae | ||
| Rubulavirus | Parainfluenza virus type 5* | |
| Mumps virus | ||
| Human parinfluenza virus types 2, 4a and 4b | ||
| Menangle Virus | ||
| Respirovirus | Sendai virus | |
| Human parinfluenza virus types 1 and 3 | ||
| Bovine parinfluenza virus type 3 | ||
| Avulavirus | Newcastle disease virus | |
| Avian paramyxovirus types 2–9 | ||
| Morbillivirus | Measles virus | |
| Canine distemper virus | ||
| Rinderpest virus | ||
| Henipavirus | Hendra virus | |
| Nipah virus | ||
| Cedar virus | ||
| Pneumovirinae | ||
| Pneumovirus | Human respiratory syncytial virus | |
| Bovine respiratory syncytial virus | ||
| Pneumovirus of mice | ||
| Metapneumovirus | Human metapneumovirus | |
| Avian pneumovirus** | ||
| Proposed | ||
| TPMV-like viruses*** | Tupaia virus | |
| Jeilongvirus*** | J-virus | |
| Beilong virus | ||
| Ferlavirus*** | Fer-de-lance virus | |
| Unclassified | ||
| Nariva virus | ||
| Mossman virus | ||
| Salem virus | ||
formerly known as simian virus 5 (SV5)
formerly known as turkey rhinotracheitis virus
proposed genus within the subfamily Paramyxovirinae
Over the past few decades, a number of new paramyxoviruses have been identified, and in spite of the increase in the number of genera in the Paramyxovirinae, several members remain unclassified. J-virus (J-V) and the newly discovered Beilong virus (BeV) have been shown to be closely related to one another, yet neither can be placed in any of the existing genera and a new genus, Jeilongvirus, has been proposed.12 Fer-de-Lance virus (FDLV), a new reptilian paramyxovirus, also has a unique genome, and a further new genus, Ferlavirus, has also been proposed.13 Most recently, a new paramyxovirus, Cedar virus (CedPV), was isolated from urine samples of flying foxes in Australia and was shown to be genetically and antigenically related to HeV and NiV.14 CedPV also appeared to utilize the same entry receptor, ephrin-B2, that both HeV and NiV employ, and CedPV is the first new proposed member of the Henipavirus genus. Other paramyxoviruses, such as Salem virus, Mossman virus and Nariva virus, have also been described but cannot be placed in any existing genera nor have new genera been proposed; therefore, these viruses remain unclassified.
Until recently, the genomes of paramyxoviruses as a group were generally considered to cluster in the range 15.1–15.9 kb. With the discovery and molecular characterization of HeV, NiV, BeV and CedPV, and the genome sequencing of the previously described Tupaia virus and J-V, the genome size range has significantly increased. BeV, with a 19,212-nt genome, now represents the largest genome among all known non-segmented negative-strand RNA viruses, longer than the 19,151-nt genome of Marburg virus.12 Indeed, aided by discovery and/or sequencing, the genetic diversity within the family Paramyxoviridae has rapidly increased within the past decade, and research efforts focused on their molecular and biochemical characterization have revealed an extraordinary breadth of biological diversity among this virus family.
Paramyxovirus entry
Fusion of enveloped viruses with the plasma membrane of a receptive host cell is a prerequisite for viral entry and infection. As a group, most paramyxoviruses contain two membrane-anchored glycoproteins that are required for the entry process, and these glycoproteins appear as spikes projecting from the envelope membrane of the viral particle when viewed under the electron microscope. Several examples are provided in Figure 1. One glycoprotein is required for virion attachment to the host cell, and depending on the particular virus, has been designated as either the hemagglutinin–neuraminidase glycoprotein (HN), the hemagglutinin glycoprotein (H) or glycoprotein (G), which has neither hemagglutinating nor neuraminidase activities (reviewed in15). Paramyxovirus attachment glycoproteins are type II membrane proteins where the molecule’s amino (N)-terminus is oriented towards the cytoplasm and the protein’s carboxy (C)-terminus is extracellular. The other glycoprotein is the fusion protein (F), which is directly involved in facilitating the fusion of the viral and host cell membranes (reviewed in16). The F glycoprotein is a type I integral membrane glycoprotein with an extracellular N-terminus that shares several conserved features with other viral fusion glycoproteins and will be discussed in greater detail in the sections to follow. A cartoon diagram of an attachment and fusion glycoproteins and their important functional domains is depicted in Figure 2.
Figure 1. Negatively stained paramyxovirus virions.
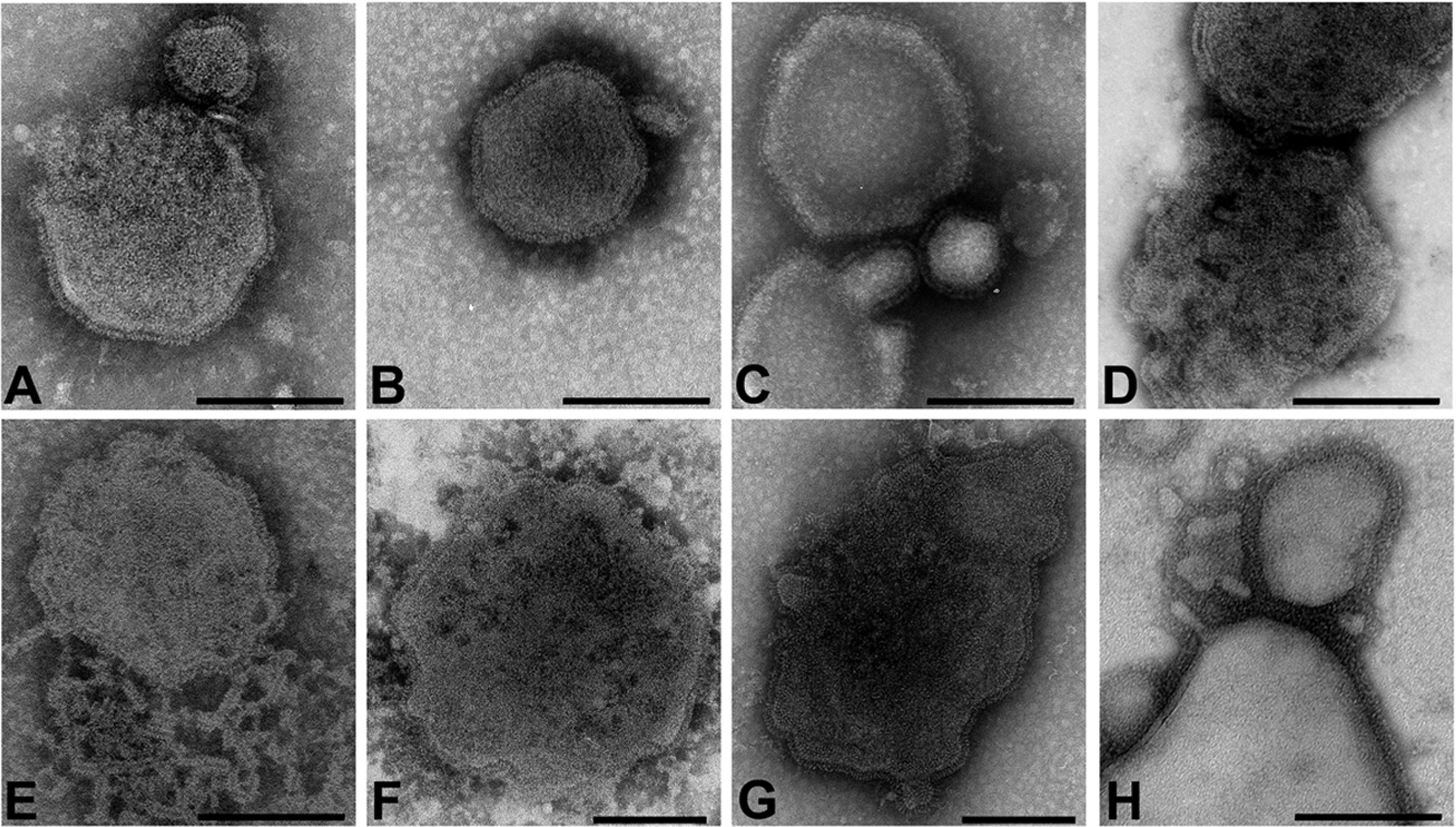
A. Newcastle disease virus (avulavirus). B. Human parainfluenza virus type 3 (respirovirus). C. Hendra virus (henipavirus). D. Canine distemper virus (morbillivirus). E. Menangle virus (proposed rubulavirus). F. Respiratory syncytial virus (pneumovirus). G. J-virus (proposed jeilongvirus). H. Mossman virus (unclassified). All micrographs were adjusted to the same magnification with exception of panels F and G. For all panels: bar, 200nm. Images courtesy of the AAHL Biosecurity Microscopy Facility, Australian Animal Health Laboratory (AAHL) Livestock Industries CSIRO, Australia.
Figure 2. The attachment and fusion envelope glycoproteins.
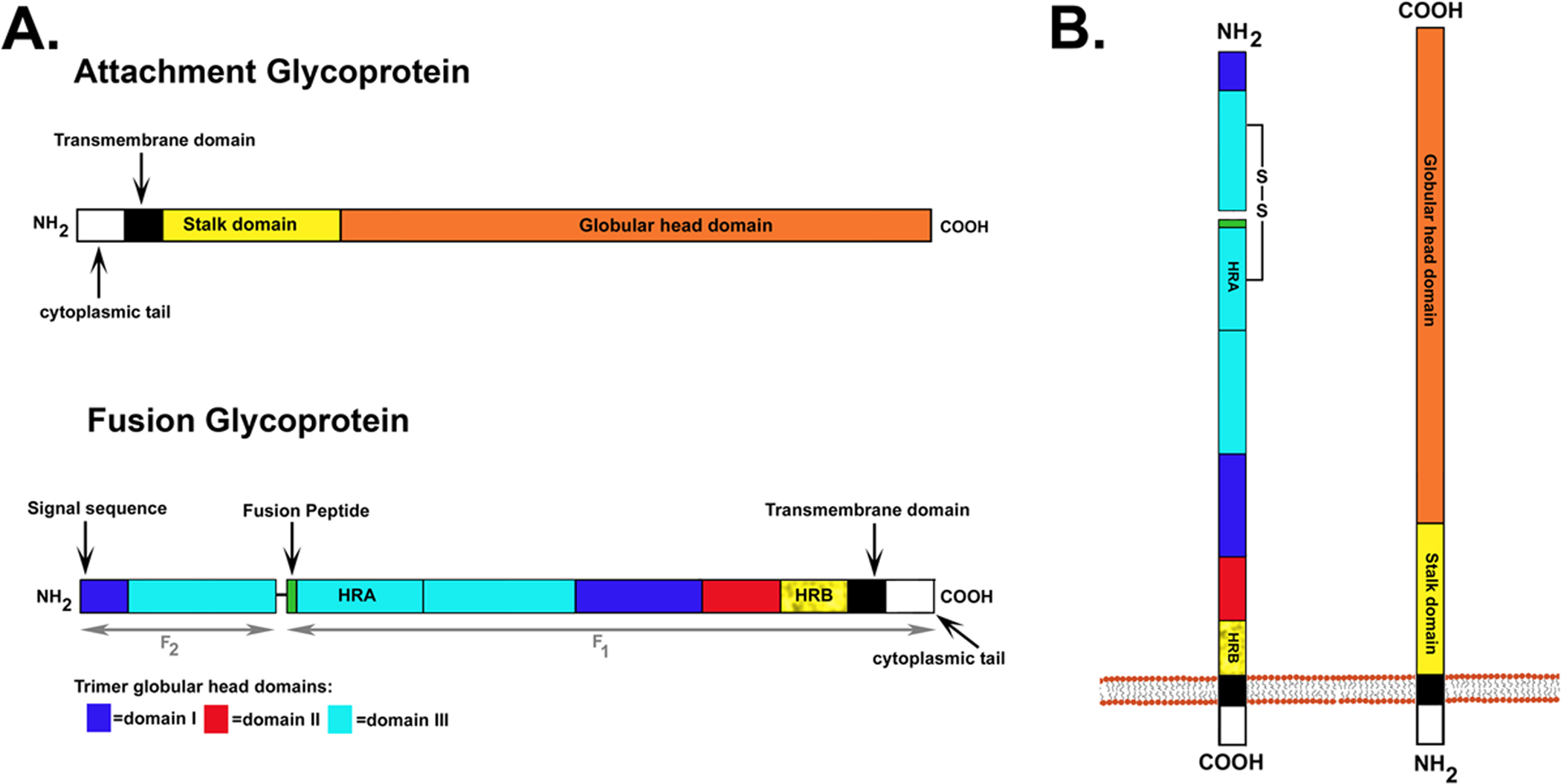
A. Important functional domains. For the attachment glycoprotein the transmembrane domain, cytoplasmic tail, proposed stalk domain and globular head are indicated. For the fusion glycoprotein the F1 and F2 subunits are depicted. F1 contains the signal sequence, transmembrane domain, cytoplasmic tail, fusion peptide, heptad repeat A (HRA) and heptad repeat B (HRB). The important domains that constitute the globular head of the F trimer, DI, DII and DIII, are represented by different shading. B. Orientation of the attachment and fusion glycoprotein in the virion membrane. F is a typical type I membrane glycoprotein with one membrane spanning domain and an extracellular N-terminus. The disulfide bond that links the F1 and F2 subunits is also shown. The attachment glycoproteins are type II membrane proteins where the molecule’s amino (N)-terminus is oriented towards the cytoplasm and the protein’s carboxy (C)-terminus is extracellular.
The attachment and fusion glycoproteins work in concert to mediate membrane fusion and particle entry into susceptible host cells. Following virus attachment to a permissive and receptor-bearing host cell, fusion of the virion and plasma membranes occurs, resulting in delivery of the viral nucleocapsid into the cytoplasm. In a related process, cells expressing attachment and fusion glycoproteins on their surface can fuse with receptor-bearing cells under physiological or cell culture conditions, leading to the formation of multinucleated giant cells (syncytia) - a hallmark of many paramyxovirus infections.
Attachment glycoproteins and their receptors
General tertiary structure of attachment glycoproteins
The paramyxovirus attachment glycoproteins consist of a stem (or stalk) and a globular head structure with the latter domain containing both the receptor binding and, if present, enzymatic activities of the molecule.17–19 The general model for the monomeric structure of HN and H is a globular head comprised of 6-folded antiparallel β-sheets (β1–6) of four strands each (S1–4), with each sheet arranged regularly around and radiating out from the central axis of the molecule.20 Although the henipavirus G glycoprotein has only limited sequence homology to HN and H glycoproteins it possesses a high structural similarity.21 The attachment glycoprotein β-propeller shape is maintained by disulfide bonds, which are highly conserved among these three types of attachment glycoproteins (reviewed in22) The 6-bladed propeller model is similar to both the earlier predicted structures of HN23 and henipavirus G21 and the now known HN structures of NDV, hPIV3 and PIV524–27, the structure of MeV H28, 29 and the structures of henipavirus G.30, 31
Earlier electron micrographs of SeV HN exhibit a box-shaped arrangement consistent with four discrete subunits32, similar to influenza virus neuraminidase (NA). The oligomeric forms of HN from different paramyxoviruses have been extensively characterized and depending on the virus consist of pairs of disulfide-linked homodimers that can come together to create noncovalently linked tetramers32–36 or also disulfide-linked tetramers (dimer of dimers).37 The disulfide-linked dimers of PIV5 HN are joined through cysteine residue 111 in the stalk domain; the exact residues responsible for tetramer association have yet to be identified although the presence of the stalk domain is critical.26 PIV526 and NDV HN38 also exist as tetramers in solution; however, significant differences exist in dimer packing.
The oligomeric organization of the MeV H glycoprotein has also been characterized by structural and functional studies identifying disulfide-linked dimers via cysteine residues at positions 139 and 15439 with a higher order tetramer configuration (dimer of dimers).40 Likewise, the biochemical characterization of native HeV G, as well as a transmembrane domain/cytoplasmic tail-deleted, soluble version of G has revealed disulfide-linked dimers and both noncovalent and disulfide-linked tetramers of G similar to the oligomeric forms of HN and H glycoproteins.41 The residues involved in the oligomerization of G are located in the stalk domain with NiV G cysteine residues 158, 162 and 146 having critical roles. Residues 158 and 162 are involved in the covalent dimer formation of NiV G and were found to be absolutely required for its fusion promotion activity, perhaps by maintaining G in a pre-receptor bound conformation, while cysteine residue 146 appeared to stabilize higher-order oligomers (tetramers)42 (reviewed in43). Additional details on the structures and functions of the types of attachment glycoproteins will be discussed in the sections below.
Hemagglutinin–neuraminidase glycoprotein (HN)
The majority of well-described paramyxoviruses, particularly those in the Respirovirus, Avulavirus and Rubulavirus genera, possess a multifunctional HN glycoprotein that attaches the virion to sialic acid receptors on host cells. In addition to binding, the HN glycoprotein cleaves sialic acid moieties from both host cell molecules and virus particles. Analogous to the role played by influenza NA, the neuraminidase activity of HN prevents re-attachment of the virion to producer cells as well as self-aggregation of progeny virions as they bud from an infected cell into the extracellular environment (reviewed in44). Because the optimal pH for paramyxovirus neuraminidases is between 4.8 and 5.5 it has been suggested that the removal of sialic acid from carbohydrate chains occurs in the acidic trans Golgi network.
Recently, the structure of the NDV HN ectodomain was reported that showed dimers of the NA domain dimers flanking the N-terminal stalk domain (Figure 3). The NDV stalk formed a parallel tetrameric coiled-coil bundle (4HB) that also permitted the classification of existing mutagenesis data, revealing broad insight into the functional roles of the HN stalk and its tetrameric configuration. Many mutations that affected only F-glycoprotein activation mapped to the 4HB surface (Figure 3A). Two of four NA domains revealed an interaction with the 4HB stalk, and residues at this interface in both the stalk and NA domain have been implicated in HN function.27 The two independent structures of PIV5 and NDV HN glycoproteins26, 27 have now been referred to as the ‘heads-up’ versus ‘heads-down’ conformations, respectively. Alternative models describing how tetrameric HN structures promote fusion have been proposed by both Zaitsev et al38 and Yuan et al26, 27 and will be discussed in a later section.
Figure 3.
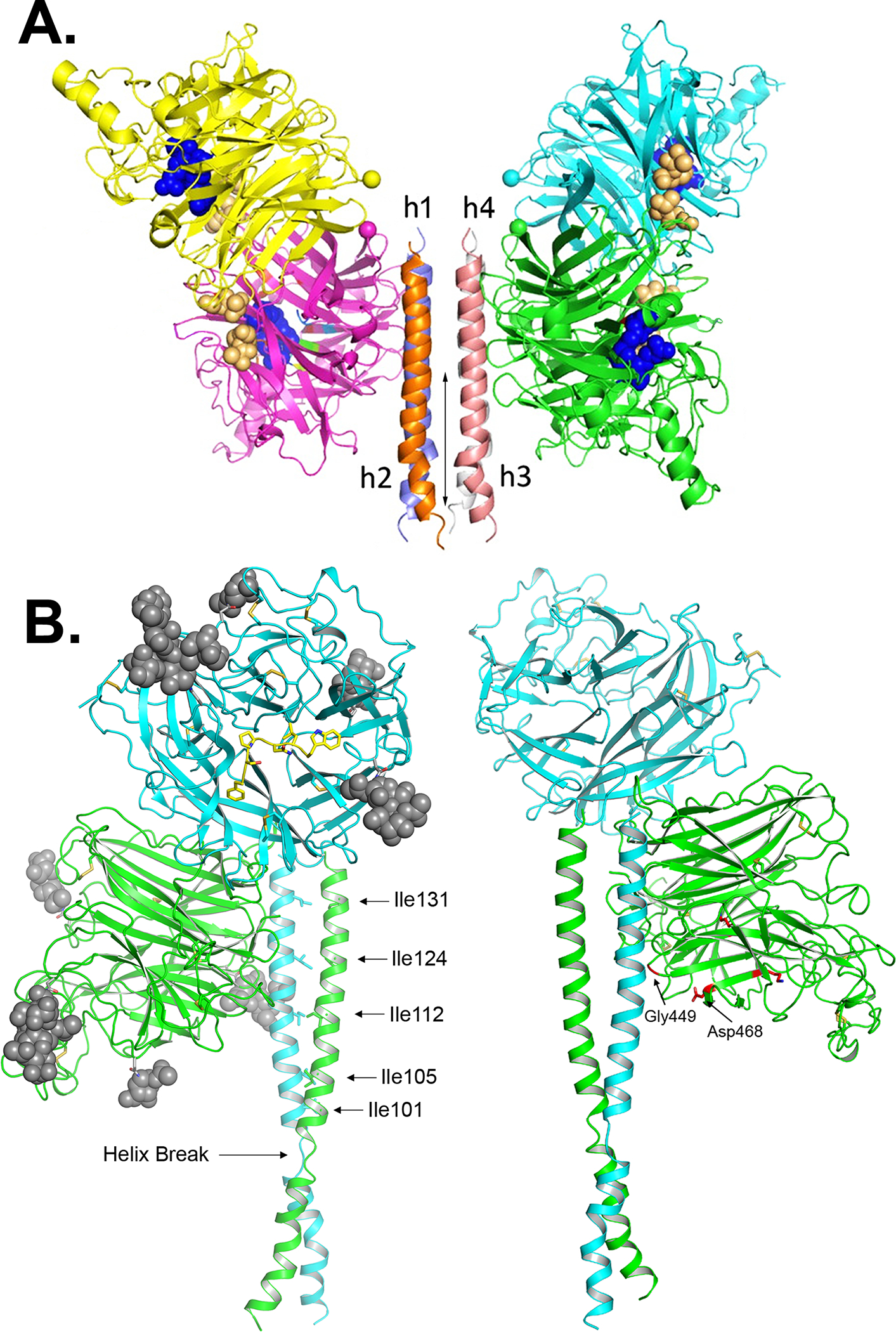
(A) Structure of the NDV HN, Australia–Victoria (AV) strain ectodomain. Two dimers of the NDV HN NA domains flank the 4HB (residues 83–114) in the stalk. The four NA active sites are shown as blue spheres. The secondary sialic acid binding sites located at the NA domain dimer interface are shown as orange spheres. Mutations of NDV HN stalk residues R83, A89, L90, L94 and L97 are known to impair F activation specifically and are implicated in forming direct contacts with the F glycoprotein. These mutations reside along the stalk region marked by the arrow in the HN tetramer structure. (B) Model of HeV G. Left: The G ectodomain is shown in the dimer conformation with the two globular head domains derived from the crystal structure, colored in green and blue, with predicted N-linked glycosylation sites shown as gray spheres. The G head domain folds as a six-bladed β-propeller with disulfide bonds illustrated as yellow sticks. The residues of the ephrin-B2 G-H loop are also shown in yellow occupying the RBS. Stalk residues 77–136 are modeled for each monomer, and the position of the HeV G head dimer and stalks are oriented based on the alignment with the NDV structure.27 Ile residues in the HeV G stalk domain that modulate G fusion promotion activity are indicated.197 Right: Model of the HeV G dimer with globular heads and stalk domains as on right and rotated with residues G449 and D468 highlighted in red showing their proximity to the stalk domain. Mutation of these residues decreases HeV fusion, suggesting they may be involved in interactions between the globular heads and stalk domains that are essential for the fusion process. Figures have been modified from original work with permission from R.A. Lamb and Proceedings of the National Academy of Sciences27 and the creative commons public license.43
HN mediates both binding to sialic acid as a receptor for viral attachment and cleavage of sialic acid via its neuraminidase enzymatic activity. An interesting question remains as to whether this is achieved through one or two separate sialic acid binding sites. Studies have demonstrated two sialic acid binding sites in the NDV HN dimer, one in the globular head domain and the second site at the dimer interface of the molecule.38 Both of these proposed sialic acid binding sites in NDV HN are shown in Figure 3A. Binding of ligand to the first active site induces a conformational change that then allows formation of the second site to which sialic acid also binds. Structural studies of hPIV3 and PIV5 HN reveal dimers very similar to those of NDV HN and suggest a similar oligomeric arrangement; however, unlike NDV the hPIV3 and PIV5 HN glycoproteins contain only one sialic acid binding site, an enzymatically active site in the globular head domain, while differences in sequence and conformation render the second sialic acid binding site implausible.25, 26 Like hPIV3 HN, PIV5 HN does not undergo conformational change upon ligand binding, highlighting further distinctions from the NDV HN structure.25, 26
Hemagglutinin glycoprotein (H)
The morbilliviruses, including MeV and CDV, have a H attachment glycoprotein, which possesses only hemagglutinating activity and does not bind to sialic acid receptors. However, H glycoproteins have significant sequence identity to HN, and similar tertiary structure models analogous to those for the HN glycoproteins of respiroviruses, avulaviruses and rubalviruses have been developed.20 MeV was also the first paramyxovirus shown to employ a cell-surface protein as a receptor45, 46, and co-immunoprecipitation experiments demonstrated an interaction between the H glycoprotein of laboratory strains of MeV and CD46.47 In addition, MeV field isolates as well as vaccine strains can utilize signaling lymphocyte activation molecule (SLAM, CD150) as a receptor48 - a receptor also employed by CDV and wild-type rinderpest virus.49, 50 Further, CD46 and SLAM expression are down-regulated in MeV-infected cells in a H glycoprotein-dependent manner.51, 52 It was hypothesized that reducing the levels of surface-expressed receptors may circumvent the need for intrinsic neuraminidase activity by MeV, and possibly morbilliviruses in general, which for HN bearing paramyxoviruses cleaves surface-associated sialic acid and prevent virus aggregation during virus budding as discussed earlier. Moreover, the use, and down-modulation, of SLAM by morbilliviruses may play a role in the general immunosuppression seen in infected hosts. Interestingly, the MeV attachment sites for both receptors appeared to overlap on the globular head domain of H53, although viruses that preferentially use either CD46 or SLAM could be selected.54 Yet a third MeV receptor, speculated to exist on epithelial cells55, 56, was recently discovered. Nectin-4, an adherens junction protein of the immunoglobulin superfamily, is the most recently identified MeV receptor.57, 58 Highly expressed in a variety of tissues including epithelial cells of the human airway, Nectin-4 supports MeV entry and is also down-regulated in infected cells. Of the three MeV receptors, Nectin-4 has the strongest affinity for MeV H58, and MeV targets Nectin-4 to emerge in tracheobronchial airways. Following the initial infection and spread of MeV within the host, facilitated by macrophages and dendritic cells, infection of epithelial cells occurs later in the disease and is important for aerosol transmission of the virus (reviewed in59).
In just the past several years a significant amount of new information on the interactions between paramyxovirus H glycoproteins and their binding partners has been obtained and the structures of MeV H alone and in complex with SLAM, CD46 and Nectin-4 have been determined.28, 29, 60–62 The most recent structure of MeV H in complex with Nectin-4 has allowed for the first time a detailed comparison of the binding of H with three different receptors, revealing overlapping but distinct binding sites for Nectin-4, CD46 and SLAM.60 Of particular interest, this latest study revealed a hydrophobic pocket centered in the MeV H β4-β5 groove involved in the binding of all three receptors, suggesting a new potential target for antivirals.
Glycoprotein (G)
There are two distinct and structurally unrelated G lineages within the family Paramyxoviridae - those described for the genus Henipavirus in the subfamily Paramyxovirinae and those described for the genera Pneumovirus and Metapneumovirus in the subfamily Pneumovirinae. Both lineages of G attachment glycoproteins lack hemagglutinin and neuraminidase activities. Only a single amino acid residue of the seven known to be critical for neuraminidase activity is conserved in HeV and NiV G, compared to at least six residues in HN or four residues in H. Additional studies demonstrated that neuraminidase treatment of Vero cells (a cell line used to propagate HeV and NiV stocks) did not inhibit HeV or NiV infection, while such treatment can abrogate their susceptibility to NDV and influenza virus A, which depend on sialic acid structures as receptors.
It was also observed that cell lines from the same species, most notably human cell lines, could be clearly positive or negative for HeV or NiV-mediated membrane fusion and that protease treatment could prevent fusion of an otherwise permissive target cell.63–65 Perhaps not unexpected in light of the observed characteristics possessed by HeV and NiV G, it was later discovered that the henipaviruses utilize a host cellular protein as a viral receptor, ephrin-B2 ligand.66, 67 Ephrin-B2 ligand is a widely-expressed and highly conserved cell surface protein across many different species, and its identification as the henipavirus receptor has aided in understanding the broad host range of HeV and NiV as well as their neurotropism. In addition, ephrin-B3 ligand was identified as a second entry receptor for NiV68 and HeV.69 Like MeV H attachment to its protein receptors, the attachment sites for both ephrin receptors in NiV G also revealed overlapping binding sites as binding to ephrin-B2 ligand can inhibit binding to ephrin-B3 ligand.68, 70 More recently, the crystal structures of both NiV and HeV G globular head domains have been determined both alone and in complex with the ephrin-B2 and -B3 receptors, revealing the exact G-receptor interactions and identical receptor binding sites.30, 31, 71–73 Also, similar to NDV HN, the henipavirus G stalk domain contains alpha helices with a predicted break from amino acids 95–98, and the stalks with the globular heads of HeV G have been modeled with the resulting structure resembling the heads-down configuration of NDV HN (Figure 3B).
An obvious difference between the G glycoproteins of henipaviruses and those of the subfamily Pneumovirinae, is size, just over 600 amino acids for HeV and NiV whereas the G glycoproteins of pneumoviruses and metapneumoviruses vary between 230 and 300 amino acids.74 There are also significant sequence differences between the G glycoprotein of pneumoviruses and metapneumonviruses, although both have a similar hydrophobicity profile, a high serine and threonine content (24–34%) and an ectodomain that also contains two mucin-like domains.75, 76 In addition, the G glycoprotein of RSV, a member of the pneumovirus genus, is heavily glycosylated with both N- and O-linked sugars that contribute greater than 50% of the weight of the mature glycoprotein - an unusual feature among viral membrane glycoproteins. The high serine and threonine content facilitates the addition of O-linked carbohydrates, and it is likely that these sugar moieties contribute to the binding of carbohydrate receptors on the cell surface.75 hMPV G has a serine/threonine content of 34%, slightly higher than RSV76, and it is predicted to have a similar profile of O-linked glycosylation and carbohydrate binding as RSV G. In addition to the membrane-bound RSV G, a soluble and secreted G glycoprotein molecule is also observed in infected cell cultures.77 It has been suggested that this soluble version of RSV G may act as an immunological decoy during infection.
Another notable characteristic of RSV, including bovine (RSV) which is being explored as a human vaccine platform, is that G-deleted viruses are still capable of replication in cell culture or in animals.77–81 These observations suggested that RSV G may function as an accessory protein that increases the efficiency of virus entry.82 Indeed, the RSV F glycoprotein has been shown to bind heparin-containing structures and the GTP-binding RhoA protein as well as interact with and subsequently signal through CD14 and toll-like receptor 4.75, 83 Clearly, RSV possesses an alternate mechanism for virus entry in which attachment and fusion can be directly mediated by F and does not strictly require G. More recently, endocytosis has been implicated as a possible route of RSV entry, either via a caveolin84 or clathrin85 mediated route(s).
Like RSV, recombinant human metapneumovirus (hMPV), which lacks G, has also been shown to be replication competent in vitro and in vivo,86, 87 and it was recently demonstrated that the first cell surface binding target for hMPV is also heparan sulfate.88 Although no definitive receptor for the hMPV F protein has been identified, integrin αvβ1 has been suggested as a host cell factor promoting entry.89 Altogether, these recent findings suggest that hMPV F glycoprotein has effectively replaced a requirement for an attachment protein with a low pH-induced triggering process,90, 91 a unique feature amongst the paramyxoviruses.
Finally, the newly characterized G genes and their encoded G glycoproteins of J-V and BeV warrant discussion, as they may represent a third lineage of G glycoproteins in the family Paramyxoviridae. The J-V and BeV G genes are 4401 and 4527-nt in length, respectively, more than double the size of most family members.12, 92 The significant increase in size is due to the presence of additional open-reading-frames (ORFs) within the G gene. To begin with, for both J-V and BeV, the 5’ half of the G mRNA contains an open reading frame (ORF-G) encoding putative proteins 709 or 734 amino acids in length, respectively. Such proteins are 105 and 130 amino acids larger, respectively, than the largest paramyxovirus attachment glycoprotein; HeV G. The putative G glycoproteins of J-V and BeV share many conserved structural elements with other paramyxovirus HN, H and G glycoproteins, and also lack any detectable hemagglutinating or neuraminidase activities.12, 92 In addition, and perhaps of greater interest, both the J-V and BeV G gene contain an additional ORF (ORF-X) within the 3’ half of their G mRNAs, encoding putative proteins, 709 and 299 amino acids in length, respectively. These additional coding regions are separated from the ORF-G by only one stop codon. BeV has another ORF downstream from, but overlapping with ORF-X which encodes another putative protein 394 amino acids in length. Probes specific for ORF-G and ORF-X in J-V both identified mRNA transcripts corresponding in size to a monocistronic G gene mRNA. However, no evidence was found for the existence of an mRNA molecule specific to ORF-X alone, nor was the protein encoded by ORF-X or a fusion protein of G-X detected93. Although the biological significance of these additional ORFs remains unknown, clearly, the G genes of J-V and BeV are unlike any other within the family Paramyxoviridae.
Fusion (F) glycoprotein
Nearly all paramyxoviruses that have been examined to date require both attachment and F glycoproteins for efficient membrane fusion to occur, although some exceptions have been noted. PIV5 F can mediate moderate levels of membrane fusion in the absence of HN94, and as described above, RSV and hMPV derivatives that lack the G gene remain fusogenic and infectious. For all paramyxoviruses the F glycoprotein is directly involved in facilitating the fusion between the virus and host cell membranes. F glycoproteins are homotrimeric oligomers with considerable hydrophobicity36, 95–100 and share several conserved features with other viral fusion proteins, including the envelope glycoprotein of retroviruses, such as gp120/gp41 of HIV-1, and the hemagglutinin (HA) of influenza virus.95, 96, 101, 102 These types of viral fusion glycoproteins have since been categorized as class I viral fusion proteins103 where from the protein’s N- to C-terminus there is a fusion peptide located just C-terminal to the cleavage site of a precursor form of the protein followed by two heptad repeat domains, a transmembrane domain (TMD) and a cytoplasmic tail. These features of the paramyxovirus F glycoprotein will be discussed below. The three prominent classes of viral fusion proteins have been recently reviewed in detail.104
Biologically active F consists of two disulfide linked subunits, F1 and F2, (Figure 2) that are generated by the proteolytic cleavage of a precursor known as F0.105, 106 Likewise, HIV-1 envelope and influenza HA are cleaved by a host cell protease, leading to the generation of a membrane distal subunit analogous to F2 and a membrane-anchored subunit analogous to F1. Cleavage of F0 is thought to play an important role that influences both infectivity and pathogenicity of paramyxoviruses. The various paramyxovirus F glycoproteins fall into two groups - those with multiple basic residues and those with a single basic residue at the cleavage site. Proteolytic activation involves two separate cleavage events mediated by host proteases. The first initially cleaves the carboxyl side of the basic residue, and the next step, mediated by a carboxypeptidase, removes the basic residue. Cleavage of F glycoproteins with multiple basic residues occurs within the cell as they traffic through the trans Golgi network of the secretion pathway and is thought to be mediated by furin, a host endoprotease1 (reviewed in106). Two different mechanisms exist for cleavage of paramyxoviruses with single basic residues at the cleavage site. The first, which has been more widely studied, has demonstrated extracellular cleavage by an exogenous protease. As an example, SeV replicates poorly in tissue culture; however, after addition of exogenous protease, productive infection significantly increases.105 When grown in eggs, SeV F is cleaved by an extracellular amniotic endoprotease.107 For NDV, virulence of the virus is directly correlated to the nature of the cleavage site, where strains with a single basic residue in the cleavage site are avirulent and are restricted to the respiratory tract, while those with multiple basic residues readily disseminate through the host.108 In general, most viruses that contain a single basic residue at the cleavage site have a more restrictive tropism and do not disseminate. Newly recognized exceptions to these general rules are HeV and NiV. Both henipaviruses have a single basic residue at the cleavage site, however, both viruses readily disseminate within the host upon infection, productively targeting a variety of organ systems. Of particular interest is the discovery that the F0 glycoprotein of HeV and NiV is cleaved in a novel process that occurs after transportation of the uncleaved molecule to the surface of infected cells. Following re-internalization of F0, cleavage occurs within the endosomal compartment and is mediated by the endoprotease cathepsin L.109, 110
Nevertheless, in all cases following F0 cleavage, the membrane-anchored subunit F1 remains linked to F2 by a disulfide bond and contains a new N-terminus, referred to as the fusion peptide (Figure 2), which is hydrophobic and conserved in its location across virus families.111, 112 The fusion peptides of paramyxoviruses, as well as other viruses including HIV-1 and influenza, are thought to intercalate into target membranes and initiate the fusion process.113 Although hydrophobic in nature, the absolute conservation of many of the residues within the fusion peptide of paramyxoviruses suggests an as yet unidentified additional function independent of actual membrane insertion.1
The paramyxovirus F glycoproteins, like those of retroviruses, contains 2 α-helical domains referred to as heptad repeats that are involved in the formation of a trimer-of-hairpins structure or 6-helix bundle (6-HB) during or immediately following fusion of virus and cell membranes.111, 114–117 For paramyxoviruses, one heptad is located adjacent to the fusion peptide in F1 and is referred to as the N-terminal heptad or heptad repeat A (HRA). The second heptad is proximal to the transmembrane domain and is referred to as the C-terminal heptad or heptad repeat B (HRB) (Figure 2). As first noted with the gp41 subunit of HIV-1 envelope glycoprotein,118, 119 peptides corresponding to either of these domains from several paramyxovirus F glycoproteins can inhibit the activity of the fusion glycoprotein when present during the fusion process.63, 64, 120–126 It has been generally accepted that significant conformational change occurs during activation of paramyxovirus F fusogenic activity. Differential antibody binding reactivity of precursor and proteolytically processed forms of PIV5 F127, in conjunction with the structure of the ‘postfusion’ 6-HB of PIV5 F111, strongly supported the conformational change model102). The postfusion structure of the hPIV3 F core is likely conserved across other paramyxoviruses and has been observed in the F core structures of RSV128, MeV129, mumps virus130 and the henipaviruses.131 A cartoon illustrating how the heptad repeats mediate 6-HB formation and membrane fusion is shown in Figure 4.
Figure 4. Model showing how paramyxovirus F heptad repeats mediate membrane merger.
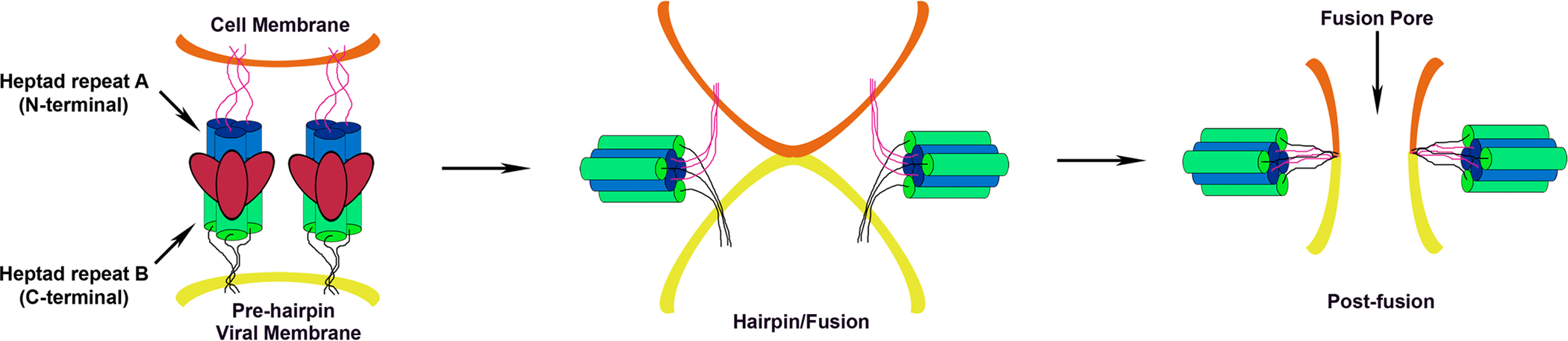
For simplicity, the attachment glycoprotein and its proposed role in triggering or promoting F activity is not shown. Upon activation, significant conformational changes in F lead to HRA forming a three-stranded α-helical coiled coil and the translocation of the fusion peptide and its intercalation into the target cell membrane. This fusion intermediate conformation of F is referred to as the prehairpin structure. Further conformational changes lead to the packing of α-helical HRB domains into the grooves of the HRA coiled coil in an antiparallel orientation giving rise to six-helix bundle formation. As the six-helix bundle forms, the two membranes are drawn closer together, and the energy released as F transitions to its most stable conformation is thought to drive membrane merger.
More recently, structural studies on the F glycoprotein of NDV reveal a trimer structure that differs from the classic influenza HA structure, principally in the manner in which HRA is oriented. In the NDV F trimer, the HRA segment is located with its C terminus directed towards the head of the molecule; this is the opposite orientation to the observed central coiled coil formed by HRA in the influenza HA trimer.98, 132 Understanding the cascade of conformational changes leading up to 6-HB formation102, 121, 127 was hindered by the spontaneous re-arrangement of secreted F glycoproteins to a conformation resembling a postfusion configuration98, 99, 132, recently reviewed in133. However, the first prefusion, metastable structure of a paramyxovirus F glycoprotein has recently been solved by appending a trimerization motif from GCN4 onto the C terminus of secreted PIV5 F, an addition thought to mimic the transmembrane domain of the glycoprotein.100 The structures of uncleaved and cleaved metastable prefusion PIV5 F glycoprotein and postfusion hPIV3 F conformations are shown in Figure 5. The differences in overall conformations between the pre- and postfusion F glycoproteins merit explanation here.
Figure 5. Structural changes between the pre- and postfusion F glycoprotein conformations derived from PIV5 and hPIV3.
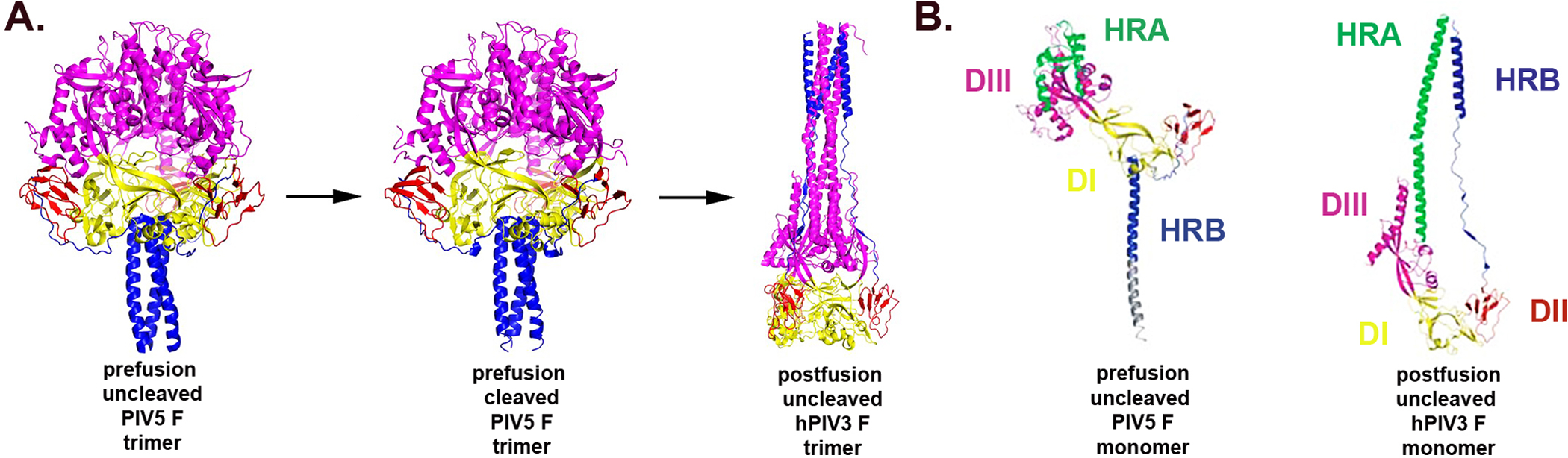
A. Ribbon diagrams of uncleaved and cleavage-activated prefusion forms and postfusion F trimers are shown side by side. The three domains, DI, DII and DIII, are depicted by different shading (DI yellow, DII red and DIII pink). B. Ribbon diagrams of pre- and postfusion F monomers similarly oriented by DI as shown in Panel A. Monomers are shown side by side to demonstrate more clearly the different domains, DI, DII and DIII, which are depicted by different shading as in A. Adapted with permission from R.A. Lamb and Macmillan Publishers Ltd: Nature100, and Proceedings of the National Academy of Sciences.136
The first metastable prefusion conformation of a PIV5 F was determined using an uncleaved version of the glycoprotein and was shown to contain a globular head connected to a trimeric coiled-coil stalk formed by HRB.100 The globular head contains three previously identified domains per subunit, referred to as DI, DII and DIII100, 132, and in DIII two sets of 6 helices form rings that cover the top of the globular head, while the HRB three-helix bundle seals the bottom. Perhaps the most intriguing aspect of the structure is that DIII undergoes major refolding between the pre- and postfusion conformations of F. In the prefusion conformation HRA is actually divided into four helices, five turn segments and two β–stands and is folded around the core of DIII. Such a conformation suggests that HRA is trapped within DIII as monomeric subunits. Further, in the prefusion conformation, the fusion peptide is wedged between the DII and DIII domains of adjacent F subunits in the trimer. Upon triggering, a total of 11 distinct segments in the prefusion HRA DIII domain refold to generate a single extended α-helical conformation necessary for translocation of the fusion peptide towards the target membrane and pre-hairpin formation. In solution, the prefusion, metastable PIV5 F molecule does not form the 6-HB structure; rather the three fold axis of the HRB three-helix bundle is aligned along the three-fold axis of the globular head with only a slight tilt. The junction of the base of the globular head and the HRB region appear to form an interactive network between trimer subunits. Previously identified residues near HRB that were hypothesized to play a role in conformational switching134 are now known to reside within this region. The presence of the additional trimeric coiled-coil domain from GCN4135 in the crystallized metastable PIV5 F is hypothesized to stabilize the trimeric coiled-coil stalk formed by HRB. The transmembrane domain and cytoplasmic tail are hypothesized to perform a similar role in native metastable F and help explain why in their absence secreted, anchorless hPIV3 and NDV F glycoproteins converted to the postfusion F conformation when crystallized.26
Very recently, the crystal structure of the cleaved prefusion form of a truncated version of the PIV5 F has been reported.136 As before; the truncated soluble form of the PIV5 F glycoprotein was appended with a trimeric coiled-coil domain (GCNt) that was able to stabilize the ectodomain of the F glycoprotein in its prefusion conformation. Unlike previous studies, the purified metastable F glycoprotein was cleaved in vitro with trypsin, re-purified and analyzed by electron microscopy, revealing its characteristic prefusion form. Interestingly, when heated briefly in vitro, the cleaved metastable F converted to a postfusion conformation.136 It was notable that other than the newly exposed N-terminus of the trypsin cleaved F0 precursor, the conformational differences near the cleavage site exhibited no net burying or exposure of hydrophobicity or charge changes136, and the conformational changes between the uncleaved to cleaved prefusion forms of the PIV5 F were not as dramatic as the prior observations between similar versions of the influenza HA glycoprotein.
Similar soluble forms of the NiV and HeV F glycoproteins have also been recently reported along with the ability to cleave purified prefusion soluble F and generate postfusion forms137, and a NiV prefusion soluble F crystal structure has been determined (K. Xu, C. Broder and D. Nikolov, unpublished findings). The structure of the cleaved prefusion PIV5 F glycoprotein, together with the previously reported structures of uncleaved prefusion PIV 5 F-GCNt and the postfusion structures of hPIV3 F, NDV F and RSV F, have now provided a detailed high-resolution view of the various static forms of paramyxovirus F glycoproteins (Figure 5).
Upon appropriate triggering of native F, it is now hypothesized that opening or “melting” of the HRB three-helix bundle stalk triggers the conformational changes in the HRA DIII domain and gives rise to the pre-hairpin fusion intermediate.100 Indeed, a multi-step process would be consistent with the early inhibition of fusion by HRA-derived peptides but not HRB-derived peptides.121, 138 Accordingly, the model depicting heptad repeat dependent membrane merger (Figure 4) requires slight adjustment. Specifically, the pre-hairpin conformation of F needs to be altered to portray HRB as three unassociated segments instead of a three-helix bundle. Overall, both prefusion conformations of the cleaved and uncleaved PIV5 F and the postfusion conformation of hPIV3 F suggest how small refolding intermediates can be coupled to the activation of F and ultimately membrane fusion.139
It has also long been recognized that truncated or cytoplasmic tail deleted versions of fusion envelope glycoproteins; particularly the human and simian immunodeficiency viruses (HIV-1, HIV-2 and SIV), could often possess an enhanced ability to mediate cell-cell fusion (Aguilar et al140 and references therein). This feature was also recorded in paramyxovirus SV5 strains where individual isolates possess an F glycoprotein with either a short (20-residue) or long (42-residue) cytoplasmic tail. It was noted earlier that an SV5 strain (W3A) possessing an F glycoprotein with a short tail could mediate syncytium formation in the absence of its HN glycoprotein partner, essentially a hyperfusogenic feature, whereas other strains with the longer tail required HN coexpression for fusion.141 Further experiments revealed that when the W3A F glycoprotein (short tail) is expressed as the longer tail (42 residues) either by mutation to remove a translational stop codon or by extension using additional sequences, the hyperfusogenic activity of the F glycoprotein is reduced. Additionally, the longer cytoplasmic tail of F modulated the F ectodomain conformation as detected by specific mAb binding, suggesting that the cytoplasmic tail could influence the conformation and function of the protein’s ectodomain.142 Likewise, the cytoplasmic tail of the NiV F glycoprotein has been shown to contain an amino acid motif (KKR) that when mutated can affect the conformation and subsequent fusion activity of the F glycoprotein ectodomain,140 a feature termed as an inside-out signaling event.
In addition, the potential role(s) of the TMD of a paramyxovirus F glycoprotein in its structure and functional features has also been under investigation. The TMD appears to influence protein folding, prefusion structure stability and the membrane fusion activity of a variety of viral fusion proteins (Smith et al143 and references therein). However, details on the TMD role have remained poorly understood among the paramyxoviruses. Recent experiments have shown that TMD-TMD interactions within the F glycoprotein trimer of HeV affect protein stability and its fusogenicity, and elements within the C terminus of the HeV F TMD appear to play an important role in the F trimer’s TMD-TMD interactions and its membrane fusion activity.143
For most paramyxoviruses, the fusion triggering event initiated by the F glycoprotein’s attachment glycoprotein partner appears to serve as an effective replacement of the acidification event required by influenza virus HA. However, the conformational changes in the transition from pre- to postfusion PIV5 F are quite different from those observed for the pre- and postfusion influenza virus HA144; nevertheless, certain trends do appear. In both viruses, the HRA is prevented from assembling, the fusion peptide is initially buried at the subunit interface, HRA projects the fusion peptide away from the viral membrane and transmembrane domain and finally the transition of HRB to its final state cannot occur due to the absence of an HRA coiled-coil and other structural barriers.
Altogether, understanding the conformational changes that occur in metastable F in its transition to a “fusogenic” and 6-HB structure has now greatly aided our understanding of how the energy required to mediate membrane fusion is captured. However, for paramyxoviruses, the precise trigger that initiates the metastable to “fusogenic” F transition continues to be investigated.
Attachment and fusion glycoproteins work together to facilitate entry
With few exceptions, the fusion activities of most paramyxovirus F glycoproteins are dependent on the activity and availability of their specific partner attachment glycoprotein (reviewed in22), and the co-expression of the attachment and fusion glycoproteins is required for virus infectivity for most members of the subfamily Paramyxovirinae. For the most part, the attachment glycoprotein-F interaction is also virus type-specific, and fusion mediated by co-expression of the F and attachment glycoproteins of different paramyxoviruses (heterotypic mixing) is rarely seen.145 Although some examples have been noted, the potency of the fusion process from heterotypic mixing is considerably reduced in comparison to that mediated by the F and attachment glycoproteins from the same virus (homotypic mixing).47, 146 HeV and NiV are closely related henipaviruses and uniquely, heterotypic combinations of the F and G glycoproteins are as potent in mediating fusion as homotypic combinations.63 Although heterotypic function of the envelope glycoproteins of the morbilliviruses MeV and CDV are not as efficient as the homotypic equivalents, like HeV and NiV, heterotypic activity is bidirectional, and fusion occurs with either heterotypic combination. These bidirectional examples are unlike the heterotypic results observed with the respiroviruses SeV and hPIV1. Here SeV F combined with hPIV1 HN functions efficiently, whereas in the reverse combination, SeV HN is unable to complement hPIV1 F.147 Given the percent amino acid similarities of F and HN from hPIV1 and SeV, which are greater than those for MeV and CDV, this would not have been expected and may represent not only the need for type-specific interactions but also the possibility that there may be genus-specific factors involved in the interaction between F and attachment glycoproteins. Although the mechanism underlying this process remains obscure, the domains of the attachment glycoprotein necessary to promote fusion have been mapped using functional assays and indicate that regions in the globular head and stalk domain are critical.148–152 It has now become increasingly clear that following receptor engagement, the attachment glycoprotein somehow signals and/or induces the required conformational changes in F leading to virion/cell fusion.100, 138, 153 However, the precise molecular details of how the fusion and attachment glycoproteins function in concert in mediating fusion continue to be gradually elucidated.
Presently there are two widely appreciated models of paramyxovirus glycoprotein-mediated membrane fusion which describe the interactions between the oligomers of an F and of an attachment glycoprotein as they relate to the role of the receptor. Model 1 suggests receptor binding to the attachment glycoprotein induces a subsequent association and triggering of F-mediated fusion. In model 2, however, receptor binds to an attachment glycoprotein that is already in complex with its partner F glycoprotein and induces the dissociation of F and the attachment glycoprotein, initiating F-mediated membrane fusion. These models were first diagrammed by McGinnes et al154 based on the available data at that time, which focused primarily on HN and F glycoproteins. A comprehensive review of the literature on the paramyxovirus fusion process by Iorio et al22 has summarized the findings on the interactions between a varied array of paramyxovirus attachment and fusion glycoprotein species and the role of their particular entry receptors. Model 1 is also referred to as the association model and model 2 as the dissociation model.22, 104
The first model proposes that the F glycoprotein and the attachment glycoprotein are not necessarily physically associated in the membrane and that following receptor binding to the attachment glycoprotein there is some alteration in the receptor-bound complex that facilitates the association of the attachment glycoprotein with its partner F glycoprotein in a manner often referred to as activation of its ‘fusion-promotion’ activity. This specific association triggers or induces the fusion activity of F, which through its subsequent conformational change drives the membrane fusion process.153, 155. This association model, also recently termed the ‘provocateur’ model156, was recently supported by key data indicating that the prefusion conformation of F (PIV5) was maintained in the absence of HN co-expression. This model is also supported by extensive functional and structural studies on the HN and F glycoproteins from hPIV3, NDV and PIV527, 94, 151, 152, 156–160 in which the overall theme indicates a positive correlation between HN and F interaction and fusion promotion or triggering activity.
In model 2, F and its partner attachment glycoprotein are pre-associated in some oligomeric complex, and a conformational alteration in the latter following receptor engagement induces some conformational change which facilitates its dissociation or release of F, thereby allowing F to undergo its conformational alterations driving the membrane merger process. Although this model was originally put forth as an alternative possibility based on studies of the HN and F glycoproteins, it was later supported by extensive studies with the MeV-H and -F glycoproteins161–164 and also with the G and F glycoproteins of the henipaviruses.69, 140, 165, 166 Here, the overall theme suggests a negative correlation between the attachment and fusion glycoprotein interaction and the membrane fusion activity of the viral species; that is, alterations in H that enhance H-F association adversely affect fusion and those that weaken the H-F interaction yield an enhanced fusion feature. The model 2, or dissociation model, has more recently been referred to as the clamp model, whereby the pre-association of complexes suggests that the attachment glycoprotein maintains the prefusion metastable conformation of its partner F glycoprotein until encountering receptor.156
The pre-association of attachment and fusion glycoproteins before receptor binding versus the post-receptor bound inducement of their association has also been addressed by other experiments. For example, as an alternative means to address these models, it has been demonstrated with several attachment glycoproteins and F glycoprotein partners, including those from MeV, NDV and hPIV2, that they interact early during biosynthesis in the endoplasmic reticulum (ER).161, 167–169 However, in other viruses (PIV5 (SV5) and hPIV3) a F and HN interaction prior to fusion was not strong.94 Additionally, the henipavirus G and F glycoproteins have a more complex biosynthesis and maturation pathway in comparison to other paramyxoviruses, and the G glycoprotein takes longer to traffic through the ER and Golgi. This longer trafficking time of G together with the complex pattern of F maturation suggests that G-F interaction does not occur until both glycoproteins are expressed on the cell membrane.167, 170
Nevertheless, either model recognizes an interaction between the attachment and fusion glycoproteins that is regulated by receptor binding (recently reviewed171, 172), and the triggering mechanisms between and HN-F pair and H/G-F pair may essentially be the same but with each differing in their general propensity to associate in their pre-receptor bound states. Indeed, recent experiments with NiV employed the addition of N-linked glycosylation modification sites as probes to examine the specific interactions between the G attachment and F glycoproteins.173 These studies with NiV revealed contrasting findings from two earlier studies with NDV and MeV that demonstrated the NDV HN, with N-glycan additions in the stalk, was defective in both its fusion promotion and F interacting capacity152 and that N-glycan additions in an implicated F-interactive site of the MeV H blocked H-F complex formation.174 Rather, the NiV G and its F interaction was not affected by most N-glycan additions and does not appear to be solely mediated by the stalk domain of G.173 Thus, although the preponderance of data to date have implicated the attachment glycoprotein stalk in the interacting and triggering process with F, these data reveal that there is another level of G-F interaction, which suggests a natural propensity for a specific association between the two glycoproteins. However, this association is not required for maintaining F in a prefusion conformation (clamp) or a trigger for F fusogenic triggering. In either case it also seems likely that upon F glycoprotein triggering, the initiation of its conformational changes leading to 6-HB formation and the driving of the membrane merger process, F would need to be free of any association with its large oligomeric partner (HN, H or G).
The recently characterized soluble forms of trimeric henipavirus F glycoprotein discussed earlier137 have also been valuable in assessing the themes within the paramyxovirus fusion models. For example, a murine mAb (5B3), which is specific for the prefusion form of henipavirus F glycoprotein, is capable of binding F on the surface of expressing cells and also from cellular lysates containing F glycoprotein in the absence of the co-expression of its partner G glycoprotein, indicating that the clamp model as defined is not accurate.137 It appears that although receptor-induced G glycoprotein triggering of the F-mediated fusion process likely takes place, the requirement of G association with its partner F glycoprotein in order to maintain F in its prefusion and metastable state is not necessary. Rather, this data is more in line with the provocateur model of fusion discussed earlier, where the prefusion conformation of F (PIV5) is maintained in the absence of the coexpression of HN.156 Further, the independent trafficking and maturation patterns of HeV and NiV F reviewed above are also in an agreement with such a scenario.
In addition, two MeV F specific mAbs (186A and 19GD) specific for either the prefusion/pretriggered versus fusion-triggered conformation of F, respectively, were recently used as probes to address the MeV F triggering process, and here as well the prefusion MeV F glycoprotein could be recognized strongly by the 186A mAb in absence of H co-expression, revealing that MeV F does not require a physical association with H to maintain its prefusion conformation.175 Similarly, a second study further explored the morbillivirus F glycoprotein triggering process with similar mAb binding techniques - identifying antibodies to both MeV and CDV F that were specific for either their prefusion, triggered or postfusion conformations.176 In these studies it was demonstrated that prefusion F-specific mAbs could bind to F in absence of H co-expression, that conversion of F from a prefusion to a triggered conformation by higher temperature revealed loss of binding by those mAbs and that antibodies specific for postfusion forms of F acquired enhanced binding upon heating. Essentially identical antibody binding profiles were observed under physiologic conditions of H triggered F activation.176 Together these studies support the conclusion recently suggested by Ader et al176 that the attachment glycoprotein, such as H, serves to lower the F triggering energy barrier rather than to maintain the prefusion conformation of F through a binding mechanism, a function more in line with a provocateur type of model. There is now evidence with at least four different species of F glycoproteins (PIV5, NiV, MeV and CDV), which together span the varied attachment glycoprotein types (HN, H or G), that demonstrate the prefusion metastable F conformation can be maintained in the absence of any ‘clamp’ or physical association with its attachment glycoprotein partner.
In summary, these findings suggest that neither a clamp model nor a provocateur model as presently defined can fully account for all the experimental observations to date on the mechanism of fusion. In fact, a recent report by Porotto et al177 describes a variation of the hPIV3 fusion mechanism that incorporates features of both the clamp and provocateur models. Here it is suggested that hPIV3 HN must continually engage receptor to activate F as interruption of hPIV3 HN and receptor blocks F-mediated membrane fusion. Although no direct HN-F interactions were assessed in this study, an approach to examine hPIV3 fusion using bimolecular fluorescence complementation to follow the dynamics of HN and F in live cells was conducted178 The authors demonstrate that HN and F associate prior to receptor engagement, that HN drives the formation of HN and F interacting clusters at the site of membrane fusion and that the interaction of the HN-F pairs of oligomers modulate the fusion process. In sum, it appears that measurable pre-association of paramyxovirus attachment and fusion glycoproteins, prior to fusion triggering by receptor, is dependent on the particular protein pair of viral species proteins and possibly due to a requirement for maintenance of the prefusion F conformation (clamp model). Nevertheless, the fusion process for most paramyxoviruses is dependent on a receptor mediated binding event by the attachment glycoprotein and this will be discussed in the next section.
Receptor induced conformational changes in attachment glycoproteins
The influence of receptor binding on the fusion-triggering process has been a major focus of research and is a factor that can potentially differentiate the most recently proposed models of the mechanism of paramyxovirus membrane fusion. The large amount of recent structural information, particularly in the many comparisons that have been made between the receptor bound and unbound structures of H and G, has revealed that major conformational changes in the attachment glycoprotein heads are not observed upon receptor binding.31, 60–62, 73 Rather, much attention has more recently focused on the role of receptor-induced conformational changes that occur in the higher-order oligomeric structures of the attachment glycoprotein tetramer.
Data derived from NDV HN structural studies suggested that receptor binding leads to conformational changes in HN and a model was suggested in which receptor (sialic acid) engagement would facilitate dimer formation or even tetramer formation in a ligand dependent manner24 and HN oligomerization would be the trigger for F activation. This model of receptor induced conformational change in HN for triggering F fusion was first proposed by Sergel et al179 and then diagramed by Lamb153 which was essentially the first ‘association’ or ‘provocateur’ model discussed earlier. The receptor-induced conformation changes in HN were first detailed at the molecular level using mutagenesis analysis.180, 181 These studies revealed differences in the structure of receptor-bound HN and non-receptor-bound HN, although importantly, the structure of HN in complex with its receptor was not then solved. Subsequently, a second sialic acid binding site was identified at the dimer interface of NDV HN38, and it was hypothesized to play a role in facilitating membrane fusion. The specific steps of paramyxovirus fusion that were proposed in this revised model38 are as follows: HN and F may exist in a complex on the cell surface and this complex holds both glycoproteins in the “off states”. Upon binding to sialic acid, conformational changes occur in HN that converts the glycoprotein to its “on state,” which leads to cleavage and release of the sialic acid from the sialic acid-containing receptor. The release of sialic-acid induces further conformational changes in HN at the dimer interface. These changes were hypothesized to alter the dimeric or tetrameric properties of HN that lead to changes in the stalk domain and in so doing trigger the fusion glycoprotein. Concurrently, the second sialic acid binding site is formed by the conformational change induced by the release of sialic acid from site 1. The existence of a second sialic acid receptor binding site for several different HN glycoproteins has also been functionally identified.182–184 Recently, using glycan array assays, it was demonstrated that the HN of hPIV1 has a second site for receptor binding masked by an N-linked glycan and that sialic acid receptor binding to the first site triggers the exposure of the second site.185 The significance of the second sialic acid binding site has been hypothesized to bind cell-surface sialic acid residues and maintain a close proximity of the virion to the cell surface to aid in efficient targeting of the fusion peptide for membrane fusion mediated by the F glycoprotein.186 This model was recently tested using a series of HN dimer interface mutants, and it was demonstrated that binding of receptor to site 1 triggered HN interaction with F and that site 2 appeared to maintain binding with the target cell membrane during the fusion process.187 A summary of this model is shown in Figure 6. However, it has also been suggested that receptor binding to the NDV HN site 2 plays an active role in transmitting the fusion activation signal to the stalk region of HN.188 This function could also be demonstrated with chimeric proteins composed of the globular head of NDV HN and the stalk region of hPIV3 or NiV where receptor binding to site 2 led to the activation of heterotypic F glycoproteins.188
Figure 6. Multiple conformational changes in NDV HN trigger F from metastable to fusogenic.
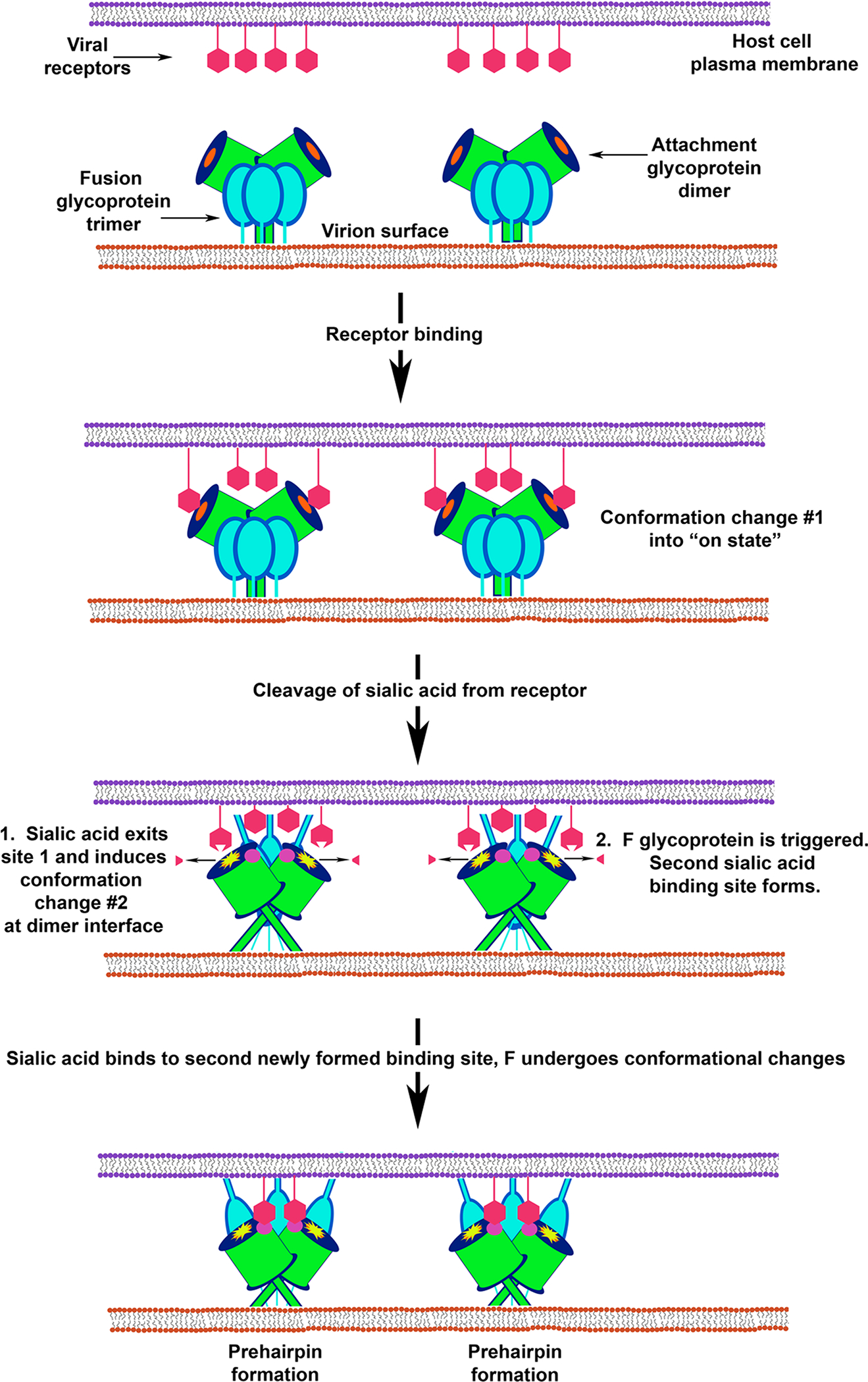
Fusion model as described in Zaitsev et al38 depicting the receptor-triggered mechanism of NDV fusion. Receptor binding induces conformational changes that convert HN to its “on state”. Subsequent cleavage and release of sialic acid leads to conformational changes at the HN dimer interface that not only are critical for triggering F but also generate a second sialic acid binding site. Binding cellular receptors via the newly formed second sialic acid site is hypothesized to keep the virion in close proximity to the target cell membrane. For simplicity F is shown as a trimer with limited conformational changes and membrane merger has been excluded.
A variation of the NDV fusion model described above was later suggested based on the solved structures of PIV5 and hPIV3 HN alone or in complex with receptor.25, 26 Several differences exist between the solved HN structures; however, a key distinction leading to the proposal of this modified model is that for both PIV5 and hPIV3 there are no major conformational changes in HN upon receptor binding. Additionally, neither PIV5 nor hPIV3 HN contain a second sialic acid binding site. Furthermore, dimers and tetramers (PIV5) were evident in the solved structures in the absence of receptor, thus a ligand induced oligomerization of HN was not at play. NDV, hPIV3 and PIV5 HN all form a similar dimer and likely tetrameric conformations. The dimer interactions are formed over a large surface, and it has been hypothesized they most likely are of high affinity, if not disulfide bond linked, requiring significant amounts of energy to destabilize the interaction.26 The tetramers, that is the dimer of dimers, by comparison appeared to be less conserved and one of weaker association. A revised model proposed that the HN dimer/tetramer is present in the absence of receptor and that receptor binding destabilizes the tetramer and it may partially dissociate. This tetrameric conformational change was suggested to lead to changes in the stalk domain of HN, the proposed site of F interaction, thus providing the necessary trigger for activating F to its fusogenic state.26
When the crystal structure of NDV HN alone and in complex with sialic acid (beta-anomer) or a neuraminidase inhibitor was reported, comparisons of the structures also suggested that the catalytic site was activated by a conformational switch in the head, providing roles for both sialic acid binding and hydrolysis activity.24 It was postulated that significant conformational change in the HN dimer essential for fusion could occur as HN transitions from an initial structure possessing minimal inter-monomeric contacts to a structure containing an extensive dimer interface. This proposed mechanism of oligomeric conformational change and its role in the fusion triggering process was later tested by mutational insertions of inter-monomeric disulfide bonds in the globular head domain of NDV HN. The insertion of disulfide bonds prevented the formation of the minimal interface configuration of HN, however, rather than inhibiting its fusion promotion activity the mutated HN possessed enhanced receptor-binding and fusion promotion activity.189 This study, using novel disulfide bond engineering to stabilize the HN dimers, showed that neither the minimal interface form of HN nor the proposed conformational changes were required for fusion. In contrast, using the extensive available structural information on H and HN, Navaratnarajah et al190 modeled and tested the role of conformational changes within the MeV H dimer. Here, the notion of a requirement for a conformational rearrangement of the head domains relative to each other within an individual dimer was also examined by the mutational insertion of strategically placed disulfide bonds. In this instance their placement prevented a required movement of the heads and subsequent F triggering and fusion, and the authors suggested a model in which H and HN transmit the fusion triggering signal in alternative ways, perhaps due to the different locations of the receptor binding site.
As discussed earlier, a tetrameric configuration of the native paramyxovirus attachment glycoprotein has been widely described, and there has recently been a considerable amount of new data on their structure and the role of the tetramer along with the receptor induced conformational changes as they relate to the F triggering fusion process. For a paramyxovirus using a protein receptor, the triggering of MeV F by H is the most extensively explored and understood system. As discussed above, MeV F and H associate intracellularly prior to any role of receptor161, and through the use of a bimolecular complementation assay it has been shown that receptor binding to H, as well as the elements in H required for F interaction174, 191 versus F triggering163, are distinct, whereby mutants of H in these various domains when expressed together can effectively restore MeV F/H fusion40 (reviewed in192)
Structural conformational changes in a central region of either the MeV or CDV H stalk domain were identified by Ader et al193 as critical in the transfer of the fusion triggering signal. Here, engineered disulfide bonds were introduced within the stalk domain central region spanning residues 91–115, which also contains the F-interacting domain mentioned above.174, 191 Several of these inserted disulfide bridges could block H fusion promotion activity and upon their reduction fusion activity was restored. Altogether, with modeling, the data suggested that the H stalk is a tetramer of subunits which undergo structural rearrangement following receptor binding, promoting an interaction of a specific stalk element of H with the associated F trimer, triggering its fusogenic activity.
Brindley et al175 later addressed the MeV fusion process by targeting the MeV H stalk through the manipulation of the existing head proximal disulfide bonds in the context of the H tetramer model together with bimolecular complementation and the use of both soluble and membrane associated MeV receptors. These studies found that the H tetramer structure is maintained in that dimers of dimers do not dissociate but that the central stalk region does require flexibility. Furthermore, receptor binding to only one dimer within the context of the tetramer was sufficient to trigger fusion. The triggering of conformational changes in the context of the H tetramer could be accomplished by soluble receptor (SLAM) and also initiate the F triggering and refolding process as detected by pre- and postfusion F specific mAbs described earlier, but cell-cell fusion pore formation required the triggering of H by membrane-anchored receptor.175
In another examination of the MeV F/H triggering process using the disulfide bridging approach, Navaratnarajah et al194 analyzed the H stalk using a comprehensive cysteine residue substitution mutagenesis process. These studies revealed three stalk regions of varying importance in which two of the three stalk segments possessed a tendency for the formation of tetrameric configurations. Some disulfide linked H stalk mutants that were fusion-triggering defective could be chemically reduced with a concomitant restoration of its fusion-promotion activity, whereas another segment of the stalk when covalently linked into H tetramers had no effect on the protein’s fusion-promoting activity. A third stalk domain, globular head-proximal, could not be readily disulfide linked and stabilized. In total, this study identified an F-triggering (interacting) domain of the H stalk as residues ~75–127, similar to the CDV stalk residues (91–115). In a companion report, the MeV F glycoprotein was analyzed by modeling to predict surface exposed residues in regions that could be predicted to interact with other glycoproteins, namely H. A large panel of some 50 possible residues were noted and by conducting an iterative mutagenesis and functional analysis, a set of specific mutants were identified that inhibited fusion with four mutants lining a cavity flanked by two monomers of the F trimer model. It was suggested that the stalk region of the H tetramers could be lodged within the sides of their companion F trimers at the site of the modeled cavity with two helices of an H tetramer contacting one side groove of an F trimer, suggesting that one H tetramer could possibly transmit the F-trigging signal to at least two opposing F trimers.195
Finally, a recent intriguing study reported on the application of a headless PIV5 HN glycoprotein in triggering PIV5 F-mediated fusion.196 Here, this study also demonstrated that essentially the entire stalk (PIV5 HN residues 1–117) was required, and it was proposed to fold into its 4-helix bundle or otherwise receptor-bound conformation that could associate with and trigger its F glycoprotein partner. The study also revealed that the F glycoprotein of PIV5 also maintained its metastable prefusion conformation in absence of HN co-expression. Further, and importantly, the 4 helix headless protein structure also displayed and maintained its viral species specificity for triggering F-mediated membrane fusion and could not trigger other F glycoproteins. The globular heads were found clearly dispensable, and the roles of the heads appear to define cell tropism and also perhaps to mask and/or maintain the stalk domains in a pre-receptor bound and non-F triggering form. In hindsight, it seems surprising that this approach was never before tested, and examining this possibility with other paramyxoviruses, particularly the morbilliviruses or henipaviruses, may yield interesting results. Remarkably, in the case of HeV, mutations in the heads that model to the head stalk interface in a heads-down structure have been identified and shown to block fusion promotion activity but allow competent receptor binding.69 Perhaps such mutants prevent the receptor-induced movement of the heads that would allow access to the henipavirus G stalk domain required for F triggering.43
Transfer of the F-mediated fusion triggering signal appeared to involve an opening and repositioning of the dimeric interface of the H head domains190, which is then followed by conformational changes of a central domain of the H stalk.193 It was suggested that receptor binding and pulling de-stabilizes this H-dimer interface190, which would subsequently elicit the conformational change in the central stalk segment of H191, 193 required for the fusion triggering signal. Indeed, such a receptor-pulling process might certainly be envisioned during the cell-cell or virus-cell contact step at the beginning of the process.
Fusion models
In consideration of the large amount of structural and functional data on the paramyxovirus fusion process, with particular attention to the recent reports over just the past few years from leaders in the field, the current modeled scenarios of the receptor binding and fusion triggering steps are shown in a refinement of the originally proposed models 1 and 2 (association and dissociation models) (Figure 7), and an attempt has been made to include elements of both the non-pre-associated and pre-associated states of a fusion and attachment glycoprotein and the heads-up and heads-down features of the attachment glycoproteins in relation to the fusion triggering process. Initial expression of the tetrameric attachment (HN/H/G) glycoprotein, with each dimer pair distinguished by color, depicts the fusion glycoprotein as not being pre-associated with the attachment glycoprotein, which is in a heads-down configuration (Figure 7A). This model has most recently been refined and proposed based on the extensive data on the function of F and HN glycoproteins.196 Here, this model recognizes that a metastable prefusion F can exist without a required association of its attachment glycoprotein partner, the importance of the F-triggering role of the stalk domain and its membrane distal elements, the receptor-mediated movement of the head domains upwards and the provocateur association and activation of F. This model does not necessarily preclude other paramyxovirus species, such as the henipaviruses that possess an F and G glycoprotein pair, because until receptor binding triggers tetrameric conformational changes in G, an F/G oligomeric pair of glycoproteins may simply have an ability to associate in membranes without inducing additional conformational changes. Likewise, the MeV F/H pair could also potentially fit a similar model recognizing that H does not have to maintain F in a prefusion state and that contact between F and H could occur with either a heads-up or heads-down position as discussed by Navaratnarajah et al194 Alternatively, in Figure 7B, a tetrameric heads-up attachment glycoprotein oligomer is pre-associated with its F glycoprotein partner, a scenario recently refined and modeled with the MeV F and H glycoprotein pairs.175, 193 Here, receptor binding triggers a conformational change in the tetramer, the central features of which are the movement to the heads-down configuration in conjunction with a twist of the stalk region, which all together facilitates a targeted association of elements within the attachment glycoprotein’s stalk to its partner F glycoprotein resulting in the disassociation and fusogenic conformational changes in F. However, it is not yet clear whether the heads-up versus heads-down attachment glycoprotein configuration is mechanistically linked to the fusion triggering process.
Figure 7. Models of paramyxovirus membrane fusion involving heads-up and heads-down conformations.
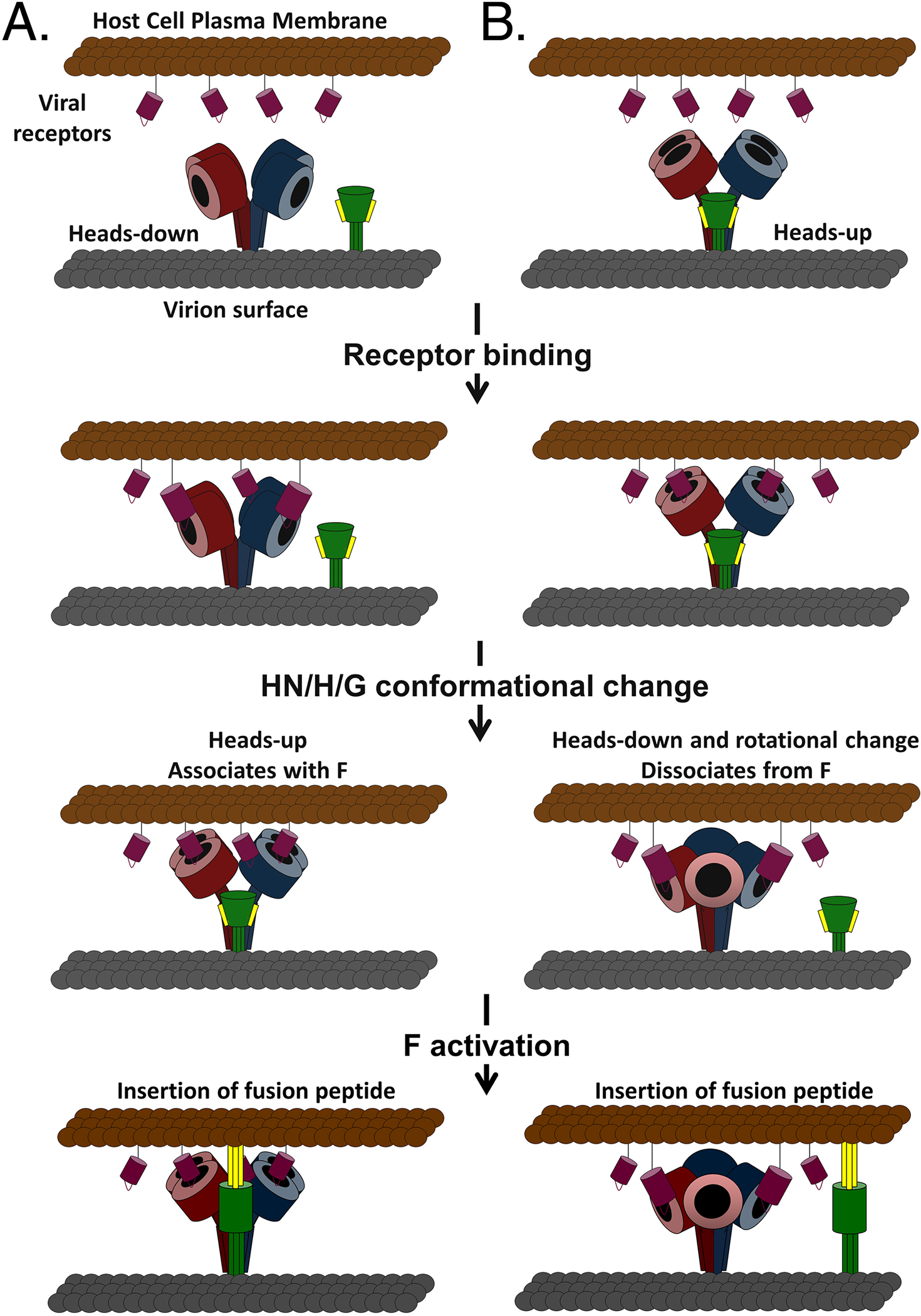
Initial expression of the tetrameric attachment (HN/H/G) glycoprotein (dimers colored red and blue) and the fusion (F) glycoprotein (green) is depicted in the (A) heads-down, non-F-associated or (B) heads-up, F-associated conformations. In both models, HN/H/G binds receptor (maroon) and undergoes receptor-induced conformational changes, switching from heads-up to heads-down or vice versa. The change in the position of the globular heads allows for (A) association (provocateur model) or (B) dissociation (clamp model) with F, leading to the fusion activation of F and the beginning of membrane fusion by the insertion of the fusion peptide (yellow) into the target cell membrane.
Conclusions and Remaining Questions
Paramyxoviruses have evolved a more complex mechanism of attachment and membrane fusion that facilitates delivery of the genome into the host cell, one that for most members requires two independent glycoproteins. The overall structural similarity of the attachment glycoproteins of viruses within the subfamily Paramyxovirinae, coupled to the highly conserved functional domains of the fusion glycoproteins in all paramyxoviruses reveal their common ancestry, while their differences in receptor engagement and the events that promote fusion most likely reflect individual adaptations, perhaps as a result of varying hosts and varying tissues within a host. Years of research by many have uncovered important facets of paramyxovirus entry and led to changing hypotheses and the proposal of alternative models of the paramyxovirus entry process. The two current and most favored models that have emerged combine old and new data with the solved structures of the attachment glycoproteins with and without their receptors. Nevertheless, the most recent revised models discussed here that have been derived from extensive data from viruses possessing an HN, H or G, are all in fair agreement and allow for subtle differences that have been experimentally explored. They can also fairly accommodate the differences in whether a sialic acid versus a proteinaceous receptor is employed as well as the locations of the receptor binding site in the attachment glycoprotein. Additionally, the recent structures of both uncleaved and cleaved metastable F glycoprotein forms have provided insights into the significant conformational changes that provide the necessary energy for a paramyxovirus fusion glycoprotein to mediate membrane merger. Although many questions relating to entry have been answered, additional information is still missing, such as the pre- and postfusion F structures from the same viral species. Equally intriguing would be to determine structures of a tetrameric attachment glycoprotein in complex with a receptor(s), particularly a receptor known to trigger fusion promotion activity with one that does not. Also, changes in the biochemical interaction of the fusion and attachment glycoproteins as fusion proceeds have yet to be adequately profiled. Finally, and perhaps difficult to achieve, the structural solution of any F trimer in complex with a stalk containing attachment glycoprotein or perhaps the stalk domain itself would be a significant advance.
Acknowledgements.
We would like to thank Alex Hyatt for generously providing the paramyxovirus electron micrographs and are indebted to Alex, Sandra Crameri and Rosemary Van Driel for their assistance in scanning and scaling all EM negatives. We thank Robert Lamb for generously allowing us to use the NDV HN, uncleaved and cleaved metastable PIV5 F and postfusion hPIV3 F structures in this chapter. Research in the author’s laboratories is supported in part by NIH research grants AI077995 and AI054715 (C.C.B.).
Footnotes
The views expressed in the manuscript are solely those of the authors, and they do not represent official views or opinions of the Department of Defense or the Uniformed Services University of the Health Sciences.
References.
- 1.Lamb RA, and Parks GD. Paramyxoviridae: The viruses and their replication. In Knipe DM and Howley PM ed., Fields Virology, 5 ed, Philadelphia. Lippincott Williams & Wilkins, 2007:1449–1496. [Google Scholar]
- 2.Alexander DJ. Newcastle disease and other avian paramyxoviruses. Rev Sci Tech 2000; 19(2):443–462. [DOI] [PubMed] [Google Scholar]
- 3.Tambi EN, Maina OW, Mukhebi AW, et al. Economic impact assessment of rinderpest control in Africa. Rev Sci Tech 1999; 18(2):458–477. [DOI] [PubMed] [Google Scholar]
- 4.Eaton BT, Broder CC, Middleton D, et al. Hendra and Nipah viruses: different and dangerous. Nat Rev Microbiol 2006; 4(1):23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eaton BT, Mackenzie JS, and Wang L-F. Henipaviruses. In Knipe DM and Howley PM ed., Fields Virology, 5 ed, Philadelphia. Lippincott Williams & Wilkins, 2007:1587–1600. [Google Scholar]
- 6.Lamb RA. Mononegavirales. In Knipe DM and Howley PM ed., Fields Virology, Fifth ed, Philadelphia. Lippincott Williams and Wilkins, 2007:1357–1361. [Google Scholar]
- 7.Mayo MA. A summary of taxonomic changes recently approved by ICTV. Arch Virol 2002; 147(8):1655–1663. [DOI] [PubMed] [Google Scholar]
- 8.de Leeuw O, and Peeters B. Complete nucleotide sequence of Newcastle disease virus: evidence for the existence of a new genus within the subfamily Paramyxovirinae. J Gen Virol 1999; 80 (Pt 1):131–136. [DOI] [PubMed] [Google Scholar]
- 9.Seal BS, Crawford JM, Sellers HS, et al. Nucleotide sequence analysis of the Newcastle disease virus nucleocapsid protein gene and phylogenetic relationships among the Paramyxoviridae. Virus Res 2002; 83(1–2):119–129. [DOI] [PubMed] [Google Scholar]
- 10.Seal BS, King DJ, and Meinersmann RJ. Molecular evolution of the Newcastle disease virus matrix protein gene and phylogenetic relationships among the paramyxoviridae. Virus Res 2000; 66(1):1–11. [DOI] [PubMed] [Google Scholar]
- 11.Wang LF, Yu M, Hansson E, et al. The exceptionally large genome of Hendra virus: support for creation of a new genus within the family Paramyxoviridae. J Virol 2000; 74(21):9972–9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Yu M, Zhang H, et al. Beilong virus, a novel paramyxovirus with the largest genome of non-segmented negative-stranded RNA viruses. Virology 2006; 346(1):219–228. [DOI] [PubMed] [Google Scholar]
- 13.Kurath G, Batts WN, Ahne W, et al. Complete genome sequence of Fer-de-Lance virus reveals a novel gene in reptilian paramyxoviruses. J Virol 2004; 78(4):2045–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsh GA, de Jong C, Barr JA, et al. Cedar virus: a novel Henipavirus isolated from Australian bats. PLoS Pathog 2012; 8(8):e1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrison TG. The three faces of paramyxovirus attachment proteins. Trends Microbiol 2001; 9(3):103–105. [DOI] [PubMed] [Google Scholar]
- 16.Lamb RA, and Jardetzky TS. Structural basis of viral invasion: lessons from paramyxovirus F. Curr Opin Struct Biol 2007; 17(4):427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parks GD, and Lamb RA. Folding and oligomerization properties of a soluble and secreted form of the paramyxovirus hemagglutinin-neuraminidase glycoprotein. Virology 1990; 178(2):498–508. [DOI] [PubMed] [Google Scholar]
- 18.Scheid A, Caliguiri LA, Compans RW, et al. Isolation of paramyxovirus glycoproteins. Association of both hemagglutinating and neuraminidase activities with the larger SV5 glycoprotein. Virology 1972; 50(3):640–652. [DOI] [PubMed] [Google Scholar]
- 19.Thompson SD, and Portner A. Localization of functional sites on the hemagglutinin-neuraminidase glycoprotein of Sendai virus by sequence analysis of antigenic and temperature-sensitive mutants. Virology 1987; 160(1):1–8. [DOI] [PubMed] [Google Scholar]
- 20.Langedijk JP, Daus FJ, and van Oirschot JT. Sequence and structure alignment of Paramyxoviridae attachment proteins and discovery of enzymatic activity for a morbillivirus hemagglutinin. J Virol 1997; 71(8):6155–6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu M, Hansson E, Langedijk JP, et al. The attachment protein of Hendra virus has high structural similarity but limited primary sequence homology compared with viruses in the genus Paramyxovirus. Virology 1998; 251(2):227–233. [DOI] [PubMed] [Google Scholar]
- 22.Iorio RM, Melanson VR, and Mahon PJ. Glycoprotein interactions in paramyxovirus fusion. Future Virol 2009; 4(4):335–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colman PM, Hoyne PA, and Lawrence MC. Sequence and structure alignment of paramyxovirus hemagglutinin-neuraminidase with influenza virus neuraminidase. J Virol 1993; 67(6):2972–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crennell S, Takimoto T, Portner A, et al. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol 2000; 7(11):1068–1074. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence MC, Borg NA, Streltsov VA, et al. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J Mol Biol 2004; 335(5):1343–1357. [DOI] [PubMed] [Google Scholar]
- 26.Yuan P, Thompson TB, Wurzburg BA, et al. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure (Camb) 2005; 13(5):803–815. [DOI] [PubMed] [Google Scholar]
- 27.Yuan P, Swanson KA, Leser GP, et al. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc Natl Acad Sci U S A 2011; 108(36):14920–14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashiguchi T, Kajikawa M, Maita N, et al. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci U S A 2007; 104(49):19535–19540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colf LA, Juo ZS, and Garcia KC. Structure of the measles virus hemagglutinin. Nat Struct Mol Biol 2007; 14(12):1227–1228. [DOI] [PubMed] [Google Scholar]
- 30.Bowden TA, Aricescu AR, Gilbert RJ, et al. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat Struct Mol Biol 2008; 15(6):567–572. [DOI] [PubMed] [Google Scholar]
- 31.Xu K, Rajashankar KR, Chan YP, et al. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc Natl Acad Sci U S A 2008; 105(29):9953–9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson SD, Laver WG, Murti KG, et al. Isolation of a biologically active soluble form of the hemagglutinin-neuraminidase protein of Sendai virus. J Virol 1988; 62(12):4653–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markwell MA, and Fox CF. Protein-protein interactions within paramyxoviruses identified by native disulfide bonding or reversible chemical cross-linking. J Virol 1980; 33(1):152–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrison TG, McQuain C, O’Connell KF, et al. Mature, cell-associated HN protein of Newcastle disease virus exists in two forms differentiated by posttranslational modifications. Virus Res 1990; 15(2):113–133. [DOI] [PubMed] [Google Scholar]
- 35.Ng DT, Randall RE, and Lamb RA. Intracellular maturation and transport of the SV5 type II glycoprotein hemagglutinin-neuraminidase: specific and transient association with GRP78-BiP in the endoplasmic reticulum and extensive internalization from the cell surface. J Cell Biol 1989; 109(6 Pt 2):3273–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell R, Paterson RG, and Lamb RA. Studies with cross-linking reagents on the oligomeric form of the paramyxovirus fusion protein. Virology 1994; 199(1):160–168. [DOI] [PubMed] [Google Scholar]
- 37.Ng DT, Hiebert SW, and Lamb RA. Different roles of individual N-linked oligosaccharide chains in folding, assembly, and transport of the simian virus 5 hemagglutinin-neuraminidase. Mol Cell Biol 1990; 10(5):1989–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaitsev V, von Itzstein M, Groves D, et al. Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. J Virol 2004; 78(7):3733–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plemper RK, Hammond AL, and Cattaneo R. Characterization of a region of the measles virus hemagglutinin sufficient for its dimerization. J Virol 2000; 74(14):6485–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brindley MA, and Plemper RK. Blue native PAGE and biomolecular complementation reveal a tetrameric or higher-order oligomer organization of the physiological measles virus attachment protein H. J Virol 2010; 84(23):12174–12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bossart KN, Crameri G, Dimitrov AS, et al. Receptor binding, fusion inhibition and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. J Virol 2005; 79(11):6690–6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maar D, Harmon B, Chu D, et al. Cysteines in the stalk of the Nipah virus G glycoprotein are located in a distinct subdomain critical for fusion activation. J Virol 2012; 86(12):6632–6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steffen DL, Xu K, Nikolov DB, et al. Henipavirus Mediated Membrane Fusion, Virus Entry and Targeted Therapeutics. Viruses 2012; 4 doi:(2):280–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choppin PW, and Scheid A. The role of viral glycoproteins in adsorption, penetration, and pathogenicity of viruses. Rev Infect Dis 1980; 2(1):40–61. [DOI] [PubMed] [Google Scholar]
- 45.Dorig RE, Marcil A, Chopra A, et al. The human CD46 molecule is a receptor for measles virus (Edmonston strain). Cell 1993; 75(2):295–305. [DOI] [PubMed] [Google Scholar]
- 46.Naniche D, Varior-Krishnan G, Cervoni F, et al. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J Virol 1993; 67(10):6025–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nussbaum O, Broder CC, Moss B, et al. Functional and structural interactions between measles virus hemagglutinin and CD46. J Virol 1995; 69(6):3341–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tatsuo H, Ono N, Tanaka K, et al. SLAM (CDw150) is a cellular receptor for measles virus. Nature 2000; 406(6798):893–897. [DOI] [PubMed] [Google Scholar]
- 49.Tatsuo H, Ono N, and Yanagi Y. Morbilliviruses use signaling lymphocyte activation molecules (CD150) as cellular receptors. J Virol 2001; 75(13):5842–5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baron MD. Wild-type Rinderpest virus uses SLAM (CD150) as its receptor. J Gen Virol 2005; 86(Pt 6):1753–1757. [DOI] [PubMed] [Google Scholar]
- 51.Galbraith SE, Tiwari A, Baron MD, et al. Morbillivirus downregulation of CD46. J Virol 1998; 72(12):10292–10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welstead GG, Hsu EC, Iorio C, et al. Mechanism of CD150 (SLAM) down regulation from the host cell surface by measles virus hemagglutinin protein. J Virol 2004; 78(18):9666–9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masse N, Ainouze M, Neel B, et al. Measles virus (MV) hemagglutinin: evidence that attachment sites for MV receptors SLAM and CD46 overlap on the globular head. J Virol 2004; 78(17):9051–9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vongpunsawad S, Oezgun N, Braun W, et al. Selectively receptor-blind measles viruses: Identification of residues necessary for SLAM- or CD46-induced fusion and their localization on a new hemagglutinin structural model. J Virol 2004; 78(1):302–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leonard VH, Sinn PL, Hodge G, et al. Measles virus blind to its epithelial cell receptor remains virulent in rhesus monkeys but cannot cross the airway epithelium and is not shed. J Clin Invest 2008; 118(7):2448–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tahara M, Takeda M, Shirogane Y, et al. Measles virus infects both polarized epithelial and immune cells by using distinctive receptor-binding sites on its hemagglutinin. J Virol 2008; 82(9):4630–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noyce RS, Bondre DG, Ha MN, et al. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog 2011; 7(8):e1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muhlebach MD, Mateo M, Sinn PL, et al. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 2011; 480(7378):530–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noyce RS, and Richardson CD. Nectin 4 is the epithelial cell receptor for measles virus. Trends Microbiol 2012; 20(9):429–439. [DOI] [PubMed] [Google Scholar]
- 60.Zhang X, Lu G, Qi J, et al. Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4. Nat Struct Mol Biol 2012. [DOI] [PubMed] [Google Scholar]
- 61.Santiago C, Celma ML, Stehle T, et al. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat Struct Mol Biol 2010; 17(1):124–129. [DOI] [PubMed] [Google Scholar]
- 62.Hashiguchi T, Ose T, Kubota M, et al. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat Struct Mol Biol 2011; 18(2):135–141. [DOI] [PubMed] [Google Scholar]
- 63.Bossart KN, Wang LF, Flora MN, et al. Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. J Virol 2002; 76(22):11186–11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bossart KN, Wang LF, Eaton BT, et al. Functional expression and membrane fusion tropism of the envelope glycoproteins of Hendra virus. Virology 2001; 290(1):121–135. [DOI] [PubMed] [Google Scholar]
- 65.Eaton BT, Wright PJ, Wang LF, et al. Henipaviruses: recent observations on regulation of transcription and the nature of the cell receptor. Arch Virol Suppl 2004(18):122–131. [PubMed] [Google Scholar]
- 66.Bonaparte MI, Dimitrov AS, Bossart KN, et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci U S A 2005; 102(30):10652–10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Negrete OA, Levroney EL, Aguilar HC, et al. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 2005; 436(7049):401–405. [DOI] [PubMed] [Google Scholar]
- 68.Negrete OA, Wolf MC, Aguilar HC, et al. Two Key Residues in EphrinB3 Are Critical for Its Use as an Alternative Receptor for Nipah Virus. PLoS Pathog 2006; 2(2):e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bishop KA, Stantchev TS, Hickey AC, et al. Identification of Hendra virus G glycoprotein residues that are critical for receptor binding. J Virol 2007; 81(11):5893–5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bossart KN, McEachern JA, Hickey AC, et al. Neutralization assays for differential henipavirus serology using Bio-Plex Protein Array Systems. J Virol Methods 2007; 142(1–2):29–40. [DOI] [PubMed] [Google Scholar]
- 71.Bowden TA, Crispin M, Harvey DJ, et al. Dimeric architecture of the Hendra virus attachment glycoprotein: evidence for a conserved mode of assembly. J Virol 2010; 84(12):6208–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bowden TA, Crispin M, Harvey DJ, et al. Crystal structure and carbohydrate analysis of Nipah virus attachment glycoprotein: a template for antiviral and vaccine design. J Virol 2008; 82(23):11628–11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu K, Chan YP, Rajashankar KR, et al. New insights into the hendra virus attachment and entry process from structures of the virus g glycoprotein and its complex with ephrin-b2. PLoS One 2012; 7(11):e48742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Collins PL, and Crowe J, J. E. Respiratory Syncytial Virus and Metapneumovirus. In Knipe DM and Howley PM ed., Fields Virology, 5 ed, Philadelphia. Lippincott Williams & Wilkins, 2007:1601–1646. [Google Scholar]
- 75.Harris J, and Werling D. Binding and entry of respiratory syncytial virus into host cells and initiation of the innate immune response. Cell Microbiol 2003; 5(10):671–680. [DOI] [PubMed] [Google Scholar]
- 76.van den Hoogen BG, Bestebroer TM, Osterhaus AD, et al. Analysis of the genomic sequence of a human metapneumovirus. Virology 2002; 295(1):119–132. [DOI] [PubMed] [Google Scholar]
- 77.Teng MN, Whitehead SS, and Collins PL. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 2001; 289(2):283–296. [DOI] [PubMed] [Google Scholar]
- 78.Karron RA, Buonagurio DA, Georgiu AF, et al. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci U S A 1997; 94(25):13961–13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karron RA, Thumar B, Schappell E, et al. Evaluation of two chimeric bovine-human parainfluenza virus type 3 vaccines in infants and young children. Vaccine 2012; 30(26):3975–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karger A, Schmidt U, and Buchholz UJ. Recombinant bovine respiratory syncytial virus with deletions of the G or SH genes: G and F proteins bind heparin. J Gen Virol 2001; 82(Pt 3):631–640. [DOI] [PubMed] [Google Scholar]
- 81.Techaarpornkul S, Barretto N, and Peeples ME. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J Virol 2001; 75(15):6825–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Teng MN, and Collins PL. Identification of the respiratory syncytial virus proteins required for formation and passage of helper-dependent infectious particles. J Virol 1998; 72(7):5707–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Feldman SA, Hendry RM, and Beeler JA. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J Virol 1999; 73(8):6610–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cantin C, Holguera J, Ferreira L, et al. Newcastle disease virus may enter cells by caveolae-mediated endocytosis. J Gen Virol 2007; 88(Pt 2):559–569. [DOI] [PubMed] [Google Scholar]
- 85.Kolokoltsov AA, Deniger D, Fleming EH, et al. Small interfering RNA profiling reveals key role of clathrin-mediated endocytosis and early endosome formation for infection by respiratory syncytial virus. J Virol 2007; 81(14):7786–7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Biacchesi S, Skiadopoulos MH, Yang L, et al. Recombinant human Metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: deletion of G yields a promising vaccine candidate. J Virol 2004; 78(23):12877–12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Biacchesi S, Pham QN, Skiadopoulos MH, et al. Infection of nonhuman primates with recombinant human metapneumovirus lacking the SH, G, or M2–2 protein categorizes each as a nonessential accessory protein and identifies vaccine candidates. J Virol 2005; 79(19):12608–12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chang A, Masante C, Buchholz UJ, et al. Human metapneumovirus (HMPV) binding and infection are mediated by interactions between the HMPV fusion protein and heparan sulfate. J Virol 2012; 86(6):3230–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cseke G, Maginnis MS, Cox RG, et al. Integrin alphavbeta1 promotes infection by human metapneumovirus. Proc Natl Acad Sci U S A 2009; 106(5):1566–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schowalter RM, Smith SE, and Dutch RE. Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. J Virol 2006; 80(22):10931–10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schowalter RM, Chang A, Robach JG, et al. Low-pH triggering of human metapneumovirus fusion: essential residues and importance in entry. J Virol 2009; 83(3):1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jack PJM, Boyle DB, Eaton BT, et al. The complete genome sequence of J-virus reveals a unique genome structure in the family Paramyxoviridae. J Virol 2005; In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jack PJ, Anderson DE, Bossart KN, et al. Expression of novel genes encoded by the paramyxovirus J virus. J Gen Virol 2008; 89(Pt 6):1434–1441. [DOI] [PubMed] [Google Scholar]
- 94.Paterson RG, Johnson ML, and Lamb RA. Paramyxovirus fusion (F) protein and hemagglutinin-neuraminidase (HN) protein interactions: intracellular retention of F and HN does not affect transport of the homotypic HN or F protein. Virology 1997; 237(1):1–9. [DOI] [PubMed] [Google Scholar]
- 95.Chan DC, Fass D, Berger JM, et al. Core structure of gp41 from the HIV envelope glycoprotein. Cell 1997; 89(2):263–273. [DOI] [PubMed] [Google Scholar]
- 96.Fass D, Harrison SC, and Kim PS. Retrovirus envelope domain at 1.7 angstrom resolution. Nat Struct Biol 1996; 3(5):465–469. [DOI] [PubMed] [Google Scholar]
- 97.Wilson IA, Skehel JJ, and Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 1981; 289(5796):366–373. [DOI] [PubMed] [Google Scholar]
- 98.Chen L, Colman PM, Cosgrove LJ, et al. Cloning, expression, and crystallization of the fusion protein of Newcastle disease virus. Virology 2001; 290(2):290–299. [DOI] [PubMed] [Google Scholar]
- 99.Yin HS, Paterson RG, Wen X, et al. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci U S A 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yin HS, Wen X, Paterson RG, et al. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 2006; 439(7072):38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cao J, Bergeron L, Helseth E, et al. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol 1993; 67:2747–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hernandez LD, Hoffman LR, Wolfsberg TG, et al. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol 1996; 12:627–661. [DOI] [PubMed] [Google Scholar]
- 103.Lescar J, Roussel A, Wien MW, et al. The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 2001; 105(1):137–148. [DOI] [PubMed] [Google Scholar]
- 104.White JM, Delos SE, Brecher M, et al. Structures and Mechanisms of Viral Membrane Fusion Proteins: Multiple Variations on a Common Theme. Crit Rev Biochem Mol Biol 2008; 43(3):189–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Scheid A, and Choppin PW. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity of proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology 1974; 57(2):475–490. [DOI] [PubMed] [Google Scholar]
- 106.Klenk HD, and Garten W. Host cell proteases controlling virus pathogenicity. Trends Microbiol 1994; 2(2):39–43. [DOI] [PubMed] [Google Scholar]
- 107.Gotoh B, Yamauchi F, Ogasawara T, et al. Isolation of factor Xa from chick embryo as the amniotic endoprotease responsible for paramyxovirus activation. FEBS Lett 1992; 296(3):274–278. [DOI] [PubMed] [Google Scholar]
- 108.Nagai Y, and Klenk HD. Activation of precursors to both glycoporteins of Newcastle disease virus by proteolytic cleavage. Virology 1977; 77(1):125–134. [DOI] [PubMed] [Google Scholar]
- 109.Pager CT, and Dutch RE. Cathepsin L is involved in proteolytic processing of the hendra virus fusion protein. J Virol 2005; 79(20):12714–12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pager CT, Craft WW Jr., Patch J, et al. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Baker KA, Dutch RE, Lamb RA, et al. Structural basis for paramyxovirus-mediated membrane fusion. Mol Cell 1999; 3(3):309–319. [DOI] [PubMed] [Google Scholar]
- 112.Hunter E Viral entry and receptors. In Coffin SH, Hughes SH, and Varmus HE ed., Retroviruses, New York. Cold Spring Harbor Laboratory Press, 1997:71–119. [PubMed] [Google Scholar]
- 113.Novick SL, and Hoekstra D. Membrane penetration of Sendai virus glycoproteins during the early stages of fusion with liposomes as determined by hydrophobic photoaffinity labeling. Proc Natl Acad Sci U S A 1988; 85(20):7433–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dutch RE, Jardetzky TS, and Lamb RA. Virus membrane fusion proteins: biological machines that undergo a metamorphosis. Biosci Rep 2000; 20(6):597–612. [DOI] [PubMed] [Google Scholar]
- 115.Singh M, Berger B, and Kim PS. LearnCoil-VMF: computational evidence for coiled-coil-like motifs in many viral membrane-fusion proteins. J Mol Biol 1999; 290(5):1031–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hughson FM. Enveloped viruses: a common mode of membrane fusion? Curr Biol 1997; 7(9):R565–569. [DOI] [PubMed] [Google Scholar]
- 117.Plemper RK, and Compans RW. Mutations in the putative HR-C region of the measles virus F2 glycoprotein modulate syncytium formation. J Virol 2003; 77(7):4181–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wild CT, Shugars DC, Greenwell TK, et al. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci U S A 1994; 91(21):9770–9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jiang S, Lin K, Strick N, et al. HIV-1 inhibition by a peptide. Nature 1993; 365:113. [DOI] [PubMed] [Google Scholar]
- 120.Lambert DM, Barney S, Lambert AL, et al. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci U S A 1996; 93(5):2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Joshi SB, Dutch RE, and Lamb RA. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology 1998; 248(1):20–34. [DOI] [PubMed] [Google Scholar]
- 122.Wild TF, and Buckland R. Inhibition of measles virus infection and fusion with peptides corresponding to the leucine zipper region of the fusion protein. J Gen Virol 1997; 78(Pt 1):107–111. [DOI] [PubMed] [Google Scholar]
- 123.Young JK, Li D, Abramowitz MC, et al. Interaction of peptides with sequences from the Newcastle disease virus fusion protein heptad repeat regions. J Virol 1999; 73(7):5945–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Young JK, Hicks RP, Wright GE, et al. Analysis of a peptide inhibitor of paramyxovirus (NDV) fusion using biological assays, NMR, and molecular modeling. Virology 1997; 238(2):291–304. [DOI] [PubMed] [Google Scholar]
- 125.Rapaport D, Ovadia M, and Shai Y. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. Embo J 1995; 14(22):5524–5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bossart KN, Mungall BA, Crameri G, et al. Inhibition of Henipavirus fusion and infection by heptad-derived peptides of the Nipah virus fusion glycoprotein. Virol J 2005; 2:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dutch RE, Hagglund RN, Nagel MA, et al. Paramyxovirus fusion (F) protein: a conformational change on cleavage activation. Virology 2001; 281(1):138–150. [DOI] [PubMed] [Google Scholar]
- 128.Zhao X, Singh M, Malashkevich VN, et al. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc Natl Acad Sci U S A 2000; 97(26):14172–14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhu J, Zhang CW, Qi Y, et al. The fusion protein core of measles virus forms stable coiled-coil trimer. Biochem Biophys Res Commun 2002; 299(5):897–902. [DOI] [PubMed] [Google Scholar]
- 130.Liu Y, Xu Y, Zhu J, et al. Crystallization and preliminary X-ray diffraction analysis of central structure domains from mumps virus F protein. Acta Crystallograph Sect F Struct Biol Cryst Commun 2005; 61(Pt 9):855–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xu Y, Lou Z, Liu Y, et al. Crystallization and preliminary crystallographic analysis of the fusion core from two new zoonotic paramyxoviruses, Nipah virus and Hendra virus. Acta Crystallogr D Biol Crystallogr 2004; 60(Pt 6):1161–1164. [DOI] [PubMed] [Google Scholar]
- 132.Chen L, Gorman JJ, McKimm-Breschkin J, et al. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure (Camb) 2001; 9(3):255–266. [DOI] [PubMed] [Google Scholar]
- 133.Lamb RA, Paterson RG, and Jardetzky TS. Paramyxovirus membrane fusion: Lessons from the F and HN atomic structures. Virology 2006; 344(1):30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Russell CJ, Kantor KL, Jardetzky TS, et al. A dual-functional paramyxovirus F protein regulatory switch segment: activation and membrane fusion. J Cell Biol 2003; 163(2):363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Harbury PB, Kim PS, and Alber T. Crystal structure of an isoleucine-zipper trimer. Nature 1994; 371(6492):80–83. [DOI] [PubMed] [Google Scholar]
- 136.Welch BD, Liu Y, Kors CA, et al. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc Natl Acad Sci U S A 2012; 109(41):16672–16677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chan YP, Lu M, Dutta S, et al. Biochemical, conformational, and immunogenic analysis of soluble trimeric forms of henipavirus fusion glycoproteins. J Virol 2012; 86(21):11457–11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Russell CJ, Jardetzky TS, and Lamb RA. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. Embo J 2001; 20(15):4024–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Steinhauer DA, and Plemper RK. Structure of the primed paramyxovirus fusion protein. Proc Natl Acad Sci U S A 2012; 109(41):16404–16405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Aguilar HC, Matreyek KA, Choi DY, et al. Polybasic KKR motif in the cytoplasmic tail of Nipah virus fusion protein modulates membrane fusion by inside-out signaling. J Virol 2007; 81(9):4520–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Paterson RG, Russell CJ, and Lamb RA. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 2000; 270(1):17–30. [DOI] [PubMed] [Google Scholar]
- 142.Waning DL, Russell CJ, Jardetzky TS, et al. Activation of a paramyxovirus fusion protein is modulated by inside-out signaling from the cytoplasmic tail. Proc Natl Acad Sci U S A 2004; 101(25):9217–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Smith EC, Culler MR, Hellman LM, et al. Beyond anchoring: the expanding role of the hendra virus fusion protein transmembrane domain in protein folding, stability, and function. J Virol 2012; 86(6):3003–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Skehel JJ, and Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 2000; 69:531–569. [DOI] [PubMed] [Google Scholar]
- 145.Hu XL, Ray R, and Compans RW. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses [published erratum appears in J Virol 1992 Aug;66(8):5176]. J Virol 1992; 66(3):1528–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bagai S, and Lamb RA. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between simian virus 5 and human parainfluenza virus 3 or Newcastle disease virus. J Virol 1995; 69(11):6712–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bousse T, Takimoto T, Gorman WL, et al. Regions on the hemagglutinin-neuraminidase proteins of human parainfluenza virus type-1 and Sendai virus important for membrane fusion. Virology 1994; 204(2):506–514. [DOI] [PubMed] [Google Scholar]
- 148.Deng R, Wang Z, Mirza AM, et al. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology 1995; 209(2):457–469. [DOI] [PubMed] [Google Scholar]
- 149.Tanabayashi K, and Compans RW. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J Virol 1996; 70(9):6112–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tsurudome M, Kawano M, Yuasa T, et al. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology 1995; 213(1):190–203. [DOI] [PubMed] [Google Scholar]
- 151.Melanson VR, and Iorio RM. Amino acid substitutions in the F-specific domain in the stalk of the newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J Virol 2004; 78(23):13053–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Melanson VR, and Iorio RM. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J Virol 2006; 80(2):623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lamb RA. Paramyxovirus fusion: A hypothesis for changes. Virology 1993; 197:1–11. [DOI] [PubMed] [Google Scholar]
- 154.McGinnes LW, Gravel K, and Morrison TG. Newcastle disease virus HN protein alters the conformation of the F protein at cell surfaces. J Virol 2002; 76(24):12622–12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Morrison T, McQuain C, and McGinnes L. Complementation between avirulent Newcastle disease virus and a fusion protein gene expressed from a retrovirus vector: requirements for membrane fusion. J Virol 1991; 65(2):813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Connolly SA, Leser GP, Jardetzky TS, et al. Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion: implications for the mechanism of fusion triggering. J Virol 2009; 83(21):10857–10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yao Q, Hu X, and Compans RW. Association of the parainfluenza virus fusion and hemagglutinin-neuraminidase glycoproteins on cell surfaces. J Virol 1997; 71(1):650–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.McGinnes LW, and Morrison TG. Inhibition of Receptor Binding Stabilizes Newcastle Disease Virus HN and F Protein-Containing Complexes. J Virol 2006; 80(6):2894–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li J, Quinlan E, Mirza A, et al. Mutated form of the Newcastle disease virus hemagglutinin-neuraminidase interacts with the homologous fusion protein despite deficiencies in both receptor recognition and fusion promotion. J Virol 2004; 78(10):5299–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Deng R, Wang Z, Mahon PJ, et al. Mutations in the Newcastle disease virus hemagglutinin-neuraminidase protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology 1999; 253(1):43–54. [DOI] [PubMed] [Google Scholar]
- 161.Plemper RK, Hammond AL, and Cattaneo R. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J Biol Chem 2001; 276(47):44239–44246. [DOI] [PubMed] [Google Scholar]
- 162.Plemper RK, Hammond AL, Gerlier D, et al. Strength of envelope protein interaction modulates cytopathicity of measles virus. J Virol 2002; 76(10):5051–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Corey EA, and Iorio RM. Mutations in the stalk of the measles virus hemagglutinin protein decrease fusion but do not interfere with virus-specific interaction with the homologous fusion protein. J Virol 2007; 81(18):9900–9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Corey EA, and Iorio RM. Measles virus attachment proteins with impaired ability to bind CD46 interact more efficiently with the homologous fusion protein. Virology 2009; 383(1):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Aguilar HC, Matreyek KA, Filone CM, et al. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol 2006; 80(10):4878–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Aguilar HC, Ataman ZA, Aspericueta V, et al. A novel receptor-induced activation site in the Nipah virus attachment glycoprotein (G) involved in triggering the fusion glycoprotein (F). J Biol Chem 2009; 284(3):1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Whitman SD, Smith EC, and Dutch RE. Differential rates of protein folding and cellular trafficking for the Hendra virus F and G proteins: implications for F-G complex formation. J Virol 2009; 83(17):8998–9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stone-Hulslander J, and Morrison TG. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J Virol 1997; 71(9):6287–6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tong S, and Compans RW. Alternative mechanisms of interaction between homotypic and heterotypic parainfluenza virus HN and F proteins. J Gen Virol 1999; 80(Pt 1):107–115. [DOI] [PubMed] [Google Scholar]
- 170.Whitman SD, and Dutch RE. Surface density of the Hendra G protein modulates Hendra F protein-promoted membrane fusion: role for Hendra G protein trafficking and degradation. Virology 2007; 363(2):419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lee B, and Ataman ZA. Modes of paramyxovirus fusion: a Henipavirus perspective. Trends Microbiol 2011; 19(8):389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chang A, and Dutch RE. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses 2012; 4(4):613–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhu Q, Biering SB, Mirza AM, et al. Individual N-glycans added at intervals along the stalk of the Nipah virus G protein prevent fusion, but do not block the interaction with the homologous F protein. J Virol 2013; Jan 2. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Paal T, Brindley MA, St Clair C, et al. Probing the spatial organization of measles virus fusion complexes. J Virol 2009; 83(20):10480–10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Brindley MA, Takeda M, Plattet P, et al. Triggering the measles virus membrane fusion machinery. Proc Natl Acad Sci U S A 2012; 109(44):E3018–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ader N, Brindley M, Avila M, et al. Mechanism for active membrane fusion triggering by morbillivirus attachment protein. J Virol 2013; 87(1):314–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Porotto M, Devito I, Palmer SG, et al. Spring-loaded model revisited: paramyxovirus fusion requires engagement of a receptor binding protein beyond initial triggering of the fusion protein. J Virol 2011; 85(24):12867–12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Porotto M, Palmer SG, Palermo LM, et al. Mechanism of fusion triggering by human parainfluenza virus type III: communication between viral glycoproteins during entry. J Biol Chem 2012; 287(1):778–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sergel T, McGinnes LW, Peeples ME, et al. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology 1993; 193(2):717–726. [DOI] [PubMed] [Google Scholar]
- 180.Takimoto T, Taylor GL, Crennell SJ, et al. Crystallization of Newcastle disease virus hemagglutinin-neuraminidase glycoprotein. Virology 2000; 270(1):208–214. [DOI] [PubMed] [Google Scholar]
- 181.Takimoto T, Taylor GL, Connaris HC, et al. Role of the hemagglutinin-neuraminidase protein in the mechanism of paramyxovirus-cell membrane fusion. J Virol 2002; 76(24):13028–13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mishin VP, Watanabe M, Taylor G, et al. N-linked glycan at residue 523 of human parainfluenza virus type 3 hemagglutinin-neuraminidase masks a second receptor-binding site. J Virol 2010; 84(6):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bousse T, and Takimoto T. Mutation at residue 523 creates a second receptor binding site on human parainfluenza virus type 1 hemagglutinin-neuraminidase protein. J Virol 2006; 80(18):9009–9016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Porotto M, Fornabaio M, Kellogg GE, et al. A second receptor binding site on human parainfluenza virus type 3 hemagglutinin-neuraminidase contributes to activation of the fusion mechanism. J Virol 2007; 81(7):3216–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Alymova IV, Portner A, Mishin VP, et al. Receptor-binding specificity of the human parainfluenza virus type 1 hemagglutinin-neuraminidase glycoprotein. Glycobiology 2012; 22(2):174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bousse TL, Taylor G, Krishnamurthy S, et al. Biological significance of the second receptor binding site of Newcastle disease virus hemagglutinin-neuraminidase protein. J Virol 2004; 78(23):13351–13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mahon PJ, Mirza AM, and Iorio RM. Role of the two sialic acid binding sites on the newcastle disease virus HN protein in triggering the interaction with the F protein required for the promotion of fusion. J Virol 2011; 85(22):12079–12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Porotto M, Salah Z, DeVito I, et al. The second receptor binding site of the globular head of the Newcastle disease virus hemagglutinin-neuraminidase activates the stalk of multiple paramyxovirus receptor binding proteins to trigger fusion. J Virol 2012; 86(10):5730–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Mahon PJ, Mirza AM, Musich TA, et al. Engineered intermonomeric disulfide bonds in the globular domain of Newcastle disease virus hemagglutinin-neuraminidase protein: implications for the mechanism of fusion promotion. J Virol 2008; 82(21):10386–10396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Navaratnarajah CK, Oezguen N, Rupp L, et al. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat Struct Mol Biol 2011; 18(2):128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lee JK, Prussia A, Paal T, et al. Functional interaction between paramyxovirus fusion and attachment proteins. J Biol Chem 2008; 283(24):16561–16572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Plemper RK, Brindley MA, and Iorio RM. Structural and mechanistic studies of measles virus illuminate paramyxovirus entry. PLoS Pathog 2011; 7(6):e1002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ader N, Brindley MA, Avila M, et al. Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger F protein refolding for membrane fusion. J Biol Chem 2012; 287(20):16324–16334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Navaratnarajah CK, Negi S, Braun W, et al. Membrane fusion triggering: three modules with different structure and function in the upper half of the measles virus attachment protein stalk. J Biol Chem 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Apte-Sengupta S, Negi S, Leonard VH, et al. Base of the measles virus fusion trimer head receives the signal that triggers membrane fusion. J Biol Chem 2012; 287(39):33026–33035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Bose S, Zokarkar A, Welch BD, et al. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc Natl Acad Sci U S A 2012; 109(39):E2625–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bishop KA, Hickey AC, Khetawat D, et al. Residues in the stalk domain of the hendra virus g glycoprotein modulate conformational changes associated with receptor binding. J Virol 2008; 82(22):11398–11409. [DOI] [PMC free article] [PubMed] [Google Scholar]


