Abstract
Two Salmonella enterica serovar Typhimurium strains from different clonal origins, both producing an extended-spectrum β-lactamase (TEM-52), were isolated from a patient. This enzyme was encoded on a single plasmid and was found at very low levels in one strain, while being encoded on multiple plasmids and in multiple different EcoRI fragments in the other strain.
Class A extended-spectrum β-lactamases (ESBLs) are mostly distributed in hospitals among Klebsiella and Escherichia coli strains (7). ESBLs are extremely rare among Salmonella strains, and so detection of these resistance genes among salmonellae is of significance. Recently, Salmonella strains having positive double-disk synergy tests were isolated from a 1-year-old patient who had been transferred from a Yugoslavian hospital to Hungary for cardiac surgery. Strains were obtained from stool and tracheal aspirate and were isolated multiple times.
Because ESBLs are rare in Salmonella, these strains attracted our interest. Upon obtaining the isolates, we detected two resistance phenotypes. One of them (MS-II) was susceptible to the expanded-spectrum cephalosporins, and the other (MS-I) was highly resistant. However, both displayed an apparent synergy between clavulanate and expanded-spectrum cephalosporins, indicating that they were producing ESBLs. Isolation of two phenotypically different ESBL-producing salmonellae from one patient attracted more interest. Here, we characterize the relatedness of the Salmonella isolates and the resistance genes for different resistance phenotypes.
The 1-year-old patient was a Yugoslavian citizen with multiple congenital heart defects. He had had a surgical intervention in 1997 in Yugoslavia, and in 1998, he was sent to the National Institute of Cardiology in Budapest, Hungary. The patient was carrying salmonellae in the gut at the admission. Despite antibiotic therapy, salmonellae were isolated twice from stool and once from tracheal aspirate of the patient. The pathogen was successfully eradicated by combination therapy with imipenem and amikacin subsequent to the surgical intervention.
MICs of antibiotics were determined according to the recommendations of the National Committee for Clinical Laboratory Standards (3). Powder forms of antibiotics were obtained from the following sources: ampicillin, chloramphenicol, and clavulanate, Deva, Kocaeli, Turkey; ceftazidime, Glaxo Wellcome; cefepime and aztreonam, Bristol-Myers Squibb; and ciprofloxacin, Bayer.
Isoelectric focusing was performed as described in detail elsewhere (9). Extracts were run on acrylamide gels supplemented with ampholines in pI ranges 3 to 10 and 5 to 8.
Probes for ribotyping and for blaTEM detection were prepared with a digoxigenin labeling kit (Boehringer Mannheim, Mannheim, Germany). For ribotyping, 16S-23S rRNA was from E. coli, while the TEM probe was a PCR product (861 bp) obtained with the primers TEM-A (5′-ATG AGT ATT CAA CAT TTC CGT G-3′) and TEM-D (5′-TTA CCA ATG CTT AAT CAG TGA G-3′). Detection of these digoxigenin-labeled probes was achieved with a digoxigenin detection kit (Boehringer Mannheim) according to the instructions of the manufacturer.
Extraction and purification of total bacterial DNA, isolation of plasmids, and the Southern blot assay have been described previously (1). The restriction digestions of total DNAs were accomplished in a 30-μl volume with 5 U of EcoRI or PvuII for 4 h at 37°C. Digested DNAs were later run on a 0.9% agarose gel at a constant voltage overnight. Digested fragments were transferred to a positively charged nylon membrane (Boehringer Mannheim) by the capillary transfer method. Hybridizations were achieved at 55°C by overnight incubation. Detection was carried out subsequent to high-stringency washes as described earlier (1).
Plasmid DNAs were isolated by an alkali lysis method. The sizes of the plasmids were estimated on a logarithmic scale relative to the migration of the known plasmids from NCTC 50192 (14). Transconjugation trials were carried out at 37°C with J-53 2 (rifampin-resistant E. coli strain) as the recipient (8). Mating experiments were tried on membranes placed on Mueller-Hinton agars, while selection of the recipients was tried on Mueller-Hinton agars supplemented with 100 mg of rifampin per liter combined with either 2 mg of ceftazidime or 8 mg of ampicillin per liter.
The sequence analyses of TEM genes were performed with PCR products of the primers TEM-A and TEM-D. PCR was accomplished with 40 cycles of 1 min of annealing at 55°C, 2 min of extension at 72°C, and 1 min of denaturation at 95°C. After gel purification of the products, the sequencing was performed with the same primers. The method was dye terminator cycle sequencing with the ABI Prism BigDye Terminator kit (Applied Biosystems, Foster City, Calif.). The assay was carried out according to the standard protocol. Data were collected on an ABI 377 automated fluorescence sequencer.
Both isolates were identified as Salmonella enterica serovar Typhimurium. The 16S-23S ribosomal patterns obtained by the digestion of PvuII enzyme were almost identical. However, the restriction patterns obtained with EcoRI digestion were clearly dissimilar, as seen in Fig. 1. This result indicates that these strains, despite having the same antigenic type, differ in clonal origin. The dissimilarity was also apparent in resistance phenotypes to β-lactam antibiotics (Table 1).
FIG. 1.
Restriction fragment patterns obtained by ribosomal DNA probe with EcoRI (A) and PvuII (B) digestions. Lanes: M, lambda DNA HindIII marker; 1, restriction patterns of MS-I; 2, restriction patterns of MS-II.
TABLE 1.
MICs of antibiotics with increasing concentrations of TEM-52-producing Salmonella inoculum
| Antibiotic(s) | MIC (mg/liter) at inoculum:
|
|||||
|---|---|---|---|---|---|---|
| 104 CFU/μl
|
105 CFU/μl
|
105 CFU/μl
|
||||
| MS-I | MS-II | MS-I | MS-II | MS-I | MS-II | |
| Ampicillin | >512 | >512 | >512 | >512 | >512 | >512 |
| Ampicillin-clavulanatea | 64 | 128 | 512 | 512 | 512 | >512 |
| Ceftazidime | 128 | 16 | 256 | 16 | 512 | 32 |
| Ceftazidime-clavulanate | 2 | <1 | 4 | 2 | 4 | 4 |
| Cefepime | 32 | 8 | 32 | 8 | 512 | 32 |
| Cefepime-clavulanate | <1 | <1 | 2 | 2 | 16 | 16 |
| Aztreonam | 128 | 8 | 128 | 16 | 512 | 16 |
| Aztreonam-clavulanate | <1 | <1 | <1 | <1 | <1 | <1 |
| Ciprofloxacin | <0.125 | <0.125 | 0.125 | <0.125 | 0.125 | <0.125 |
| Chloramphenicol | >512 | >512 | >512 | >512 | >512 | >512 |
Clavulanate was supplemented at a fixed proportion of 4 mg/liter.
Both isolates produce an enzyme that is focused at pI 6.0. The enzyme from MS-I was more intense and rapid than that of the enzyme from MS-II, which was very weak and appeared very late on the gel. This was a consequence of the small quantity of the enzyme in MS-II.
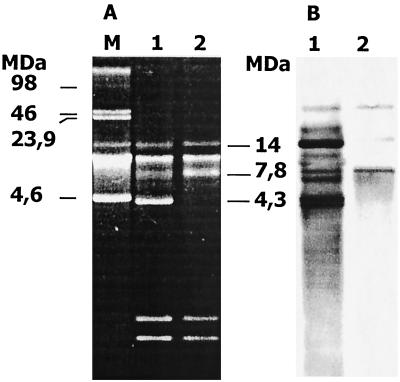
The plasmid profiles of the strains were similar, with the exception of the 4.3-MDa plasmid (Fig. 2A). This plasmid was highly intense with ethidium bromide staining, indicating a high copy number, which was further supported by a strong signal with the TEM probe (Fig. 2B). Hybridization signals with the TEM probe were distinct in either localization or intensity with the plasmids of the strains. The EcoRI restriction fragments when hybridized with TEM probe displayed a single band in MS-II, while three bands were seen in MS-I (Fig. 3). These all indicated the existence of blaTEM in MS-I with high copy number. Trasnsconjugation experiments failed to transfer this enzyme to the recipient.
FIG. 2.
Plasmid profiles of MS-I and MS-II (A) and hybridization with TEM probe of the membrane transfer of this gel (B). Lane M, plasmids with known length from NCTC 50192. In panel A, lanes 1 and 2 represent plasmids from MS-I and MS-II, respectively; lanes 1 and 2 in panel B are the corresponding lanes after hybridization with the TEM probe of the membrane transfer from the same gel.
FIG. 3.
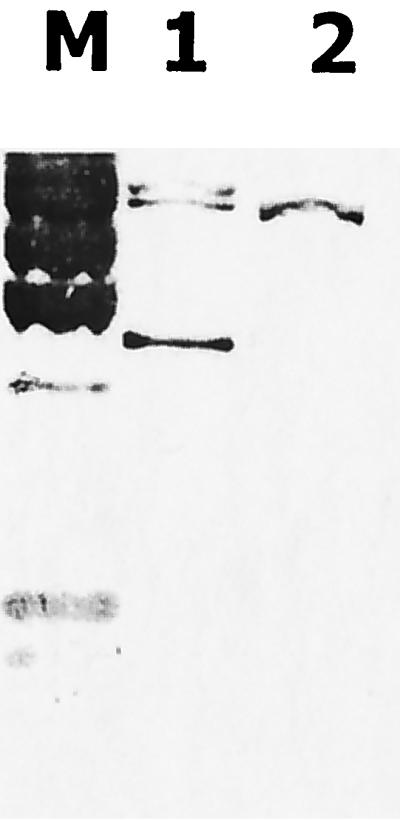
Hybridization patterns with the TEM probe after EcoRI digestion of total DNAs. Lanes: M, lambda DNA HindIII digest; 1, EcoRI fragments from MS-I; 2, EcoRI fragments from MS-II.
The sequence analysis of blaTEM showed that both strains carry exactly the same TEM gene. This gene differed from blaTEM-1 by five bases. Three of these mutations caused the following amino acid substitutions: Glu 104 to Lys, Met 182 to Thr, and Gly 238 to Ser (Ambler numbering). The gene carrying these substitutions in the TEM-1 gene was named previously TEM-52 (5). The isoelectric focusing data (enzyme with pI of 6.0) further confirmed the presence of an active TEM-52 gene in both isolates. Other base differences were silent and located at the positions Gly 78 (GGC to GGT) and Ala 134 (GCG to GCT).
Ribotypes of Salmonella enterica serovar Typhimurium strains differ by multiple fragments with EcoRI digestion. This piece of data provides reliable evidence that MS-I and MS-II differ in clonal origin. The carriage of different Salmonella clones at the same time is highly interesting and is not a well-known condition. However, if the resistance phenotypes were not apparently different, we would have failed to notice this.
Depending on the exact similarity of the silent mutations in the sequences of TEM-52 genes, one can speculate that both isolates might have acquired the gene from a common source. The patient was a carrier for these strains at the time of admission. Therefore, Yugoslavia must be the country of origin of these pathogens.
Another interesting question was how TEM-52 disseminated among these clones. Apart from clonal spread, ESBLs often disseminate by the transmission of plasmids between bacteria (2, 10). ESBL genes are typically found on transposons (4, 6; I. Casin, B. Hanau, P. Mugnier, H. Vahaboglu, and E. Collatz, Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-122, p. 67, 1997). Transposons cannot replicate themselves, and thus they depend on self-replicating genetic elements, such as plasmids, which also serve as their vehicles of transmission. However, transposons can also jump between plasmids and between chromosomes. The data obtained with the strains of this study do not support plasmid dissemination for TEM-52: first, the 4.3-MDa plasmid is not present in MS-II; second, the intensities of the hybridization signals of the MS-I plasmids were distinct from those of MS-II; and, third, the EcoRI fragments carrying blaTEM-52 in MS-I are multiple and so differ at least in number from the single fragment of MS-II. The unsuccessful transconjugation experiments between the two clones indicate that these plasmids are not readily transmissible, and so this might be a further argument against a hypothesis of plasmid dissemination.
Accordingly, we believe that our findings support the view that this TEM-52 gene was not spreading via plasmids but behaved as a jumping gene moving between various genetic elements. If this hypothesis is correct, then TEM-52 must be located on a highly mobile transposon. However, the investigation of this issue was beyond the scope of the study.
This study has demonstrated that TEM-52, which had never been reported in Europe before, is being disseminated at least among Salmonella enterica serovar Typhimurium strains. This fact and the jumping nature of the ESBL gene should keep us alert for signs of further dissemination.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 2.Liu P Y-F, Tung J-C, Ke S-C, Chen S-L. Molecular epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates in a district hospital in Taiwan. J Clin Microbiol. 1998;36:2759–2762. doi: 10.1128/jcm.36.9.2759-2762.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. Approved standard M7–A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 4.Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000;44:622–632. doi: 10.1128/aac.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poyart C, Mugnier P, Quesne G, Berche P, Trieu-Cuot P. A novel extended-spectrum TEM-type β-lactamase (TEM-52) associated with decreased susceptibility to moxalactam in Klebsiella pneumoniae. Antimicrob Agents Chemother. 1998;42:108–113. doi: 10.1128/aac.42.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preston K E, Radomski C C, Venezia R A. The cassettes and 3′ conserved segment of an integron from Klebsiella oxytoca plasmid pACM1. Plasmid. 1999;42:104–114. doi: 10.1006/plas.1999.1418. [DOI] [PubMed] [Google Scholar]
- 7.Sirot D L, Goldstein F W, Soussy C J, Courtieu A L, Husson M O, Lemozy J, Meyran M, Morel C, Perez R, Quentin Noury C, Reverdy M E, Scheftel J M, Rosembaum M, Rezvani Y. Resistance to cefotaxime and seven other β-lactams in members of the family Enterobacteriaceae: a 3-year survey in France. Antimicrob Agents Chemother. 1992;36:1677–1681. doi: 10.1128/aac.36.8.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vahaboglu H, Hall L M, Mulazimoglu L, Dodanli S, Yildirim I, Livermore D M. Resistance to extended-spectrum cephalosporins, caused by PER-1 beta-lactamase, in Salmonella typhimurium from Istanbul, Turkey. J Med Microbiol. 1995;43:294–299. doi: 10.1099/00222615-43-4-294. [DOI] [PubMed] [Google Scholar]
- 9.Vahaboglu H, Öztürk R, Aygün G, Coşkunkan F, Yaman A, Kaygusuz A, Leblebicioglu H, Balik İ, Aydin K, Otkun M. Widespread detection of PER-1-type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob Agents Chemother. 1997;41:2265–2269. doi: 10.1128/aac.41.10.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan J-J, Wu S-M, Tsai S-H, Wu J-J, Su I-J. Prevalence of SHV-12 among clinical isolates of Klebsiella pneumoniae producing extended-spectrum β-lactamases and identification of a novel AmpC enzyme (CMY-8) in Southern Taiwan. Antimicrob Agents Chemother. 2000;44:1438–1442. doi: 10.1128/aac.44.6.1438-1442.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]