Abstract
Owing to climate change impacts, waterlogging is a serious abiotic stress that affects crops, resulting in stunted growth and loss of productivity. Cassava (Manihot esculenta Grantz) is usually grown in areas that experience high amounts of rainfall; however, little research has been done on the waterlogging tolerance mechanism of this species. Therefore, we investigated the physiological responses of cassava plants to waterlogging stress and analyzed global gene transcription responses in the leaves and roots of waterlogged cassava plants. The results showed that waterlogging stress significantly decreased the leaf chlorophyll content, caused premature senescence, and increased the activities of superoxide dismutase (SOD), catalase (CAT) and peroxidase (POD) in the leaves and roots. In total, 2538 differentially expressed genes (DEGs) were detected in the leaves and 13364 in the roots, with 1523 genes shared between the two tissues. Comparative analysis revealed that the DEGs were related mainly to photosynthesis, amino metabolism, RNA transport and degradation. We also summarized the functions of the pathways that respond to waterlogging and are involved in photosynthesis, glycolysis and galactose metabolism. Additionally, many transcription factors (TFs), such as MYBs, AP2/ERFs, WRKYs and NACs, were identified, suggesting that they potentially function in the waterlogging response in cassava. The expression of 12 randomly selected genes evaluated via both quantitative real-time PCR (qRT-PCR) and RNA sequencing (RNA-seq) was highly correlated (R2 = 0.9077), validating the reliability of the RNA-seq results. The potential waterlogging stress-related transcripts identified in this study are representatives of candidate genes and molecular resources for further understanding the molecular mechanisms underlying the waterlogging response in cassava.
1. Introduction
Waterlogging, or soil flooding, is estimated to affect more than 17 million km2 of land area per year worldwide. Waterlogging events are expected to increase in frequency, severity, and unpredictability in the future because of global climate change [1]. It has been reported that the intensification of rainfall and evaporation in response to global warming will cause wet regions such as most tropical and subtropical zones to experience waterlogging [2]. Approximately 10% of irrigated farmlands suffer from frequent waterlogging, resulting in substantial yield losses (from 40 to 80%) [3, 4]. In China, losses in crop production due to flooding were second to those due to drought in 2013, accounting for more than RMB 300 billion Yuan [5]. This abiotic stress also causes problems for agricultural production in Australia, North America and Central Europe, especially in regions with heavy-textured soils [6].
The gas diffusion rate in water is much lower than that in air, which is a major determinant of the adverse effects of waterlogging. This low diffusion rate leads to reduced concentrations of oxygen in the root zone, limiting mitochondrial aerobic respiration, supplying energy for nutrient uptake and transport, and causing energy loss [7]. Waterlogging stress further decreases plant shoot metabolism, stomatal conductance, hydraulic conductance, transpiration, respiration and photosynthesis, which manifest as stunted growth and reduced biomass accumulation [8]. Obtaining sufficient knowledge about the mechanisms that drive waterlogging tolerance in plants to develop stress-tolerant crops and anticipate ecosystem changes is an enormous challenge for the plant science research community.
To cope with anaerobiosis due to waterlogging and to regulate different adaptive responses, plants modulate various transcriptional and metabolic changes [9–12]. A major plant response to soil waterlogging is the metabolic switch from aerobic respiration to anaerobic fermentation [13]. This switch involves metabolic adaptations such as induced expression of fermentation pathway enzymes, which leads to a rapid reduction in cellular adenosine triphosphate (ATP) levels. Plants have also developed a series of antioxidant mechanisms to defend themselves against oxidative stress. Antioxidant enzymes such as superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), and glutathione reductase (GR) have been reported to remove toxic oxygen substances and prevent or reduce cell damage in many plant species [14]. These reports strongly suggest that the regulation of the waterlogging response in plants involves more than a simple adaptation and is far more complex than anticipated for many years.
Cassava (Manihot esculenta Grantz) is considered an important cash and biofuel crop species in Asia, Latin America and Africa because of its starchy roots, making it critical for food security and economic development [15]. This species has been reported to be very drought tolerant; moreover, it can also use light and water resources efficiently and is tolerant to heat [16]. However, existing cassava cultivars that are injured by waterlogging can sometimes never fully recover, and there have been few studies on the adaptations of cassava to waterlogging stress. Globally, although cassava is grown across a wide range of environments, the majority of this species is cultivated in areas where the rainfall is more than 700 mm per year [17]; therefore, cassava is subjected to excess rainfall during the summer rainy season. RNA sequencing (RNA-Seq) has been used as an efficient approach to understanding transcriptome profiles [18]. To better understand the molecular mechanisms underlying the response of cassava to soil waterlogging, the transcriptional profiles of waterlogged cassava roots were analyzed via RNA-seq. Genes expressed in the control and treatment groups were compared to determine the species-specific responses of cassava and to identify genes or strategies associated with waterlogging resistance.
2. Materials and methods
2.1 Plant growth and waterlogging treatment
The cassava cultivar South China 6068 (SC6068), which is a common cultivar, was used in this study. Cassava plants were propagated clonally from cuttings of parental stems that had at least two nodes and were approximately 8 cm in length. The plants were grown in plastic pots (12 cm in height, 15 cm in diameter) containing a 2:1 mixture (V/V) of potting soil and vermiculite. One plant was grown in each pot. The plants were grown in a glasshouse under natural lighting and temperature (average daily temperature of 28–30°C) at Hainan University (20° 50’ N and 108° 38’ E). At 45 days after the cuttings were planted, those at the same developmental stage were waterlogged. Briefly, the plants were transplanted into plastic containers (62 cm × 36 cm × 42 cm) that were filled with water until the water level was approximately 3–4 cm above the soil surface. Waterlogging treatments were maintained for 6 days. The plants were separated into two groups. Three plants were randomly selected for each group, the leaves and roots of each plant were harvested for RNA extraction, and the remaining samples were used for physiological measurements. Four different types of samples were taken: leaves under waterlogged conditions (WL), roots under waterlogged conditions (WR), leaves under nonwaterlogged conditions (CL) and roots under nonwaterlogged conditions (CR). All the samples were immediately frozen in liquid nitrogen and stored at -80°C until use. All of the waterlogging treatments were performed with three independent biological replicates.
2.2 Measurements of chlorophyll content and maximum quantum efficiency of photosystem II (PSII) (Fv/Fm)
To estimate the relative chlorophyll content in the leaves of waterlogged plants, the chlorophyll contents were measured using a SPAD-502 Plus portable chlorophyll meter (Konica Minolta, Japan), which calculates a relative chlorophyll content value (SPAD) from the ratio of optical absorbance at 650 nm to that at 940 nm; major veins and areas of obvious visual damage were avoided. An average relative chlorophyll content was obtained for each leaf sample. The Fv/Fm values were measured using a handheld fluorometer (FluorPen FP100, Photon Systems Instruments, Czech Republic).
2.3 Measurements of enzyme activity
Colorimetric assays were used to measure the contents of peroxidase (POD), CAT and SOD (the kit was purchased from Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). A sample of leaves (0.1 g) without the midrib was thoroughly ground with a cold mortar and pestle in an ice bath. The grinding medium consisted of 1 mL of phosphate buffer plus homogenizing glass beads. The homogenate was centrifuged for 10 min at 8500 rpm and 4°C. The supernatant in turn constituted the crude enzyme extract and was used to determine enzyme activity. The absorbance of the reaction mixture was determined by using an Infinite M200 PRO instrument (Tecan, Switzerland).
2.4 Procedures of RNA-sequencing
Library construction, quality detection and Illumina sequencing were carried out by Beijing Biomarker Cloud Technology Co., Beijing, China (www.bmkcloud.com). In total, 1 μg of RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using a NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, USA) according to the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. The adaptor sequences and low-quality sequence reads were removed from the data sets. Raw sequences were transformed into clean reads after data processing. Clean data (clean reads) were obtained from the raw data by removing reads containing adapters, reads containing poly-N sequences and low-quality reads and then mapped to the reference genome sequence. Only reads with a perfect match or one mismatch were further analyzed and annotated based on the reference genome. Hisat2 software was used to map the reads to the reference (https://www.ncbi.nlm.nih.gov/assembly/GCA_013618965.1). The accession numbers of the transcriptome data was PRJNA699429, which were deposited in the NCBI Sequence Read Archive (SRA) database.
2.5 Gene annotation and pathway analysis
For annotations, all unigenes, which were proven to be longer than 200 bp, were subjected to a BLAST search (E-value < 1e-5) against the NCBI nonredundant (NR) protein database [19], manually annotated and reviewed protein sequence database Swiss-Prot [20], Gene Ontology (GO) [21], Clusters of Orthologous Groups of proteins (KOG/COG) [22, 23], Protein family (Pfam) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases [24, 25].
2.6 Differential expression analysis
Gene expression levels in each sample was analyzed by using FPKM (fragments per kilobase of exons model per million mapped reads) method [26]. Differential expression analyses of genes between two groups of comparison were performed using the DESeq2 package for FPKM data with biological replicates [27]. The p values were corrected using the Benjamini and Hochberg’s method to control the false discovery rate (FDR). Genes differentially expressed with at least 2-fold change (i.e., the absolute value of log2 Fold change ≥ 1.0) and a FDR corrected p-value of < 0.01 found by DESeq were considered as differentially expressed genes (DEGs). KOBAS software was used to test the statistical enrichment of differentially expressed genes in KEGG pathways [28]. The KEGG pathway with a FDR corrected p-value of < 0.05 were considered as enrichment in DEGs.
2.7 Quantitative real-time PCR (qRT-PCR) analysis
The expression patterns of twelve genes were analyzed via qRT-PCR. A pair of primers for each gene was designed on the basis of content from the NCBI database (https://www.ncbi.nlm.nih.gov/). The primer pairs are listed in S1 Table. Approximately 1 μg of isolated total RNA was used to generate cDNA using a reverse transcriptase kit (Thermo, USA). qRT-PCR was then performed using a 7500 Real Time PCR System with a total reaction volume of 20 μL, which consisted of 2 μL of cDNA template, 0.4 μL of forward and reverse primers (10 μM each), 10 μL of qPCR Master Mix, 0.4 μL of Rox and 6.8 μL of sterilized ddH2O. The PCR conditions were as follows: 95°C for 30 s; 40 cycles of 95°C for 5 s, 60°C for 34 s, and 95°C for 15 s; and 60°C for 1 min. The expression abundances of 12 genes were determined based on the ΔΔCT method described by Schmittgen and Livak [29], and relative changes in gene expression from the qRT-PCR experiments were calculated using elongation factor 1 alpha (EF1α) as a reference gene. EF1α was validated as one of two most stably expressed genes across different tissues and developmental stages in cassava [30]. Three biological replicates and three technical replicates of each group were assessed.
2.8 Statistical analysis
All of the experiments used for data comparisons were repeated three times. The statistics were analyzed via analysis of variance (ANOVA) followed by Duncan’s new multiple range test with SPSS version 20.0. The significance level was set to P < 0.05.
3. Results
3.1 Morphological and physiological changes in cassava in response to waterlogging stress
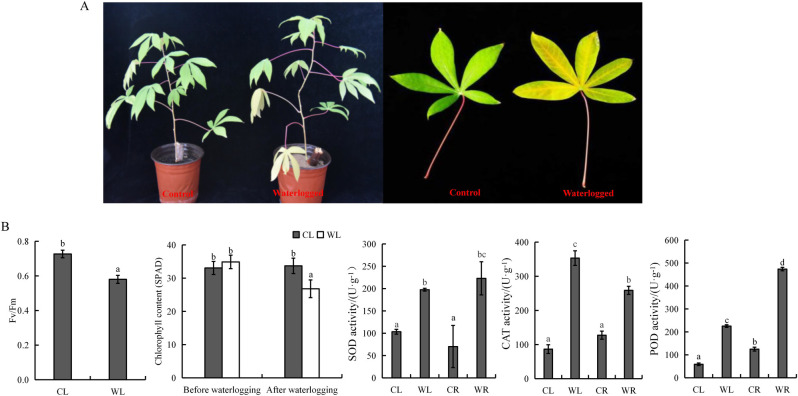
We compared the morphological changes in control and waterlogged plants after 6 days of waterlogging stress. No morphological changes were observed in the control plants, which had green leaves and upright stems. However, the leaves of the stem base of the waterlogged plants were withered and yellow, although the young leaves remained green (Fig 1A).
Fig 1. Morphological and physiological changes of cassava in response to waterlogging stress.
(A) Changes of phenotypes and stem base leaves in control and waterlogged seedlings. (B) Determination of photosynthetic indexes and antioxidant enzymes in the leaves and roots under waterlogging stress. The photosynthetic indexes included maximum quantum yield (Fv/Fm) and chlorophyll content (SPAD). The antioxidant enzymes are superoxide dismutase (SOD), catalase (CAT) and peroxidase (POD). Different letters indicate significant differences from the control (P < 0.05). Bars indicate standard error (n = 3).
To investigate the photosynthetic ability of cassava plants under waterlogging stress, the chlorophyll content and Fv/Fm were determined. The chlorophyll content decreased by 20% under the waterlogging treatment (Fig 1B). The Fv/Fm values of the waterlogged samples also significantly decreased compared with those of the CL samples. In the WL samples, the Fv/Fm ratio was 0.5–0.6, while in the CL samples, the Fv/Fm ratio was approximately 0.7 or higher (Fig 1B). The observed decrease in Fv/Fm values may be associated with the sensitivity of the photosynthetic apparatus to waterlogging stress.
The activities of SOD, POD and CAT were measured for the CL, WL, and CR (roots under nonwaterlogged conditions) samples and the WR (roots under waterlogged conditions) samples. The activity of three antioxidant enzymes increased in the WL and WR samples. The SOD activity in the WL and WR samples reached values that were 1.9-fold and 3.2-fold higher than those in the CL and CR samples, respectively (Fig 1B). The activity of CAT in the WL samples reached 353 U·g-1, which was more than four times that in the CL samples (Fig 1B). The POD activity in the WR samples was 3.8 times higher than that in the CR samples (Fig 1B).
3.2 Waterlogging leads to extensive transcriptomic reprogramming
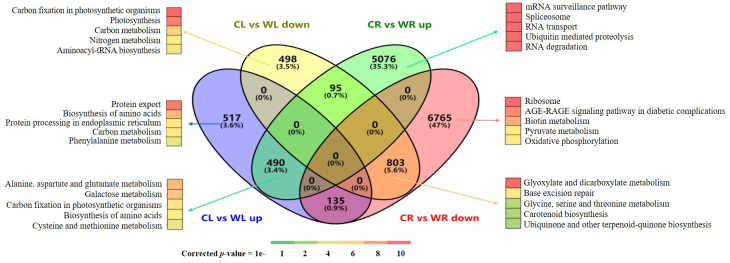
After removing the unknown reads (those whose proportion of undetermined bases was > 10%), low-quality reads and reads that contained adapters and at least 39.4 million clean reads were obtained for each sample (Table 1). The clean reads were subsequently mapped to the reference genome, with the mapping ratio varying from 79.4% to 86.1%. More than 78% of the reads were uniquely mapped (Table 1). With a fold-change ≥ 2 and a FDR corrected p-value of < 0.01 used as screening criteria, 15902 genes were identified as differentially expressed in at least one tissue between the nonwaterlogged conditions and waterlogged conditions (Fig 2, S2 Table). Among them, 13364 differentially expressed genes (DEGs) were found in the CR vs WR pairwise comparison, and 2538 DEGs were found in the CL vs WL comparison. We discovered that there were many more DEGs in the CR vs WR comparison than in the CL vs WL comparison, suggesting that the waterlogging response is more complex in the roots than in the leaves. This is consistent with findings for Taxodium ‘Zhongshansa’ [31]. A total of 1523 genes were found to be shared between the two tissues, of which 625 showed opposite expression responses. Additionally, 11841 genes (5076 upregulated and 6765 downregulated) were exclusively differentially expressed between the CR and WR samples, and the expression levels of 1015 genes (517 upregulated and 498 downregulated) exclusively changed between the CL and WL samples (Fig 2, S2 Table).
Table 1. Total number of sequencing reads obtained from each sample.
| Sample name | Total Reads | Mapped Reads | Uniq Mapped Reads | Multiple Map Reads |
|---|---|---|---|---|
| CL1 | 40,918,904 | 35,153,610(85.91%) | 34,569,539(84.48%) | 584,071(1.43%) |
| CL2 | 47,888,734 | 41,180,289(85.99%) | 40,468,518(84.51%) | 711,771(1.49%) |
| CL3 | 45,846,222 | 39,494,069(86.14%) | 38,830,063(84.70%) | 664,006(1.45%) |
| CR1 | 39,610,692 | 33,168,267(83.74%) | 32,547,312(82.17%) | 620,955(1.57%) |
| CR2 | 40,630,974 | 33,601,548(82.70%) | 32,933,270(81.05%) | 668,278(1.64%) |
| CR3 | 43,169,994 | 34,980,971(81.03%) | 34,327,845(79.52%) | 653,126(1.51%) |
| WL1 | 39,464,384 | 33,934,945(85.99%) | 33,361,618(84.54%) | 573,327(1.45%) |
| WL2 | 40,027,610 | 34,443,486(86.05%) | 33,897,841(84.69%) | 545,645(1.36%) |
| WL3 | 41,557,772 | 35,653,680(85.79%) | 35,037,818(84.31%) | 615,862(1.48%) |
| WR1 | 40,983,672 | 32,810,482(80.06%) | 32,218,760(78.61%) | 591,722(1.44%) |
| WR2 | 47,124,748 | 37,432,143(79.43% | 36,741,146(77.97% | 690,997(1.47%) |
| WR3 | 42,694,908 | 34,396,57(80.56%) | 33,762,811(79.08%) | 633,346(1.48%) |
Note: CL: leaves under nonwaterlogged conditions; CR: roots under nonwaterlogged conditions; WL: leaves under waterlogged conditions; WR: roots under waterlogged conditions.
Fig 2. Venn diagrams of the differentially expressed genes under waterlogging treatment in the leaf and root samples (FDR corrected p-value of ≤ 0.01 and fold change ≥ 2) and their respective top five most significantly enriched KEGG pathways.
The numbers of DEGs exclusively up- or down-regulated in one tissue are shown in each circle. The numbers of DEGs with a common or opposite tendency of expression changes between the two tissues are shown in the overlapping regions. Only the top 5 enriched pathway terms are shown.
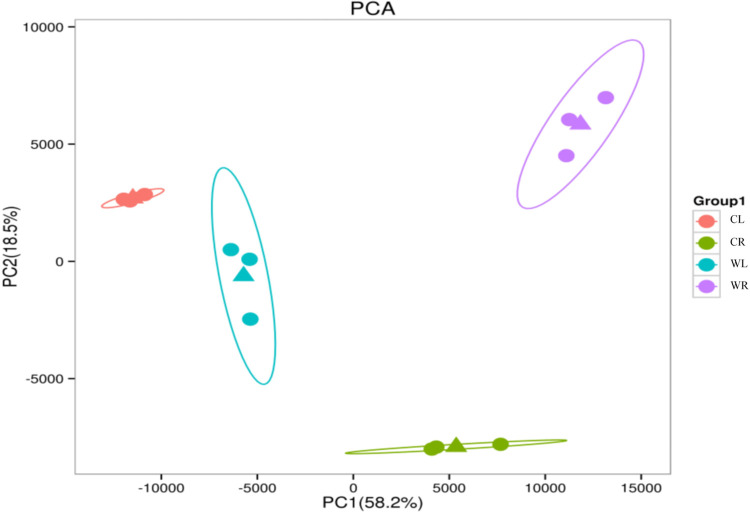
Principal component analysis (PCA) was used to visualize the overall changes in gene expression in the different treatments. The first two principal components, which explained 76.7% of the total variance (58.2% by the first component, 18.5% by the second component), showed that the waterlogged tissues and nonwaterlogged tissues were dissimilar. Moreover, the replicates showed a high degree of similarity for all four treatments (Fig 3).
Fig 3. Principal component analysis of transcriptome data.
Principal component analysis was applied to differentially expressed contigs identified in CL, CR, WL and WR. The triangles mean cluster centers.
3.3 Functional annotation of waterlogging-responsive DEGs
To further characterize the expression changes discussed above, we compared the enriched KEGG pathways for DEGs between the two tissues, the results of which indicated some tissue-specific or highly performed functions (Fig 2). The following pathways were highly enriched in the DEGs whose expression was upregulated: ‘alanine, aspartate and glutamate metabolism’; ‘galactose metabolism’; and ‘carbon fixation in photosynthetic organisms’. However, other pathways were highly enriched in the DEGs whose expression was typically downregulated in both tissues, including ‘glyoxylate and dicarboxylate metabolism’; ‘base excision repair’; and ‘glycine, serine and threonine metabolism’. The pathways ‘protein export’, ‘biosynthesis of amino acids’, and ‘protein processing in endoplasmic reticulum’ were highly enriched in the DEGs whose expression was upregulated specifically in the CL and WL samples, while the ‘mRNA surveillance pathway’, ‘spliceosome’ and ‘RNA transport’ pathways were dramatically enriched in DEGs whose expression was specifically increased in the CR and WR samples. The DEGs involved in all aspects of photosynthesis, which were associated with ‘carbon fixation in photosynthetic organisms’, ‘photosynthesis’, ‘carbon metabolism’ and ‘nitrogen metabolism’, were listed as the top four enriched pathways in the CL and WL samples. The ‘ribosome’ pathway was the pathway most enriched by DEGs whose expression was downregulated specifically in the CR and WR samples.
3.4 Analysis of DEGs related to the photosynthesis pathway
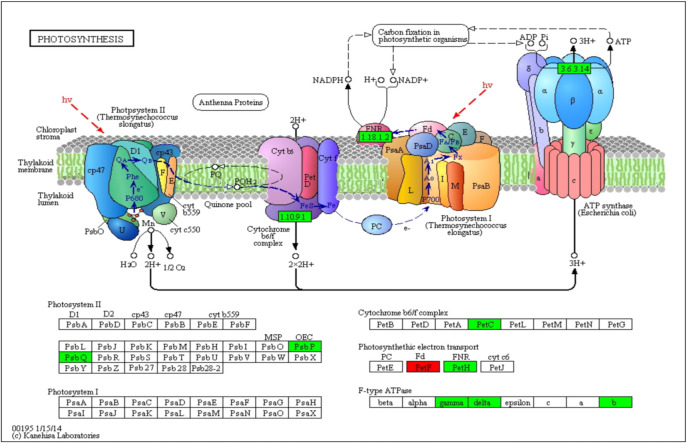
PSI, PSII, the cytochrome b6/f complex, photosynthetic electron transport, and F-type ATPase are key components in the photosynthetic pathway. The comparison between the CL and WL libraries revealed 12 DEGs related to the photosynthesis pathway, including three genes related to photosystem II (PSII), four related to photosynthetic electron transport, one related to the cytochrome b6/f complex, and four related to the F-type ATPase. DEGs involved in PSII, the cytochrome b6/f complex and F-type ATPase were downregulated in the leaves when cassava was under waterlogging stress, two upregulated and two downregulated DEGs were related to photosynthetic electron transport (Fig 4, S3 Table). The malfunction of PSII reduced the efficiency of electron transport for photosynthetic reactions, which could result in substantive accumulation of reactive oxygen species (ROS) and further reduced PN under WL. Additionally, 12 genes involved in photosynthetic activities may be associated with the differences in leaf color and may cause a decrease in the Fv/Fm ratio.
Fig 4. Differentially expressed genes mapped to the photosynthesis pathway in the leaves.
The known pathways were obtained from the KEGG database. Red squares denote upregulated genes, and green squares denote downregulated genes.
3.5 Analysis of DEGs related to the glycolysis pathway
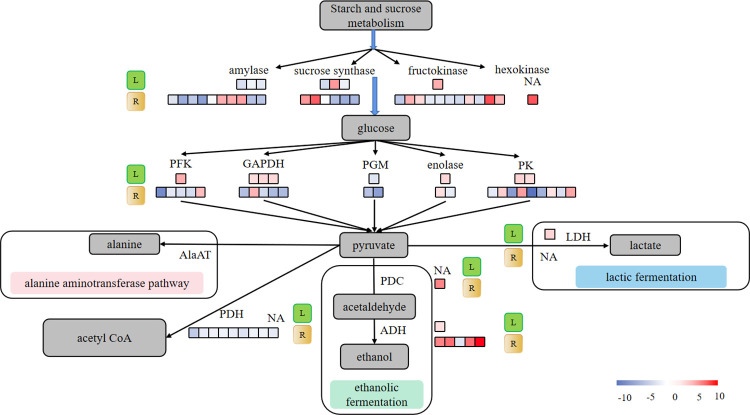
In both the roots and leaves of waterlogged cassava, many genes with potential roles in glycolysis and fermentation were identified as displaying a significant transcriptional response to waterlogging stress (Fig 5, S4 Table). A total of 113 DEGs (47 upregulated and 66 downregulated) were annotated as encoding enzymes involved in the glycolysis/gluconeogenesis pathway according to their KEGG classification (S4 Table). The expression of most of the DEGs was upregulated in the WL samples compared to the CL samples and were involved in enzymes such as glyceraldehyde 3-phosphate dehydrogenase (GAPDH), phosphofructokinase (PFK), enolase, alcohol dehydrogenase (ADH), L-lactate dehydrogenase (LDH) and pyruvate kinase (PK). However, in the WR samples, the expression of major glycolysis- and fermentation-related genes in response to waterlogging stress decreased, indicating a decrease in throughput of these pathways (Fig 5).
Fig 5. Schematic diagram of glycolysis and fermentation proposed during plant waterlogging stress.
AlaAT, alanine aminotransferase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PDC, pyruvate decarboxylase; ADH, alcohol dehydrogenase; PFK, 6-phosphofructokinase; PK, pyruvate kinase; LDH, lactic dehydrogenase; PDH, pyruvate dehydrogenase; PGM, phosphoglucomutase. L, leaves; R, roots; The color of the boxes represents the fold-change value of DEGs. The number of boxes represents the number of DEGs. NA indicates that there are no DEGs encoding the enzyme. The pathway was designed based on KEGG pathways (http://www.kegg.jp/kegg/pathway.html).
3.6 Analysis of DEGs related to the galactose metabolic pathway
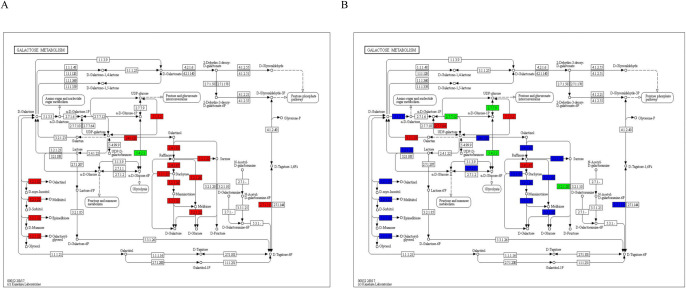
A comprehensive analysis of the DEGs in the leaves and roots involved in the galactose metabolism pathway identified two and four genes in the leaves and roots that encode glucuronosyltransferase (inositol galactoside synthase) (Fig 6, S5 Table). This protein catalyzes the synthesis of inositol galactoside from UDP-galactose and myoinositol, which is the first key step in the synthesis of raffinose family oligosaccharides (RFOs) and the most critical regulatory step in RFO synthesis. The expression difference levels of two and three genes increased successively in leaves and roots, and one gene was downregulated in the roots (Fig 6, S5 Table). The analysis identified four and five genes encoding inositol galactoside-sucrose galactosyltransferase (raffinose synthase) in leaves and roots, respectively. This protein mainly catalyzes the synthesis of raffinose from inositol galactoside and sucrose. One and three genes encoding β-D-fructofuranoside were identified in the leaves and roots and mainly catalyze the decomposition of stachyose and raffinose into melibiose and the decomposition of sucrose into glucose and fructose. We also found two genes in the roots encoding β-galactosidase (β-GAL), which mainly catalyzes the decomposition of lactose and raffinose into melibiose (Fig 6B, S5 Table). Two of the main pectin deglycosylating enzymes that participate in this process are alpha galactosidases (α-GAL) (EC:3.2.1.22) and β-GAL [32]. In this study, four α-GAL DEGs were identified, two of which were upregulated after waterlogging in the roots, while two of them were downregulated. One β-GAL gene was identified and downregulated in the roots. Two α-GAL DEGs were upregulated in the leaves (Fig 6). One alpha glucosidase (α-Glu) gene, which is involved in cellulose degradation, was downregulated in the roots (S5 Table).
Fig 6. Analysis of DEGs involved in the galactose metabolic pathway in the leaves and roots.
Pattern diagram of the lactose metabolic pathway in the (A) leaves and (B) roots. The red squares indicate upregulated genes, the green squares indicate downregulated genes, and the blue squares indicate both upregulated and downregulated genes. The pathway was designed based on KEGG pathways (http://www.kegg.jp/kegg/pathway.html).
3.7 Responses of transcription factors (TFs) to waterlogging stress
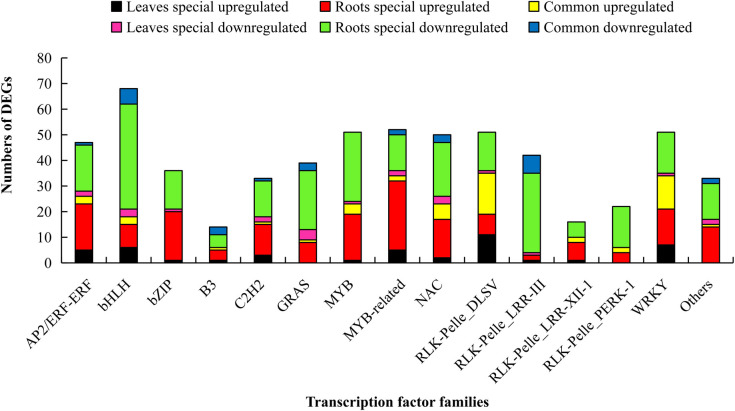
TFs are key regulators of target gene expression in response to various biotic or abiotic stresses by binding to specific cis-acting elements in these gene promoters [33]. In this study, a total of 605 waterlogging-regulated TFs were identified according to their assigned gene families (Fig 7, S6 Table). Of these, 84 TFs (55 upregulated and 29 downregulated) were commonly expressed in the leaves and roots, whereas 66 TFs (43 upregulated and 23 downregulated) were expressed in the leaves only, and 455 TFs (179 upregulated and 276 downregulated) were expressed only in the roots. These data strongly suggest that transcriptional regulation occurs in response to waterlogging. Remarkably, genes belonging to the MYB, WRKY, NAC, and AP2/ERF families encode most of the differentially expressed TFs, implying that these genes have important roles in waterlogging stress responses (Fig 7, S6 Table).
Fig 7. Response of transcription factors to waterlogging.
Graphical representations of waterlogging-regulated transcription factors based on their assigned protein families. Different colors represent the different sources of DEGs, and the length of the bar represents the number of DEGs.
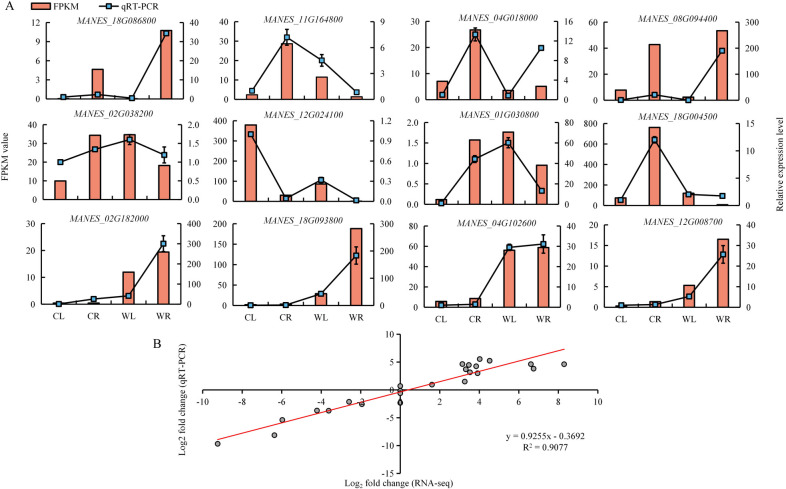
3.8 RNA-seq results validated by qRT-PCR
To verify the reliability of the RNA-seq data, we selected twelve genes for further investigation using qRT-PCR methods (Fig 8A, S1 Table). The results showed that the expression patterns determined via qRT-PCR were highly consistent with the RNA-seq data, with a relative R2 of 0.9077 (Fig 8B).
Fig 8. The expression patterns of 12 selected genes were verified by qRT-PCR, and the correlation between RNA-seq and qRT-PCR in the leaves and roots was verified.
(A) The broken line represents the qRT-PCR results, and the bar graph represents the FPKM of RNA-seq. (B) The gene fold change log2 values (Y-axis) were plotted against the qRT-PCR fold change log2 values (X-axis). Each value denotes the mean relative level of expression of three biological replicates.
4. Discussion
Substantial progress has been made in understanding waterlogging or low-oxygen-stress response mechanisms at the transcriptional level in Arabidopsis [34, 35] and in various crop species, such as rice [36], rape [37], cucumber [38, 39] and maize [40], using transcriptomic approaches. These studies indicated that genes regulated by low oxygen availability are extremely diverse among species and are involved in ethylene synthesis, glycolysis, ethanol fermentation, carbohydrate catabolism, photosynthesis and galactose metabolism.
4.1 Comparisons of transcriptomes between leaves and roots
Waterlogging stress is different from complete submergence in that only the lower and subsoil portions of affected plants are subjected directly to the stress. In this study, comparison of a transcriptional response to waterlogging stress between the aboveground tissues (leaves) and belowground tissues (roots) revealed a high number of DEGs during the same period. These data can be used to discern among the enriched gene function categories, which can be reasonably explained by the functional differences between the two tissues. A previous report showed that the ability to tolerate hypoxic stress in the leaves and roots could be genetically based [31, 41], and the anaerobic induction of most known anaerobic proteins (ANPs) was root specific.
Genes associated with functional categories related to mRNA surveillance, RNA transport and degradation were most highly enriched in the roots, but they were also dramatically enhanced in the WR samples, which indicated a large scale of energy savings under hypoxic conditions. However, few studies have analyzed the role of these genes in the waterlogging response. In addition, the genes that were involved with ribosomes and whose expression was downregulated were the most enriched in the WR samples, suggesting that waterlogging decreased ribosome biosynthesis. Ribosomes are large and complex molecular machines found within all living cells in plants and serve as the primary sites of biological protein synthesis or translation [42]. Downregulation of ribosomal genes has been found in several plant species exposed to heavy metal stress [43, 44]. Our results suggest that there is a common ribosome biosynthesis network between waterlogging and heavy metal stress in cassava.
Moreover, the disruption in aerobic respiration caused by waterlogging may inhibit the tricarboxylic acid (TCA) cycle and activate glycolysis and fermentation pathways at the whole-plant level, resulting in the accumulation of amino acids closely derived from glycolysis intermediates (glycine, serine, threonine, etc.) and a decrease in TCA cycle intermediate-derived amino acids (asparagine, aspartic acid, glutamine, glutamic acid, etc.) [45]. In this study, as expected, the genes related to ‘alanine, aspartate and glutamate metabolism’ and ‘glycine, serine and threonine metabolism’ were highly enriched in both tissues, suggesting that pathways related to carbohydrate and amino metabolism were activated when cassava was exposed to hypoxic conditions. This finding was consistent with those in alfalfa [46].
4.2 Effects of waterlogging stress on photosynthesis
Chlorosis is a typical symptom that inevitably occurs after severe waterlogging. Because waterlogging causes water saturation and subsequent rapid closure of stomata, a high concentration of O2 cannot be released out, and photosynthetic electron transportation is blocked in chloroplasts [47, 48].
Waterlogging at the plant stage of cassava plants caused significant decreases in chlorophyll content and the Fv/Fm ratio and caused premature senescence of leaves (Fig 1). These results were consistent with those of previous studies in which waterlogging-induced chlorophyll loss was associated with a decrease in photosynthetic activity [49]. Interestingly, the Fv/Fm ratio was inconsistent with the findings of Guide and Soldatini [50], who observed that after 6 days of waterlogging, the Fv/Fm ratio remained unchanged and photosynthesis continued to decrease in the absence of stomatal closure in soybean. DEGs relevant to photosynthesis were abundant in the leaves compared with the roots. In this study, the top two enriched pathways by genes whose differential expression was specifically downregulated in the WL samples were associated with photosynthesis pathways, indicating a possible reduction in the photosynthesis rate (Fig 4, S3 Table). These DEGs were involved in PSII and the photosynthetic electron transport chain (cytochrome b6-f complex, ferredoxin, transporting ATPase subunit, etc.). The reaction centers of PSII in chloroplast thylakoids are also the major generation sites of ROS and are largely affected by abiotic factors such as low oxygen concentrations [37, 51]. In addition, the expression levels of genes encoding rubisco and rubisco activase, which are involved in carbon-fixing reactions, were also downregulated, suggesting that waterlogging stress not only inhibits photosynthetic reactions but also reduces the efficiency of CO2 fixation in cassava. Similar waterlogging responses in other crop species have been previously reported [37, 52].
4.3 Effects of waterlogging stress on glycolysis pathway
Generally, plants receive their essential energy supply through glycolysis and ethanol fermentation when facing energy shortages caused by waterlogging stress [53]. In this study, many genes, including well-known hypoxia-related genes associated with glycolysis and fermentative processes, were identified as having a significant transcriptional response to waterlogging stress in both tissues, which indicated that the pathway was activated to maintain ATP production under hypoxic conditions. Most ANPs have been identified as enzymes involved in the glycolysis or sugar phosphate pathways that are needed to maintain energy production under waterlogging conditions [54, 55].
Fermentation is a process of energy conversion necessary during waterlogging stress, during which the expression of some anaerobic genes, such as PDC and ADH, is upregulated [56, 57]. Overexpression of PDC1 and PDC2 in Arabidopsis improves survival under low-oxygen conditions [58]. A similar result was obtained by Rivoal et al. in rice [59]. ADH activity has been reported to increase under anoxia, and compared with wild-type plants, maize mutants deficient in ADH activity are more sensitive to waterlogging stress [60]. In this study, the expression of four out of five ADH genes was significantly upregulated in the roots under waterlogging stress (S4 Table), and the expression of one PDC gene was also significantly upregulated. These results suggest that energy was produced under hypoxic conditions by the activation of alcoholic fermentation. The expression of none of the genes encoding PDC changed in the CL vs WL comparison. This finding was consistent with findings for Taxodium ‘Zhongshansa’ [31] and gray poplar [45]. In addition to PDC and ADH, LDH is also involved in the response to low-oxygen conditions and is activated in the initial stages of root hypoxia in many plant species. In a study on gray poplar, LDH transcripts were also rather abundant during the initial reaction to oxygen deprivation but decreased after approximately 5 h due to the decrease in cytosolic pH caused by lactic acid [45]. In this study, the expression of an LDH gene was significantly upregulated in the WL samples, while in the WR samples, it was unchanged. Additional quantitative real-time PCR experiments are needed to confirm the importance of LDH in the roots of cassava during waterlogging tolerance.
4.4 Effects of waterlogging stress on galactose metabolism
The energy metabolism pathway is not only related to the alcohol dehydrogenase-catalyzed step of glycolysis but also associated with starch and sucrose metabolism and the galactose metabolism pathway. Accumulating data suggest that low oxygen concentrations play a key role in the induction of hypoxia metabolism, such as the expression of genes triggering anaerobic fermentation, sugar utilization and antioxidant defense [61, 62]. We found that after waterlogging treatment, the expression levels of major genes related to galactose metabolism in the leaves and roots were upregulated. Raffinose and galactitol, as ROS scavengers, can reduce oxidative damage under abiotic stress conditions [63]. Additionally, raffinose can be transported to chloroplasts to protect thylakoids and stabilize PSⅡ [64]. Both galactinol and raffinose accumulate at higher levels in plants in response to abiotic stresses [65]. They play a novel role in the protection of cellular metabolism from oxidative damage caused by salinity, chilling, or drought [66]. Under the condition of long-term waterlogging, the cell wall of plants will disintegrate before the whole cell disintegrates [67, 68]. Degradation of cell wall polysaccharides is a consequence of synergistic action among several key cell wall modifying enzymes, including polygalacturonase (PG), pectin methyl esterase (PME), β-GAL and cellulase (CEL) [69]. In our study, two β-GAL genes in the roots and one β-GAL gene in the leaves were downregulated, suggesting that the cell wall degrades slowly under waterlogging stress (S5 Table).
4.5 Changes in the expression of TFs
Previous studies have reported that MYB TFs are closely related to the primary and secondary products of plant morphogenesis and to the metabolic regulation of plant resistance [55, 70–72]. The members of this TF family are also known to trigger ADH gene. For instance, the induction of AtADH1 expression is tightly coupled to the initial increase in AtMYB2 transcripts [73]. Research on wheat has shown that the expression of TaMyb1 is strongly induced in the roots under hypoxic conditions [74]. We found 23 MYB- and 34 MYB-related TFs in the upregulated group, indicating that these genes have potential roles in waterlogging stress responses and tolerance. ERF TFs belong to a plant-specific TF superfamily related to the stress response. Previous studies have shown that AP2/ERF TFs such as Sub1A, HRE1, HRE2, Snorkel1, Snorkel2, and RAP2.2 are important for tolerance to plant hypoxia caused by waterlogging [75–77]. For instance, overexpression of SK1 and SK2 significantly enhances the waterlogging tolerance of rice and is characterized by excessive elongation of internodes [75]. Hinz reported that constitutive overexpression of HRE1 or HRE2 could rapidly induce the expression of hypoxia-responsive genes, such as PDC and ADH, to enhance waterlogging tolerance in Arabidopsis [76]. Under hypoxia, the presence of RAP2.12 in the nucleus may lead to not only relocalization of the existing protein but also de novo synthesis [78]. In this study, the expression of 26 and 21 AP2/ERF genes was upregulated and downregulated, respectively, under waterlogging stress. Whether the TF changes are related to cassava waterlogging tolerance requires further demonstration. Additionally, members of the WRKY and NAC TF families may also be associated with the transcriptional regulation of genes involved in the waterlogging response of cassava, as these TFs were overrepresented in the list of DEGs. These results were similar to those for soybean [79] and kiwifruit [11].
5. Conclusion
In this study, we used high-throughput sequencing to characterize the transcriptome responses in the roots and leaves of partially submerged cassava plants. In total, 15902 transcripts were identified as being differentially expressed. KEGG enrichment analysis provided comparisons of iCL vs WL and of CR vs WR. The results suggested that waterlogging stress mainly represses photosynthesis reactions in the leaves and improves energy savings in the roots. Amino acid metabolism also greatly changed in both tissues, and a nitrate-producing pathway may be induced to help maintain ATP levels. Furthermore, complex interactions between energy production and the antioxidant enzyme system were observed, suggesting that they have important roles in the waterlogging response. The changes in the expression of these genes involved in the waterlogging response might be regulated by the synthesis and perception of some TFs, such as ERFs, MYBs, WRKYs and NACs.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We appreciate the reviewers and editors for the patience to the work.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the Natural Science Foundation of Hainan Province (320QN193) and the China Agriculture Research System (CARS-11). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Roscoe PA. Changing Climate for Anthropological and Archaeological Research? Improving the Climate‐Change Models. Am Anthropol. 2014; 116(3): 535–548. 10.1111/aman.12115 [DOI] [Google Scholar]
- 2.Durack PJ, Wijffels SE, Matear RJ. Ocean Salinities Reveal Strong Global Water Cycle Intensification During 1950 to 2000. Science. 2012; 336(6080): 455–458. doi: 10.1126/science.1212222 [DOI] [PubMed] [Google Scholar]
- 3.Voesenek LACJ, Sasidharan R, Weber A. Ethylene—and oxygen signalling—drive plant survival during flooding. Plant Biol. 2013; 15(3): 426–435. doi: 10.1111/plb.12014 [DOI] [PubMed] [Google Scholar]
- 4.Shabala S, Shabala L, Barcelo J, Poschenrieder C. Membrane transporters mediating root signalling and adaptive responses to oxygen deprivation and soil flooding. Plant Cell Environ. 2014; 37(10):2216–2233. doi: 10.1111/pce.12339 [DOI] [PubMed] [Google Scholar]
- 5.Huo ZG, Fan YX, Yang JY, Shang Y. Review on Agricultural Flood Disaster in China. J Appl Meteor Sci. 2017; 28(6): 641–653. 10.11898/1001-7313.20170601 [DOI] [Google Scholar]
- 6.Bailey-Serres J, Fukao T. Plant responses to hypoxia-is survival a balancing act? Trends Plant Sci. 2004; 9(9): 449–456. doi: 10.1016/j.tplants.2004.07.005 [DOI] [PubMed] [Google Scholar]
- 7.Arbona V, Hossain Z, López-Climent MF, Pérez-Clemente RM, Gómez-Cadenas A. Antioxidant enzymatic activity is linked to waterlogging stress tolerance in citrus. Physiol Plant. 2008; 132(4): 452–466. doi: 10.1111/j.1399-3054.2007.01029.x [DOI] [PubMed] [Google Scholar]
- 8.Visser EJW, Voesenek LCJ, Vartapetian BB, Jackson MB. Flooding and plant growth: Preface. Ann Bot. 2003; 91(2): 107–109. 10.1093/aob/mcg014 [DOI] [Google Scholar]
- 9.Bailey-Serres J, Voesenek LACJ. Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol. 2008; 59: 313–339. doi: 10.1146/annurev.arplant.59.032607.092752 [DOI] [PubMed] [Google Scholar]
- 10.Kim YH, Hwang SJ, Waqas M, Khan AL, Lee JH, Lee JD, et al. Comparative analysis of endogenous hormones level in two soybean (Glycine max L.) lines differing in waterlogging tolerance. Front Plant Sci. 2015; 6, 714. doi: 10.3389/fpls.2015.00714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang JY, Huang SN, Mo ZH, Xuan JP, Jia XD, Wang G, et al. De novo transcriptome sequencing and comparative analysis of differentially expressed genes in kiwifruit under waterlogging stress. Mol Breed. 2015; 35(11): 208–220. 10.1007/s11032-015-0408-0 [DOI] [Google Scholar]
- 12.Phukan UJ, Jeena GS, Tripathi V, Shukla RK. MaRAP2-4 a waterlogging-responsive ERF from Mentha regulates bidirectional sugar transporter AtSWEET10 to modulate stress response in Arabidopsis. Plant Biotechnol J. 2017; 16(1): 221–233. doi: 10.1111/pbi.12762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parent C, Capelli N, Berger A, Cre`vecoeur M, Dat JF. An overview of plant responses to soil waterlogging. Plant Stress. 2008; 2(1): 20–27. https://hal.archives-ouvertes.fr/hal-00374639 [Google Scholar]
- 14.Lin KR, Weng C, Lo H, Chen J. Study of the root antioxidative system of tomatoes and eggplants under waterlogged conditions. Plant Sci. 2004; 167(2): 355–365. 10.1016/j.plantsci.2004.04.004 [DOI] [Google Scholar]
- 15.FAO. Food and Agricultural Organization, World meat markets at a glance (FAO World Food Outlook 2014). 2016.
- 16.Yu XY, Yao Y, Hong YH, Hou PY, Li CX, Xia ZQ, et al. Differential expression of the Hsf family in cassava under biotic and abiotic stresses. Genomics. 2019; 62(8): 563–569. doi: 10.1139/gen-2018-0163 [DOI] [PubMed] [Google Scholar]
- 17.El-Sharkawy MA, Cock JH. Response of cassava to water stress. Plant Soil. 1987; 100(28): 345–360. 10.1007/BF02370950 [DOI] [Google Scholar]
- 18.Luo K, Wu F, Zhang D, Dong R, Fan Z, Zhang R, et al. Transcriptomic profiling of Melilotus albus near-isogenic lines contrasting for coumarin content. Sci Rep. 2017. Jul 4;7(1):4577. doi: 10.1038/s41598-017-04111-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007; 35: 61–65. doi: 10.1093/nar/gkl842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2004; 32(1): D115–D119. doi: 10.1093/nar/gkh131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashburner M, Ball CA, Blake JA. Gene ontology: tool for the unification of biology. Nature Genet. 2000; 25(1): 25–29. doi: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koonin EV, Fedorova ND, Jackson JD. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004; 5(2): R7. doi: 10.1186/gb-2004-5-2-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome scale analysis of protein functions and evolution. Nucleic Acids Res. 2000; 28(1): 33–36. doi: 10.1093/nar/28.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finn RD, Tate J, Mistry J. The Pfam protein families database. Nucleic Acids Res. 2008; 36: D281–D288. doi: 10.1093/nar/gkm960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Araki M, Goto S. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008; 36: D480–D484. doi: 10.1093/nar/gkm882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortazavi A.; Williams B.A.; McCue K.; Schaeffer L.; Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. doi: 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 27.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao X, Cai T, Olyarchuk J G. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005; 21(19): 3787–3793. doi: 10.1093/bioinformatics/bti430 [DOI] [PubMed] [Google Scholar]
- 29.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008; 3: 1101–1108. doi: 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 30.Salcedo A, Zambrana C, Siritunga D. Comparative expression analysis of reference genes in field-grown cassava. Tropical Plant Biol. 2014; 7: 53–64. http://doi.org/ 10.1007/s12042-014-9137-5 [Google Scholar]
- 31.Qi B, Yang Y, Yin YL, Xu M, Li HG. De novo sequencing, assembly, and analysis of the Taxodium ’Zhongshansa’ roots and shoots transcriptome in response to short-term waterlogging. BMC Plant Biol. 2014; 14(1): 201–213. 10.1186/s12870-014-0201-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goulao LF, Oliveira CM. Cell wall modifications during fruit ripening: when a fruit is not the fruit. Trends Food Sci. Tech. 2008; 19: 4–25. 10.1016/j.tifs.2007.07002 [DOI] [Google Scholar]
- 33.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006; 57: 781–803. doi: 10.1146/annurev.arplant.57.032905.105444 [DOI] [PubMed] [Google Scholar]
- 34.Liu FL, Tara VT, Moy LP, Bock G, Lara D, John L. Global Transcription Profiling Reveals Comprehensive Insights into Hypoxic Response in Arabidopsis. Plant Physiol. 2005; 137(3): 1115–1129. doi: 10.1104/pp.104.055475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Juntawong P, Girke T, Bazin J, Bailey-Serres J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. P Natl Acad Sci. 2014; 111(1): E203–E212. doi: 10.1073/pnas.1317811111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narsai R, Rocha M, Geigenberger P, Whelan J, Dongen JTV. Comparative analysis between plant species of transcriptional and metabolic responses to hypoxia. New Phytol. 2011; 190(2): 472–487. doi: 10.1111/j.1469-8137.2010.03589.x [DOI] [PubMed] [Google Scholar]
- 37.Lee YH, Kim KS, Jang YS, Hang JH., Lee DH, Choi IH. Global gene expression responses to waterlogging in leaves of rape seedlings. Plant Cell Reports. 2014; 33(2): 289–299. doi: 10.1007/s00299-013-1529-8 [DOI] [PubMed] [Google Scholar]
- 38.Qi XH, Xu XW, Lin XJ, Zhang WJ, Chen XH. Identification of differentially expressed genes in cucumber (Cucumis sativus L.) root under waterlogging stress by digital gene expression profile. Genomics. 2012; 99(3): 160–168. doi: 10.1016/j.ygeno.2011.12.008 [DOI] [PubMed] [Google Scholar]
- 39.Xu X, Chen M, Ji J, Xu Q, Qi XH, Chen XH. Comparative RNA-seq based transcriptome profiling of waterlogging response in cucumber hypocotyls reveals novel insights into the de novo adventitious root primordia initiation. BMC Plant Biol. 2017; 17(1): 129–142. doi: 10.1186/s12870-017-1081-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malachy TC, Christopher AP, Dou YC, Aaron JS, Piyaporn P, Greg RK, et al. Genetic and molecular characterization of submergence response identifies subtol6 as a major submergence tolerance locus in maize. Plos One. 2016; 110(3): e0120385. 10.1371/journal.pone.0120385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellis MH, Dennis ES, Peacock WJ. Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol. 1999; 119(1): 57–64. doi: 10.1104/pp.119.1.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell Mol Life Sci. 2007; 64(1): 29–49. doi: 10.1007/s00018-006-6278-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Planello R, Martinez-Guitarte J, Morcillo G. Ribosomal genes as early targets of cadmium-induced toxicity in Chironomus riparius larvae. Sci Total Environ. 2007; 373(1): 113–121. doi: 10.1016/j.scitotenv.2006.10.038 [DOI] [PubMed] [Google Scholar]
- 44.Zhu QL, Guo SN, Wen F, Zhang XL, Wang CC, Si LF, et al. Transcriptional and physiological responses of dunaliella salina to cadmium reveals time-dependent turnover of ribosome, photosystem, and ros-scavenging pathways. Aquat Toxicol. 2018; 207: 153–162. doi: 10.1016/j.aquatox.2018.12.007 [DOI] [PubMed] [Google Scholar]
- 45.Kreuzwieser JUR, Hauberg J, Howell KA, Carroll A, Rennenberg H, Millar AH, et al. Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiol. 2009; 149(1): 461–473. doi: 10.1104/pp.108.125989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng N, Yang ZJ, Zhang ZF, Hu LX, Chen L. Comparative Transcriptome Combined with Proteome Analyses Revealed Key Factors Involved in Alfalfa (Medicago sativa) Response to Waterlogging Stress. Int J Mol Sci. 2019; 20(6): 1359–1383. doi: 10.3390/ijms20061359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Musgrave ME. Waterlogging Effects on Yield and Photosynthesis in Eight Winter Wheat Cultivars. Crop Sci. 1994; 34(5): 1314–1318. 10.2135/cropsci1994.0011183X003400050032x [DOI] [Google Scholar]
- 48.Smethurst CF, Shabala S. Screening methods for waterlogging tolerance in lucerne: comparative analysis of waterlogging effects on chlorophyll fluorescence, photosynthesis, biomass and chlorophyll content. Funct Plant Biol. 2003; 30: 335–343. doi: 10.1071/FP02192 [DOI] [PubMed] [Google Scholar]
- 49.Qian H, Li J, Sun L, Chen W, Sheng GD, Liu W, et al. Combined effect of copper and cadmium on Chlorella vulgaris growth and photosynthesis-related gene transcription. Aquat Toxicol. 2009; 94(1): 56–61. doi: 10.1016/j.aquatox.2009.05.014 [DOI] [PubMed] [Google Scholar]
- 50.Guidi L, Soldatini F. Chlorophyll fluorescence and gas exchanges in flooded soybean and sunflower plants. Plant Physiol Biochem. 1997; 35: 713–717. [Google Scholar]
- 51.Production Asada K. and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006; 141(2): 391–396. doi: 10.1104/pp.106.082040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Christianson JA, Llewellyn DJ, Dennis ES, Wilson IW. Global Gene Expression Responses to Waterlogging in Roots and Leaves of Cotton (Gossypium hirsutum L.). Plant Cell Physiol. 2010; 51(1): 21–37. doi: 10.1093/pcp/pcp163 [DOI] [PubMed] [Google Scholar]
- 53.Caruso P, Baldoni E, Mattana M, Paolo DP, Genga A, Coraggio I, et al. Ectopic expression of a rice transcription factor, Mybleu, enhances tolerance of transgenic plants of Carrizo citrange to low oxygen stress. Plant Cell Tiss Org. 2012; 109(2): 327–339. 10.1007/s11240-011-0098-1 [DOI] [Google Scholar]
- 54.Ahsan N, Lee DG, Lee SH, Kang KY, Bahk JD, Choi MS, et al. A comparative proteomic analysis of tomato leaves in response to waterlogging stress. Physiol Plant. 2007; 131(4): 555–570. doi: 10.1111/j.1399-3054.2007.00980.x [DOI] [PubMed] [Google Scholar]
- 55.Zhang YF, Cao GY, Qu LJ, Gu HY. Characterization of Arabidopsis MYB transcription factor gene AtMYB17 and its possible regulation by LEAFY and AGL15. J Genet Genomics. 2009; 36(2): 99–107. doi: 10.1016/S1673-8527(08)60096-X [DOI] [PubMed] [Google Scholar]
- 56.Kato-Noguchi H. Pyruvate metabolism in rice coleoptiles under anaerobiosis. Plant Growth Regul. 2006; 50(1): 41–46. 10.1007/s10725-006-9124-4 [DOI] [Google Scholar]
- 57.Takahashi H, Greenway H, Matsumura H, Tsutsumi N, Nakazono M. Rice alcohol dehydrogenase 1 promotes survival and has a major impact on carbohydrate metabolism in the embryo and endosperm when seeds are germinated in partially oxygenated water. Ann Bot. 2014; 113(5): 851–859. doi: 10.1093/aob/mct305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ismond KP, Dolferus R, De PM, Dennis ES, Good AG. Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiol. 2003; 132(3): 1292–1302. doi: 10.1104/pp.103.022244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rivoal J, Thind S, Ricard AP. Differential induction of pyruvate decarboxylase subunits and transcripts in anoxic rice seedlings. Plant Physiol. 1997; 114(3): 1021–1029. doi: 10.1104/pp.114.3.1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roberts JK, Callis J, Wemmer D, Walbot V, Jardetzky O. Mechanism of Cytoplasmic pH Regulation in Hypoxic Maize Root Tips and Its Role in Survival under Hypoxia. P Natl Acad Sci. 1984; 81(11): 3379–3383. 10.1073/pnas.81.11.3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olga B, Eija V, Kurt VF. Antioxidants, Oxidative Damage and Oxygen Deprivation Stress: a Review. Ann Bot. 2003; 91(2): 179–194. 10.1093/aob/mcf118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang WWP, Huang L, Shen M, Webster C, Burlingame AL, Roberts JKM. Patterns of protein synthesis and tolerance of anoxia in root tips of maize seedlings acclimated to a low-oxygen environment, and identification of proteins by mass spectrometry. Plant Physiol. 2000; 122: 295–317. doi: 10.1104/pp.122.2.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nishizawa A, Yabuta Y, Shigeoka S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008; 147: 1251–1263. doi: 10.1104/pp.108.122465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneider T, Keller F. Raffinose in Chloroplasts is Synthesized in the Cytosol and Transported across the Chloroplast Envelope. Plant Cell Physiol. 2009; 50(12): 2174–2182. doi: 10.1093/pcp/pcp151 [DOI] [PubMed] [Google Scholar]
- 65.Gu L, Zhang Y, Zhang M, Li T, Dirk LM, Downie B, et al. ZmGOLS2, a target of transcription factor ZmDREB2A, offers similar protection against abiotic stress as ZmDREB2A. Plant Mol Biol. 2016; 90(1–2): 157–170. doi: 10.1007/s11103-015-0403-1 [DOI] [PubMed] [Google Scholar]
- 66.Ayako N, Yukinori Y, Shigeru S. Galactinol and Raffinose Constitute a Novel Function to Protect Plants from Oxidative Damage. Plant Physiol. 2008; 147(3): 1251–1263. doi: 10.1104/pp.108.122465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen YH, Yan QQ, Xiao GY. Research Progress of Rice Tolerance to Flooding. Chinese Agricultural Science Bulletin. 2005; 21(12): 151–153, 159. 10.3969/j.issn.1007-7731.2008.18.102 [DOI] [Google Scholar]
- 68.Zhu GC, Luo CM. Research progress on the anatomical structure of gramineous plants under flooding stress. Anhui Agricultural Science Bulletin. 2008; 14(18): 166–167, 195. 10.3969/j.issn.1007-7731.2008.18.102 [DOI] [Google Scholar]
- 69.Phetsirikoon S, Paull RE, Chen N, Ketsa S, Doorn WGV. Increased hydrolase gene expression and hydrolase activity in the abscission zone involved in chilling-induced abscission of Dendrobium flowers. Postharvest Biol Technol. 2016; 117: 217–229. 10.1016/j.postharvbio.2016.03.002 [DOI] [Google Scholar]
- 70.Cone KC, Burr FA, Burr B. Molecular analysis of the maize anthocyanin regulatory locus C1. Proc Natl Acad Sci USA. 1986; 83(24): 9631–9635. doi: 10.1073/pnas.83.24.9631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, et al. The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell. 2011; 3(3): 1000–1013. doi: 10.1105/tpc.111.083089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ruan MB, Guo X, Wang B, Yang YL, Li WQ, Yu XL, et al. Genome-wide characterization and expression analysis enables identification of abiotic stress-responsive MYB transcription factors in cassava (Manihot esculenta). J Exp Bot. 2017; 68(13): 3657–3672. doi: 10.1093/jxb/erx202 [DOI] [PubMed] [Google Scholar]
- 73.Hoeren FU, Dolferus R, Wu YR, Peacock WJ, Dennis ES. Evidence for a Role for AtMYB2 in the Induction of the Arabidopsis Alcohol Dehydrogenase Gene (ADH1) by Low Oxygen. Genetics. 1998; 149(2): 479–490. doi: 10.1093/genetics/149.2.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee TG, Jang CS, Kim JY, Seong RC, Kim IG, Kim DS, et al. Expressed Sequence Tags from Wheat Roots under Hypoxia. Russ J Plant Physiol. 2007; 54: 659–668. 10.1134/S1021443707050147 [DOI] [Google Scholar]
- 75.Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature. 2009; 460(7258): 1026–1030. doi: 10.1038/nature08258 [DOI] [PubMed] [Google Scholar]
- 76.Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, et al. Arabidopsis RAP2.2, an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010; 153(2): 757–772. doi: 10.1104/pp.110.155077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gibbs DJ, Lee SC, Isa NM, Gramuglia S, Fukao T, Bassel GW, et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature. 2011; 479(7373): 415–418. doi: 10.1038/nature10534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kosmacz M, Parlanti S, Schwarzländer M, Kragler F, Licausi F, Dongen JT. The stability and nuclear localization of the transcription factor RAP2.12 are dynamically regulated by oxygen concentration. Plant Cell Environ. 2015; 38(6): 1094–1103. doi: 10.1111/pce.12493 [DOI] [PubMed] [Google Scholar]
- 79.Nanjo Y, Maruyama K, Yasue H, Yamaguchi-Shinozaki K, Shinozaki K, Komatsu S. Transcriptional responses to flooding stress in roots including hypocotyl of soybean seedlings. Plant Mol Biol. 2011; 77(1–2): 129–144. doi: 10.1007/s11103-011-9799-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.