Abstract
Access to electron acceptors supports oxidized biomass synthesis and can be limiting for cancer cell proliferation, but how cancer cells overcome this limitation in tumors is incompletely understood. Nontransformed cells in tumors can help cancer cells overcome metabolic limitations, particularly in pancreatic cancer, where pancreatic stellate cells (PSCs) promote cancer cell proliferation and tumor growth. However, whether PSCs affect the redox state of cancer cells is not known. By taking advantage of the endogenous fluorescence properties of reduced nicotinamide adenine dinucleotide and oxidized flavin adenine dinucleotide cofactors we use optical imaging to assess the redox state of pancreatic cancer cells and PSCs and find that direct interactions between PSCs and cancer cells promote a more oxidized state in cancer cells. This suggests that metabolic interaction between cancer cells and PSCs is a mechanism to overcome the redox limitations of cell proliferation in pancreatic cancer.
Interactions between pancreatic stromal cells and pancreatic cancer cells affect the redox state of the cancer cells.
INTRODUCTION
Tumors are a heterogeneous collection of cell types, and cancer cells must adapt their metabolism to use available nutrients and proliferate (1). A prominent feature of pancreatic ductal adenocarcinoma (PDAC) is that cancer cells are a relatively rare cell type in tumors, with interactions between cancer cells and noncancer cells affecting many properties of this cancer including disease progression and drug sensitivity (2–6). Diverse populations of stromal and immune cells within the tumor microenvironment can also influence nutrient utilization, and there is evidence for metabolic competition and cooperation between cancer cells and other cell types in pancreatic cancer (2, 3, 5, 7–9). Efforts to target the desmoplastic stroma, including pancreatic stellate cells (PSCs), have been challenging because stromal cells can both promote and restrain tumor progression (10–14). Thus, identifying specific ways that stromal cells such as PSCs interact with cancer cells to promote tumor growth is necessary to target the stroma and improve pancreatic cancer treatment.
Oncogenic activation of KRAS is an early event in pancreatic tumor progression that promotes metabolic reprogramming of cancer cells and the retention of stromal cells within the tumor (15). Moreover, activation of PSCs, a major resident cell type within the pancreas, is observed before tumor formation, suggesting that cooperative interactions between PSCs and cancer cells influence early disease progression (16). Cancer-associated fibroblasts (CAFs), including activated PSCs, can also affect therapy response. CAFs and associated desmoplasia can account for up to 90% of the tumor, leading to a high interstitial pressure that limits drug delivery (17). CAFs can also produce metabolites that compete for uptake of nucleoside analog chemotherapy (2). As a result, the presence of CAFs, including PSCs, can promote pancreatic tumor growth and affect response to therapy (9, 18, 19).
Pancreatic cancer organoids derived from genetically engineered mouse models mimic some aspects of human PDAC and can be used to study tumor and stromal cell interactions in a coculture setting where morphology-based identification of cancer cells and PSCs is feasible (4, 9, 20, 21). Another advantage of pancreatic cancer organoid cocultures is that metabolic interactions between two defined cell types can be studied in an in vitro setting that is more tractable for metabolic studies than analysis of whole tumors (4, 9, 22). PSCs can support the growth of tumor organoids in minimal media conditions that are not conducive to culture of organoids derived only from cancer cells (4, 9). This argues that coculture of cancer cells and PSCs in organoid cultures is a system where metabolic interactions between cell types are present. However, exactly which media conditions can be replaced by PSCs to support cancer cell proliferation are not known.
Access to electron acceptors to support regeneration of oxidized nicotinamide adenine dinucleotide (NAD+) as a cofactor for oxidized biomass synthesis can be limiting for cancer cell proliferation (23–31). This problem is particularly apparent in PDAC tumors, where levels of oxygen, an important biological electron acceptor, are extremely low (32). Thus, how pancreatic cancer cells dispose of excess electrons to regenerate NAD+ is unknown, and one hypothesis is that interactions with PSCs allow cancer cells to overcome this metabolic limitation of tumor growth.
The rapid time scales of metabolic processes can be an impediment to studying metabolic interactions between cell types in a mixed population. The time needed to dissociate either organoids or tumors and isolate individual cell types is long relative to the turnover of many metabolites, and dissociating and sorting cells can affect metabolic pathways known to be responsive to cell stress (9, 33–35). One approach that circumvents these limitations is imaging metabolic changes in intact systems such as organoids and tumors. A parameter that is amenable to imaging with minimal perturbation of cell-cell interactions is assessment of cell redox state, which can be assessed at the single-cell level even when multiple cell types are present. An optical redox ratio can be measured using label-free, high-resolution multiphoton microscopy of redox cofactor autofluorescence (36–39). This technique takes advantage of the fact that only the reduced form of NAD+ (NADH) and nicotinamide adenine dinucleotide phosphate (NADPH) and the oxidized form of flavin adenine dinucleotide (FAD) are fluorescent.
NADPH has overlapping spectral properties with NADH; hence, the autofluorescence measured is derived from both NADH and NADPH. However, while the NADH/NAD+ ratio in cells is less than the NADPH/NADP+ ratio, these ratios are coupled, suggesting that changes in NAD(P)H fluorescence can be a surrogate for changes in the reduced state of these cofactors (40–44). Likewise, the FAD/FADH2 ratio is coupled to redox reactions in pathways that also involve NAD(P)H, enabling changes in FAD fluorescence to reflect changes in oxidized cofactors in oxidation-reduction reactions (37, 45, 46). Thus, the optical redox ratio, defined as the ratio of fluorescence intensity of NAD(P)H to FAD, provides a quantitative measurement of the relative oxidation-reduction state of individual cells (36, 38, 47). This optical redox measurement has been used to study the metabolic state of myriad cell types including cell populations found in tumors such as cancer cells (48–50) and immune cells (51–53). Multiphoton microscopy also allows optical sectioning to accommodate metabolic imaging of three-dimensional (3D) biological samples including organoids and intravital imaging of mouse tumors (36, 54, 55). Last, the nondestructive nature of the technique allows for measurements of redox state over time at a single-cell level without dissociating cells.
In this study, we use optical redox imaging to show that PSCs can affect the redox state of pancreatic cancer cells in organoid culture. Physical interactions between these cell types promote a more oxidized state in cancer cells, arguing that a metabolic symbiosis exists whereby fibroblasts facilitate oxidation reactions in cancer cells to support proliferation.
RESULTS
Coculture with PSCs supports pancreatic cancer cell proliferation in the absence of exogenous pyruvate
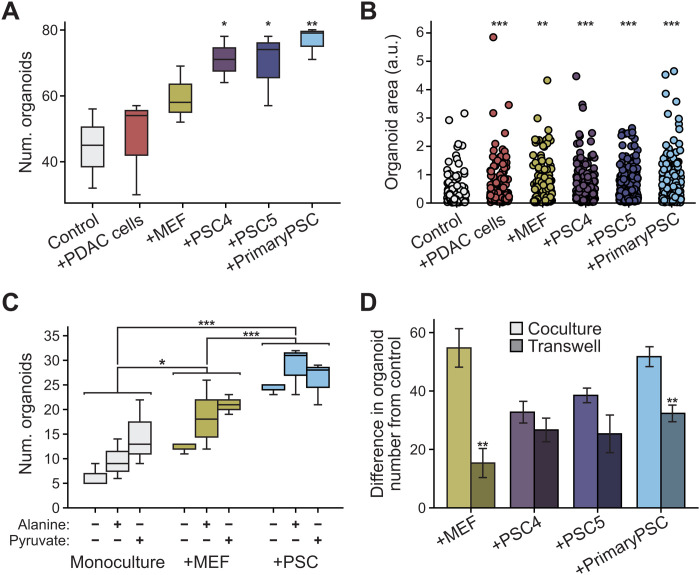
Prior studies have found that PSCs can secrete metabolites and other factors that affect pancreatic cancer cell metabolism and proliferation in vivo and in organoid culture (2, 4, 7, 9, 56). To further examine how PSCs might support pancreatic cancer cell metabolism and enable organoid growth, we cocultured primary or immortalized murine PSCs (4, 9) with PDAC cells from tumors arising in the LSL-KrasG12D; Trp53fl/fl; Pdx1-Cre (KP−/−C) autochthonous mouse PDAC model (57) as 3D organoids. Because prior work suggested that coculture with PSCs can enable organoid growth in minimal media (4, 9), we first examined whether contact between PSCs and cancer cells was necessary for this phenotype using a transwell system. In this experiment, media is shared between PDAC cancer cells grown as 3D organoids and 2D cells grown on the transwell inserts, but organoids are not in physical contact with PSCs or other cell types included in the transwell system. Specifically, PDAC cells cultured as 3D organoids in minimal media were seeded in the bottom chamber in Matrigel domes, and murine PSCs, mouse embryonic fibroblasts (MEFs), or PDAC cells were seeded in the upper chamber of the transwell system (fig. S1). We found that culturing MEFs or PSCs in the upper chamber of the transwells increased the number of organoids formed in the lower chamber when compared with the number of organoids observed when cultured without cells added to the upper chamber of the transwell or when PDAC cells were seeded in the upper chamber (Fig. 1A). This increase in organoid number was similar when either primary or immortalized PSCs were added to the upper well of the transwell, suggesting that these two cell types have a similar effect on organoid growth in this context. Given the similar findings observed with both primary and immortalized PSCs, we chose to focus our studies on immortalized PSCs for two reasons. First, immortalized PSCs allow access to sufficient material for repeated experiments evaluating cancer cell–PSC cocultures. Second, the use of immortalized PSCs obviates the need to rederive fresh primary cells for each experiment, which may exhibit variability across isolates. To capture the desmoplasia observed in pancreatic tumors, PSCs were cultured in 5- to 10-fold excess to cancer cells in all coculture studies. Although culturing PDAC cell organoids with any other cell type added to the upper chamber of the transwells increased the area of a subset of the organoids that formed (Fig. 1B), organoid number was specifically enhanced when cultured with PSCs (Fig. 1A). These results are in agreement with previous observations that both cocultures and transwell cultures involving PSCs and PDAC cells can increase cancer cell proliferation (4, 7, 9). Moreover, these data argue that interactions with other cell types can affect organoid size and number differently.
Fig. 1. Pancreatic stellate cells support pancreatic cancer cell organoid growth.
(A and B) Murine PDAC cells were cultured as 3D organoids in Matrigel domes in the bottom transwell chamber alone (control) or with PDAC cells (+PDAC cells), MEFs (+MEF), immortalized PSCs (+PSC4 and +PSC5), or primary PSCs (+primary PSC) in standard 2D culture in the upper transwell chamber. (A) Organoid number (num. organoids) and (B) relative area of organoids, quantified from bright-field images. At least three images from three independent wells were assessed per condition; significance was determined using one-way analysis of variance (ANOVA) with post hoc Dunnett’s test. (***P < 0.001; *P < 0.05). Error bars, 95% confidence interval. (C) PDAC cells cultured as 3D organoids alone (monoculture) or cocultured in Matrigel with MEFs (+MEF) or immortalized PSCs (+PSC). Cells exposed to DMEM without pyruvate, supplemented with dialyzed serum and 1 mM alanine or 1 mM pyruvate. Organoid number quantified from bright-field images from three independent wells per condition; significance determined using one-way ANOVA with post hoc Tukey’s test. (***P < 0.001; *P < 0.05). Error bars, 95% confidence interval. (D) Difference in organoid number between PDAC cells as 3D organoids alone (control) or in coculture with MEF, PSC4, PSC5, or primary PSC (light-colored bars, calculated from data in fig. S2E). Also plotted are differences in organoid number for PDAC cells cultured as 3D organoids (control, no cells) or cultured in a transwell bottom chamber with indicated cells in the upper chamber [darker-colored bars, calculated from data in (A)]. For all conditions, cultures were grown in media without exogenous pyruvate or alanine. n = 4 images (cocultures), n = 3 images (transwells) analyzed per condition. Significance assessed using t test **P < 0.01. Error bars, SD. a.u., arbitrary units.
To examine whether specific metabolites could partly explain the observed phenotypic differences, we next explored which metabolites in PSC- and cancer cell–conditioned media could be promoting the increase in pancreatic cancer organoid number. Because alanine has been shown to be secreted by PSCs (7) and pyruvate has been shown to promote proliferation of multiple cancer cell types including pancreatic cancer (23, 24, 58–60), we examined whether the organoid growth-promoting role of PSCs could be replaced by addition of exogenous pyruvate or alanine to the media of organoid monocultures (fig. S1). Pyruvate addition had a larger effect than alanine, and notably, when grown in monoculture, the presence of exogenous pyruvate approximately doubled pancreatic cancer cell organoid number (Fig. 1C and fig. S2, A to C). The increase in organoid number was similar to that observed with MEF coculture, although not as great an increase as that observed with PSC coculture. A possible trend toward a slight increase in organoid size was observed when either exogenous alanine or pyruvate is added to cancer cell organoid monocultures, although this difference was not significant (fig. S2D). These results indicate that exogenous pyruvate can promote increased numbers of pancreatic cancer-derived organoids and that including pyruvate in the medium can partially replace access to PSCs to increase the number of organoids.
PSCs can alter cancer cell growth and proliferation in many ways, independent of media conditioning. To further examine whether the growth-promoting effects of stromal cells supersedes the effects of pyruvate or alanine addition, PDAC cells grown as organoids were cocultured with immortalized PSCs or MEFs (fig. S1), with and without addition of these exogenous metabolites (Fig. 1C and fig. S2, A to D). Compared to organoid monocultures containing only PDAC cells that were supplemented with pyruvate or alanine, we observed a further increase in the total number of organoids when cocultured with MEFs (P < 0.05), and this effect was further increased when the PDAC cells were cocultured with PSCs (P < 0.001) (Fig. 1C and fig. S2C). Of note, this effect was observed regardless of whether alanine or pyruvate was supplemented in the media (P < 0.001) (Fig. 1C and fig. S2B). Coculture with MEFs and PSCs may also cause a slight increase in the size of some organoids compared to monoculture; however, the overall difference in size observed was not significant (fig. S2D). Of note, exogenous pyruvate has a greater effect on organoid number in monoculture than it does in coculture with PSCs (Fig. 1C and fig. S2A), suggesting that exogenous pyruvate provides less of an added growth benefit in the presence of PSCs. Similarly, organoid number was increased when organoids were in coculture with stromal cells to a greater degree than that observed when pyruvate is added to monocultures (Fig. 1C and fig. S2E). Notably, coculture with MEFs or PSCs increased organoid size and number more than was observed following pyruvate addition to monocultures or when including stromal cells in transwell cultures (Fig. 1D and fig. S2F). Moreover, these data suggest that PSCs are not unique in their ability to promote organoid growth, in agreement with previous work suggesting that both PSCs and MEFs may share some metabolic properties (2). We next examined whether coculture of organoids with PSCs also enhances the growth of PSCs relative to PSCs cultured alone. Consistent with prior work (4), we observed that coculture with PDAC cells grown as 3D organoids enhanced the growth of two independently derived PSC cell lines (fig. S2, G and H). Together, these data argue that interactions between cancer cells and stromal cells can reduce the requirement for pyruvate or alanine addition to the media. Furthermore, physical contact between cell types may enhance both pancreatic cancer cell and stromal cell proliferation, with physical interactions mitigating the benefits of exogenous pyruvate for cancer cell proliferation.
Optical redox imaging reveals redox differences between cell types in PDAC cancer cell–PSC cocultures
Both pyruvate and alanine can contribute carbon to central carbon metabolism, but unlike alanine, which has a redox state that is equivalent to lactate (61), exogenous pyruvate can serve as an electron acceptor to support biomass production, which can be limiting for proliferation in some contexts (23, 24, 26, 58). To study whether metabolic interactions between PDAC cells and PSCs can overcome a requirement for pyruvate as an electron acceptor to support cancer cell proliferation, we first asked whether PDAC cells and PSCs have different redox states when cultured individually in standard cell culture conditions. We measured the NADH/NAD+ ratio in murine PDAC cells derived from the KP−/−C or LSL-KrasG12D; Trp53LSL-R172H; Pdx1-Cre (KPC) mouse models and in independently derived immortalized mouse PSCs that did or did not express green fluorescent protein (GFP), which were cultured in media with or without pyruvate. We found that each cell line had a distinct redox ratio that was variably affected by pyruvate addition (fig. S3A). These data suggest that in monoculture, PDAC cells and PSCs do not exhibit consistent differences in redox state.
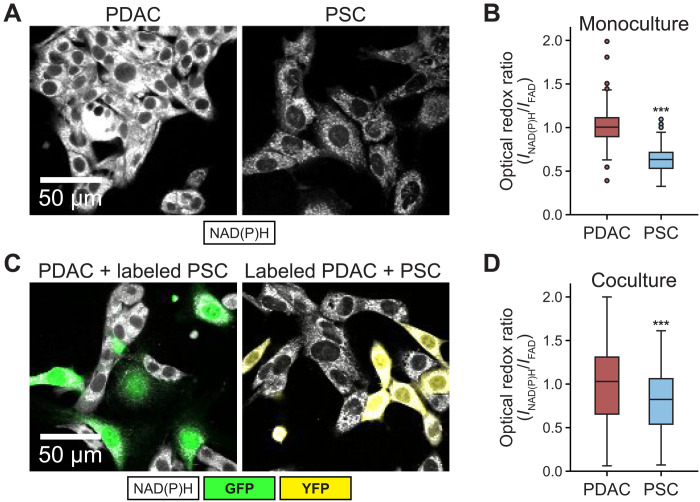
To determine whether coculture affects redox state in PDAC cells or PSCs, we took advantage of the fact that the optical redox ratio, measured using autofluorescence of NAD(P)H and FAD (i.e., INAD(P)H/IFAD), allows assessment of redox state in different cell types in a mixed population. First, we confirmed that the optical redox ratio reflected the NADH/NAD+ ratio observed in PDAC cells and PSCs in 2D monoculture (Fig. 2, A and B, and fig. S3A). Next, we performed optical redox imaging of PDAC cell–PSC cocultures. We find that the optical redox ratios of the PDAC cells and PSCs in 2D coculture were closer to each other than the optical redox ratio observed for each cell type in monoculture (Fig. 2, A to D, and fig. S3B). The optical redox ratio of PDAC cells was more oxidized in coculture with PSCs than it was in monoculture (fig. S3b), while the optical redox ratio of PSCs, even when imaged after 48 hours, was less affected by coculture (fig. S3, B to D). A difference in optical redox ratio was also observed between PSCs and PDAC cells in coculture when the cancer cells were derived from the widely used KPC autochthonous PDAC mouse model (fig. S3, E and F). Together, these data suggest that PSCs can affect the redox state of PDAC cancer cells in coculture conditions.
Fig. 2. PDAC cells and PSCs in coculture exhibit different redox states.
(A) Representative NAD(P)H intensity images of PDAC cells and PSCs in monoculture. (B) Quantification of optical redox ratio of PDAC cells and PSCs in monoculture (two independent cultures per cell type were analyzed), normalized to PDAC cells, at single-cell level (PDAC cells: n = 227; PSCs: n = 265). (C) For imaging cocultures, either PDAC cells or PSCs were fluorescently labeled with YFP or GFP, respectively, to distinguish each cell type. Two distinct cocultures were prepared with one labeled and one unlabeled cell type, and optical redox ratios were assessed in the unlabeled cell type to circumvent interference of fluorescent labels with FAD signal due to spectral overlap. For both cultures, PDAC cells were mixed with PSCs in a 1:1 ratio and cultured in standard 2D conditions for 24 hours before assessment of the optical redox ratio. Left: Representative NAD(P)H intensity (white) images of unlabeled PDAC cells in coculture with GFP+ PSCs (labeled PSC, green). Right: Representative NAD(P)H intensity (white) images of PDAC cells labeled with YFP (labeled PDAC, yellow) in coculture with unlabeled PSCs. (D) Quantification of optical redox ratio of PDAC cells and PSCs from two independent cultures per condition as shown in (C). Optical redox ratio was computed for only unlabeled cells, at single-cell level (PDAC cells: n = 91; PSCs: n = 119) and normalized to PDAC cells. Error bars in (B) and (D) are the 95% confidence interval. Statistical significance for (B) and (D) were assessed using one-way ANOVA with Tukey’s post hoc test (***P < 0.001).
PDAC cells and PSCs exhibit redox differences in 3D organoid cocultures
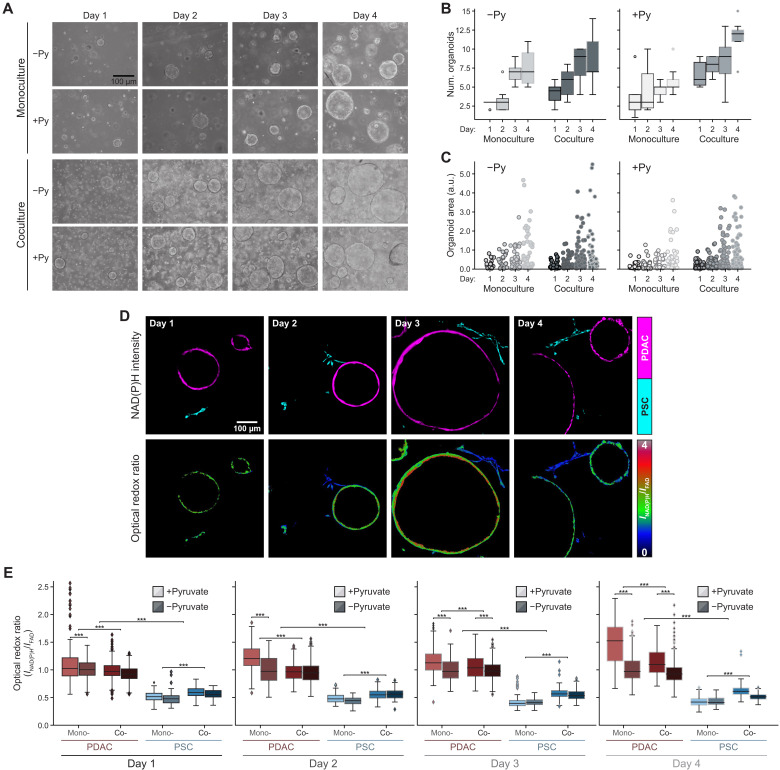
We next asked whether the differences in redox state between PDAC cells and PSCs in coculture were retained in 3D organoid cocultures where PSCs can replace pyruvate addition to the media to support growth. In addition, because optical redox ratio imaging is nondestructive and amenable to analysis of 3D samples at single-cell level, we assessed the redox states of different cells in organoid cocultures over time, with and without pyruvate addition to the culture media. Quantification of organoid number and size shows an increase in both parameters over time in all the culture conditions evaluated (Fig. 3, A to C, and fig. S4, A and B). In organoid cultures, PDAC cells exhibit a more reduced optical redox ratio compared to PSCs at all time points regardless of whether the different cell types were grown in monoculture or coculture or whether pyruvate was included in the medium (Fig. 3, D and E; see also table S1). These data argue that cancer cells in organoid cultures are in a more reduced metabolic state than PSCs.
Fig. 3. Differences in redox state between PDAC cells and PSCs are diminished in organoid coculture.
(A) Representative bright-field images of PDAC cells cultured as 3D organoids in monoculture or in coculture with immortalized PSCs (PSC) in DMEM supplemented with or without 10 mM exogenous pyruvate (Py) as indicated. Images are shown from days 1 through 4 after plating. The number of organoids (num. organoids) formed (B) and the area for each organoid (C) were quantified from bright-field images of three independent cultures per condition on each day as indicated. Data shown is the average value per image per condition (n = 4 images analyzed per condition). Statistical significance for (B) and (C) was assessed using one-way ANOVA with Tukey’s post hoc test, and these same data are presented differently in fig. S4 (A and B) for all the conditions. (D) Representative NAD(P)H fluorescence intensity images of PDAC cells cultured as 3D organoids (shown in magenta) and PSCs (shown in cyan) from cocultures obtained on days 1 through 4 after plating as indicated (top). The corresponding optical redox ratio from the same images is also shown (bottom). (E) Quantification of optical redox ratios of PDAC cells cultured as 3D organoids or PSCs cultured in 3D either in monoculture (mono-) or coculture (co-) for days 1 through 4. The cultures were grown in DMEM supplemented with or without 10 mM exogenous pyruvate (Py) indicated with light and dark colors, respectively. Optical redox ratio is analyzed for n = 6 images from the three independent cultures per condition per day. The data are computed at the single-cell level and normalized to PDAC cells in monoculture, grown without exogenous pyruvate for each day. The number of cells segmented per condition is presented in table S1. Statistical significance was accessed using one-way ANOVA with Tukey’s post hoc test (***P < 0.001;). All error bars are the 95% confidence interval.
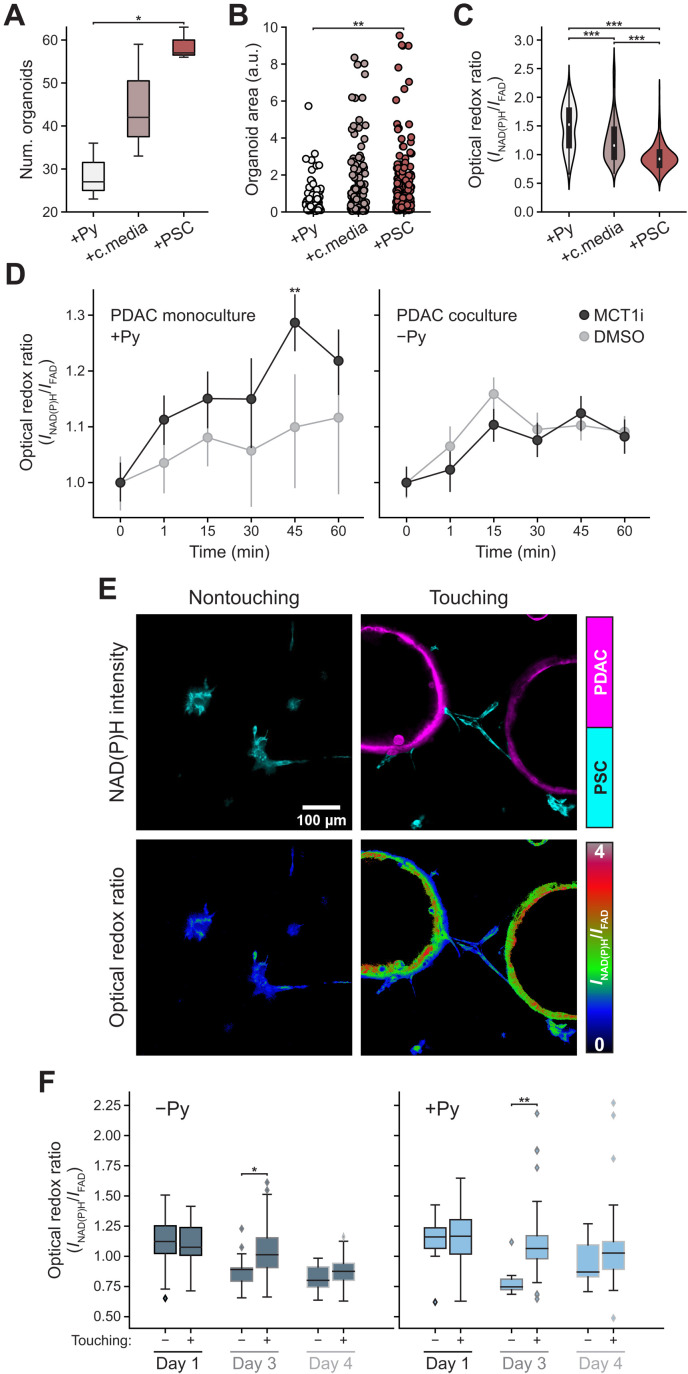
The difference in optical redox ratio between mono- and coculture conditions with pyruvate shows that cancer cells in organoid coculture with PSCs become more oxidized by day 4 compared to cancer cells in organoid monoculture (Fig. 3E). Conversely, PSCs in coculture conditions with pyruvate become more reduced compared to the monoculture condition (Fig. 3E). The difference in optical redox ratio between PDAC cells and PSCs is also greater in monoculture than in coculture for each day (fig. 4C), suggesting that PSCs may promote a more oxidized state in cancer cells.
Fig. 4. Physical interactions between PSCs and PDAC cells influence the redox state of both cell types and organoid growth.
(A and B) Organoid number (A) and area (B) of PDAC cells in media with 10 mM pyruvate (+Py) for 4 days, DMEM media for 2 days and then PSC conditioned media (+c.media) added for 2 days, or coculture with PSCs (+PSC) for 4 days. Each data point is one organoid (n = 6 images per condition from three independent cultures). One-way ANOVA with post hoc Tukey’s test. (C) Optical redox ratio from (A) for each PDAC cell segmented from organoids and normalized to PDAC cells without pyruvate. n (+Py) = 201; n (+c.media) = 329; n (+PSC) = 347. Some data are subset of data in Fig. 3E. One-way ANOVA with post hoc Tukey’s test. (D) PDAC cells as 3D organoids in media with 10 mM pyruvate (PDAC monoculture +Py) or coculture with PSCs without pyruvate (PDAC coculture −Py) both for 3 days, followed by MCT1 inhibitor [AZD 3965 (100 nM)] or dimethyl sulfoxide (DMSO). Optical redox ratio at pretreatment (time 0) or indicated time. Optical redox ratio for each PDAC cell (min n = 300 cells) segmented from organoids normalized to 0 min; n = 6 images per condition per time point from three independent cultures. ANOVA and Tukey’s post hoc test. (E) NAD(P)H intensity images of PSCs (cyan) of nontouching (left) or touching (right) reflecting proximity to PDAC cells as organoids (magenta) in cocultures (top) or corresponding optical redox images (bottom). PSCs within 454-μm field-of-view of each organoid assigned to touching group, rest to nontouching group. (F) Optical redox ratio of nontouching or touching PSCs with or without 10 mM pyruvate (Py) for indicated time. n = 6 images from three independent cultures. Computed at the single-cell level; normalized to PDAC cells in monoculture without pyruvate for each day. Cells per condition, table S2. T test. (***P < 0.001; **P < 0.01; *P < 0.05).
The optical redox differences between PDAC cells and PSCs observed were similar when, instead of imaging the same dish daily for 4 days as shown in Fig. 3, the cells were plated as 3D organoid mono- and cocultures every 24 hours, and all the cultures were imaged on day 4 (fig. S4, D and E; see also table S2). These data suggest that the measured differences in redox ratio in cocultures are present across multiple biological replicates. The pyruvate and lactate in the 3D culture media were also assessed on days 3 and 4. Of note, the optical redox ratio of 3D PDAC cells in monoculture correlated with the pyruvate to lactate ratio present in the culture media, while the optical redox ratio of both cell types was decoupled from the pyruvate to lactate ratio in coculture (fig. S4F). Select additional metabolites in the 3D culture media were also measured on days 3 and 4, but no observed changes were sufficient to explain how PSCs might affect the redox state of cancer cells (fig. S5 and table S3). These data support a model in which media metabolite changes alone are insufficient to fully explain the effect of PSCs on organoid growth.
Organoid coculture with PSCs alters cancer cell redox state despite inhibition of pyruvate uptake
We hypothesized that perturbing the redox state of organoids could also improve organoid growth in media without pyruvate. Expression of the NADH oxidase LbNox has previously been used to modulate redox state following response to mitochondrial electron transport inhibition (26). Thus, we tested whether LbNox expression affects PDAC cancer cells in monoculture without pyruvate and found that it did not alter cancer cell redox state or organoid growth (fig. S6, A to C). In addition, LbNox expression in PSCs did not further enhance the ability of PSCs to promote PDAC organoid growth in media without pyruvate (fig. S6, E and E). To test whether the enhanced organoid growth observed in coculture can be explained by exchange of pyruvate or another metabolite between the organoids and the PSCs, we cultured the organoids alone in either pyruvate, PSC-conditioned media, or in coculture with PSCs. We observe that while conditioned media does enhance both organoid number and area, the physical presence of PSCs provides an additional growth advantage to organoids, and this is also accompanied by a more oxidized cancer cell redox ratio (Fig. 4, A to C), arguing that PSC-derived factors alone are not responsible for the redox changes in cancer cells. We also examined whether pharmacological inhibition of monocarboxylate transporter 1 (MCT1), which can mediate exchange of pyruvate and other monocarboxylates across the cell membrane (62), had an effect on the optical redox ratio of cancer cells grown as organoids in monoculture or coculture with PSCs. We found that, consistent with an inhibition of pyruvate uptake, acute MCT1 inhibition with AZD 3965 makes the optical redox ratio of cancer cells more reduced in monoculture but does not change the optical redox ratio of cancer cells in coculture with PSCs (Fig. 4D). Together, these data support a model where PSCs affect the redox state of PDAC cancer cells in coculture via mechanisms other than by excretion of pyruvate or another factor into the media.
Physical contact with PSCs facilitates redox changes in cancer cells
In considering how PSCs might affect the redox state of PDAC cells, we noted that PSCs spread toward the PDAC cells grown as 3D organoid cocultures, suggesting that PSCs wrap around the PDAC cell organoids or form bridges between different organoids by days 3 and 4, respectively (Fig. 3D). Of note, there is evidence that mechanical interactions between PDAC cells and some populations of PSCs can provide some of the growth promoting benefits of coculture (4), although whether cell-cell interactions may be modulating a change in redox state is unknown. Thus, to investigate the possibility that physical contact with PSCs might influence the redox state of PDAC cells, PSCs were divided into “touching” and “nontouching” subsets based on their spatial proximity to the organoids in the same coculture (Fig. 4, E and F). The single-cell level optical redox ratio was calculated for PSCs in each group for days 1, 3, and 4 of coculture to capture time points that were clearly before and after the observed wrapping of PSCs around organoids. There was no difference in optical redox ratio between the two groups on day 1 (Fig. 4F). However, by day 3 the PSCs in the touching group had a significantly more reduced redox state compared to the nontouching group (Fig. 4F). On day 4, the redox differences appear to be maintained, although this difference was no longer statistically significant. Overall, these results suggest that coculture with PSCs supports PDAC cell proliferation through secretion of factors into the media as well as by cell-cell interactions such that the PSCs in contact with cancer cells assume a more reduced redox state than those not in contact with cancer cells. Together, these data suggest that PSCs engage in a metabolic interaction with PDAC cells to promote a more oxidized state in the cancer cells.
Redox differences between different cells are also found in pancreatic tumors
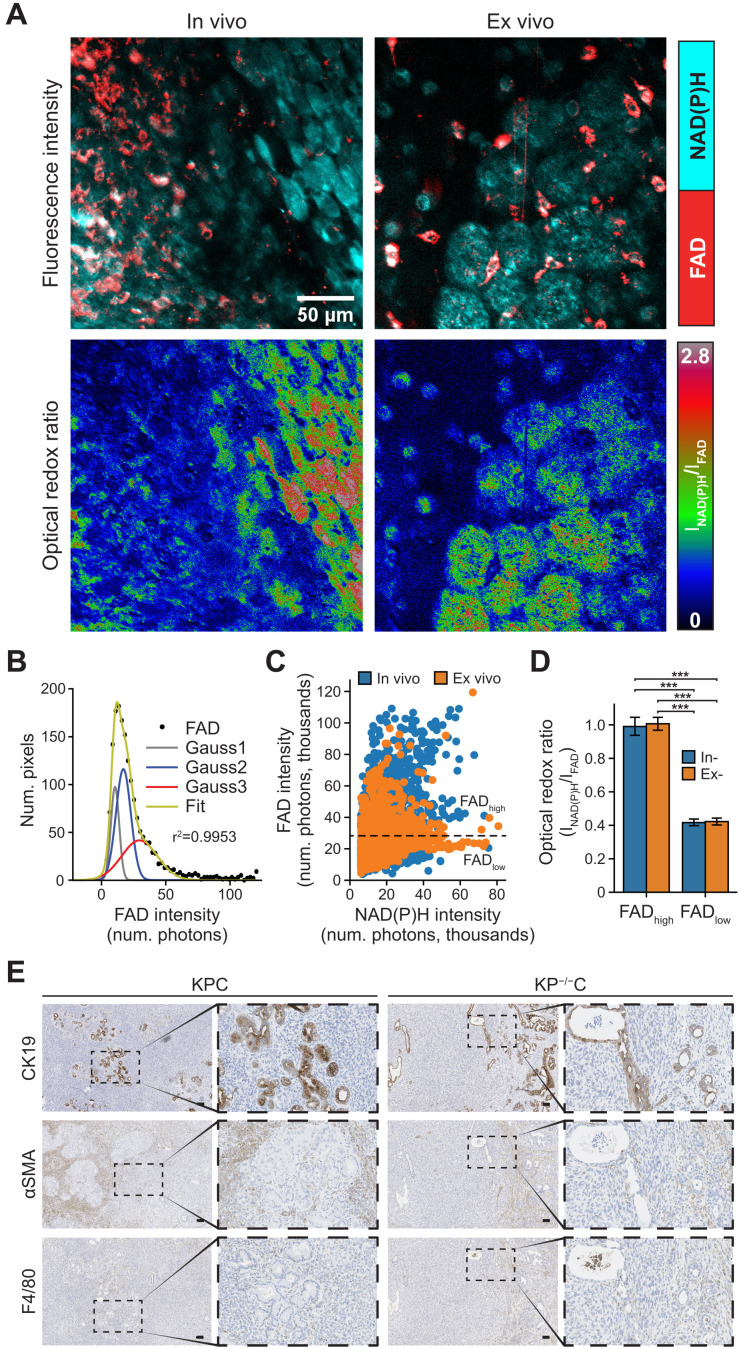
To explore whether the redox differences observed with in vitro culture systems are also reflective of the redox state of different cell types in pancreatic tumors, we used the KP−/−C and KPC pancreatic cancer mouse model to image redox state in tumor tissue in vivo (in live animals) and ex vivo (freshly excised tumor tissue). A heterogeneous FAD intensity distribution was observed across cell types within tumors imaged both in vivo and ex vivo (Fig. 5A). To identify the number of different cell populations with heterogeneous FAD intensity, Gaussian fitting was performed on FAD intensity histograms (Fig. 5B), which resulted in a three Gaussian fit. The first Gaussian (Gauss1, gray) was attributed to low-intensity pixels from background, while the second (Gauss2, blue) and third (Gauss3, red) Gaussians indicated the presence of two cell populations. A scatterplot of FAD versus NAD(P)H intensity shows two populations of cells, one with high FAD intensity (FADhigh) and one with low FAD intensity (FADlow) separated at threshold (black dotted line in Fig. 5C) calculated from Gaussian fits (Fig. 5C). The optical redox ratio calculations of bulk tumor tissue showed FADlow cells and interspersed FADhigh cells with a more oxidized optical redox ratio, with similar observations made when tumor tissue was imaged in vivo or ex vivo (Fig. 5D).
Fig. 5. Cells in pancreatic tumors exhibit distinct redox states.
(A) Representative NAD(P)H (cyan) and FAD (red) intensity images (top) and corresponding optical redox ratio images (bottom), obtained from imaging in vivo and freshly excised (ex vivo) pancreatic tumor tissue. Imaging was performed on tumors and tumor tissue from both LSL-KRasG12D; Trp53 R172H/+; Pdx1-Cre (KPC) and LSL-KRasG12D; Trp53 fl/fl; Pdx1-Cre (KP−/−C) mice. (B) Gaussian fit (green curve) was performed on FAD intensity (black solid circles) with the goodness of fit of (r2 = 0.9953). Gray (Gauss1), blue (Gauss2), and red (Gauss3) curves represent the best three-component Gaussian fits. The intersecting point between Gauss2 and Gauss3 was computed to be 25.3 × 103 photons. (C) Scatterplot of FAD versus NAD(P)H intensity computed from nine in vivo and eight ex vivo tissue images as indicated (n = 4 mice). Each data point is regions of interest (ROIs) segmented from NAD(P)H and FAD intensity images, n (in vivo) = 3990, n (ex vivo) = 4647. Black dotted line is drawn at 25.3 × 103 photons, the intersecting point calculated in (B) and serves as the threshold between high FAD intensity regions (FADhigh) and low FAD intensity regions (FADlow). (D) Optical redox ratio from in vivo and ex vivo images for regions within FADlow (bulk tissue, low FAD intensity) and FADhigh (interspersed high FAD intensity cells) (n = 17 images analyzed). The optical redox ratio for each image was normalized to its FADhigh regions. Statistical significance was tested using one-way ANOVA with post hoc Tukey’s test. (***P < 0.001). Error bars are the SD. (E) Representative immunohistochemistry images of PDAC tumors from KPC and KP−/−C mice stained for the PDAC cell marker CK19 (top), fibroblast marker αSMA (middle), or macrophage marker F4/80 (bottom). Scale bars, 100 μm.
While fluorescent markers can be used to identify cell populations within the tumor and enable cell type–specific studies, these markers also interfere with imaging. Moreover, mouse models were not available with a specific label for PSCs in the far red so as not to interfere with the weak signals from NAD(P)H and FAD fluorescence, and fluorescent antibodies against PSCs are difficult to deliver in vivo and achieve distribution throughout tumor tissue (63). Therefore, we processed parallel unlabeled tumors from the same mouse model to compare how different cell populations are distributed in tumors for comparison to the cell redox distributions observed upon redox imaging of tumor tissue. These studies were done to examine overall heterogeneity across cell populations within the tumor. While we cannot definitively categorize the redox states of specific cells, we can use histological analysis to confirm heterogeneous tumors including cell populations that stain positive for markers of macrophages, PSCs, and epithelial cells (Fig. 5E). Of note, stromal cells are distributed among nests of cancer cells. This is consistent with the dispersed FAD high-cell population representing a stromal cell population in the tumors. When considered along with the findings in organoid cocultures, these data suggest that pancreatic tumors are composed of cell types with distinct redox states and support the hypothesis that stromal cells are more oxidized than cancer cells in pancreatic tumors.
DISCUSSION
The tumor microenvironment affects pancreatic tumor development and progression. Pancreatic tumor organoid model systems that incorporate stromal cells can be used to study the cooperative interactions between cancer cells and the supporting stromal cells (2, 4, 9). Here, we take advantage of these systems and optical redox imaging and find that PSCs assume a more oxidized redox state than cancer cells. This interaction could help pancreatic cancer cells carry out oxidation reactions despite limited access to oxygen as an electron acceptor and direct contact between cancer cells and PSCs appears necessary to maximize this effect.
Optical redox imaging has been previously used to study metabolic interactions between HeLa cells and human skin fibroblasts (64). Consistent with our findings, the fibroblasts were more oxidized compared to cancer cells in 2D monoculture. Optical redox imaging also revealed a more oxidized state for the human MIA PaCa-2 PDAC cell line when cocultured in 3D with patient-derived CAFs compared to monoculture conditions but did not assess the redox state of the CAFs (65). Nevertheless, these studies further support a hypothesis where interactions between cancer cells and stromal cells can promote a more oxidized state in cancer cells.
Access to electron acceptors can limit cancer cell proliferation, as NAD+ regeneration supports oxidized biomass production (23–25, 27, 28, 31, 66). Consistent with this notion, we find cancer cells in both organoid and mouse tumor PDAC models are more reduced than surrounding noncancer cells. Gaining access to exogenous electron acceptors, such as pyruvate, can overcome this redox limitation (23, 24), and secretion of pyruvate by PSCs is one mechanism by which stromal cells might help cancer cells proliferate (59). However, the concentration of pyruvate in interstitial fluid isolated from pancreatic tumors is lower than the concentration of pyruvate found in plasma, suggesting that some cells in pancreatic tumors may not have extensive access to this metabolite in vivo (67). Nevertheless, local exchange of pyruvate and lactate between cells is possible and would not be evident in bulk fluid measurements. It is also conceivable that the NADH/NAD+ ratio varies among different cancer cell populations within tumors and over different stages of disease progression, in alignment with nutrient fluctuations within the tumor microenvironment or based on interactions between cancer cells and stromal cells that vary in both time and space.
In addition to secretion of pyruvate or other metabolites, our findings that the PDAC cancer cell redox ratio in coculture with PSCs is unaltered in the absence of pyruvate and is not affected by MCT1 inhibition suggests that pyruvate exchange alone cannot entirely explain PSCs’ support of organoid growth. Moreover, these data argue that physical interactions between cancer cells and stellate cells also help cancer cells overcome redox limitations of proliferation.
As previously reported, physical interactions between PSCs and PDAC organoids can support proliferation (4) and induce a proinflammatory stromal cells state. How physical interaction with PSCs might alter cancer cell redox state is unknown. The coupling of hydrophobic molecules in plasma cell membranes to transfer electrons is a possibility for how cell contact might mediate redox changes in different cell types, and there is evidence of electron transport in cell membranes other than the mitochondrial membrane, including the plasma membrane (68). Plasma membrane electron transfer has been reported in pancreatic cells (69) and may be involved in how physical interaction with PSCs maintains a more oxidized state in pancreatic cancer cells. Furthermore, recent studies found mitochondrial transfer between nonmalignant cells and cancer cells can promote tumor progression in prostate cancer and glioblastoma (70, 71). It is possible that PSCs modulate the redox state of cancer cells in PDAC through mitochondrial sharing via direct cell-cell contact.
Mechanical forces including those produced by cell-cell interactions and interactions with a more or less stiff extracellular matrix (ECM) can also affect cell metabolism, and fibroblasts can synthesize the ECM proteins in the pancreatic tumor stroma (72, 73). The ECM can influence nutrient uptake and affect cell signaling pathways that regulate metabolism (72). Metabolite exchange has also been reported between cell types in pancreatic cancer cells based on physical contact. For example, cell-cell interactions between the glutamatergic presynaptic protein Netrin G1 in CAFs and its receptor NGL-1 in PDAC cells can play a role glutamate/glutamine sharing between cell types (74). Regardless of the mechanism, when taken together, these data argue that direct interactions between PSCs and cancer cells can promote a more oxidized state in cancer cells and suggest that redox sharing is another way by which cell-cell interactions between cancer cells and stromal cells can support pancreatic cancer cell metabolism and tumor growth.
MATERIALS AND METHODS
Experimental design
Cell culture
To generate PDAC cell lines or organoids for in vitro experiments, cancer cells were isolated from pancreatic tumors arising in the Kras+/LSL-G12D; Trp53+/LSL-R172H; Pdx1-Cre (KPC) or Kras+/LSL-G12D; Trp53−/−; Pdx1-Cre (KP−/−C) mouse models bred onto a C57BL/6J background using established protocols (9). Briefly, PDAC tumors were harvested, minced, and digested in 5 ml of phosphate-buffered saline (PBS) containing cold collagenase I (1 mg/ml; Worthington Biochemical Corporation, LS004194), dispase II (3 mg/ml; Roche, 04942078001) and deoxyribonuclease I (DNase I) (0.1 mg/ml; Sigma-Aldrich, D4527) rotating at 37°C for 30 min. After digestion, EDTA was added to a final concentration of 10 mM, and the solution was filtered through a 70-μm cell strainer and then incubated at room temperature for 5 min before spinning at 200g RCF (Relative Centrifugal Force) for 5 min at 4°C and washed with PBS. For cell lines, cells were resuspended in Dulbecco’s minimum essential medium medium (DMEM) containing pyruvate with 10% heat-inactivated serum and penicillin-streptomycin.
To isolate and immortalized PSCs, GFP+ PSCs were isolated from β-actin-GFP mice on a C57BL6/J background (006567) as previously described (9, 75, 76). Briefly, 3 ml of cold collagenase P (1.3 mg/ml; Sigma-Aldrich, 11213865001) and DNase (0.01 mg/ml; Sigma-Aldrich, D5025) in GBSS (Gey’s Balanced Salt Solution) (Sigma-Aldrich, G9779) were injected into the pancreas. The tissue was then placed into 2 ml of collagenase P solution on ice. Cells were then placed in a 37°C water bath for 15 min. The digested pancreas was filtered through a 250-μm strainer and washed with GBSS with 0.3% bovine serum albumin (BSA). A gradient was created by resuspending the cells in Nycodenz (VWR, 100356-726) and layering in GBSS with 0.3% BSA. Cells were then centrifuged at 1300g for 20 min at 4°C. The layer containing PSCs was removed, filtered through a 70-μm strainer, washed in GBSS with 0.3% BSA, and plated for cell culture in DMEM with 10% FBS and penicillin-streptomycin. GFP+ PSCs were immortalized with TERT and SV40 large T antigen after several passages. Unlabeled, immortalized PSCs were a gift from R. Hynes and D. Tuveson’s laboratories (4). Immortalized MEFs were created as described previously (77). Cell lines were regularly tested for mycoplasma contamination using the MycoAlert Plus Kit (Lonza) or the Mycoprobe Mycoplasma Detection Kit (R&D Systems).
Organoid culture
To generate organoids for in vitro experiments, cells were isolated from pancreatic tumors arising in Kras+/LSL-G12D; Trp53−/−; Pdx1-Cre (KP−/−C) mouse models bred onto the C57BL/6J background using established protocols (9, 20). Cancer cells were isolated directly from mice bearing PDAC tumors and cultured as organoids, referred to here as PDAC cells cultured as 3D organoids. Organoids were never exposed to standard 2D culture conditions and were propagated as organoids as described previously (9, 20). PDAC cells were resuspended in 50 μl of growth factor–reduced (GFR) Matrigel (Corning, 356231) and propagated in organoid feeding media. Specifically, organoid domes were cultured in 500 μl of complete feeding media that consisted of Advanced DMEM/F12 (Thermo Fisher Scientific,12634) containing GlutaMAX (Thermo Fisher Scientific, 35050), penicillin-streptomycin, HEPES (Thermo Fisher Scientific,15630), 0.5 μM transforming growth factor–β (TGF-β) inhibitor A-83-01 (Tocris, 2939), epidermal growth factor (0.05 μg/ml; Thermo Fisher Scientific, PMG8041), fibroblast growth factor (0.1 μg/ml; Peprotech, 100-26), 0.01 μM Gastrin I (Tocris, 3006), Noggin (0.1 μg/ml; Peprotech 250-38), 10.5 μM Rho Kinase Inhibitor Y-27632 (Sigma-Aldrich, Y0503), 1.25 mM N-acetylcysteine (Sigma-Aldrich, A9165), 10 mM nicotinamide (Sigma-Aldrich, N0636), 1× B-27 supplement (Thermo Fisher Scientific,17504), and R-spondin (1 μg/ml). R-spondin was purified from 293 T cells engineered to produce it using the Protein A Antibody Purification Kit (Sigma-Aldrich, PURE1A). For PDAC cells grown as 3D organoids in coculture, cocultures were grown in DMEM without pyruvate (Corning, 10-017-CV) supplemented with 10% dialyzed fetal bovine serum (FBS) and penicillin-streptomycin (minimal media). PDAC cells grown as 3D organoids were regularly tested for mycoplasma contamination using the MycoAlert Plus Kit (Lonza) or the Mycoprobe Mycoplasma Detection Kit (R&D Systems).
For passaging and assessment of cell numbers, PDAC cells grown as 3D organoids were digested to single cells by incubating with dispase (2 mg/ml) in advanced DMEM/F12 with penicillin-streptomycin, HEPES, and GlutaMAX at 37°C for 20 min. PDAC cells grown as 3D organoids were then triturated with a fire-polished glass pipette and enzymatically digested with 1 ml of TrypLE Express (Thermo Fisher Scientific,12605-010) for 10 min rotating at 37°C, followed by addition of 1 ml of dispase-containing media and 10 μl of DNase (10 mg/ml; Sigma-Aldrich, 4527) and digested rotating at 37°C for 20 min or until single cells were visible under a microscope. PDAC cells were counted and plated in GFR Matrigel at a concentration of 2000 cells per well.
For coculture experiments using transwells, 2000 PDAC cells grown as 3D organoids were plated on the bottom of the plate in a GFR Matrigel dome, and 10,000 pancreatic cancer cells, MEFs, or PSCs were plated on the top of the transwell insert in 24-well plates. Organoids were imaged on day 6 after plating.
For coculture experiments of PDAC cells cultured as 3D organoids and stromal cells including addition of exogenous pyruvate or alanine, 2000 PDAC cells and 10,000 MEFs or PSCs were mixed together and plated in a GFR Matrigel dome in 24-well plates. A total of 1 mM alanine or 1 mM or 10 mM pyruvate was added to the culture media where indicated, and cells were imaged on day 6 after plating.
To assess PSC growth in monoculture or in coculture with PDAC cells cultured as organoids, 10,000 GFP+ PSCs or 2000 PDAC cells and 10,000 GFP+ PSCs were mixed together in 70 μl of a 1:1 mixture of GFR Matrigel and media in black 96-well plates on ice. A total of 200 μl of prewarmed media was then added to the top of the wells, and GFP fluorescence was read on a Tecan M200 plate reader daily for 7 days. Background fluorescence of Matrigel was subtracted using Matrigel-only wells as a control. DMEM without pyruvate supplemented with 10% dialyzed FBS was used for these experiments.
Sample preparation for multiphoton imaging
For multiphoton imaging of 2D monocultures, either PDAC cells isolated from the KP−/−C or KPC mouse models or PSCs were seeded (seeding density: 200,000) on 35-mm glass-bottom dishes (#P35G-1.5-14-C, MatTek) with 2 ml of media (DMEM with 10% FBS and 1% penicillin-streptomycin), either 24 or 48 hours before imaging. For 2D cocultures, either fluorescently labeled PDAC cells from the KP−/−C model [labeled with yellow fluorescent protein (YFP)], PDAC cells from the KPC model (labeled with tdTomato), or PSCs (labeled with GFP) were used to distinguish each cell type where indicated during imaging. Of note, the optical redox ratio of cells labeled with GFP, YFP, and tdTomato cannot be measured because these fluorophores obscure the FAD autofluorescence signal due to spectral overlap. Thus, data were obtained from distinct sets of cocultures in this study where only one of the two cells types was labeled. For all cocultures, the two cell types were mixed in 1:1 ratio and cultured for either 24 or 48 hours before imaging.
Multiphoton imaging of 3D culture of organoids and PSCs was performed in 35-mm glass-bottom dishes. PDAC cells cultured as organoids or PSCs were embedded in a Matrigel matrix and overlaid with media (DMEM without pyruvate supplemented with 10% dialyzed FBS and 5% penicillin-streptomycin). For coculture, PSCs (seeding density: 200,000 cells per dish) were mixed with the PDAC cells. For +pyruvate conditions, 10 mM pyruvate (Sigma-Aldrich) was added to the media. For inhibition of MCT1 transporter, 100 nM AZD 3965 (Cayman Chemicals) was added to the media. At least three representative organoids were imaged near the center of the organoid per dish.
Bright-field imaging and analysis
All bright-field images were collected on a Nikon Eclipse Ti-S microscope using 2×, 0.10 numerical aperture (NA) or 4×, 0.13 NA objectives, except that data for Figs. 3 (A to C) and 4 (A and B) were collected on an EVOS XL Core Imaging microscope (Thermo Fisher Scientific) using 20×, 0.40 NA (LPlan PH2, EVOS) and 4×, 0.13 NA (LPlan PH2, EVOS) objectives, respectively. The organoids were segmented and counted manually from bright-field images. Image analysis, organoid segmentation, organoid area, and number calculations were performed using ImageJ software [National Institutes of Health (NIH)] (78).
Multiphoton imaging and optical redox ratio analysis
2D and 3D cultures
Multiphoton imaging of 2D and 3D cultures was performed on a custom-built Ultima Multiphoton Imaging System (Bruker) consisting of an inverted microscope (TI-E, Nikon) coupled to an ultrafast tunable laser source (Insight DS+, Spectra Physics). The data were acquired in photon-counting mode using time-correlated single-photon counting electronics (SPC 150, Becker & Hickl GmbH). Imaging was performed using Prairie View Software (Bruker). The tunable multiphoton laser allowed sequential excitation of NAD(P)H and FAD/YFP/GFP/tdTomato at 750 and 890 nm, respectively. All 2D culture samples were illuminated through a 40× water immersion, 1.15 NA objective (Plan Apo, Nikon) with an image scan speed of 4.8 μs per pixel, 60-s integration time, image size of 256 × 256 pixels, and field of view of 270 μm. 3D culture samples were imaged via a 20× air objective, 0.75 NA (Plan Apo, Nikon) with 1.19× digital zoom, image scan speed of 2.4 μs per pixel, 100-s integration time, image size of 512 × 512 pixels, and field of view of 454 μm. Power at the sample was 4 mW. A dichroic at 720 nm was used to separate excitation from emission signals. The emission filter used for NAD(P)H was bandpass 460/80 nm, while FAD, YFP, and GFP were collected using bandpass 550/100 nm. GaAsP photomultiplier tubes (H7422P-40, Hamamatsu, Japan) were used for detection.
For data collected on photon-counting mode (2D and 3D cultures), the fluorescence decay curve at each pixel was integrated to generate the intensity of each fluorophore using SPCImage software (Becker & Hickl GmbH). For all data, fluorescence intensity image analysis was performed using ImageJ software (NIH) (78).
Single- cell segmentation was performed using a customized CellProfiler pipeline that has been previously applied to 2D cells and 3D organoid images (79). Briefly, the nuclei of each cell was first manually segmented in the NAD(P)H fluorescence intensity image. This was set as the primary object and served as seed areas. Next, the boundary for each cell was automatically detected using the Voronoi-based segmentation (80). This is a propagation method that identifies boundaries between cells based on intensity gradients and proximity to the primary object. Last, the cytoplasm masks for each cell were generated by subtracting the cell nucleus from the cell boundary. Figure S7 shows representative NAD(P)H intensity images and corresponding nuclei, cell boundary, and cytoplasm masks for 2D and 3D samples. Cytoplasm masks were applied to all images to determine single-cell optical redox ratio using Eq. 1 where I represents the intensity of the fluorophore at each pixel
| (1) |
For 2D cocultures, the optical redox ratio from only unlabeled cells in cocultures was computed by eliminating cells with GFP, YFP, or tdTomato fluorescence signal in the imaged region.
In vivo and ex vivo samples
All animal work was done in accordance with the guidelines and procedures approved by Massachusetts Institute of Technology Committee on Animal Care. For in vivo imaging, the mice were anesthetized by intraperitoneal injection of Avertin. A small vertical incision on the left side at the level of the pancreas was made, followed by pancreatic tumor exteriorization. The exposed organ was orientated so that the tissue was underneath the animal, pressed gently on a glass slide dish for imaging. For ex vivo imaging, pancreatic tumor tissue was excised and placed on the glass coverslip of 35-mm glass-bottom dishes. All ex vivo imaging was performed within 1 hour after excision.
In vivo and ex vivo PDAC tumors from mice were imaged on an Olympus multiphoton laser scanning microscope (FV1000MPE) with nondescanned detectors, coupled to an ultrafast Ti:Sapphire laser (DeepSea, Spectral Physics) using 25× water immersion, 1.05 NA objective (Olympus) in analog scanning mode. Image acquisition speed was 12.5 μs per pixel, and the total integration time was 118 s. The image size was 512 × 512 pixels with a field of view of 254 μm. The emission filters used for NAD(P)H and FAD were bandpass 472/30 and 525/45 nm, respectively.
Image analysis for ex vivo and in vivo experiments was performed using ImageJ software. Regions of interest (ROIs) were segmented from NAD(P)H intensity images, and masks were generated using “analyze particles” function. Similarly, masks for FADhigh ROI were generated from thresholded FAD intensity images. FADlow ROI masks were generated by applying a XOR function between original NAD(P)H ROI and the FADhigh ROI masks. To generate consistent number of ROIs per image, the images were subsampled into 1024 images of size 16 by 16 pixels. The optical redox ratio for each segmented ROI was calculated using Eq 1.
Biochemical assessment of NADH/NAD+ ratio
NADH/NAD+ measurements were done using a modified NADH/NAD+ Glo Assay (Promega) as described previously (23). Briefly, 200,000 cells were plated on a 35-mm dish in 2 ml of media in DMEM. Twenty-four hours later, cells were washed three times in PBS, and cells were maintained in 2 ml of DMEM with or without pyruvate in dialyzed serum for 48 hours. For extraction, cells were washed three times in ice-cold PBS and extracted in 100 μl of lysis buffer (1% dodecyltrimethylammonium bromide in 0.2 N NaOH diluted 1:1 with PBS) and frozen in −80°C. After establishing the linear range of detection with sample volume and measurement time, NADH was measured by incubating samples at 75°C for 30 min using 5 μl of the lysate and 15 μl of the lysis buffer. An equal sample volume of 5 μl of lysate and 15 μl of lysis buffer was added to 20 μl of lysis buffer and 20 μl of 0.4 N HCl and incubated at 60°C for 15 min to measure NAD+. Samples were quenched and neutralized with 20 μl of 0.25 M tris in 0.2 N HCl (NADH) or 20 μl 0.5 M tris base (NAD+), and bioluminescence was measured using a plate reader.
GC-MS analysis of metabolites
Metabolites were analyzed by gas chromatography–mass spectrometry (GC-MS) as described previously (9, 81). Briefly, 10 μl of each media sample was extracted with 600 μl of cold methanol, vortexed at 4°C for 10 min, and spun down at maximum speed on a table top centrifuge for 10 min at 4°C. A total of 450 μl of each sample was then dried under nitrogen gas and stored at −80°C until GC-MS analysis. Dried metabolite extracts were dissolved in 16 μl of methoxamine reagent (Thermo Fisher Scientific, TS-45950) and incubated at 37°C for 90 min, followed by addition of 20 μl of N-methyl-N-(tert-butyldimethylsilyl)trifluoroacetamide + 1% tert-butyldimethylchlorosilane (Sigma-Aldrich, 375934) and incubated at 60°C for 1 hour. Following derivatization, samples were analyzed using a DB-35MS column (Agilent Technologies) in an Agilent 7890 gas chromatograph coupled to an Agilent 5975C mass spectrometer. Helium was used as the carrier gas at a flow rate of 1.2 ml/min. One microliter of sample was injected at 270°C. After injection, the GC oven was held at 100°C for 1 min and increased to 300°C at 3.5°C/min. The oven was then ramped to 320°C at 20°C/min and held for 5 min at this 320°C. The MS system operated under electron affect ionization at 70 eV, and the MS source and quadrupole were held at 230° and 150°C, respectively. The detector was used in scanning mode, and the scanned ion range was 100 to 650 mass/charge ratio.
To generate metabolite heatmaps (fig. S5), the total ion count (TIC) value of each metabolite was first normalized to norvaline TIC, followed by normalization of each metabolite to that of PDAC cells cultured as 3D organoids without pyruvate. The heatmap score for each metabolite per culture condition was calculated using Eq. 2
| (2) |
where observed is the normalized TIC of the metabolite present in conditioned media from 3D culture, DMEM is the TIC of the metabolite present in fresh culture media [DMEM without pyruvate (Corning, 10-017-CV) supplemented with 10% dialyzed FBS and 5% penicillin-streptomycin], and σrow is the SD of the metabolite level of all culture conditions (i.e., heatmap row for the metabolite).
Immunohistochemistry
Five-micron sections from formalin-fixed, paraffin-embedded mouse tumors were stained with antibodies against cytokeratin-19 (1:500 dilution; Abcam, #ab133496), α-smooth muscle actin (αSMA) (1:125 dilution; Cell Signaling Technologies, #19245), or F4/80 (1:125 dilution; Cell Signaling Technologies, #70076) using standard techniques.
Statistical analysis
Data analysis was performed in Python, and graphs and heatmaps for all the figures were plotted in Python using the Seaborn data visualization library. Statistical tests were performed in R. Data were compared using either Student’s t test or one-way analysis of variance (ANOVA) with a Dunnett’s or Tukey’s post hoc test for multiple comparisons, and P values have been indicated as follows: ***P < 0.001; **P < 0.01; *P < 0.05. Gaussian curve fitting for Fig. 5B was performed using MATLAB.
Acknowledgments
We would like to thank the Koch Institute Swanson Biotechnology Center for assistance with microscopy and histology, as well as the members of the Vander Heiden laboratory and Skala laboratory for discussions. We would also like to acknowledge J. Eickhoff for advice on statistical analysis, S. Trier for guidance with in vivo imaging, and M. Stefely for input on design of scientific diagrams and data presentation.
Funding: S.S. is supported by the Damon Runyon Cancer Research Foundation (DRG-2367-19). A.N.L. was a Robert Black Fellow of the Damon Runyon Cancer Research Foundation (DRG-2241-15) and is supported by an NIH Pathway to Independence Award (K99CA234221). M.G.V.H. and M.C.S. received substantial support for this work from SU2C, a division of the Entertainment Industry Foundation. M.G.V.H. acknowledges additional support from the Lustgarten Foundation, a Faculty Scholar grant from the Howard Hughes Medical Institute, the MIT Center for Precision Cancer Medicine, the Ludwig Center at MIT, the Emerald Foundation, and the NCI (R35CA242379, R01CA201276, and P30CA14051). M.C.S. acknowledges additional support from the NCI (R21CA224280, R01CA211082, R01CA205101, and R01CA185747), the NSF (CBET-1642287), the Morgridge Institute for Research, and the University of Wisconsin Carbone Cancer Center (P30CA014520).
Author contributions: R.D.: Conceptualization, methodology, software, formal analysis, investigation, writing (original draft and review and editing), and visualization. S.S.: Conceptualization, methodology, investigation, and writing (original draft and review and editing). A.N.L.: Conceptualization, methodology, investigation, and writing (original draft and review and editing). L.V.F.: Formal analysis. A.M.B.: Investigation and writing (review and editing). J.W.: Investigation and methodology. M.C.S.: Conceptualization, supervision, funding acquisition, methodology, and writing (original draft and review and editing). M.G.V.H.: Conceptualization, supervision, funding acquisition, methodology, and writing (original draft and review and editing).
Competing interests: A.N.L. is a current employee at Pfizer; however, all work was performed while affiliated with MIT. M.G.V.H. is a scientific advisor for Agios Pharmaceuticals, Aeglea Biotherapeutics, iTeos Therapeutics, Faeth Therapeutics, Sage Therapeutics, and Auron Therapeutics. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S3
REFERENCES AND NOTES
- 1.Kim J., DeBerardinis R. J., Mechanisms and implications of metabolic heterogeneity in cancer. Cell Metab. 30, 434–446 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalin S., Sullivan M. R., Lau A. N., Grauman-Boss B., Mueller H. S., Kreidl E., Fenoglio S., Luengo A., Lees J. A., Vander Heiden M. G., Lauffenburger D. A., Hemann M. T., Deoxycytidine release from pancreatic stellate cells promotes gemcitabine resistance. Cancer Res. 79, 5723–5733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halbrook C. J., Pontious C., Kovalenko I., Lapienyte L., Dreyer S., Lee H.-J., Thurston G., Zhang Y., Lazarus J., Sajjakulnukit P., Hong H. S., Kremer D. M., Nelson B. S., Kemp S., Zhang L., Chang D., Biankin A., Shi J., Frankel T. L., Crawford H. C., Morton J. P., di Magliano M. P., Lyssiotis C. A., Macrophage-released pyrimidines inhibit gemcitabine therapy in pancreatic cancer. Cell Metab. 29, 1390–1399.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Öhlund D., Handly-Santana A., Biffi G., Elyada E., Almeida A. S., Ponz-Sarvise M., Corbo V., Oni T. E., Hearn S. A., Lee E. J., Chio I. I. C., Hwang C.-I., Tiriac H., Baker L. A., Engle D. D., Feig C., Kultti A., Egeblad M., Fearon D. T., Crawford J. M., Clevers H., Park Y., Tuveson D. A., Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 214, 579–596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherman M. H., Yu R. T., Tseng T. W., Sousa C. M., Liu S., Truitt M. L., He N., Ding N., Liddle C., Atkins A. R., Leblanc M., Collisson E. A., Asara J. M., Kimmelman A. C., Downes M., Evans R. M., Stromal cues regulate the pancreatic cancer epigenome and metabolome. Proc. Natl. Acad. Sci. U.S.A. 114, 1129–1134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhim A. D., Oberstein P. E., Thomas D. H., Mirek E. T., Palermo C. F., Sastra S. A., Dekleva E. N., Saunders T., Becerra C. P., Tattersall I. W., Westphalen C. B., Kitajewski J., Fernandez-Barrena M. G., Fernandez-Zapico M. E., Iacobuzio-Donahue C., Olive K. P., Stanger B. Z., Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell 25, 735–747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sousa C. M., Biancur D. E., Wang X., Halbrook C. J., Sherman M. H., Zhang L., Kremer D., Hwang R. F., Witkiewicz A. K., Ying H., Asara J. M., Evans R. M., Cantley L. C., Lyssiotis C. A., Kimmelman A. C., Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auciello F. R., Bulusu V., Oon C., Tait-Mulder J., Berry M., Bhattacharyya S., Tumanov S., Allen-Petersen B. L., Link J., Kendsersky N. D., Vringer E., Schug M., Novo D., Hwang R. F., Evans R. M., Nixon C., Dorrell C., Morton J. P., Norman J. C., Sears R. C., Kamphorst J. J., Sherman M. H., A stromal lysolipid–autotaxin signaling axis promotes pancreatic tumor progression. Cancer Discov. 9, 617–627 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau A. N., Li Z., Danai L. V., Westermark A. M., Darnell A. M., Ferreira R., Gocheva V., Sivanand S., Lien E. C., Sapp K. M., Mayers J. R., Biffi G., Chin C. R., Davidson S. M., Tuveson D. A., Jacks T., Matheson N. J., Yilmaz O., Vander Heiden M. G., Dissecting cell-type-specific metabolism in pancreatic ductal adenocarcinoma. eLife 9, e56782 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhim A. D., Mirek E. T., Aiello N. M., Maitra A., Bailey J. M., McAllister F., Reichert M., Beatty G. L., Rustgi A. K., Vonderheide R. H., Leach S. D., Stanger B. Z., EMT and dissemination precede pancreatic tumor formation. Cell 148, 349–361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Özdemir B. C., Pentcheva-Hoang T., Carstens J. L., Zheng X., Wu C. C., Simpson T. R., Laklai H., Sugimoto H., Kahlert C., Novitskiy S. V., de Jesus-Acosta A., Sharma P., Heidari P., Mahmood U., Chin L., Moses H. L., Weaver V. M., Maitra A., Allison J. P., LeBleu V. S., Kalluri R., Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell 28, 831–833 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Lonardo E., Frias-Aldeguer J., Hermann P. C., Heeschen C., Pancreatic stellate cells form a niche for cancer stem cells and promote their self-renewal and invasiveness. Cell Cycle 11, 1282–1290 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Ikenaga N., Ohuchida K., Mizumoto K., Cui L., Kayashima T., Morimatsu K., Moriyama T., Nakata K., Fujita H., Tanaka M., CD10+ pancreatic stellate cells enhance the progression of pancreatic cancer. Gastroenterology 139, 1041–1051.e8 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Jiang H., Torphy R. J., Steiger K., Hongo H., Ritchie A. J., Kriegsmann M., Horst D., Umetsu S. E., Joseph N. M., McGregor K., Pishvaian M. J., Blais E. M., Lu B., Li M., Hollingsworth M., Stashko C., Volmar K., Yeh J. J., Weaver V. M., Wang Z. J., Tempero M. A., Weichert W., Collisson E. A., Pancreatic ductal adenocarcinoma progression is restrained by stromal matrix. J. Clin. Invest. 130, 4704–4709 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ying H., Kimmelman A. C., Lyssiotis C. A., Hua S., Chu G. C., Fletcher-Sananikone E., Locasale J. W., Son J., Zhang H., Coloff J. L., Yan H., Wang W., Chen S., Viale A., Zheng H., Paik J. H., Lim C., Guimaraes A. R., Martin E. S., Chang J., Hezel A. F., Perry S. R., Hu J., Gan B., Xiao Y., Asara J. M., Weissleder R., Wang Y. A., Chin L., Cantley L. C., DePinho R. A., Oncogenic kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Won J. H., Zhang Y., Ji B., Logsdon C. D., Yule D. I., Phenotypic changes in mouse pancreatic stellate cell Ca2+ signaling events following activation in culture and in a disease model of pancreatitis. Mol. Biol. Cell 22, 421–436 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Provenzano P., Cuevas C., Chang A. E., Goel V. K., Von Hoff D. D., Hingorani S. R., Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer cell 21, 418–429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neesse A., Michl P., Frese K. K., Feig C., Cook N., Jacobetz M. A., Lolkema M. P., Buchholz M., Olive K. P., Gress T. M., Tuveson D. A., Stromal biology and therapy in pancreatic cancer. Gut 60, 861–868 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Helms E., Onate M. K., Sherman M. H., Fibroblast heterogeneity in the pancreatic tumor microenvironment. Cancer Discov. 10, 648–656 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boj S. F., Hwang C. I., Baker L. A., Chio I. I. C., Engle D. D., Corbo V., Jager M., Ponz-Sarvise M., Tiriac H., Spector M. S., Gracanin A., Oni T., Yu K. H., van Boxtel R., Huch M., Rivera K. D., Wilson J. P., Feigin M. E., Öhlund D., Handly-Santana A., Ardito-Abraham C. M., Ludwig M., Elyada E., Alagesan B., Biffi G., Yordanov G. N., Delcuze B., Creighton B., Wright K., Park Y., Morsink F. H. M., Molenaar I. Q., Borel Rinkes I. H., Cuppen E., Hao Y., Jin Y., Nijman I. J., Iacobuzio-Donahue C., Leach S. D., Pappin D. J., Hammell M., Klimstra D. S., Basturk O., Hruban R. H., Offerhaus G. J., Vries R. G. J., Clevers H., Tuveson D. A., Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biffi G., Oni T. E., Spielman B., Hao Y., Elyada E., Park Y., Preall J., Tuveson D. A., IL1-induced JAK/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. 9, 282–301 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muir A., Danai L. V., Heiden M. G. V., Microenvironmental regulation of cancer cell metabolism: Implications for experimental design and translational studies. Dis. Model. Mech. 11, dmm035758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan L. B., Gui D. Y., Hosios A. M., Bush L. N., Freinkman E., Vander Heiden M. G., Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birsoy K., Wang T., Chen W. W., Freinkman E., Abu-Remaileh M., Sabatini D. M., An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diehl F. F., Lewis C. A., Fiske B. P., Vander Heiden M. G., Cellular redox state constrains serine synthesis and nucleotide production to impact cell proliferation. Nat. Metab. 1, 861–867 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Titov D. V., Cracan V., Goodman R. P., Peng J., Grabarek Z., Mootha V. K., Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science 352, 231–235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwörer S., Berisa M., Violante S., Qin W., Zhu J., Hendrickson R. C., Cross J. R., Thompson C. B., Proline biosynthesis is a vent for TGFβ-induced mitochondrial redox stress. EMBO J. 39, e103334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bajzikova M., Kovarova J., Coelho A. R., Boukalova S., Oh S., Rohlenova K., Svec D., Hubackova S., Endaya B., Judasova K., Bezawork-Geleta A., Kluckova K., Chatre L., Zobalova R., Novakova A., Vanova K., Ezrova Z., Maghzal G. J., Novais S. M., Olsinova M., Krobova L., An Y. J., Davidova E., Nahacka Z., Sobol M., Cunha-Oliveira T., Sandoval-Acuña C., Strnad H., Zhang T., Huynh T., Serafim T. L., Hozak P., Sardao V. A., Koopman W. J. H., Ricchetti M., Oliveira P. J., Kolar F., Kubista M., Truksa J., Dvorakova-Hortova K., Pacak K., Gurlich R., Stocker R., Zhou Y., Berridge M. V., Park S., Dong L., Rohlena J., Neuzil J., Reactivation of dihydroorotate dehydrogenase-driven pyrimidine biosynthesis restores tumor growth of respiration-deficient cancer cells. Cell Metab. 29, 399–416.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Bermudez J., Baudrier L., la K., Zhu X. G., Fidelin J., Sviderskiy V. O., Papagiannakopoulos T., Molina H., Snuderl M., Lewis C. A., Possemato R. L., Birsoy K., Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat. Cell Biol. 20, 775–781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan L. B., Luengo A., Danai L. V., Bush L. N., Diehl F. F., Hosios A. M., Lau A. N., Elmiligy S., Malstrom S., Lewis C. A., Vander Heiden M. G., Aspartate is an endogenous metabolic limitation for tumour growth. Nat. Cell Biol. 20, 782–788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu M., Wang Y., Yang C., Ruan Y., Bai C., Chu Q., Cui Y., Chen C., Ying G., Li B., Inhibiting both proline biosynthesis and lipogenesis synergistically suppresses tumor growth. J. Exp. Med. 217, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koong A. C., Mehta V. K., le Q. T., Fisher G. A., Terris D. J., Brown J. M., Bastidas A. J., Vierra M., Pancreatic tumors show high levels of hypoxia. Int. J. Radiat. Oncol. Biol. Phys. 48, 919–922 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Llufrio E. M., Wang L., Naser F. J., Patti G. J., Sorting cells alters their redox state and cellular metabolome. Redox Biol. 16, 381–387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roci I., Gallart-Ayala H., Schmidt A., Watrous J., Jain M., Wheelock C. E., Nilsson R., Metabolite profiling and stable isotope tracing in sorted subpopulations of mammalian cells. Anal. Chem. 88, 2707–2713 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Binek A., Rojo D., Godzien J., Rupérez F. J., Nuñez V., Jorge I., Ricote M., Vázquez J., Barbas C., Flow cytometry has a significant impact on the cellular metabolome. J. Proteome Res. 18, 169–181 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Skala M. C., Riching K. M., Gendron-Fitzpatrick A., Eickhoff J., Eliceiri K. W., White J. G., Ramanujam N., In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl. Acad. Sci. U.S.A. 104, 19494–19499 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinn K. P., Sridharan G. V., Hayden R. S., Kaplan D. L., Lee K., Georgakoudi I., Quantitative metabolic imaging using endogenous fluorescence to detect stem cell differentiation. Sci. Rep. 3, 3432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Georgakoudi I., Quinn K. P., Optical imaging using endogenous contrast to assess metabolic state. Annu. Rev. Biomed. Eng. 14, 351–367 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Stringari C., Abdeladim L., Malkinson G., Mahou P., Solinas X., Lamarre I., Brizion S., Galey J. B., Supatto W., Legouis R., Pena A. M., Beaurepaire E., Multicolor two-photon imaging of endogenous fluorophores in living tissues by wavelength mixing. Sci. Rep. 7, 3792 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williamson D. H., Lund P., Krebs H. A., The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem. J. 103, 514–527 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veech R. L., Eggleston L. V., Krebs H. A., The redox state of free nicotinamide–adenine dinucleotide phosphate in the cytoplasm of rat liver. Biochem. J. 115, 609–619 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao W., Wang R.-S., Handy D. E., Loscalzo J., NAD(H) and NADP(H) redox couples and cellular energy metabolism. Antioxid. Redox Signal. 28, 251–272 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodman R. P., Calvo S. E., Mootha V. K., Spatiotemporal compartmentalization of hepatic NADH and NADPH metabolism. J. Biol. Chem. 293, 7508–7516 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang S., Heikal A. A., Webb W. W., Two-photon fluorescence spectroscopy and microscopy of NAD(P)H and flavoprotein. Biophys. J. 82, 2811–2825 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.B. Chance, G. R. Williams, The respiratory chain and oxidative phosphorylation, in Advances in Enzymology and Related Areas of Molecular Biology (John Wiley & Sons Ltd, 2006), pp. 65–134. [DOI] [PubMed] [Google Scholar]
- 46.Heikal A. A., Intracellular coenzymes as natural biomarkers for metabolic activities and mitochondrial anomalies. Biomark. Med. 4, 241–263 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chance B., Schoener B., Oshino R., Itshak F., Nakase Y., Oxidation-reduction ratio studies of mitochondria in freeze-trapped samples. NADH and flavoprotein fluorescence signals. J. Biol. Chem. 254, 4764–4771 (1979). [PubMed] [Google Scholar]
- 48.Li L. Z., Xu H. N., Ranji M., Nioka S., Chance B., Mitochondrial redox imaging for cancer diagnostic and therapeutic studies. J. Innov. Opt. Health Sci. 02, 325–341 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shah A. T., Demory Beckler M., Walsh A. J., Jones W. P., Pohlmann P. R., Skala M. C., Optical metabolic imaging of treatment response in human head and neck squamous cell carcinoma. PLOS ONE 9, e90746 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ostrander J. H., McMahon C. M., Lem S., Millon S. R., Brown J. Q., Seewaldt V. L., Ramanujam N., Optical redox ratio differentiates breast cancer cell lines based on estrogen receptor status. Cancer Res. 70, 4759–4766 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smokelin I. S., Mizzoni C., Erndt-Marino J., Kaplan D., Georgakoudi I., Optical changes in THP-1 macrophage metabolism in response to pro- and anti-inflammatory stimuli reported by label-free two-photon imaging. J. Biomed. Opt. 25, 014512 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heaster T. M., Humayun M., Yu J., Beebe D. J., Skala M. C., Autofluorescence imaging of 3D tumor-macrophage microscale cultures resolves spatial and temporal dynamics of macrophage metabolism. bioRxiv 2020.03.12.989301 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walsh A. J., Mueller K. P., Tweed K., Jones I., Walsh C. M., Piscopo N. J., Niemi N. M., Pagliarini D. J., Saha K., Skala M. C., Classification of T-cell activation via autofluorescence lifetime imaging. Nat. Biomed. Eng. 5, 77–88 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones J. D., Ramser H. E., Woessner A. E., Quinn K. P., In vivo multiphoton microscopy detects longitudinal metabolic changes associated with delayed skin wound healing. Commun. Biol. 1, 198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walsh A. J., Cook R. S., Sanders M. E., Aurisicchio L., Ciliberto G., Arteaga C. L., Skala M. C., Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer. Cancer Res. 74, 5184–5194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Omary M. B., Lugea A., Lowe A. W., Pandol S. J., The pancreatic stellate cell: A star on the rise in pancreatic diseases. J. Clin. Invest. 117, 50–59 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bardeesy N., Aguirre A. J., Chu G. C., Cheng K. H., Lopez L. V., Hezel A. F., Feng B., Brennan C., Weissleder R., Mahmood U., Hanahan D., Redston M. S., Chin L., DePinho R. A., Both p16Ink4a and the p19Arf-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc. Natl. Acad. Sci.U.S.A. 103, 5947–5952 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gui D. Y., Sullivan L. B., Luengo A., Hosios A. M., Bush L. N., Gitego N., Davidson S. M., Freinkman E., Thomas C. J., Vander Heiden M. G., Environment dictates dependence on mitochondrial complex I for NAD+ and aspartate production and determines cancer cell sensitivity to metformin. Cell Metab. 24, 716–727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kerk S. A., Lin L., Myers A. L., Chen B., Sajjakulnukit P., Robinson A., Thurston G., Nelson B. S., Kemp S. B., Steele N. G., Hoffman M. T., Wen H.-J., Long D., Ackenhusen S. E., Ramos J., Gao X., Zhang L., Andren A., Nwosu Z. C., Galbán S., Halbrook C. J., Lombard D. B., Ying H., Crawford H. C., Pasca di Magliano M., Shah Y. M., Lyssiotis C. A., The pancreatic tumor microenvironment buffers redox imbalance imposed by disrupted mitochondrial metabolism. bioRxiv 2020.08.07.238766 (2020). [Google Scholar]
- 60.Luengo A., Li Z., Gui D. Y., Sullivan L. B., Zagorulya M., Do B. T., Ferreira R., Naamati A., Ali A., Lewis C. A., Thomas C. J., Spranger S., Matheson N. J., Heiden M. G. V., Increased demand for NAD+ relative to ATP drives aerobic glycolysis. Mol. Cell 81, 691–707.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hosios A. M., Vander Heiden M. G., The redox requirements of proliferating mammalian cells. J. Biol. Chem. 293, 7490–7498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Halestrap A. P., Meredith D., The SLC16 gene family-from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflugers Arch. 447, 619–628 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Jain R. K., Physiological barriers to delivery of monoclonal antibodies and other macromolecules in tumors. Cancer Res. 50, 814s–819s (1990). [PubMed] [Google Scholar]
- 64.Druzhkova I. N., Shirmanova M. V., Lukina M. M., Dudenkova V. V., Mishina N. M., Zagaynova E. V., The metabolic interaction of cancer cells and fibroblasts—Coupling between NAD(P)H and FAD, intracellular pH and hydrogen peroxide. Cell Cycle 15, 1257–1266 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Broekgaarden M., Anbil S., Bulin A. L., Obaid G., Mai Z., Baglo Y., Rizvi I., Hasan T., Modulation of redox metabolism negates cancer-associated fibroblasts-induced treatment resistance in a heterotypic 3D culture platform of pancreatic cancer. Biomaterials 222, 119421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bao X. R., Ong S. E., Goldberger O., Peng J., Sharma R., Thompson D. A., Vafai S. B., Cox A. G., Marutani E., Ichinose F., Goessling W., Regev A., Carr S. A., Clish C. B., Mootha V. K., Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife 5, e10575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sullivan M. R., Danai L. V., Lewis C. A., Chan S. H., Gui D. Y., Kunchok T., Dennstedt E. A., Vander Heiden M. G., Muir A., Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. eLife 8, e44235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun I. L., Crane F. L., Grebing C., Löw H., Properties of a transplasma membrane electron transport system in HeLa cells. J. Bioenerg. Biomembr. 16, 583–595 (1984). [DOI] [PubMed] [Google Scholar]
- 69.Gray J. P., Eisen T., Cline G. W., Smith P. J. S., Heart E., Plasma membrane electron transport in pancreatic β-cells is mediated in part by NQO1. Am. J. Physiol. Endocrinol. Metab. 301, E113–E121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ippolito L., Morandi A., Taddei M. L., Parri M., Comito G., Iscaro A., Raspollini M. R., Magherini F., Rapizzi E., Masquelier J., Muccioli G. G., Sonveaux P., Chiarugi P., Giannoni E., Cancer-associated fibroblasts promote prostate cancer malignancy via metabolic rewiring and mitochondrial transfer. Oncogene 38, 5339–5355 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Salaud C., Alvarez-Arenas A., Geraldo F., Belmonte-Beitia J., Calvo G. F., Gratas C., Pecqueur C., Garnier D., Pérez-Garcià V., Vallette F. M., Oliver L., Mitochondria transfer from tumor-activated stromal cells (TASC) to primary glioblastoma cells. Biochem. Biophys. Res. Commun. 533, 139–147 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Pickup M. W., Mouw J. K., Weaver V. M., The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 15, 1243–1253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tian C., Clauser K. R., Öhlund D., Rickelt S., Huang Y., Gupta M., Mani D. R., Carr S. A., Tuveson D. A., Hynes R. O., Proteomic analyses of ECM during pancreatic ductal adenocarcinoma progression reveal different contributions by tumor and stromal cells. Proc. Natl. Acad. Sci. U.S.A. 116, 19609–19618 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Francescone R., Vendramini-Costa D. B., Franco-Barraza J., Wagner J., Muir A., Lau A. N., Gabitova L., Pazina T., Gupta S., Luong T., Rollins D., Malik R., Thapa R. J., Restifo D., Zhou Y., Cai K. Q., Hensley H. H., Tan Y., Kruger W. D., Devarajan K., Balachandran S., Klein-Szanto A. J., Wang H., El-Deiry W. S., Heiden M. G. V., Peri S., Campbell K. S., Astsaturov I., Cukierman E., Netrin G1 promotes pancreatic tumorigenesis through cancer associated fibroblast driven nutritional support and immunosuppression. Cancer Discov. 11, 446–479 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.M. Apte, Isolation of quiescent pancreatic stellate cells from rat and human pancreas. Pancreapedia: Exocrine pancreas knowledge base. 10.3998/panc.2011.1 (2011).
- 76.Danai L. V., Babic A., Rosenthal M. H., Dennstedt E. A., Muir A., Lien E. C., Mayers J. R., Tai K., Lau A. N., Jones-Sali P., Prado C. M., Petersen G. M., Takahashi N., Sugimoto M., Yeh J. J., Lopez N., Bardeesy N., Fernandez-del Castillo C., Liss A. S., Koong A. C., Bui J., Yuan C., Welch M. W., Brais L. K., Kulke M. H., Dennis C., Clish C. B., Wolpin B. M., Vander Heiden M. G., Altered exocrine function can drive adipose wasting in early pancreatic cancer. Nature 558, 600–604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lunt S. Y., Muralidhar V., Hosios A. M., Israelsen W. J., Gui D. Y., Newhouse L., Ogrodzinski M., Hecht V., Xu K., Acevedo P. N. M., Hollern D. P., Bellinger G., Dayton T. L., Christen S., Elia I., Dinh A. T., Stephanopoulos G., Manalis S. R., Yaffe M. B., Andrechek E. R., Fendt S. M., Vander Heiden M. G., Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol. Cell 57, 95–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.A. J. Walsh, M. C. Skala, An automated image processing routine for segmentation of cell cytoplasms in high-resolution autofluorescence images, in Multiphoton Microscopy in the Biomedical Sciences XIV (International Society for Optics and Photonics, 2014), vol. 8948 89481M.
- 80.T. R. Jones, A. Carpenter, P. Golland, Voronoi-based segmentation of cells on image manifolds, in International Workshop on Computer Vision for Biomedical Image Applications (Springer, 2005), pp. 535–543. [Google Scholar]
- 81.Lewis C. A., Parker S. J., Fiske B. P., McCloskey D., Gui D. Y., Green C. R., Vokes N. I., Feist A. M., Vander Heiden M. G., Metallo C. M., Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell 55, 253–263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S3