Abstract
Hemophilia is a hereditary disease that remains incurable. Although innovative treatments such as gene therapy or bispecific antibody therapy have been introduced, substantial unmet needs still exist with respect to achieving long-lasting therapeutic effects and treatment options for inhibitor patients. Antithrombin (AT), an endogenous negative regulator of thrombin generation, is a potent genome editing target for sustainable treatment of patients with hemophilia A and B. In this study, we developed and optimized lipid nanoparticles (LNPs) to deliver Cas9 mRNA along with single guide RNA that targeted AT in the mouse liver. The LNP-mediated CRISPR-Cas9 delivery resulted in the inhibition of AT that led to improvement in thrombin generation. Bleeding-associated phenotypes were recovered in both hemophilia A and B mice. No active off-targets, liver-induced toxicity, and substantial anti-Cas9 immune responses were detected, indicating that the LNP-mediated CRISPR-Cas9 delivery was a safe and efficient approach for hemophilia therapy.
We showed that lipid nanoparticle–packed CRISPR- mediated antithrombin gene editing offers a sustainable and safe hemophilia therapy.
INTRODUCTION
Hemophilia is a representative genetic disease with spontaneous bleeding caused by a loss of gene function related to the intrinsic, extrinsic, and common coagulation pathway (1). A fundamental treatment has not yet been developed, and hemophilia A and B are among the most prominent targets for gene therapy (2). The most commonly used treatment for hemophilia is prophylaxis, wherein deficient clotting factors are supplemented depending on the type of hemophilia. Adeno-associated virus (AAV) gene therapy has been clinically tested and has demonstrated efficacy in restoring deficient clotting factors. However, this approach is not suitable for patients with inhibitors for factor VIII (FVIII) or FIX, which comprise approximately 30 and 10% of cases for hemophilia A and hemophilia B, respectively (3). The inhibitor is neutralizing antibodies, and 95% of patients with severe hemophilia develop inhibitor within the first 75 days of FVIII replacement therapy (4).
Recently, various treatment strategies have been suggested for patients with inhibitors. Emicizumab, a U.S. Food and Drug Administration (FDA)–approved bispecific antibody, has been used clinically for treating patients with inhibitors and involves the bypassing strategy to mimic the FVIII function linking FIX and FX for the downstream clotting pathway (5). However, this approach requires the patients to undertake repeated injections over their lifetime. Fitusiran is an RNA interference therapeutic that targets antithrombin (AT) (6), and it has been used in a treatment strategy that covers all patients with hemophilia A and B. AT is an endogenous negative regulator of thrombin generation that is encoded by the serpin family C member 1 (SERPINC1) gene. Inhibition of AT using fitusiran has been demonstrated to be a potent bypassing method that restores the balance in the coagulation system without the direct expression of clotting factors. This approach is an attractive and versatile strategy as it is applicable to most patients with hemophilia regardless of the factors responsible for the disease or the occurrence of inhibitors. However, a limitation of this strategy is the short-term therapeutic effects of fitusiran (6, 7).
To date, all the strategies that have been approved for treating hemophilia are known to exert a temporary therapeutic effect. However, repeated treatments are known to negatively affect the quality of life of patients with hemophilia. In recent times, the life expectancy of patients with hemophilia has increased, resulting in the need for a longer duration of periodic treatment (8, 9). Hence, attaining a long-term effect that is sustainable and safe has been one of the most critical considerations in the treatment of hemophilia. In this regard, gene therapy is suitable for maintaining long-term therapeutic effects. Notably, the therapeutic efficacy of AAVs has been reported for several years (10), and gene therapy studies using various AAV serotypes are being conducted. However, use of AAVs has been limited owing to immunogenicity and random integration (11, 12).
To date, genome editing is widely considered as an effective strategy for the long-term cure of several hereditary and some chronic diseases (13). Viral and nonviral vectors that deliver genome editing tools have been applied in preclinical models, and some pioneering in vivo genome editing approaches are being clinically tested (14–16). The use of nonviral vectors for the delivery of genome editing tools has relative advantages in terms of safety when compared with the viral vector approach (17). Notably, the nonviral approach is not hampered by the size of the payload, preexisting immunity, or long-term Cas9 expression–associated immunogenicity, which are some of the main limitations encountered when using viral delivery systems. Cas9 protein could induce innate and cellular immune responses (18), and such immunogenicities have been associated with safety issues and prevents long-term therapeutic outcomes because of the lysis of edited cells, which are more threatening in cells having an antigen-presenting function such as hepatocytes (19). In comparison, the nonviral delivery approach has reduced off-target effects because of the transient expression of CRISPR-Cas9. Although the expression of CRISPR-Cas9 is relatively brief, it is sufficient to edit on-target genomic sites. Therefore, recent studies have prominently focused on the nonviral delivery systems such as polymer- or lipid-based vehicles as more suitable delivery tools for in vivo genome editing therapeutics (20).
In this study, we hypothesized that CRISPR-Cas9–mediated AT editing may represent a long-term and versatile therapeutic option for treating hemophilia A and B. We developed and optimized lipid nanoparticles (LNPs) to deliver the Cas9 mRNA along with a potent single guide RNA (sgRNA) targeting mouse AT (mAT) to the mouse liver. Next, mAT gene editing–mediated thrombin generation was evaluated in hemophilia A and B mouse models to confirm the therapeutic efficacy of the developed system. Here, we demonstrate that in vivo delivery of CRISPR-Cas9 using LNPs enables AT gene editing for sustainable hemophilia A and B therapy.
RESULTS
Potent sgRNA selection in the SERPINC1 gene by in vitro screening
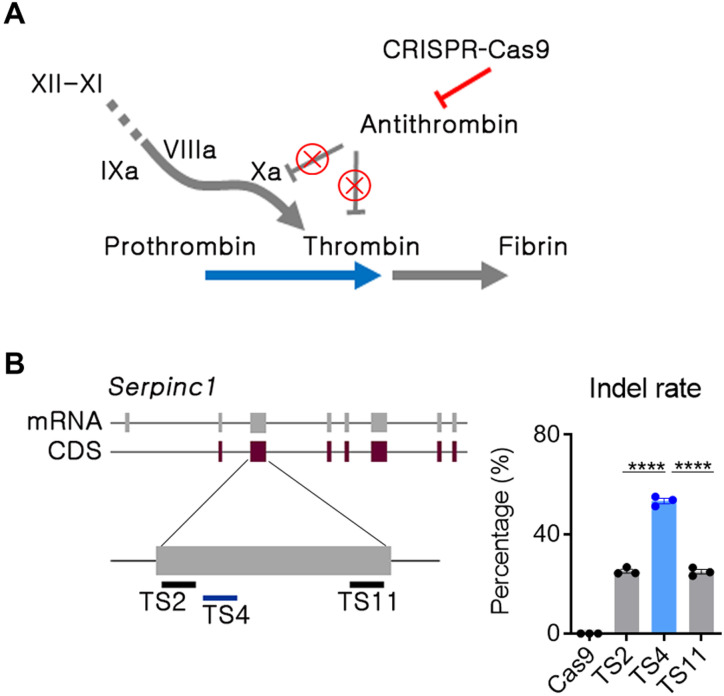
For hemophilia therapy by mAT genome editing (Fig. 1A), sgRNA candidates targeting Serpinc1 exon 3 were selected on the basis of minimal off-target risk (fig. S1A). The 11 sgRNAs that did not contain up to 2–base pair (bp) mismatches were transfected with a ribonucleoprotein (RNP) formulation into the mouse C2C12 cell line. Insertion or deletion (indel) frequency was analyzed by targeted deep sequencing. Three sgRNAs were chosen by primary screening (fig. S1B), and the TS4 sgRNA was shown to be most potent in a subsequent comparison test (Fig. 1B). Although the C2C12 cell line was well transfected and convenient for sgRNA activity screening, it was not close to our target cell in the context of AT regulation. Thus, we proceeded to test the three sgRNAs in mouse primary hepatocytes, and TS4 sgRNA consistently led to the highest indels (fig. S1C). Accordingly, the TS4 sgRNA was selected for further LNP-mediated in vivo studies after the in vitro studies.
Fig. 1. The AT locus was selected as the target gene for rebalancing.
(A) Strategy for CRISPR-Cas9–mediated in vivo Serpinc1 (encoding AT) gene editing. The red line and “X” symbols indicate inhibition of gene expression or its function. (B) Single guide RNAs (sgRNAs) were selected in the second exon of the Serpinc1 gene, and double-stranded breakage potential was evaluated by deep sequencing after transfection of C2C12 cells with an RNP formulation. Transfection and sequencing were conducted in triplicates, and each dot indicates the double-stranded breakage frequency from each experiment. TS, target site. Data were presented as mean ± SEM. ****P < 0.001.
LNP optimization for in vivo hepatocyte transfection by buffer modification
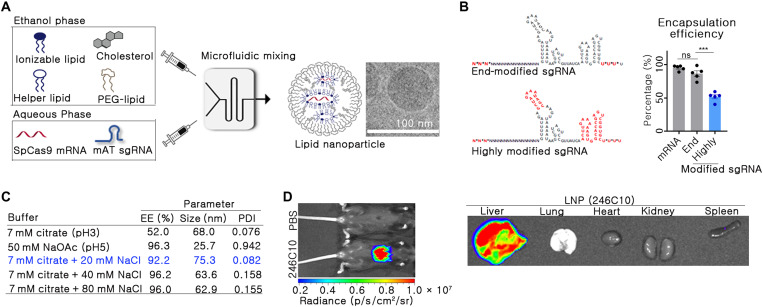
For the in vivo delivery of Cas9 mRNA and sgRNA, LNPs were formulated using 246C10, dioleoylphosphatidylethanolamine (DOPE), cholesterol, and polyethylene glycol (PEG)–ceramide. A highly modified sgRNA was used in this study to induce targeted genome editing at desired doses efficiently. Phosphorothioate bond was introduced at the 5′- and 3′-end of an sgRNA. In addition, 2′-O-methyl was introduced at the stem loops of an sgRNA as previously described (16). A rapid microfluidic mixing system was used to formulate the LNPs with defined particle size and distribution (Fig. 2A). In the aqueous phase, Cas9 mRNA and mAT-targeted sgRNA were dissolved in a 1:1 weight ratio, in either sodium citrate (pH 3) or sodium acetate buffer (pH 5). Coencapsulation of Cas9 mRNA and sgRNA was performed using a microfluidic system, and various buffer conditions were tested to confirm the encapsulation of the RNA molecules. We observed that the highly modified sgRNA contributed to poor encapsulation efficiency of RNA into the LNPs (fig. S2). In addition, depending on the extent of chemical modification in the sgRNA, the encapsulation efficiency of the sgRNA was quite different (Fig. 2B). A highly modified sgRNA demonstrated poor encapsulation efficiency (~50%), while an end-modified sgRNA resulted in an encapsulation efficiency of nearly 85% (P = 0.0002).
Fig. 2. LNPs for CRISPR-Cas9–mediated in vivo gene editing were prepared.
(A) Schematic image of LNP formulation using a microfluidic mixing system. (B) Encapsulation efficiency (EE) was analyzed between end- and highly modified sgRNA at 7 mM citrate buffer (n = 5). In the sgRNA structure, the asterisks and red font indicate the phosphorothioate bond and 2′-O-methyl ribonucleotide, respectively. Data are exhibited as means ± SEM. ***P < 0.001. (C) EE, size, and PDI of LNPs were analyzed with different buffer conditions. The condition selected in this study has been indicated in blue. (D) Biodistribution was analyzed by in vivo bioluminescence imaging. mFlucLNPs were formulated using the Luc mRNA (0.1 mpk dose) and injected intravenously into C57BL/6 mice. Three hours after the injection, luminescence signal was detected in the live mice and in their organs.
Previous studies have demonstrated that using a highly modified sgRNA results in superior genome editing than an end-modified sgRNA (16, 21). Hence, we attempted to optimize the buffer condition to ensure the encapsulation of highly modified sgRNA in the LNPs. Different buffer conditions, either citrate or acetate, were evaluated to optimize the encapsulation of sgRNA in the presence of varying concentrations of sodium chloride (NaCl). The ionic buffer strength and pH are important parameters during microfluidic mixing. Citrate buffer at low pH (10 mM, pH 3) is often used to prepare RNA-encapsulated LNPs to decrease base hydrolysis of RNA. Moreover, most ionizable lipid amines in the citrate buffer are positively charged (pKa of 246C10 ionizable lipid is 6.9), giving ionizable lipids the advantage of electrostatically interacting with the negatively charged RNA. However, we sought to examine the effect of acetate buffer at high pH (50 mM, pH 5). Accordingly, we determined that highly modified sgRNA-encapsulated LNPs produced using citrate buffer had low encapsulation efficiency (68%) and were monodisperse [polydispersity index (PDI) < 0.1]. Contrastingly, highly modified sgRNA-encapsulated LNPs produced using acetate buffer had high encapsulation efficiency (96%) and were relatively more heterodisperse (PDI > 0.9) (Fig. 2C). Given that using citrate buffer during microfluidic mixing produced monodispersed LNPs, we selected citrate buffer during microfluidic mixing. In addition, we added NaCl during microfluidic mixing to increase ionic strength and encapsulation efficiency. Previously, Finn et al. (16) have incorporated NaCl during microfluidic mixing, giving high encapsulation efficiency of highly modified sgRNA. On the basis of the study, we optimized the amount of NaCl added to the buffer.
The amount of NaCl added to the buffer systems during the microfluidic mixing distinctly affected the encapsulation of sgRNA into the formulated LNPs (Fig. 2C). This effect was negligible when working only with Cas9 mRNA; however, a highly modified sgRNA required a certain amount of ionic strength to be encapsulated. At the optimized condition (7 mM citrate with 20 mM NaCl), coencapsulation of Cas9 mRNA and mAT sgRNA demonstrated over 90% encapsulation efficiency with a narrow PDI. On the basis of these results, we selected this buffer condition with better RNA encapsulation efficiency (>90%) than the initial formulation for further studies. Next, the organ-specific delivery potential of LNP was confirmed by introducing a luciferase packed with LNP through the intravenous route. Notably, the mouse liver exhibited high transgene expression, but the other organs did not show detectable luciferase expression (Fig. 2D).
Sustained down-regulation of mAT by LNPs packed with Spcas9 and sgRNA
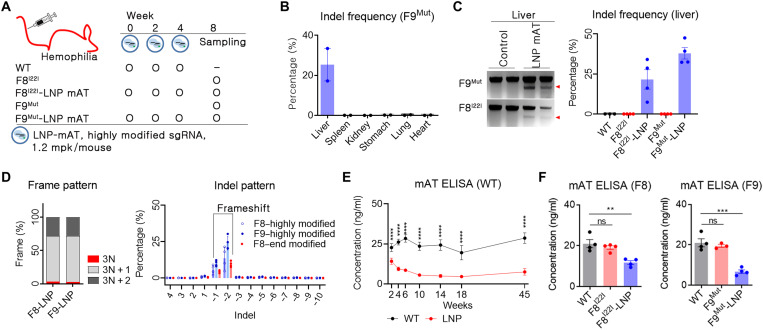
In vivo mAT targeting was attempted after optimizing the LNP-CRISPR-mAT for hepatocyte delivery. In humans, hemophilia A and B are the representative types of coagulation disorder, and down-regulation of mAT expression resulted in improved clinical symptoms in both hemophilia A and B (7, 22, 23). Hemophilia A, the most common type of hemophilia, is a severe coagulation disorder that results from an inversion of the 22nd intron of the FVIII gene (F8I22I) (24). On the other hand, hemophilia B is caused by the loss of function of the FIX gene (F9Mut). Hence, mice with F8I22I and F9Mut were used in this study; these mice were generated using CRISPR-Cas9–mediated gene editing and verified for the hemophilia phenotype. As a trial dose, we administered 1.2 mg/kg (mpk) per mouse of LNP-CRISPR-mAT via the intravenous route at an interval of 2 weeks (Fig. 3A). The bioluminescence signal was easily detected at a low luciferase mRNA dose; however, a higher dose was needed to produce therapeutically relevant Cas9 proteins in the target cells (25, 26). The LNP-CRISPR-mAT developed detectable indels only in the liver tissue (Fig. 3B and fig. S3), and an average of 22 and 38% indel frequencies were observed in the liver tissue of F8I22I and F9Mut, respectively (Fig. 3C). Also, LNP-CRISPR-mAT developed a dominant 1 or 2 bp deletion with frameshift, thereby enhancing the loss of function of mAT (Fig. 3D).
Fig. 3. LNP-CRISPR-mAT induced a prolonged down-regulation of mAT expression.
(A) Brief schematic for the in vivo gene targeting using LNP-CRISPR-mAT. C57BL6 (B6, n = 4), B6.FVIII intron 22 inversion (F8I22I, n = 4 in each group), and B6.FIX knockout (F9Mut, n = 4 in each group) mice were used in this study. (B and C) Indel frequency was calculated by deep sequencing and confirmed using T7E1 analysis (number of each group, 2 or 4). Data are shown as means ± SEM. (D) Indel pattern was analyzed according to frameshift and indel size (number of each group, 3 or 4). Each dot indicates the percentage of each sized indel from the total sequencing result. (E) Prolonged AT down-regulation was screened by ELISA using B6 mice (n = 6 per group). Blood was collected from the tail vein using a sodium citrate–coated tube, and the plasma was subjected to mAT ELISA. ****P < 0.0001. (F) Blood mAT concentration was measured and compared between the LNP-CRISPR-mAT–treated and the control hemophilia mice group. Each dot indicates the mAT concentration of an individual mouse (number of each group, 3 or 4). Data are presented as means ± SEM. **P < 0.01 and ***P < 0.001; ns, nonsignificant.
Since double-stranded DNA breakage was observed in the liver tissue, the evaluation of mAT function was performed by assessing the blood mAT concentration. As we administered LNP-CRISPR-mAT three times at an interval of 2 weeks, we analyzed the blood mAT concentration repeatedly during this duration using wild-type (WT) mice. The concentration of mAT declined with the repeated rounds of LNP-CRISPR-mAT injection but plateaued at 10 weeks after the first LNP-CRISPR-mAT injection (Fig. 3E). Thus, when highly modified sgRNA and SpCas9 mRNA were packed in LNPs and administered repeatedly, Serpinc1 gene function was down-regulated by more than 70% compared with the average mAT values in control WT mice, and Serpinc1 down-regulation was maintained stably thereafter for 10 months. While the control WT mice were useful for analyzing gene expression changes through repeated sampling, however, it is important to note that mAT expression may differ between the control WT and the hemophilia mouse model. Hence, LNP-CRISPR-mAT was administered to the mice with F8I22I and F9Mut, and the blood mAT concentration was assessed. We observed that the blood mAT concentration decreased by about 40% in F8I22I and 70% in F9Mut mice (Fig. 3F).
Rescued thrombin generation activity using LNP-CRISPR-mAT in mice with hemophilia
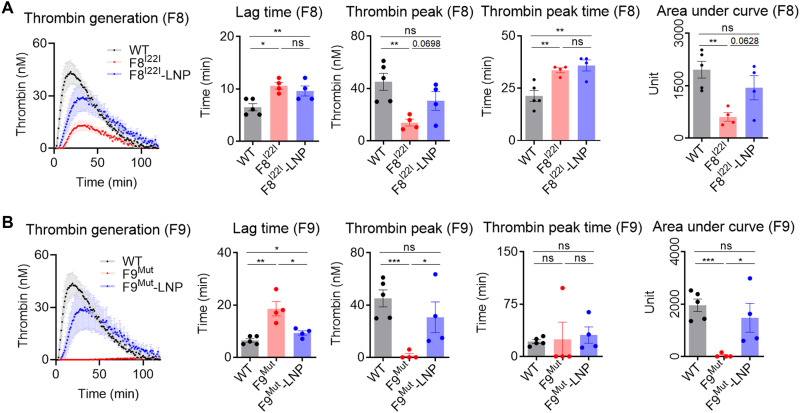
Among the various diagnostic methods that are available to assess the coagulation disorder, activated partial thromboplastin time (aPTT) test and calibrated automated thrombin (CAT) generation are widely used (27). However, aPTT is suitable for evaluating the initial phase of the clotting potential, and hence, it is not useful for analyzing the therapeutic effect of rebalancing (28). On the other hand, CAT can measure the thrombin generation over 100 min and is applicable for evaluating the rebalancing efficacy (6). Notably, the LNP-CRISPR-mAT treatment restored the thrombin generation potential in both the F8I22I and F9Mut mice. LNP-CRISPR-mAT reduced lag time, enhanced thrombin peak, and increased total thrombin generation (area under curve) in F8I22I and F9Mut. In addition, the LNP-CRISPR-mAT treatment resulted in thrombin peak enhancement of approximately up to 65% that of WT. On the basis of the thrombin peak and total thrombin formation ability identified using the thrombogram, we noted that LNP-CRISPR-mAT treatment enhanced thrombin generation ability in the mice with F8I22I and F9Mut (Fig. 4, A and B).
Fig. 4. In vivo mAT targeting rescued thrombogenesis.
(A and B) Plasma from F8I22I and F9Mut mice were collected from the inferior vena cava (control WT, n = 5; F8I22I, F8I22I-LNP, F9Mut, and F9Mut-LNP, n = 4). The thrombogenesis potential was calculated and analyzed for lag time, thrombin peak, thrombin peak time, and area under the curve. Each dot indicates data from one mouse and represents means ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001. Thrombogenesis in WT mice was assessed once and used for comparison with the F8I22I and F9Mut mice.
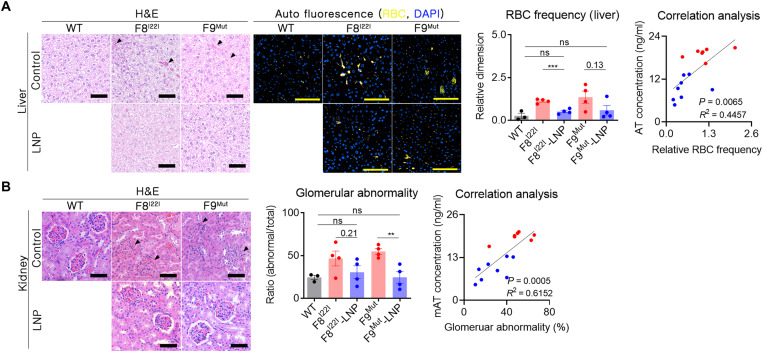
Reduced spontaneous bleeding and secondary hemophilia complication by enhanced thrombosis potential
The CAT analysis demonstrated the improvement in the coagulation ability due to the LNP-CRISPR-mAT–mediated rebalancing. However, it was also essential to assess for a reduction in spontaneous bleeding, which is a substantial complication of hemophilia. The most common spontaneous bleeding sites found in patients with hemophilia are the brain and joints (29, 30). However, we did not observe bleeding lesions in the brain and joints in the hemophilic mice (fig. S4). The histopathological analysis revealed spontaneous bleeding in the interstitial liver region in both groups of mice with hemophilia (Fig. 5A). The frequency or severity of bleeding was verified by measuring the total number of red blood cells (RBCs) detected in the tissue section. As RBCs exhibit autofluorescence (31), the false-colored yellow signal that produced by coexpression at 470- and 530-nm wavelengths was defined as autofluorescence produced by RBC. Ten areas of the liver were randomly selected from each mouse, and the dimension ratio of the yellow fluorescence signal was quantified. As expected, treatment with LNP-CRISPR-mAT reduced the frequency of RBCs in the liver tissue, indicating a reduction in spontaneous bleeding. In particular, there was a significant correlation between the mAT concentration and RBC frequency in each mouse (P = 0.0065) (Fig. 5A).
Fig. 5. Enhanced thrombosis reduced spontaneous bleeding and secondary hemophilia complication.
(A) Paraffin-embedded liver tissues were prepared without perfusion to prevent the loss of evidence of spontaneous bleeding (WT, n = 3; F8I22I, F8I22I-LNP, F9Mut, and F9Mut-LNP, n = 4). RBCs in the interstitial liver tissue [marked by black triangles in the liver tissue using hematoxylin and eosin (H&E) staining] were measured by immunofluorescence staining by detecting the autofluorescence signal at 470 and 540 nm. The intensity of the coexpressions (yellow signals) were then calculated. Ten areas of the liver were randomly selected from each mouse, and each measured autofluorescence signal was subjected to the RBC frequency analysis. Each dot indicates a signal from one selected area. Correlation analysis was conducted using the mean yellow fluorescence values obtained from each mouse. Blue dot, LNP-CRISPR-mAT groups; red dot, control group. Scale bars, 100 μm. (B) The whole kidney was formalin fixed, and gross histology was examined using H&E staining. The number of abnormally shaped glomerular capsules (black triangles) was calculated from three randomly selected regions of the cortex (1 mm2) per mouse and analyzed. **P < 0.01 and ***P < 0.001.
Renal lesions are not common in patients with hemophilia (32), but unexpected histological changes in the kidney were observed in this study. Both the F8I22I and F9Mut hemophilic mice exhibited characteristic edema symptoms in the Bowman’s capsule (Fig. 5B). However, these symptoms were not detected in the control WT mice. Hence, we assessed the therapeutic effect of LNP-CRISPR-mAT on the kidney as well. For this purpose, we analyzed the shape of the Bowman’s capsule by randomly selecting three sections from the kidney of each mouse. The LNP-CRISPR-mAT treatment reduced edema symptoms, and the overall ratio of the regular glomerular capsule in the F8I22I and F9Mut mice was similar to those of the control WT mice (Fig. 5B). Together, these observations confirmed that rebalancing through LNP-CRISPR-mAT treatment resulted in sustained and reduced mAT concentration and consequentially improved the clinical symptoms of hemophilia.
No detectable CRISPR-LNP–mediated side effect
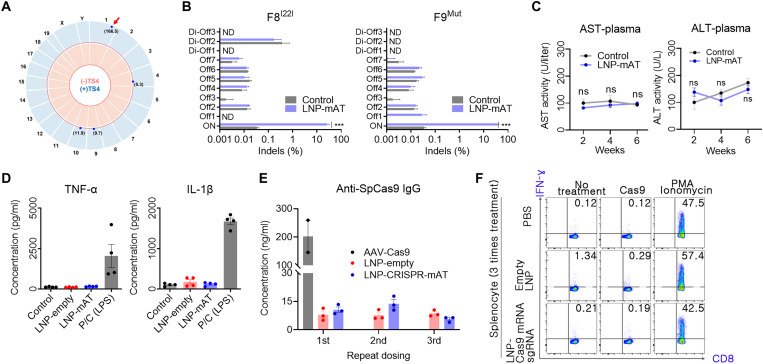
The most critical complication in in vivo gene editing is the occurrence of undesirable off-target effects. Thus, we first used the in silico method to select the seven highest potential off-target sites that differed from the on-target sgRNA sequence by up to three nucleotides (table S1). In addition, a genome-wide detection using Digenome-seq (33) was performed, and we found three more potential sites in which cleavage scores were substantially lower than that of the on-target site (Fig. 6A and table S1). Next the total 10 off-target candidates were subsequently validated in mouse liver treated with LNP-CRISPR-mAT. Targeted deep sequencing analysis using liver genomic DNA from the control WT, F8I22I, and F9Mut (n = 3, each) showed no meaningful indels at the potential off-target sites in both hemophilic models (Fig. 6B).
Fig. 6. Safety-related assessments of the LNP-CRISPR-Cas9.
(A) Genome-wide Circos plot for in vitro cleavage sites in the absence (pink) or presence (blue) of the TS4 sgRNA. Numbers in bracket: cleavage scores. Red arrow: on-target cleavage (B) Targeted deep sequencing results of the top seven homologous candidates (Offs) and the three candidates detected by the Digenome-seq analysis (Di-Offs) (n = 3). ND, not detected; ***P < 0.001. (C) Serum AST and ALT concentrations after injection with 1.2 mpk of LNP-CRISPR-mAT thrice to WT at an interval of 2 weeks (n = 6). (D) Serum TNF-α and IL-1β concentration after injection with 1.2 mpk of LNP or LNP-CRISPR-mAT thrice. Positive control group was injected 20 mpk of lipopolysaccharide (n = 4). (E) Serum anti-SpCas9 IgG concentration after repeated injection with LNP-CRISPR-mAT (n = 3). Mouse intravenously injected with AAV9-EFS-SpCas9 (5 × 1013 vg/kg) was also tested after 6 weeks of the treatment. The concentrations were calculated from the standard curve from ELISA (R2 = 0.989). (F) Representative flow cytometry plots illustrating IFN-γ expression in CD8+ T cells. IFN-γ expression in CD8+ T cells was evaluated after repeated injections of PBS, empty LNPs, and sgRNA/Cas9 mRNA–encapsulated LNPs to mice (n = 3). Detailed results are supplied in fig. S5.
Another substantial safety issue by LNP-CRISPR is an immune response or inflammation. Plasma was applied to aspartate transaminase (AST) and alanine transferase (ALT) analysis, and there was little difference between the groups. Thus, LNP-CRISPR-mAT administration did not induce liver injury (Fig. 6C). Next, inflammatory response to LNP-CRISPR was evaluated by measuring serum tumor necrosis factor–α (TNF-α) and interleukin-1β (IL-1β) concentrations. There was no detectable increase in inflammatory cytokine production following LNP-CRISPR injection (Fig. 6D). In addition, we investigated systemic anti-Cas9 antibody response through enzyme-linked immunosorbent assay (ELISA) using sera from the LNP-injected mouse. Compared with the LNP empty treatment, anti-Cas9 immunoglobulin G (IgG) was not evoked by repeated LNP-CRISPR-mAT injection (Fig. 6E). However, when we treated the mice with AAV-Cas9 intravenously, systemically elevated anti-Cas9 IgG was detected after 6 weeks of treatment (Fig. 6E). This suggests that repeated LNP-CRISPR injection was a relatively less immunogenic approach than the persistent AAV-mediated Cas9 expression. Next, we evaluated whether repeated administration of sgRNA/Cas9 mRNA–encapsulated LNPs would induce a cellular immune response against cells expressing Cas9 protein. Since a cellular immune response is specifically mediated by CD8+ cytotoxic T lymphocytes, we confirmed the existence of anti-Cas9 T cells in mice splenocytes (34) by determining interferon-γ (IFN-γ) level after Cas9 protein challenge to mice splenocytes. Repeated LNP injection did not induce cellular immune responses in mice (Fig. 6F and fig. S5).
DISCUSSION
Hemophilia is an X-linked recessive genetic disease mainly caused by loss-of-function mutations in the FVIII or FIX gene (35). Although prophylaxis by supplying the deficient clotting factor proteins is widely used in the clinic, the need for frequent administration, high costs, and inhibitor incidence negatively affect the quality of life of patients (36). Hence, advanced therapeutic strategies are currently being developed to address this issue. In this study, we assessed whether genome editing of SERPINC1 encoding AT can be used as a potential advanced therapeutic option for treating patients with hemophilia. We demonstrated that LNP-mediated delivery of CRISPR-Cas9 resulted in the inhibition of mAT and improved thrombin generation and bleeding-associated phenotypes in both hemophilia A and B mouse models. Notably, the administration of LNP-CRISPR-mAT could provide flexible opportunities to treat patients with or without inhibitors, exerting long-term therapeutic effects even with a limited number of doses (7). Moreover, we also demonstrated that non–virally delivered SpCas9 and sgRNA effectively targeted liver Serpinc1 without notable off-targeting effects. Hence, such a CRISPR-Cas9–compatible delivery tool may have potential clinical applicability as well. A similar approach using LNP-CRISPR-Cas9 recently underwent human trials using patients with transthyretin (ATTR) amyloidosis (NCT04601051). The results showed that LNP-CRISPR-Cas9 effectively inhibited pathogenic TTR expression by approximately 90% with only mild adverse events (37), demonstrating the translational feasibility of this approach.
The therapeutic application of rebalancing the clotting pathway is strongly encouraged because of the observation that bleeding phenotypes are substantially ameliorated in individuals with hemophilia who co-inherit the deficiency in anticoagulation factors, such as AT and tissue factor pathway inhibitor (TFPI) (38, 39). AT inhibits various coagulation-related genes such as Factor XIIa, XIa, IXa, Xa, VIIa, and thrombin (IIa) (40), while TFPI reversibly inhibits Factor VIIa and Xa (41). Thus, the loss of function of AT or TFPI could recover the impaired balancing by reducing their anticoagulant activity. TFPI-targeting approaches using monoclonal antibodies, including concizumab and marstacimab, have been clinically tested. However, the clinical regimens involved are daily and once-weekly injections for concizumab and marstacimab, respectively (42). Therefore, genome editing of TFPI appears to be a more long-lasting effective strategy, and some potent sgRNAs targeting TFPI were observed in the human genome (fig. S6). Notably, TFPI is expressed by diverse cell types (43), and development of a delivery method targeting multiple tissues may facilitate the clinical translation of the TFPI-editing approach. Instead, we suggest that translation of the SERPINC1-editing strategy may be a quicker approach, because the delivery tools developed in this study are very potent at targeting the liver, which is the primary source of AT secretion. Some of the sgRNAs targeting the human SERPINC1 resulted in effective indels and out-of-frame portions inducing a knockout through the nonsense-mediated decay mechanism (fig. S6). Hence, these sgRNA candidates can be considered for the next lead optimization stage.
Enormous efforts directed toward the development of new technology have contributed in curing patients suffering from diverse refractory diseases (44, 45). Specifically, AAV-mediated gene replacement therapy is now widely accepted as an option for long-term therapy rather than protein replacement or antibody-mediated treatment. In hemophilia, clinical trials of AAV-FVIII have similarly demonstrated a decline in FVIII levels within a few years after dosing, while trials of AAV-FIX have shown a persistent effect for more than 10 years (46, 47). The reason for such decline is not proven, but expression or secretion of FVIII in the hepatocyte, not an endogenous source of FVIII, may be involved. Genome editing strategies that directly correct the common mutations such as inversion of intron 22 or intron 1 of FVIII have been proposed (48). However, requirements of a sinusoidal endothelial cell-targeting delivery system and high reversion efficiency may limit the application of such an approach. Therefore, other bypassing strategies combined with long-term therapeutic approaches could help overcome the limitations associated with the short duration of the therapeutic effectiveness of current approaches. We hypothesize that the SERPINC1-editing strategy is an excellent therapeutic option that allows broad targeting of the patient population.
In this study, we applied LNPs to deliver CRISPR-Cas9. Three consecutive repeated dosing of LNP treatment inhibited AT expression by approximately 40 and 70% in the F8I22I and F9Mut mouse, respectively. In addition, the levels of inhibition were accompanied by phenotype recovery in both the hemophilic models. Accordingly, the therapeutic window of AT inhibition may be less than 50%; however, further studies are necessary to dissect the minimum effective regulation level in detail. One of the advantages of LNP is the ease of repeat dosing, which is not suitable for AAVs because of the high anticapsid immune response (49). Notably, onpattro, an FDA-approved LNP–short hairpin RNA inhibiting ATTR can be injected every 3 weeks (50). Several potent LNPs such as C12-200, MC3, and cKK-E12 have been reported to target the liver effectively (51), and these could be further optimized by the partial application of a biodegradable form of lipid (52). Similarly, our prototype LNPs can be further optimized with respect to the biodegradability or endosomal release of the particles, facilitating the development of a more clinically compatible delivery tool.
We did not observe any active off-target sites when assessing the candidate genomic sites selected using both the homology-based and genome-wide detection methods. Although we used Digenome-seq, other methods, including Circle-, Change-, and Guide-seq, are also applicable methods that involve in vitro or cell-based genome-wide cleavage together with next-generation sequencing (53). The combinations of the biased and unbiased methods to capture potential off-target loci have been used for IND (Investigational New Drug Application) approval in recently conducted clinical programs using CRISPR-Cas9 (15, 37). Accordingly, site-specific validation using targeted deep-sequencing combined with more than one off-target detection method could be a standard protocol to validate the translational applicability of genome editing strategies. Meanwhile, considering the off-target reducing strategy, some engineered high-fidelity versions of Cas9 such as HFCas9, eCas9, HypaCas9, and SniperCas9 can be used as alternatives to WT SpCas9 (53–55).
In conclusion, we provide a new, CRISPR-Cas9–based approach for treating hemophilia. To the best of our knowledge, no previous reports have demonstrated CRISPR-Cas9–mediated AT gene editing with nonviral vectors and a therapeutic effect on hemophilia. Our genome editing approach presents a versatile option for solving diverse unmet needs that persist in major advanced therapies, including gene replacement therapies or other bypassing therapies.
MATERIALS AND METHODS
sgRNA screening
Mouse C2C12 [American Type Culture Collection (ATCC), CRL-1722] cells or human Jurkat (ATCC, TIB-152) cells were maintained in Dullbecco’s Modified Essential Medium supplemented with 4 mM glutamine (4.5 g/liter), 1 mM sodium pyruvate, 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 mg/ml). For initial sgRNA screening, plasmids encoding human optimized SpCas9 and sgRNA or RNP complex formed by 4 μg of Cas9 with 1 μg of sgRNA were subjected to electrical shock using 10-μl tips on the Neon Transfection (Life Technologies, Carlsbad CA, USA) at 1150 V for 15 ms and two pulses. For additional comparison test, mouse primary hepatocytes (4 × 105 cells) were treated by 1 μg of LNP for three selected sgRNAs (TS2, TS4, and TS11). After 3 days of transfection, the cells were harvested, and extracted genomic DNA using a DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) was subjected to targeted deep sequencing measuring indel frequencies.
LNP preparation
LNPs were prepared using NanoAssemblr Benchtop Instrument (Precision Nanosystems Inc., Vancouver, Canada) according to a previously described method (26). The lipid components (ionizable lipid, DOPE, cholesterol, and PEG lipid at 26.5:20:52:1.5 molar ratio) were dissolved in ethanol, and RNAs (Cas9 mRNA/sgRNA at 1:1 weight ratio) were dissolved in 10 mM citrate buffer (pH 3). The final ionizable lipid:RNA weight ratio was 10:1, and the final volume ratio was 1:3. Then, LNPs were formulated by microfluidic mixing of the prepared solutions at a 12 ml/min flow rate. The resulting LNPs were dialyzed against 1X phosphate-buffered saline (PBS) with 3500–molecular weight cutoff dialysis cassettes (Life Technologies) for 16 hours to exchange buffer. To characterize the prepared LNPs, dynamic light scattering was used to confirm the size, PDI, and zeta potential of LNPs. The encapsulation efficiency of RNAs was measured by Quant-iTTM Ribogreen Assay (Life Technologies).
In vivo biodistribution analysis of LNPs
C57BL/6 mice with weights of 18 to 20 g were purchased from Orient Bio (Seongnam, Gyeonggi, Korea). LNPs with firefly luciferase encoding mRNA (dose of 0.1 mpk) were injected intravenously into mice. Three hours later, mice were injected with d-luciferin intraperitoneally and incubated for 15 min. Bioluminescence imaging of the whole body and ex vivo organ was performed using an IVIS (Perkin Elmer, Waltham, MA, USA). All animal study procedures were approved by the Institutional Animal Care and Use Committee at the Ewha Woman’s University (Institutional Animal Care and Use Committee 19-022).
Animal experiment for LNP-CRISPR
C57BL/6 (B6) mice were purchased from Koatech (PyeongTaek, Gyeonggi, Korea). B6.FVIII intron 22 inversion (F8I22I) and B6.FIX knockout (F9Mut) were generated by CRISPR-Cas9–based gene editing (fig. S7) (56). Seven- to 9-week-old male mice were subjected to further experiments. B6 and each hemophilia mice were randomly divided, and half of them were injected LNP packed SpCas9 mRNA and highly modified sgRNA (LNP-CRISPR-mAT) three times with 2-week interval. Injection solution was prepared by mixing 1.2 mpk of LNP-CRISPR-mAT and warm saline up to 600 μl and was injected via the intravenous route. The 600 μl of injection solution could not induce hydrodynamic gene delivery in the liver (fig. S8). B6 mice have been subjected to repeated blood sampling via tail vein for 18 weeks. Hemophilia mice were euthanized at 8 weeks after the first LNP-CRISPR-mAT injection. A total of 450 μl of fresh blood was collected from the inferior vena cava and mixed with 50 μl of 3.2% sodium citrate, and plasma was prepared by collecting supernatant after centrifugation. After blood sampling, each organ was collected without perfusion, a part of the tissues was fixed by formalin, and the remains were used for genomic DNA extraction. This study was approved by the Institutional Animal Care and Use Committees of Seoul National University (SNU-200715-2) and was conducted under approved guidelines.
Polymerase chain reaction, T7E1 analysis, and targeted deep sequencing
Genomic DNA was extracted from organ tissues by a G-DEX IIc genomic DNA extraction kit (Intron Biotechnology, Gyeonggi, Korea). Primers were designed for amplifying the LNP-CRISPR-mAT–targeted genomic region. Next, in T7E1 analysis, polymerase chain reaction (PCR) amplicons were subjected to hetero-duplex hybridization and 30 min of T7E1 endonuclease (New England Biolabs, Ipswich, MA, USA) incubation. The presence of the cut band in gel electrophoresis was designated as indel formation. For targeted deep sequencing, interesting genomic regions were amplified by PCR from genomic DNA extracted from transfected cells or LNP-injected tissues. The produced amplicons were barcoded during subsequent PCR with Illumina TrueSeq adaptors. The products were purified with a PCR purification kit (Geneall, Seoul, Korea) and then were pooled in an equimolar ratio. The final libraries were paired-end sequenced using Illumina Miseq v2 (PE150) (Illumina, San Diego, CA, USA). Indel frequencies were quantified using Cas-Analyzer (www.rgenome.net). Indels in the region 3-bp upstream from the protospacer-adjacent motif sequence were considered mutations resulting from Cas9.
Enzyme-linked immunosorbent assay
Blood AT concentration was measured by an ELISA using an mAT III ELISA kit (Abcam, Cambridge, UK). ELISA was conducted using plasma according to the manufacturer’s instructions. Absorbance was measured by a spectrophotometer (Tecan, Zurich, Switzerland) at 450 nm, and AT concentrations were calculated by applying the measured optical density value to the standard curve. AST and ALT were measured by an AST/ALT activity assay kit (Apexbio, Houston, Texas, USA). Plasma was diluted with provided assay buffer 1:9 and 1:3 for AST and ALT analysis, respectively. AST/ALT assay was conducted according to the manufacturer’s instructions. Absorbance was measured by a spectrophotometer (Tecan, Zurich, Switzerland) at 450 and 570 nm for AST and ALT analysis, respectively. To analyze inflammatory cytokine production, WT mice were injected with 1.2 mpk of LNP or LNP-CRISPR-mAT thrice into the tail vein at a 7-day interval. Within 24 hours after the final injection, blood was collected from the anterior vena cava, and serum TNF-α and IL-1β concentrations were measured using ELISA (Bioss, Woburn, MA, USA).
The calibrated automated thrombogram (CAT)
Thrombin generation was measured using a Technothrombin TGA kit (Diapharma, West Chester, OH, USA). Briefly, a mixture of 40 μl of plasma dilution, 10 μl of reagent C low buffer, and 50 μl of the substrate was added in a single well, and fluorescence at 360/450 nm (excitation/emission) was measured for 120 min with 1-min intervals. Fluorescence measurement of thrombin generation was performed in a 96-well plate using Cytation 5 (BioTek, Winooski, VT, USA). The thrombin generation curves were calculated by using a manufacturer’s provided software.
Histological analysis
In hematoxylin and eosin stain, deparaffinized tissues were stained by 0.1% Mayer’s H&E solution. To detect autofluorescence, deparaffinized tissues were stained by 4′,6-diamidino-2-phenylindole (DAPI), and signals at 470- and 530-nm wavelengths were detected using Cytation 5 (BioTek). After image acquisition by a microscope, images were analyzed with the ImageJ program (NIH, Rockville Pike, MD, USA). All images were converted to 8-bit grayscale, and then “Image” under “Adjust > Threshold” commands were selected. All mean value of each image was measured automatically by ImageJ software.
Off-target analysis
Potential off-target sites were first identified using an in silico tool, Off-finder (www.rgenome.net). Mouse genomic sites containing up to 3-bp mismatches were considered off-target sites and further confirmed by targeted deep sequencing. Additional off-target candidates were obtained by Digenome-seq (33). Briefly, after in vitro cleavage of human genomic DNA (8 μg) with SpCas9 (300 nM) and sgRNA (900 nM) in the proper buffer [100 mM NaCl, 50 mM tris-HCl, 10 mM MgCl2, and bovine serum albumin (100 μg/ml)], the digested DNA was purified and used to generate an initial library using a Covaris system (Life Technologies) together with an End Repair Mix (Thermo Fisher Scientific, Waltham, MA, USA). The end of the DNA fragment was ligated with adaptors, and the final library was sequenced through WGS (whole genome sequencing) using an Illumina HiSeq X Ten Sequencer (Illumina, San Diego, CA, USA). After mapping the sequenced reads to the mouse genome reference GRCm38, the produced BAM files were analyzed using Digenome-seq tools (www.rgenome.net). The loci showing cleavage scores of more than >2.5 were selected as potential off-target sites and further verified by targeted deep sequencing. Target site and primer information for off-target candidates are listed in table S1 and S2, respectively.
T cell stimulation and intracellular cytokine staining
Mice were injected thrice with empty LNPs, sgRNA/Cas9 mRNA–encapsulated LNPs, and PBS. Twenty-four hours after the last injection, single-cell suspensions were prepared from mice spleens. Cells (1 × 106) were cultured with medium alone (negative control), 2.5 μg of recombinant Cas9 (Cas9), or phorbol 12-myristate 13-acetate (PMA) (50 ng/ml) and ionomycin (1 μg/ml) (positive control) for 18 hours. To measure the IFN-γ–producing cells, splenocytes were further treated with brefeldin A (eBioscience Inc., San Diego, CA, USA) for 5 hours. Cells were then permeabilized with fluorescence-activated cell sorting (FACS) buffer containing 0.5% saponin (Sigma-Aldrich, St. Louis, MO, USA) and stained with markers. The stained cells were acquired using a CytoFLEX S flow cytometer (Beckman Coulter, Brea, CA, USA) and analyzed using FlowJo software (TreeStar Inc., Ashland, OR, USA). The antibodies used were antigen-presenting cell (APC)/Cy7 anti-mouse CD3 (BioLegend, San Diego, CA, USA, 100706), fluorescein isothiocyanate (FITC) anti-mouse CD8 (BioLegend, 100222), and phycoerythrin (PE) anti-mouse IFN-γ (BioLegend, 505808).
Measurement of anti-Cas9 antibody response
The anti-Cas9 antibody response was measured using ELISA. Briefly, a 96-well plate was coated with 0.1 μg per well of SpCas9 protein (Aldevron, 9212) for 3 hours at 37°C. Subsequently, the plate was washed four times and then blocked by adding 100 μl of assay diluent buffer (BioLegend, 421203) for 1 hour at 25°C. Sera from mice injected with LNP-CRISPR-mAT or AAV9-Cas9 were diluted 40 times, and the plate was incubated with the samples for 2 hours at 25°C. After washing, the plate was incubated with anti-mouse IgG–conjugated with horseradish peroxidase (HRP; Sigma, A9044; Sigma-Aldrich), subsequently washed, and then treated with trimethylboron (TMB) substrate (BioLegend, 421101). After stopping the HRP/TMB reaction using stop solution (BioLegend, 423001), the plate was subjected to absorbance measurement at 450 nm. Commercial mouse anti-Cas9 antibody (Abcam, 191468) was used for a standard curve test, and the standard curve (R2 = 0.989) was used for determining anti-Cas9 IgG concentrations of the tested samples.
Statistical analysis
Statistical analysis was performed with unpaired Student’s t test and correlation analysis using GraphPad Prism (Version 5.02, GraphPad, San Diego, CA, USA).
Acknowledgments
Funding: National Research Foundation of Korea (2017M3A9B4061406 and 2020R1A2C2004364), the Korea Ministry of Education (2018R1A5A2025286), and the Korea Ministry of Food and Drug Safety (21173MFDS562).
Author contributions: Conceptualization: Hyukjin Lee, D.W.S., and S.C.Y.; methodology: J.Y.L. and S.K.; investigation: J.P.H., M.J.K., B.S.C., J.H.L., G.S.L., E.-A.K., M.J., Y.L., H.-K.O., N.G., Hyerim Lee, K.J.L., U.G.K., and J.C.; visualization: J.P.H., M.K., B.S.C., Hyukjin Lee, D.W.S., and S.C.Y.; project administration: Hyukjin Lee, D.W.S., and S.C.Y.; supervision: Hyukjin Lee, D.W.S., and S.C.Y.; writing, original draft: J.P.H., M.K., B.S.C.; writing, review and editing: J.P.H., M.K., B.S.C., Hyukjin Lee, D.W.S., and S.C.Y.
Competing interests: D.W.S., B.S.C., H.-K.O., N.G., Hyerim Lee, S.K., J.Y.L., U.G.K., and K.J.L. are employed by ToolGen. D.W.S., B.S.C., U.G.K., and K.J.L. are inventors on a patent application related to the investigation of human sgRNAs filed by ToolGen Inc. (PCT/KR2019/009238, filed on 26 July 2018). M.J.K., M.J., and Hyukjin Lee are inventors on a patent related to this work filed by the Ewha Womans University (no. KR10-2198736, filed 13 August 2018, published 5 October 2020). The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 and S2
REFERENCES AND NOTES
- 1.Peters R., Harris T., Advances and innovations in haemophilia treatment. Nat. Rev. Drug Discov. 17, 493–508 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Bulcha J. T., Wang Y., Ma H., Tai P. W. L., Gao G., Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 6, 53 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valentino L. A., Cooper D. L., Goldstein B., Surgical experience with rFVIIa (NovoSeven) in congenital haemophilia A and B patients with inhibitors to factors VIII or IX. Haemophilia 17, 579–589 (2011). [DOI] [PubMed] [Google Scholar]
- 4.van den Berg H. M., Fischer K., Carcao M., Chambost H., Kenet G., Kurnik K., Konigs C., Male C., Santagostino E., Ljung R.; PedNet Study Group , Timing of inhibitor development in more than 1000 previously untreated patients with severe hemophilia A. Blood 134, 317–320 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Oldenburg J., Levy G. G., Emicizumab prophylaxis in hemophilia a with inhibitors. N. Engl. J. Med. 377, 2194–2195 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Pasi K. J., Rangarajan S., Georgiev P., Mant T., Creagh M. D., Lissitchkov T., Bevan D., Austin S., Hay C. R., Hegemann I., Kazmi R., Chowdary P., Gercheva-Kyuchukova L., Mamonov V., Timofeeva M., Soh C. H., Garg P., Vaishnaw A., Akinc A., Sorensen B., Ragni M. V., Targeting of antithrombin in hemophilia A or B with RNAi therapy. N. Engl. J. Med. 377, 819–828 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Machin N., Ragni M. V., An investigational RNAi therapeutic targeting antithrombin for the treatment of hemophilia A and B. J. Blood Med. 9, 135–140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mejia-Carvajal C., Czapek E. E., Valentino L. A., Life expectancy in hemophilia outcome. J. Thromb. Haemost. 4, 507–509 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Lovdahl S., Henriksson K. M., Baghaei F., Holmstrom M., Nilsson J. A., Berntorp E., Astermark J., Incidence, mortality rates and causes of deaths in haemophilia patients in Sweden. Haemophilia 19, 362–369 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Kattenhorn L. M., Tipper C. H., Stoica L., Geraghty D. S., Wright T. L., Clark K. R., Wadsworth S. C., Adeno-associated virus gene therapy for liver disease. Hum. Gene Ther. 27, 947–961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colella P., Ronzitti G., Mingozzi F., Emerging issues in AAV-mediated in vivo gene therapy. Mol. Ther. Methods Clin. Dev. 8, 87–104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verdera H. C., Kuranda K., Mingozzi F., AAV vector immunogenicity in humans: A long journey to successful gene transfer. Mol. Ther. 28, 723–746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shahryari A., Saghaeian Jazi M., Mohammadi S., Razavi Nikoo H., Nazari Z., Hosseini E. S., Burtscher I., Mowla S. J., Lickert H., Development and clinical translation of approved gene therapy products for genetic disorders. Front. Genet. 10, 868 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma R., Anguela X. M., Doyon Y., Wechsler T., DeKelver R. C., Sproul S., Paschon D. E., Miller J. C., Davidson R. J., Shivak D., Zhou S., Rieders J., Gregory P. D., Holmes M. C., Rebar E. J., High K. A., In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood 126, 1777–1784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maeder M. L., Stefanidakis M., Wilson C. J., Baral R., Barrera L. A., Bounoutas G. S., Bumcrot D., Chao H., Ciulla D. M., DaSilva J. A., Dass A., Dhanapal V., Fennell T. J., Friedland A. E., Giannoukos G., Gloskowski S. W., Glucksmann A., Gotta G. M., Jayaram H., Haskett S. J., Hopkins B., Horng J. E., Joshi S., Marco E., Mepani R., Reyon D., Ta T., Tabbaa D. G., Samuelsson S. J., Shen S., Skor M. N., Stetkiewicz P., Wang T., Yudkoff C., Myer V. E., Albright C. F., Jiang H., Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 25, 229–233 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Finn J. D., Smith A. R., Patel M. C., Shaw L., Youniss M. R., van Heteren J., Dirstine T., Ciullo C., Lescarbeau R., Seitzer J., Shah R. R., Shah A., Ling D., Growe J., Pink M., Rohde E., Wood K. M., Salomon W. E., Harrington W. F., Dombrowski C., Strapps W. R., Chang Y., Morrissey D. V., A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 22, 2227–2235 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Patil S., Gao Y. G., Lin X., Li Y., Dang K., Tian Y., Zhang W. J., Jiang S. F., Qadir A., Qian A. R., The development of functional non-viral vectors for gene delivery. Int. J. Mol. Sci. 20, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehta A., Merkel O. M., Immunogenicity of Cas9 protein. J. Pharm. Sci. 109, 62–67 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pien G. C., Basner-Tschakarjan E., Hui D. J., Mentlik A. N., Finn J. D., Hasbrouck N. C., Zhou S., Murphy S. L., Maus M. V., Mingozzi F., Orange J. S., High K. A., Capsid antigen presentation flags human hepatocytes for destruction after transduction by adeno-associated viral vectors. J. Clin. Invest. 119, 1688–1695 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilbie D., Walther J., Mastrobattista E., Delivery aspects of CRISPR/Cas for in vivo genome editing. Acc. Chem. Res. 52, 1555–1564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin H., Song C. Q., Suresh S., Wu Q., Walsh S., Rhym L. H., Mintzer E., Bolukbasi M. F., Zhu L. J., Kauffman K., Mou H., Oberholzer A., Ding J., Kwan S. Y., Bogorad R. L., Zatsepin T., Koteliansky V., Wolfe S. A., Xue W., Langer R., Anderson D. G., Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 35, 1179–1187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ragni M. V., Targeting antithrombin to treat hemophilia. N. Engl. J. Med. 373, 389–391 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Sehgal A., Barros S., Ivanciu L., Cooley B., Qin J., Racie T., Hettinger J., Carioto M., Jiang Y., Brodsky J., Prabhala H., Zhang X., Attarwala H., Hutabarat R., Foster D., Milstein S., Charisse K., Kuchimanchi S., Maier M. A., Nechev L., Kandasamy P., Kel’in A. V., Nair J. K., Rajeev K. G., Manoharan M., Meyers R., Sorensen B., Simon A. R., Dargaud Y., Negrier C., Camire R. M., Akinc A., An RNAi therapeutic targeting antithrombin to rebalance the coagulation system and promote hemostasis in hemophilia. Nat. Med. 21, 492–497 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Pandey G. S., Yanover C., Miller-Jenkins L. M., Garfield S., Cole S. A., Curran J. E., Moses E. K., Rydz N., Simhadri V., Kimchi-Sarfaty C., Lillicrap D., Viel K. R., Przytycka T. M., Pierce G. F., Howard T. E., Sauna Z. E.; PATH (Personalized Alternative Therapies for Hemophilia) Study Investigators , Endogenous factor VIII synthesis from the intron 22–inverted F8 locus may modulate the immunogenicity of replacement therapy for hemophilia A. Nat. Med. 19, 1318–1324 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Q., Wei T., Farbiak L., Johnson L. T., Dilliard S. A., Siegwart D. J., Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim M., Jeong M., Hur S., Cho Y., Park J., Jung H., Seo Y., Woo H. A., Nam K. T., Lee K., Lee H., Engineered ionizable lipid nanoparticles for targeted delivery of RNA therapeutics into different types of cells in the liver. Sci. Adv. 7, eabf4398 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kintigh J., Monagle P., Ignjatovic V., A review of commercially available thrombin generation assays. Res. Pract. Thromb. Haemost. 2, 42–48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duarte R. C. F., Ferreira C. N., Rios D. R. A., Reis H. J. D., Carvalho M. D. G., Thrombin generation assays for global evaluation of the hemostatic system: Perspectives and limitations. Rev. Bras. Hematol. Hemoter. 39, 259–265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peyvandi F., Garagiola I., Young G., The past and future of haemophilia: Diagnosis, treatments, and its complications. Lancet 388, 187–197 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Husseinzadeh H., Chiasakul T., Gimotty P. A., Pukenas B., Wolf R., Kelty M., Chiang E., Fogarty P. F., Cuker A., Prevalence of and risk factors for cerebral microbleeds among adult patients with haemophilia A or B. Haemophilia 24, 271–277 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Rey-Barroso L., Roldan M., Burgos-Fernandez F. J., Gassiot S., Ruiz Llobet A., Isola I., Vilaseca M., Spectroscopic evaluation of red blood cells of thalassemia patients with confocal microscopy: A pilot study. Sensors 20, 4039 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esposito P., Rampino T., Gregorini M., Fasoli G., Gamba G., Dal Canton A., Renal diseases in haemophilic patients: Pathogenesis and clinical management. Eur. J. Haematol. 91, 287–294 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Kim D., Bae S., Park J., Kim E., Kim S., Yu H. R., Hwang J., Kim J. I., Kim J. S., Digenome-seq: Genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–243 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Crudele J. M., Chamberlain J. S., Cas9 immunity creates challenges for CRISPR gene editing therapies. Nat. Commun. 9, 3497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castaman G., Matino D., Hemophilia A and B: Molecular and clinical similarities and differences. Haematologica 104, 1702–1709 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mannucci P. M., Hemophilia therapy: The future has begun. Haematologica 105, 545–553 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gillmore J. D., Gane E., Taubel J., Kao J., Fontana M., Maitland M. L., Seitzer J., O’Connell D., Walsh K. R., Wood K., Phillips J., Xu Y., Amaral A., Boyd A. P., Cehelsky J. E., McKee M. D., Schiermeier A., Harari O., Murphy A., Kyratsous C. A., Zambrowicz B., Soltys R., Gutstein D. E., Leonard J., Sepp-Lorenzino L., Lebwohl D., CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 385, 493–502 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Franchini M., Mannucci P. M., Non-factor replacement therapy for haemophilia: A current update. Blood Transfus. 16, 457–461 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sridharan G., Liu J. X., Qian K., Goel V., Huang S., Akinc A., In silico modeling of the impact of antithrombin lowering on thrombin generation in rare bleeding disorders. Blood 130, 3659 (2017). [Google Scholar]
- 40.Palta S., Saroa R., Palta A., Overview of the coagulation system. Indian J. Anaesth. 58, 515–523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crawley J. T., Lane D. A., The haemostatic role of tissue factor pathway inhibitor. Arterioscler. Thromb. Vasc. Biol. 28, 233–242 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Shapiro A. D., Angchaisuksiri P., Astermark J., Benson G., Castaman G., Chowdary P., Eichler H., Jimenez-Yuste V., Kavakli K., Matsushita T., Poulsen L. H., Wheeler A. P., Young G., Zupancic-Salek S., Oldenburg J., Subcutaneous concizumab prophylaxis in hemophilia A and hemophilia A/B with inhibitors: Phase 2 trial results. Blood 134, 1973–1982 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thyzel E., Siegling S., Brinkmann T., Kleesiek K., Gotting C., Expression and characterization of wild-type TFPI and the [P151L]TFPI mutant in insect cells. Mol. Cell. Biochem. 283, 31–38 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Lu R. M., Hwang Y. C., Liu I. J., Lee C. C., Tsai H. Z., Li H. J., Wu H. C., Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 27, 1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uddin F., Rudin C. M., Sen T., CRISPR gene therapy: Applications, limitations, and implications for the future. Front. Oncol. 10, 1387 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buchlis G., Podsakoff G. M., Radu A., Hawk S. M., Flake A. W., Mingozzi F., High K. A., Factor IX expression in skeletal muscle of a severe hemophilia B patient 10 years after AAV-mediated gene transfer. Blood 119, 3038–3041 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Batty P., Lillicrap D., Hemophilia gene therapy: Approaching the first licensed product. Hemasphere 5, e540 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Y., Hu Z., Li Z., Pang J., Feng M., Hu X., Wang X., Lin-Peng S., Liu B., Chen F., Wu L., Liang D., In situ genetic correction of F8 intron 22 inversion in hemophilia A patient-specific iPSCs. Sci. Rep. 6, 18865 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boutin S., Monteilhet V., Veron P., Leborgne C., Benveniste O., Montus M. F., Masurier C., Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: Implications for gene therapy using AAV vectors. Hum. Gene Ther. 21, 704–712 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Adams D., Gonzalez-Duarte A., O’Riordan W. D., Yang C. C., Ueda M., Kristen A. V., Tournev I., Schmidt H. H., Coelho T., Berk J. L., Lin K. P., Vita G., Attarian S., Plante-Bordeneuve V., Mezei M. M., Campistol J. M., Buades J., Brannagan T. H. III, Kim B. J., Oh J., Parman Y., Sekijima Y., Hawkins P. N., Solomon S. D., Polydefkis M., Dyck P. J., Gandhi P. J., Goyal S., Chen J., Strahs A. L., Nochur S. V., Sweetser M. T., Garg P. P., Vaishnaw A. K., Gollob J. A., Suhr O. B., Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 379, 11–21 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Witzigmann D., Kulkarni J. A., Leung J., Chen S., Cullis P. R., van der Meel R., Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv. Drug Deliv. Rev. 159, 344–363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miao L., Lin J., Huang Y., Li L., Delcassian D., Ge Y., Shi Y., Anderson D. G., Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun. 11, 2424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naeem M., Majeed S., Hoque M. Z., Ahmad I., Latest developed strategies to minimize the off-target effects in CRISPR-cas-mediated genome editing. Cell 9, 1608 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee J. K., Jeong E., Lee J., Jung M., Shin E., Kim Y. H., Lee K., Jung I., Kim D., Kim S., Kim J. S., Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 9, 3048 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen J. S., Dagdas Y. S., Kleinstiver B. P., Welch M. M., Sousa A. A., Harrington L. B., Sternberg S. H., Joung J. K., Yildiz A., Doudna J. A., Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han J. P., Song D. W., Lee J. H., Lee G. S., Yeom S. C., Novel severe hemophilia a mouse model with factor VIII intron 22 inversion. Biology 10, 704 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 and S2