Abstract
Chondrocytes secrete massive extracellular matrix (ECM) molecules that are produced, folded, and modified in the endoplasmic reticulum (ER). Thus, the ER-associated degradation (ERAD) complex—which removes misfolded and unfolded proteins to maintain proteostasis in the ER— plays an indispensable role in building and maintaining cartilage. Here, we examined the necessity of the ERAD complex in chondrocytes for cartilage formation and maintenance. We show that ERAD gene expression is exponentially increased during chondrogenesis, and disruption of ERAD function causes severe chondrodysplasia in developing embryos and loss of adult articular cartilage. ERAD complex malfunction also causes abnormal accumulation of cartilage ECM molecules and subsequent chondrodysplasia. ERAD gene expression is decreased in damaged cartilage from patients with osteoarthritis (OA), and disruption of ERAD function in articular cartilage leads to cartilage destruction in a mouse OA model.
Reduced secretory activity due to the unfolded protein accumulation in chondrocytes may worsen cartilage damage and loss in osteoa.
INTRODUCTION
One-third of all proteins are secreted or transported from the endoplasmic reticulum (ER) to other membranous cellular organelles. The ER provides a specialized environment to produce secretory and transmembrane proteins; it thus houses molecular chaperones, folding catalysts, and enzymes for posttranslational modification. The ER must organize diverse posttranslational processes, such as folding, oligomerization, and glycosylation. However, proteins in the ER are often misfolded or unfolded, and these must be corrected or removed to maintain functional ER homeostasis.
Accumulation of misfolded or unfolded proteins causes ER stress. Excess ER stress impairs ER functions and leads to apoptotic cell death (1). To prevent this, the ER performs quality control checks to remove unfolded or misfolded proteins to maintain cellular homeostasis and prevent proteotoxicity caused by misfolded protein accumulation. Disruption of protein quality control can cause pathologies, such as Alzheimer’s disease, Huntington’s disease, or cystic fibrosis (2). The ER uses two degradation pathways to remove misfolded or unfolded proteins: the ER-associated degradation (ERAD) pathway and autophagy. The ERAD pathway is a major degradation pathway (3) and facilitates proteasome-dependent degradation of unfolded or misfolded proteins in response to unfolded protein response (UPR) signaling in the ER lumen (4). The ERAD complex comprises many proteins, including Sel1L, HRD1, EDEM1, and Derlin-1/2 (Derl1/2), which participate in recognizing nonnative proteins produced because of errors in transcription, translation, or failure to form a correct oligomeric complex. These nonnative proteins are then translocated, ubiquitinated, and removed by proteasomal degradation in the cytosolic proteasomal complex (5–7).
Although ERAD functions as a downstream response to ER stress or UPR are generally understood, recent data suggest that the ERAD complex has UPR- or ER stress–independent functions (i.e., constitutive in vivo functions in uninduced conditions) (8). The evidence suggests that ERAD complex pathophysiology is dependent on the endogenous targets of specific tissues. In addition, the phenotypes of tissue-specific deletion of ERAD genes are distinct from the loss-of-function phenotypes of UPR components (8). The in vivo roles of the constitutive ERAD complex in various tissues and organs have yet to be studied.
Chondrocytes are specialized cells solely responsible for producing extracellular matrix (ECM)–rich cartilage tissue. During development, chondrocytes secrete large amounts of ECM molecules, such as collagens and proteoglycans (9, 10), that are essential to the mechanical properties and integrity of cartilage (9, 11, 12). ER homeostasis is therefore critical for cartilage construction and maintenance.
Here, we investigated the roles of the ERAD complex in cartilage synthesis and maintenance and found that ERAD complex dysfunction in developing cartilage resulted in severe chondrodysplasia. Chondrocytes from patients with osteoarthritis (OA) exhibited reduced ERAD gene expression. Furthermore, disruption of ERAD function in mouse articular cartilage resulted in OA-like cartilage damage. Last, we showed that defects in cartilage formation are derived from the abnormal accumulation of cartilage matrix proteins in the ER due to ERAD complex malfunction. Collectively, our results provide compelling evidence that impaired ERAD function is a main cause of cartilage malformation and destruction, such as seen in OA.
RESULTS
ERAD gene expression is strongly increased and necessary during chondrogenesis
The ER in chondrocyte consistently synthesizes matrix proteins, while surveying and removing misfolded or unfolded proteins to maintain proteostasis in the ER. Therefore, we considered that chondrocytes may express high levels of ERAD genes and be vulnerable to ERAD complex malfunction. To confirm this hypothesis, we measured ERAD gene expression during chondrogenesis. We first quantitively analyzed expression of several ERAD genes during in vitro chondrogenesis with ATDC5 mouse chondrogenic cell line and human primary chondrocytes. As predicted, the expression of all ERAD genes tested markedly increased 10 days after chondrogenic induction (Fig. 1, A and B). These data suggest that ERAD functions may be critical for both formation and maintenance of cartilage tissue. Chondrocytes usually differentiate into hypertrophic chondrocytes in vitro, and we further examined whether ERAD gene expression increases during chondrocyte hypertrophy. Both ATDC5 and human primary chondrocytes did not undergo hypertrophy at 10 days after chondrogenic induction (fig. S1, A and B). Then, we forced hypertrophic chondrocyte differentiation using a differentiation medium (13). Both ATDC5 and human primary chondrocytes that differentiated into hypertrophic chondrocytes did not show notable correlation in expression of ERAD genes (fig. S1, C and D). This line of evidence suggests that ERAD gene expression is specifically elevated during chondrogenic differentiation.
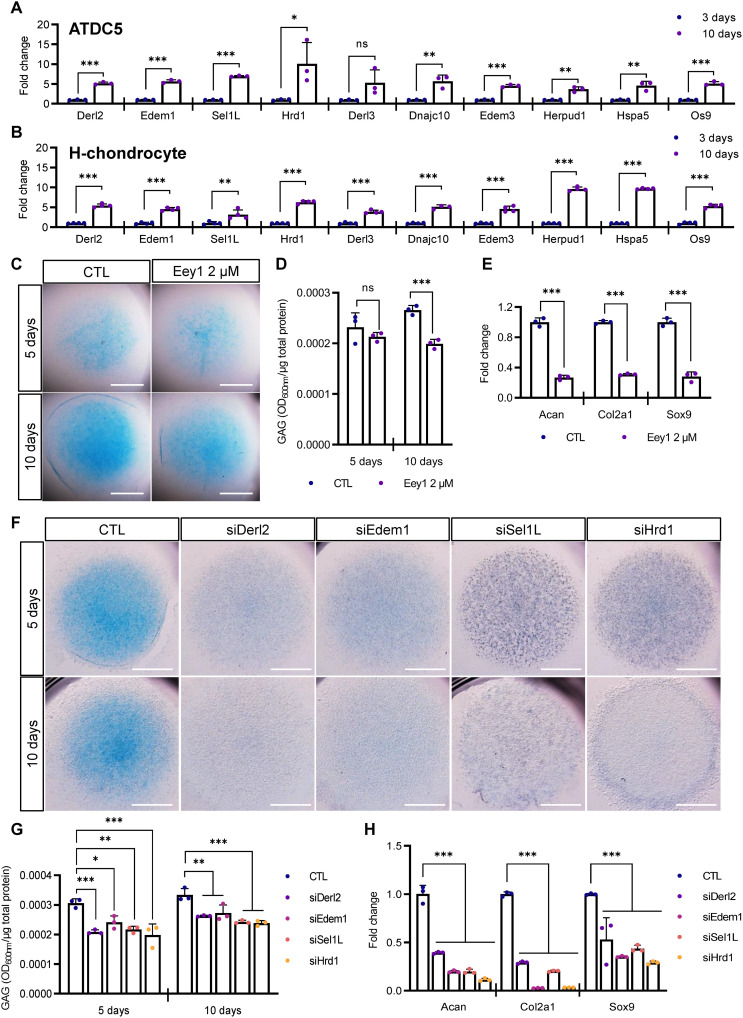
Fig. 1. ERAD genes are highly expressed in differentiating chondrocytes, and ERAD inhibition impairs chondrogenesis.
(A and B) Expression levels of ERAD genes in differentiating ATDC5 (A, n = 3) or H-chondrocyte (B, n = 4) micromasses were measured using qPCR (n = 3). ERAD gene expression was significantly increased at 10 days after induction of chondrogenesis. (C) Differentiating ATDC5 cell micromasses were treated with Eey1, and Alcian blue staining was performed at the indicated days after induction. Eey1-treated micromasses displayed sharp reductions in glycosaminoglycan (GAG) accumulation. Scale bar, 1 mm. (D) GAG levels in chondrogenic micromasses shown in (C) were measured and normalized to total protein level (n = 3). (E) Expression of cartilage anabolic factors, Acan, Col2a1, and Sox9 was measured by qPCR. Eey1 treatment significantly reduced the expression of anabolic factors (n = 3). (F) ERAD genes were depleted with siRNAs, and chondrogenesis was induced in the ATDC5 cell micromasses. Depletion of any one of the ERAD genes strongly impaired chondrogenesis. Scale bar, 1 mm. (G) GAG levels in chondrogenic micromasses shown in (F) were measured and normalized to total protein level. Depletion of any one of the ERAD genes significantly decreased GAG accumulation (n = 3). (H) Expression of cartilage anabolic factors was reduced upon depletion of any one of the ERAD genes (n = 3). Statistical analysis was performed using Student’s t test (A, B, D, and E) or one-way ANOVA (G and H). ***P < 0.001, **(0.001 < P < 0.01), *(0.01 < P < 0.05), ns (0.05 < P), OD600 = optical density at 600 nm, CTL, cytotoxic T lymphocyte.
To elucidate the functions of the ERAD complex, we treated cells with eeyarestatin 1 (Eey1), which inhibits the ATPases associated with various cellular activities (AAA +) adenosine triphosphatase (ATPase) p97 transporter in the ERAD complex during chondrogenic differentiation in micromass culture (14). As we hypothesized, Eey1-mediated inhibition of ERAD markedly reduced glycosaminoglycan (GAG) deposition as visualized by Alcian blue staining (Fig. 1C and fig. S2A). We further verified reductions in chondrogenesis by measuring total GAG levels in the micromasses (Fig. 1D and fig. S2B). Up to 5 days of differentiation, GAG deposition was comparable in the Eey1-treated and control cells; however, after 7 days of differentiation, GAG levels gradually decreased in Eey1-treated micromasses (fig. S2B). We further examined whether the inhibition of the ERAD complex function may inhibit proliferation of chondrogenic cells by performing 5-ethynyl-2’-deoxyuridine (EdU) staining. However, the inhibition of ERAD complex function did not change the proliferation rate of differentiating chondrocytes (fig. S2C).
Next, we investigated the expression of cartilage anabolic factors, such as sex-determining region Y (SRY)-box transcription factor 9 (Sox9), collagen type II alpha 1 (Col2a1), and aggrecan (Acan). During chondrogenesis, Sox9 is a master regulator of chondrogenesis that works at an early stage of differentiation (15, 16). Col2a1 is the major collagen type, and Acan is a cartilage-specific proteoglycan secreted during chondrogenesis (17). Quantitative real-time polymerase chain reaction (qRT-PCR) analysis revealed that expression of these anabolic factors was sharply decreased in Eey1-treated micromasses (Fig. 1E). These data suggest that sustained ERAD function may be necessary for chondrogenic differentiation.
Knockdown of ERAD genes impairs chondrogenesis
Although Eey1 is known to inhibit ERAD functions, it may inhibit other AAA + ATPase p97–dependent cellular functions. Therefore, we directly inhibited ERAD function by depleting ERAD genes. The ERAD complex comprises several proteins, including DERL1/2, EDEM1/2/3, SEL1L, and HRD1, that participate in recognition, translocation, ubiquitination, and proteasomal degradation (18). We designed small interfering RNAs (siRNAs) targeting the ERAD genes Derl2, Edem1, Sel1L, and Hrd1, which were shown to be strongly increased during chondrogenesis. siRNA transfection efficiently knocked down target gene expression without affecting chondrocyte proliferation (fig. S2, C and D). All the siRNA-transfected micromasses showed marked reductions in cartilage-specific matrix deposition (Fig. 1F and fig. S2E). We further measured total GAG levels of those micromasses and yielded similar data (Fig. 1G and fig. S2F). In addition, ERAD gene–depleted micromasses also showed reduced anabolic factor expression (Fig. 1H). Together, these data strongly support the critical role of the ERAD complex for efficient chondrogenesis.
Depletion of ERAD expression causes abnormal accumulation of matrix proteins
Next, we sought to elucidate why malfunction or depletion of the ERAD complex impaired chondrogenesis. Chondrocytes secrete massive amounts of cartilage-specific matrix proteins, of which Col2a1 is a major component. Therefore, we analyzed Col2a1 deposition in ERAD complex–depleted micromasses. Consistent with the reduction in GAG deposition, malfunction of the ERAD complex significantly reduced Col2a1 deposition in chondrogenic micromasses (Fig. 2, A to C). Eey1-treated micromasses showed similar results to ERAD complex–depleted micromasses (fig. S3, A to C).
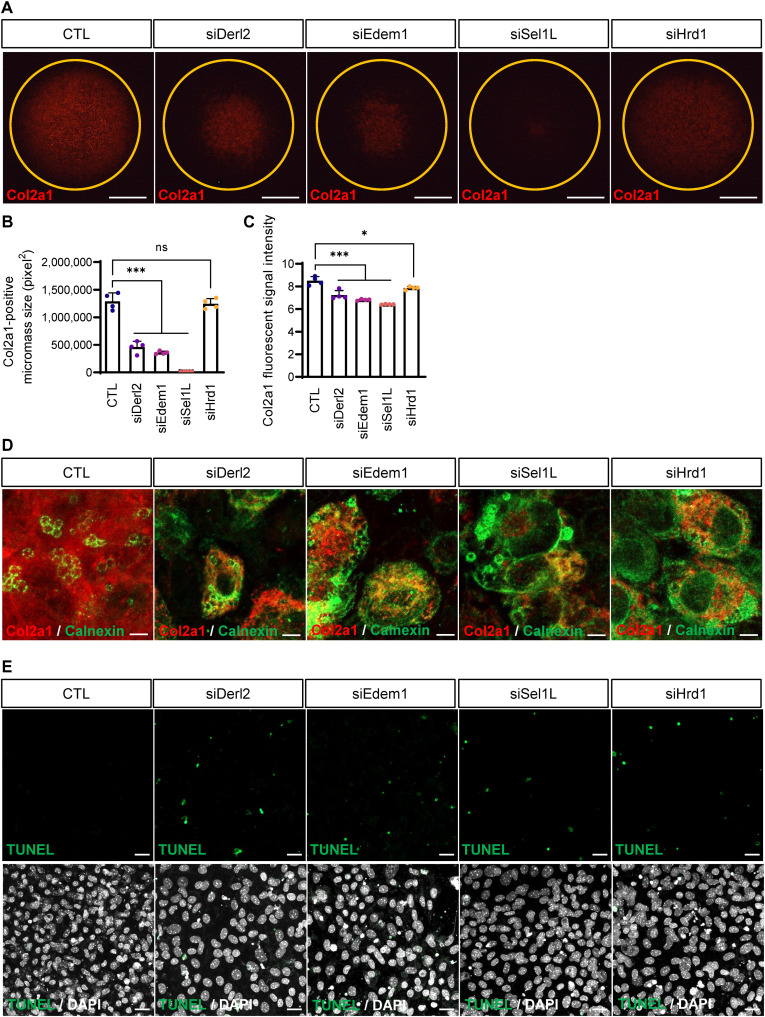
Fig. 2. Inhibition of ERAD function causes abnormal collagen accumulation in the ER lumen.
(A) Collagen deposition in chondrogenic micromasses was detected by immunofluorescence staining upon ERAD gene depletion. Depletion of ERAD genes caused severe reduction in collagen deposition compared to control micromasses. The Sel1L-depleted micromass showed a severe phenotype. Scale bar, 1 mm (B) The areas of collagen-positive micromass cultures shown in (A) were measured and plotted (n = 4). Areas of collagen positivity for Derl2- and Edem1-knockdown micromasses were significantly reduced. Sel1L depletion caused the most severe phenotypes, whereas Hrd1-depleted micromasses displayed mild reductions in collagen deposition. (C) Staining intensity of collagen-positive micromass cultures shown in (A) were measured and plotted (n = 4). Statistical analysis was performed using one-way ANOVA. ***(P < 0.001), *(0.01 < P < 0.05), ns (0.05 < P). (D) High-resolution images of micromasses shown in (A) (top). Red represents Col2a1, and green represents the ER marker calnexin. In control micromasses, collagen was secreted and deposited in the ECM. However, in ERAD-depleted micromasses, most of the collagen signal was detected in the ER lumen. Scale bars, 5 μm. (E) TUNEL staining revealed increased cell death in ERAD-depleted micromasses. The bottom panels show the merged images with DAPI staining. Scale bars, 20 μm.
During chondrogenesis, an important step in ECM biosynthesis is protein folding in the ER, which usually is dependent on posttranslational modifications (2). Secretion of misfolded or unfolded ECM proteins is attenuated, so they accumulate in the ER (2). We speculated that cartilage-specific ECM proteins may accumulate in the ER because of disrupted ERAD function. To test this hypothesis, we stained differentiating chondrocytes for Col2a1 and the ER marker calnexin to investigate collagen localization after ERAD depletion. In control micromasses, Col2a1 was well secreted to the extracellular space. Unexpectedly, in Derl2, Edem1, Sel1L, or Hrd1 knockdown micromasses, Col2a1-positive punctae were accumulated in the calnexin-positive ER lumen, rather than being secreted to the extracellular space (Fig. 2D). Eey1-treated micromasses also showed Col2a1 accumulation in the ER lumen (fig. S3D).
We observed that expression of the ER marker calnexin was noticeably increased in ERAD-depleted cells (Fig. 2D). The malfunction or depletion of the ERAD complex increases ER stress; expression of the ER chaperone protein calnexin is known to increase in response to ER stress (19). Because excess amount of the ER stress may also cause apoptotic cell death (1, 8), we measured apoptotic cell death by performing TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling) staining of ERAD complex–depleted micromasses. TUNEL-positive cells were increased in all ERAD complex–depleted micromasses and ERAD function–inhibited micromasses (Fig. 2E and fig. S3, E to G).
Sel1L knockdown causes severe chondrodysplasia
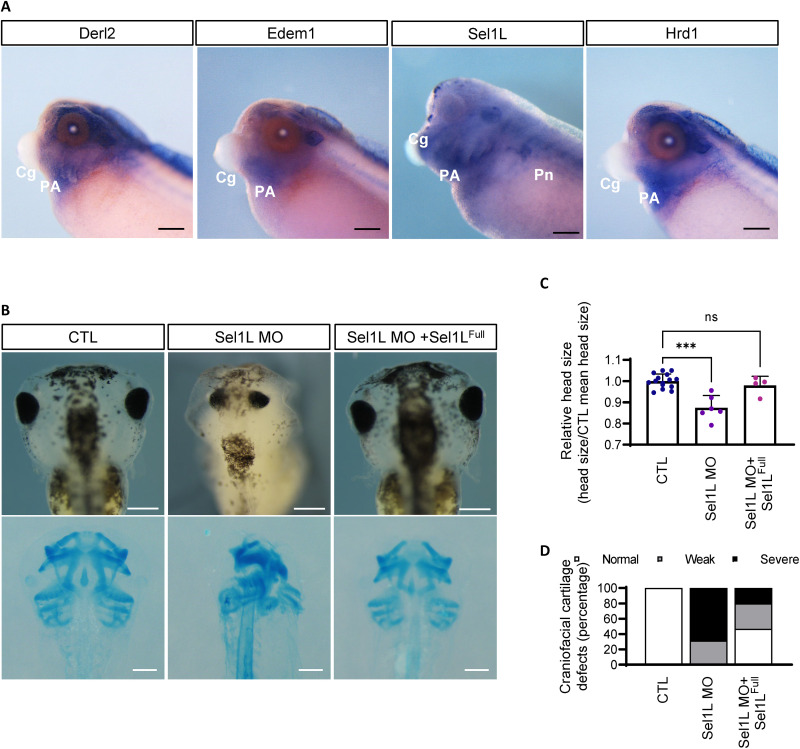
Xenopus laevis is a powerful model organism to investigate developmental processes, and we and others (20–22) previously demonstrated that the craniofacial cartilage of Xenopus would be an excellent model to study molecular mechanisms of chondrogenesis. To understand in vivo functions of the ERAD complex, we next analyzed ERAD gene expression in developing cartilage tissue by performing whole-mount in situ hybridization using Xenopus embryos. Facial cartilage develops from embryonic pharyngeal arches (PA), and whole-mount in situ hybridization analysis revealed that most ERAD genes were strongly expressed in the PAs of developing embryos (Fig. 3A and fig. S4). ERAD genes were also expressed in the kidney and head structures (Fig. 3A and fig. S4). Because Sel1L depletion in ATDC5 micromass cultures resulted in severe phenotypes, we assumed that Sel1L knockdown may also result in cartilage defects in developing embryos. To investigate Sel1L function in cartilage development, we knocked down Sel1L expression using translation-blocking morpholino antisense oligonucleotides (Sel1L-MO) and analyzed Sel1L loss-of-function phenotypes. As expected, Sel1L knockdown resulted in severe chondrodysplasia phenotypes, which were rescued by ectopic expression of full-length Sel1L (Sel1LFull) (Fig. 3, B to D).
Fig. 3. Sel1L depletion results in severe chondrodysplasia in Xenopus laevis.
(A) Whole-mount in situ hybridization analysis showed that ERAD genes are also highly expressed in developing facial cartilage tissue (PA) of X. laevis embryos. PA, pharyngeal arches; Cg, cement gland; Pn, pronephros. Scale bars, 200 μm. (B) Sel1L depletion by antisense morpholino oligos resulted in severe chondrodysplasia in craniofacial cartilage, whereas coinjection of full-length Sel1L mRNA (Sel1LFull) with the antisense morpholino rescued the phenotype. The bottom panels show Alcian blue staining of craniofacial cartilage. Scale bars, 200 μm. (C) Head sizes of Xenopus tadpoles shown in (B) were measured and plotted. The head size of Sel1L morphant was significantly reduced; however, full-length Sel1L mRNA rescued the phenotype. From left to right, n = 15, 6, and 4. (D) Craniofacial cartilage defects in Xenopus shown in (B) were counted and plotted. The morphants showed craniofacial cartilage defects that were rescued by full-length Sel1L mRNA. From left to right, n = 16, 9, and 15. Statistical analysis was performed using the Student’s t test. ***(P < 0.001), **(0.001 < P < 0.01), *(0.01 < P < 0.05), ns (0.05 < P).
To rule out nonspecific effects of Sel1L-MO in developing embryos, we further verified the loss-of-function phenotypes of Sel1L for chondrogenesis by overexpressing a dominant negative mutant in developing Xenopus embryos. Sel1L contains 11 Sel1L-like-repeat (SLR) motifs. We previously reported that 5 to 9 SLR motifs constitute the central domain of Sel1L (Sel1Lcent), the main homodimerization domain, and the carboxyl domain (Sel1Lc term), containing 10 to 11 SLR motifs, functions as an interaction domain with the E3 ligase HRD1 (23). We reasoned that Sel1Lc term overexpression may inhibit the interaction between Sel1LFull and Hrd1, and we confirmed that the Sel1Lc term effectively interrupted Sel1L-HRD1 binding (fig. S5A). Then, we assumed that overexpression of Sel1Lc term may cause defects in chondrogenesis by functioning as a dominant negative mutant. To confirm this hypothesis, we overexpressed Sel1Lc term and analyzed the chondrogenic process. Alcian blue staining of the Sel1Lc term–overexpressed embryos revealed severe craniofacial defects similar to those of Sel1L-MO–injected Xenopus embryos (fig. S5, B to D).
Depletion of Sel1L causes abnormal accumulation of matrix proteins in differentiating chondrocytes in vivo
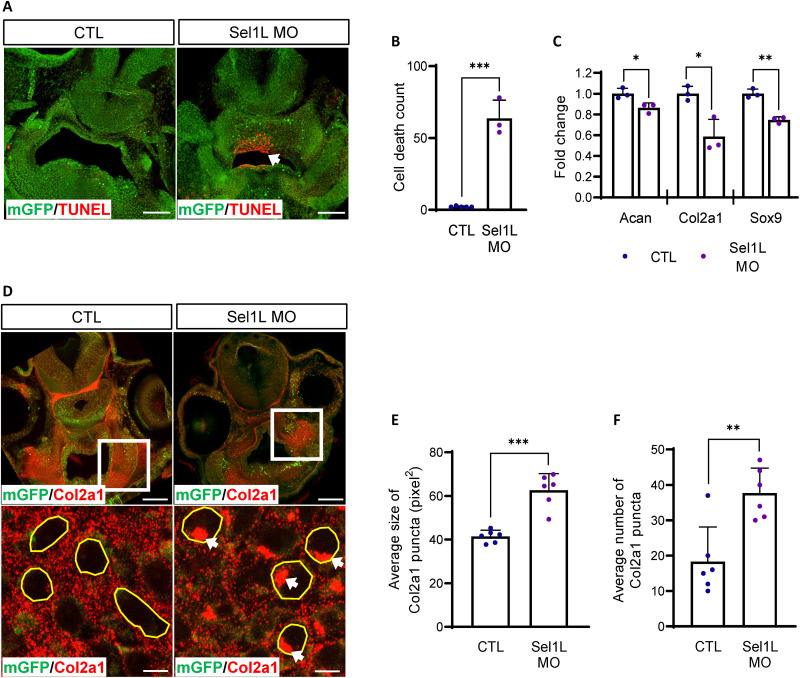
Next, we examined whether malfunction of the ERAD complex may cause cell death and abnormal collagen accumulation in differentiating chondrocytes in vivo using the Xenopus model. We sectioned craniofacial tissues of Xenopus embryos and performed TUNEL staining and Col2a1 immunostaining. Consistent with the ATDC5 micromass data, Sel1L-MO–injected embryos showed increased cell death compared with controls (Fig. 4, A and B). Expression of cartilage anabolic factors was also decreased in Sel1L-depleted embryos (Fig. 4C). Col2a1 immunostaining showed that, while most of the Col2a1 signal was exclusively found in the extracellular space in control embryos, Sel1L morphant embryos showed severe intracellular accumulation of Col2a1 in developing chondrocytes (Fig. 4, D to F). Developing chondrocytes with Sel1Lc term overexpression showed elevated cell death and accumulation of Col2a1 (fig. S5, E to I). The overexpression of Sel1Lc term did not cause significant differences in cartilage anabolic factor expression (fig. S5J). We believe that this discrepancy between Sel1L-MO and Sel1Lc term–injected embryos is probably due to the local distribution of Sel1Lc term mRNA. We further investigated Sel1Lc term overexpression phenotypes using ATDC5 mouse chondrocyte. Consistent with the ERAD gene depletion data, Alcian blue–positive areas and GAG levels were significantly decreased in Sel1Lc term–overexpressing micromasses (fig. S6, A and B), as were cartilage anabolic factors (fig. S6C), but Sel1Lc term overexpression did not alter proliferation (fig. S6D). These data strongly suggest that the ERAD complex plays a critical role in developing chondrocytes by maintaining the proteostasis of secretory pathways.
Fig. 4. Sel1L depletion causes abnormal accumulation of collagen in craniofacial cartilages in Xenopus laevis.
(A) Cell death was analyzed in Sel1L-depleted craniofacial tissues of Xenopus (green, membrane GFP; red, TUNEL-positive cells). Cell death was significantly increased in Sel1L morphants (white arrow). Scale bar, 100 μm. (B) TUNEL-positive cells in Xenopus shown in (E) were counted and plotted. Sel1L depletion increased the number of TUNEL-positive cells. From left to right, n = 5 and 3. (C) Expression of cartilage anabolic factors in Xenopus was measured using qPCR. Morpholino-injected embryos showed mild decreases. n = 3. (D) Collagen deposition was analyzed in Sel1L-depleted craniofacial tissues of Xenopus. Sel1L morphants showed intracellular collagen accumulation. White arrows indicate abnormal collagen puncta. Scale bars, 100 μm (top), 5 μm (bottom). (E) Statistical analysis of the average size of collagen-positive punctae. Collagen puncta were significantly increased in Sel1L-depleted Xenopus. n = 6. (F) Statistical analysis of number of accumulated collagen-positive punctae. Abnormal collagen-positive puncta were significantly increased in Sel1L-depleted Xenopus. n = 6. Statistical analysis was performed using the Student’s t test. ***(P < 0.001), **(0.001 < P < 0.01), *(0.01 < P < 0.05), ns (0.05 < P).
ERAD genes are down-regulated in patients with OA
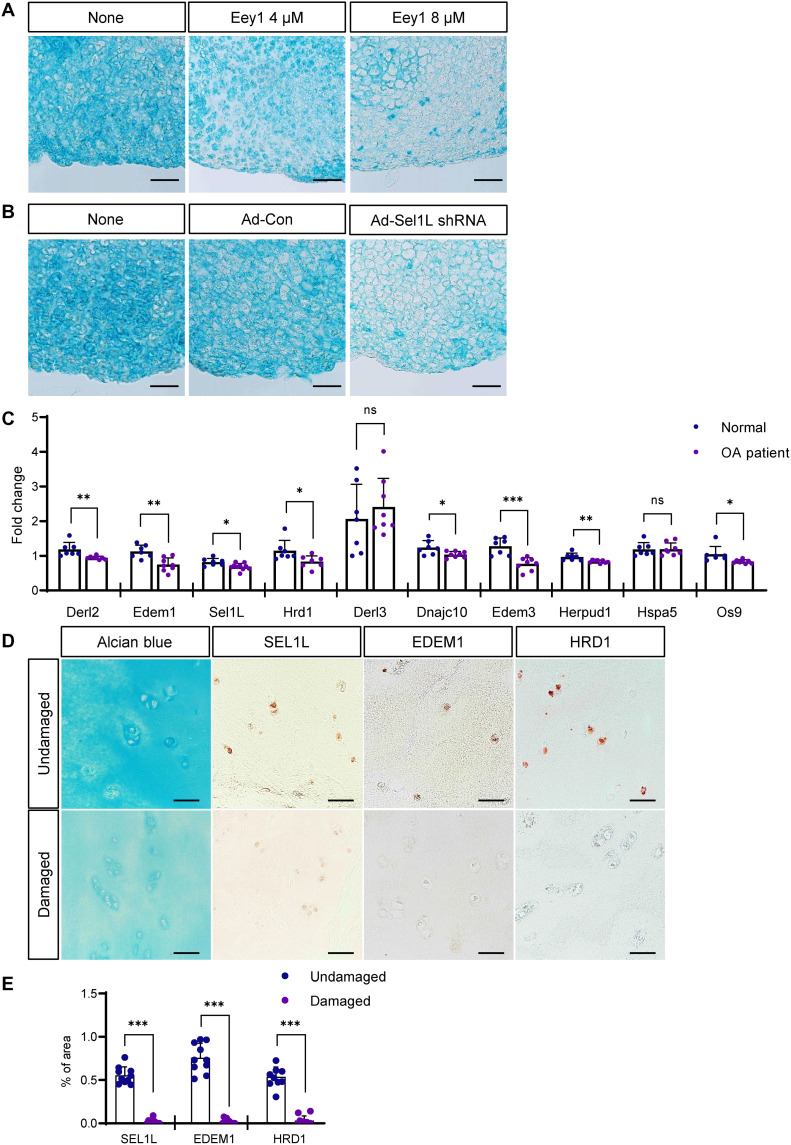
Notable increases and strong expression of ERAD genes during chondrogenesis imply that ERAD function may be necessary for both the construction and maintenance of cartilage. We treated developed cartilage micromasses with Eey1 and analyzed cartilage matrix integrity. As expected, Eey1 treatment for 7 days after micromass differentiation significantly reduced total GAG deposition and anabolic gene expression required for chondrogenesis (fig. S7, A to C). We also inhibited ERAD function in cartilage ex vivo cultures explanted from mouse knee joints by either treating with Eey1 or transducing Sel1L short hairpin–mediated RNA (shRNA) using an adenoviral gene delivery system. In both cases, cartilage matrix integrity was significantly compromised in cartilage explants (Fig. 5, A and B), which suggested that intact, active ERAD function might be critical to the maintenance of healthy cartilage in humans. Therefore, we examined the expression of ERAD genes in cartilage samples from patients with OA. qRT-PCR analysis demonstrated that ERAD genes were consistently down-regulated in patients with OA (Fig. 5C). Immunohistochemistry (IHC) also revealed that expression of ERAD proteins, such as SEL1L, EDEM1, and HRD1, were decreased in damaged cartilage identified with weaker Alcian blue staining (Fig. 5, D and E).
Fig. 5. Inhibition of ERAD function causes cartilage loss in mouse cartilage explants, and ERAD genes are down-regulated in OA chondrocytes.
(A) Mouse cartilage explants were treated with Eey1, and GAG deposition was visualized by Alcian blue staining. Eey1-treated explants displayed severe cartilage loss. Scale bars, 50 μm. (B) Sel1L expression in mouse cartilage explants was depleted by IA injection of adenoviral Sel1L-shRNA (Ad-sel1l shRNA), and GAG deposition was visualized by Alcian blue staining. Sel1L-depleted explants displayed severe cartilage loss. Scale bars, 50 μm. (C) Expression of ERAD genes in chondrocytes isolated from patients with OA was measured by qPCR. Most ERAD genes were down-regulated in OA chondrocytes. Normal, n = 7; OA patient, n = 8. (D) Immunohistochemistry (IHC) using several antibodies against ERAD genes showed decreased protein levels in OA cartilage tissue. Scale bars, 50 μm. (E) Measurement of SEL1L, EDEM, and HRD1 IHC density in OA cartilage tissues shown in (D). Undamaged, n = 10; damaged, n = 10. Statistical analysis was performed using the Student’s t test. ***(P < 0.001), **(0.001 < P < 0.01), *(0.01 < P < 0.05), ns (0.05 < P).
Knockdown of ERAD genes in articular cartilage caused destruction of cartilage matrix
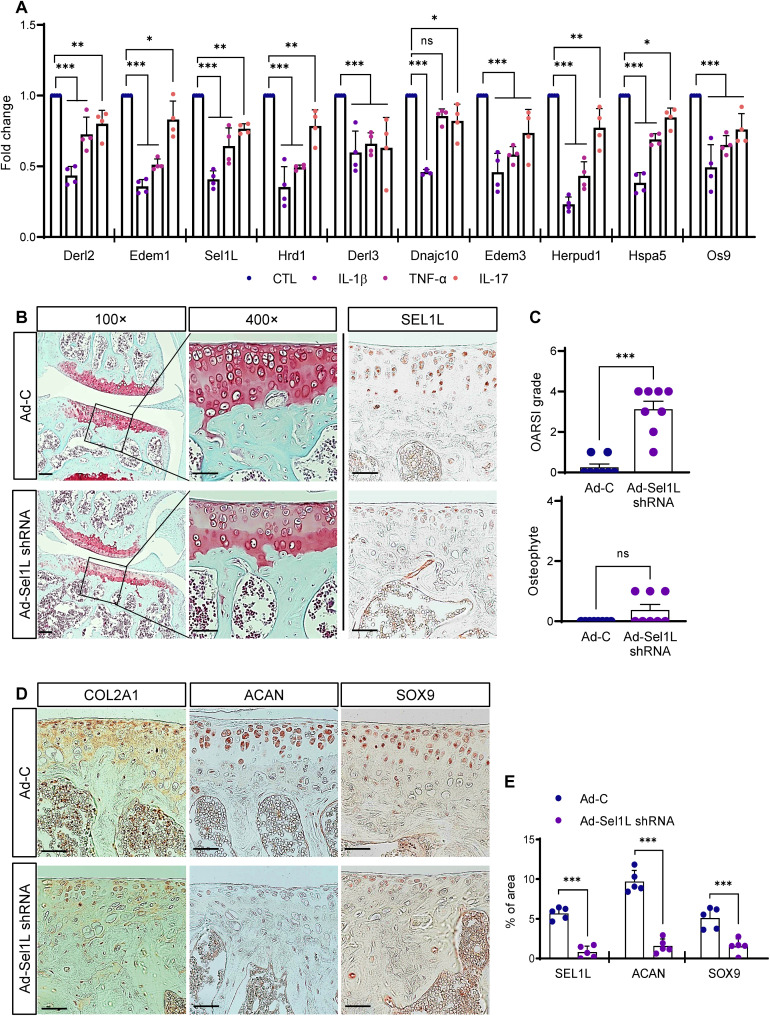
Inflammatory cytokines are key molecules in OA pathogenesis. Elevated cytokine levels increase the expression of matrix-degrading proteases [e.g., matrix metalloproteinase (MMP)] and promote cartilage destruction. Interleukin-1β (IL-1β), tumor necrosis factor–α (TNF-α), IL-6, IL-15, IL-17, and IL-18 are major proinflammatory cytokines and are thought to accelerate OA progression (24, 25). We hypothesized that proinflammatory cytokines may also reduce ERAD gene expression and promote OA development. To investigate this possibility, we treated mouse primary chondrocytes with several proinflammatory cytokines and analyzed changes in ERAD gene expression. Unexpectedly, treatment with all cytokines we examined (IL-1β, TNF-α, and IL-17) consistently reduced ERAD gene expression (Fig. 6A).
Fig. 6. Proinflammatory cytokines down-regulated ERAD gene expression in mouse primary chondrocytes, and impaired ERAD function in chondrocytes promoted OA-like cartilage damage.
(A) Mouse primary chondrocytes were treated with proinflammatory cytokines IL-1β, TNF-α, or IL-17, and ERAD gene expression was analyzed by qPCR. Proinflammatory cytokines significantly reduced ERAD gene expression (n = 4). (B) Sel1L expression in articular chondrocytes was depleted by IA injection of Ad-Sel1L-shRNA, and articular cartilage integrity was analyzed by Safranin-O staining. GAG levels were reduced in Sel1L-depleted cartilage. Scale bars, 100 μm (top), 50 μm (bottom). (C) Osteoarthritic cartilage destruction was measured by OARSI grade and osteophyte formation after IA injection of Ad-Sel1L-shRNA. n = 8. (D) Expression levels of cartilage anabolic factors Col2a1, Acan, and Sox9 in Sel1L-depleted mouse articular cartilages were visualized by immunohistochemical staining. Sel1L depletion decreased Col2a1, Acan, and Sox9 expression. Scale bars, 50 μm. (E) Measurement of SEL1L, ACAN and SOX9 IHC density in Sel1L-depleted mouse articular cartilages. n = 5. Statistical analysis was performed using the Student’s t test. ***(P < 0.001), **(0.001 < P < 0.01), *(0.01 < P < 0.05), ns (0.05 < P).
Last, we examined whether reduced ERAD gene expression may directly cause OA in vivo. To test this hypothesis, Sel1L expression in mouse articular cartilage was depleted by intra-articular (IA) injection of adenoviral Sel1L shRNA (Ad-Sel1L shRNA) into the mouse knee joint. IHC showed that this effectively depleted Sel1L expression in mouse cartilage (Fig. 6, B and E). Then, we analyzed cartilage matrix integrity by safranin O staining. Compared to the articular cartilage of adenovirus control (Ad-C)–infected mice, Ad-Sel1L shRNA–infected cartilage showed significant loss of cartilage matrix (Fig. 6, B and C). Consistent with the data obtained using micromasses and Xenopus model, Sel1L depletion in mouse articular cartilage reduced the expression of cartilage anabolic factors, such as Col2a1, Acan, and Sox9 (Fig. 6, D and E).
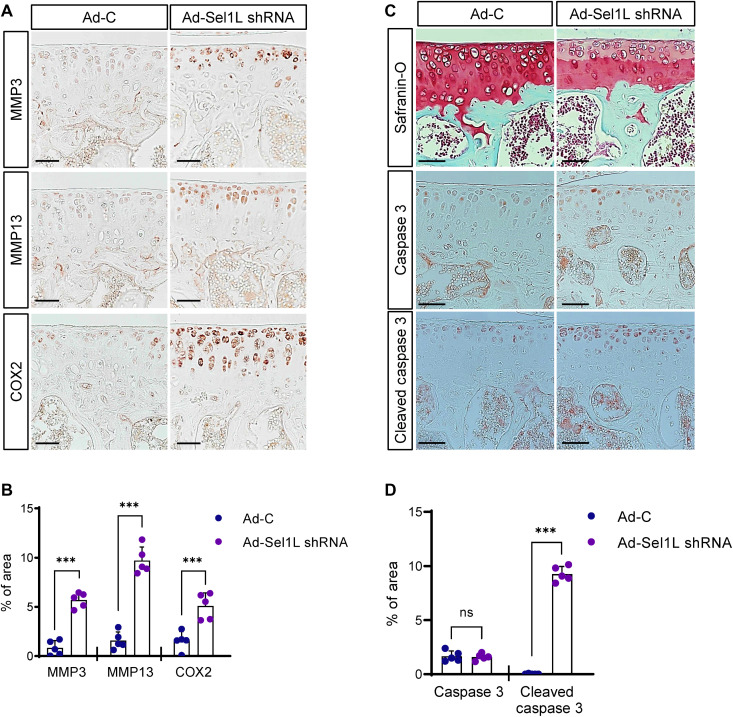
We further analyzed molecular markers known to cause cartilage destruction in Sel1L-depleted mouse articular cartilage. Immunohistochemical analysis using anti-MMP3/13 and anti–cyclooxygenase-2 (COX2) antibodies revealed strong up-regulation of cartilage catabolic factors in Sel1L-depleted cartilage (Fig. 7, A and B). Also, in vitro micromass culture studies revealed that ERAD complex malfunction induced by Eey1, siRNA, or Sel1Lc term led to increases in cartilage catabolic factors (MMP3 and MMP13), inflammatory cytokines (IL-1β and IL-6), and OA disease markers (ColX, Runx2, and Cox2) (fig. S7, C and D, and fig. S8). Furthermore, we observed increased caspase activities in Ad-Sel1L shRNA–infected cartilages (Fig. 7, C and D).
Fig. 7. Sel1L depletion in mouse articular cartilages increases the expression of cartilage catabolic factors.
(A) IHC of the cartilage catabolic factors MMP3, MMP13, and COX2 in mouse articular cartilage upon Sel1L depletion. Sel1L depletion increased MMP3, MMP13, and COX2 expression. Scale bars, 50 μm. (B) Increased MMP3, MMP13, and COX2 were quantified in Sel1L-depleted mouse articular cartilages. n = 5. (C) Sel1L expression in articular chondrocytes was depleted by IA injection of adenoviral Sel1L-shRNA (Ad-Sel1L-shRNA), and articular cartilage integrity was analyzed by Safranin-O staining and caspase 3 IHC. Scale bars, 50 μm. (D) The areas of IHC signals in (C) were quantified. n = 5. Statistical analysis was performed using the Student’s t test. ***(P < 0.001), **(0.001 < P < 0.01), *(0.01 < P < 0.05), ns (0.05 < P).
Synovial fibroblasts produce and secrete proteins into the synovial fluid for joint lubrication and impact on the joint integrity. Also, inflammation of the synovial membrane is observed in more than half of the patients with OA (26) and is known to contribute to cartilage destruction by secreting cartilage catabolic factors, such as MMP1/3/13 (27). Thus, we examined the possibility that reductions in ERAD function in synovial fibroblasts may also trigger cartilage destruction. However, although we observed mild thickening of synovial membranes in Ad-Sel1L shRNA–injected knee joints, we did not observe any noticeable synovitis (fig. S9, B and C).
Also, in contrast to chondrocytes, the presence of proinflammatory cytokines did not affect Sel1L gene expression in mouse primary synovial fibroblasts (fig. S9D), and Sel1L depletion did not increase MMP3/13 and Cox2 expression, which may directly cause cartilage destruction (fig. S9E). While we believe that ERAD function could be important for fibroblast-like synoviocytes (FLS) function, cartilage destruction is more directly affected by the malfunction of the ERAD complex in chondrocytes. These in vivo data from the mouse model imply that ERAD pathway malfunction is one of the major causes of OA.
DISCUSSION
Our results demonstrate the necessity of ERAD function for cartilage development and OA disease. ERAD genes were highly up-regulated during chondrogenesis, and inhibition of ERAD function by Eey1 treatment strongly reduced chondrogenic matrix deposition (Fig. 1, A to D). Also, depleting ERAD genes almost completely inhibited chondrogenesis in ATDC5 micromasses (Fig. 1, F and G). Furthermore, our data show that ERAD pathway malfunction induces abnormal accumulation of cartilage matrix proteins in the ER lumen (Fig. 2D).
The necessity of ERAD function during cartilage development was also confirmed using an in vivo Xenopus model. ERAD genes were expressed at the PA of developing embryos, which develop into the facial cartilage. Our data show that Sel1L depletion caused severe chondrodysplasia in craniofacial cartilage (Fig. 3B). Sel1L is one of the ERAD complex proteins that participate in the recognition and translocation of misfolded or unfolded proteins (28–30). Consistent with the in vitro micromass data, we observed abnormal Col2a1 accumulation in Sel1L-depleted cartilage in developing Xenopus embryos (Fig. 4, D to F). Collectively, these in vitro and in vivo data support the hypothesis that developing chondrocytes must maintain efficient and high-level ERAD complex activity for cartilage formation.
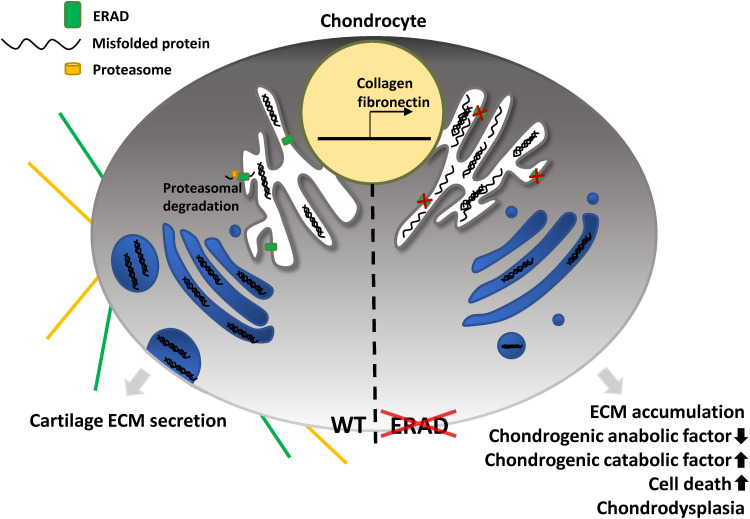
Chondrocytes secrete high amounts of ECM molecules to build and maintain cartilage tissues, and it is important to maintain the capacity of the secretory pathway. Therefore, ER performance must be deliberately monitored, and misfolded or unfolded proteins must be efficiently removed to prevent proteotoxicity. The ER performs quality control of proteins using the ERAD pathway or autophagy. The ERAD pathway is the major process that degrades unwanted proteins (3). For this reason, we believe that cartilaginous tissue is highly vulnerable to imbalanced proteostasis in the secretory pathway (Fig. 8). Our in vivo data in the Xenopus model support this idea, as facial cartilage malformation and chondrodysplasia were the most severe phenotypes caused by ERAD gene depletion.
Fig. 8. Graphical abstract.
The ERAD complex maintains proteostasis in the ER and facilitates chondrogenesis by constantly removing misfolded matrix proteins from the ER. Reduced ERAD functions result in misfolded protein accumulation and consequently cause cartilage loss in patients with OA.
A growing body of evidence suggests that elevated ER stress responses in chondrocytes cause apoptotic cell death and loss of cartilage matrix by down- and up-regulating cartilage anabolic and catabolic factors, respectively (31). The UPR pathway also seems to be critical for maintaining functionally intact articular cartilage (31). ERAD genes are major targets of UPR signaling, and the ERAD complex restores ER proteostasis by facilitating proteasome-dependent degradation of unfolded or misfolded proteins in the ER lumen (4). Although ERAD functions as downstream responses to ER stress or UPR are generally understood, the in vivo pathophysiological roles of ERAD in uninduced conditions (i.e., constitutive functions) are just beginning to be understood (8). Accumulating data suggest that ERAD complex functions in vivo are independent of the UPR or ER stress; rather, ERAD complex pathophysiology is largely dependent on the endogenous targets of specific tissues (8). For example, tissue-specific deletion of ERAD genes results in distinct phenotypes that differ from the loss-of-function phenotypes of UPR components (32). However, the in vivo roles of the ERAD complex in various tissues and organs are just beginning to be understood.
Regarding these current hypotheses for the tissue-specific function of the ERAD complex, our data strongly indicate that the ECM molecules of cartilage tissues, including Col2a1, could be the in vivo targets of the ERAD complex in developing chondrocytes. Compromising ERAD complex function, either by treating with the ERAD inhibitor Eey1 or by depleting any one of the ERAD genes, almost completely blocked normal chondrogenesis, indicating the indispensable function of the ERAD complex in cartilage tissue construction. Further investigation of the molecular bases of severe chondrodysplasia caused by malfunction of the ERAD complex revealed that the major ECM molecule of cartilage, Col2a1, was abnormally accumulated in the ER lumen. This was observed in both in vitro and in vivo models, indicating that the ERAD complex controls ER proteostasis in chondrocytes by monitoring maturation of cartilage-specific ECM molecules (Fig. 4D).
We also showed that ERAD function was consistently down-regulated in the damaged cartilage tissues of patients with OA (Fig. 5, C and D), and inhibition of ERAD function in mouse articular cartilage severely compromised cartilage matrix integrity (Fig. 6B). These data imply that ERAD function is critical for both the construction and maintenance of cartilage matrix. Furthermore, treatment with proinflammatory cytokines down-regulated most ERAD genes in mouse chondrocytes (Fig. 6A), suggesting that inflammatory environment in patients with OA may reprogram ERAD gene expression and promote the loss of cartilage matrix.
Although our data strongly suggest that augmented ERAD function is necessary to maintain the integrity of cartilage tissues, it remains unclear whether recovering ERAD functions is sufficient to ameliorate OA symptoms or to attenuate cartilage destruction. Unfortunately, because of the complexity of the ERAD pathway, it is challenging to enhance ERAD activity by treatments with either small molecules or overexpressing ERAD genes.
To further address this important issue, we believe that an in-depth study of the upstream regulation of ERAD genes in OA, such as in conditions of sustained inflammation or aging, must be performed. Also, developing therapies to enhance ERAD activity or gene expression would lead to previously unknown approaches for treating destructive cartilage disorders.
In most OA pathology research, ERAD function is considered a downstream target of ER stress or UPR signaling, but the constitutive functions of ERAD during chondrogenesis and OA development have not been systematically investigated. This study provides the first compelling evidence that ERAD function is critical not only for building cartilage tissue but also for maintaining healthy and functional cartilage matrix.
MATERIALS AND METHODS
Human OA cartilage
Human cartilage samples were obtained from individuals 45 to 65 years of age undergoing total knee arthroplasty. All patients provided written informed consent, and sample collection was approved by the Institutional Review Board of the Catholic University of Korea (UC14CNSI0150).
Plasmid construction
The Xenopus Sel1L gene was cloned into pCS108 with a 3 × HA (hemagglutinin) tag fused to the C terminus of Sel1L. The signal peptide of Xenopus Sel1L was cloned into pCS108, and the SEL1Lc term domain was fused to the C terminus of the signal peptide with the C terminus 3 × HA tag. Cloned plasmids underwent in vitro transcription using an SP6 transcription kit (AM1340, Invitrogen) for microinjection. The translation-blocking antisense morpholino was designed and synthesized by Gene Tools LLC (OR, USA).
The mouse Sel1LFull gene was cloned into pCS108 with a C terminus 3 × FLAG tag. The signal peptide of Xenopus Sel1L was cloned into pCS108, and the mouse SEL1Lc term was fused to the carboxyl terminus of the signal peptide with a C terminus 3 × FLAG and ER retention signal, KDEL. The mouse HRD1 gene was cloned into pCS108 with a C terminus 3 × HA tag.
Cell culture
For immunoprecipitation, human embryonic kidney (HEK) 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) with 10% fetal bovine serum (FBS) (Gibco) and 1% anti-antimycotic (Gibco). One day before transfection, cells were plated in a six-well plate at 2 × 105 and then transfected with plasmids using JetPRIME (Polyplus) according to the manufacturer’s manual.
For micromass culture, ATDC5 cells or human primary chondrocytes were cultured in DMEM/F12 (Gibco) with 5% FBS (Gibco) and 1% antimycotic (Gibco). One day before transfection, cells were plated in 60-mm cell culture dishes (Nunc) at 1 × 106 and then transfected with plasmids using JetPRIME (Polyplus) according to the manufacturer’s manual. One day before micromass culture, siRNAs were transfected if required.
For mouse articular chondrocyte primary cultures, articular chondrocytes were isolated from cartilage tissue obtained from mice on postnatal day 5. Cartilage tissue was subjected to consecutive enzymatic digestions with proteinase and collagenase, as previously described (33). Primary cultured mouse FLS were cultured from 8-week-old C57BL/6 mice as previously described (34). Briefly, mouse synovial tissues were dissected and digested with collagenase type I (1 mg/kg) in DMEM for 4 hours to isolate synovial cells. The synovial cells were cultured selectively from other types of cells by multiple passages and analyzed for CD90.2+ expression. More than 90% of CD90.2+ cells were used for related experiments.
Chondrocyte isolation and fluorescence-activated cell sorting analysis
To isolate chondrocyte in adult mouse cartilage, C57BL/6 mice (DBL Co. Ltd) were sacrificed, skin and muscle around the bones were discarded, and the femur and tibia were dissected. The knee joint area connecting the femur and tibia (the tip of each bone) was chopped and digested with 1% collagenase (C6885, Sigma-Aldrich) in DMEM for 3 hours at 37°C. The digested solution was filtered through a 40-μm cell strainer, and only cells that passed through were collected. The cells were washed with DMEM with 10% FBS and 1% penicillin-streptomycin. To elucidate chondrocyte proliferation in cartilage, flow cytometry was performed using fluorescein isothiocyanate (FITC)–conjugated anti–type II collagen (bs-10589R-FITC, Bioss) and Pacific blue–conjugated anti-Ki67 (652422, BioLegend) antibodies.
Micromass culture
Cells were transfected with siRNAs (when applicable) 1 day before initiation of micromass culture. Cultured ATDC5 chondrocytes were harvested using 0.25% trypsin-EDTA (Gibco). Droplets (20 μl) containing ATDC5 cell suspension (1× 107 cells/ml) were dropped into 48-well culture plates (Nunc). After cell attachment for 3 hours, 200 μl of α-MEM (Welgene) containing 100 nM dexamethasone (Sigma-Aldrich), 100 μM ascorbate-2-phosphate (Sigma-Aldrich), and human transforming growth factor–β1 (10 μg/ml; Gibco) was added. At 3, 5, 7, and 10 days of differentiation, total RNA was extracted or micromass samples were fixed using 4% paraformaldehyde (PFA).
Fixed micromasses were washed with phosphate-buffered saline (PBS). Micromasses were then incubated in 0.1% Alcian blue in 1 N HCl for a day. Stained micromasses were washed with distilled water several times. Micromasses were imaged with an SZX16 microscope (Olympus) and quantified by ImageJ software [National Institutes of Health (NIH)]. After imaging, micromasses were incubated with 200 μl of 6 M guanidine hydrochloride for a day. To detect GAG content in micromasses, 150 μl of the solution was transferred to 96-well plates, and optical density was detected with a SpectraMax microplate reader (Molecular Devices) at 660 nm. To normalize GAG content in micromasses, 10 μl of solution was mixed with 150 μl of protein assay reagent (22660, Thermo Fisher Scientific), and then optical density was detected with a SpectraMax microplate reader (Molecular Devices) at 660 nm.
For immunofluorescence analysis, droplets (20 μl) containing 1 × 107 cells/ml ATDC5 cell suspension were dropped on a coverslip. Micromass samples were fixed using 4% PFA for TUNEL staining. For Col2a1 immunofluorescence, micromass samples were fixed using Dent’s Fixative [80% methanol + 10% dimethyl sulfoxide (DMSO)].
Xenopus embryo manipulation
Adult Xenopus females were induced to ovulate by administration of human chorionic gonadotropin. One day later, eggs were collected by squeezing to perform in vitro fertilization using dissected testis from an adult male frog. Jelly layers were removed using 3% cysteine (pH 7.9) in 1/3× Marc’s modified Ringer’s (MMR) solution, and dejellied eggs were maintained in 1/3× MMR with 5% gentamycin. Antisense morpholinos or Sel1L mutant mRNAs were microinjected into two-cell stage embryos in 1/3× MMR with 0.5% Ficoll; after 4 hours, that solution was replaced with 1/3× MMR solution with 5% gentamycin, and cells were cultured until the indicated stages. All animal experiments were approved by the animal care and use committee of the Ulsan National Institute of Science and Technology (UNIST) and complied with all relevant ethical regulations (UNISTIACUC-19-22).
For Alcian blue staining, the embryos were fixed using MEMFA solution (MEM salt + 4% formaldehyde). Fixed embryos were washed using PBS + 0.1% Tween-20 (PTW) and acidic alcohol (80% ethanol + 20% acetic acid). Embryos were stained in Alcian blue solution (80% ethanol + 10 mM MgCl2 + 0.04% Alcian blue) for 2 days. Embryos were then washed in PTW, and pigment was removed using a bleach solution (1% hydrogen peroxide + 5% formamide + 0.5× saline-sodium citrate buffer). After bleaching, embryos were cleared using trypsin and imaged with an SZX16 microscope (Olympus). Small or abnormal head structures were counted as mild defects, and some part of the head structure missing or no craniofacial cartilages were considered severe defects.
For immunofluorescence analysis, embryos were fixed using Dent’s fixative solution (80% ethanol + 20% DMSO). Fixed embryos were rehydrated by serial dilution of methanol in PBS. Then, embryos were washed using PBS. Embryos were then embedded in 2% low-melt agarose (Biopure) in PBS. Embryos were sectioned using a vibratome (Leica).
Immunofluorescence analysis
Sectioned embryos were incubated in blocking solution [tris-buffered saline (TBS) + 0.1% Triton X-100 + 10% FBS + 2% DMSO] for 30 min. Immunostaining was performed using anti–green fluorescent protein (GFP, Abcam), anti-Col2a1 (DSHB), and TUNEL staining kits (Roche). Secondary antibodies were Alexa Fluor 488–, 555–, or 633–conjugated antibodies (Invitrogen), and all samples were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Molecular Probes). Samples were dehydrated using methanol and then cleared using BA:BB solution (1 volume of benzyl alcohol and 2 volumes of benzyl benzoate). Cleared embryos were mounted using 10% glycerol in PBS. Imaging was performed using an LSM780 confocal microscope (Zeiss). Images were quantified by ImageJ software (NIH). COL2A1 puncta size was quantified by measuring the size of each COL2A1 puncta larger than 10 pixels and normalized by total number of COL2A1 puncta. The number of COL2A1 puncta was quantified by counting all COL2A1 punctae larger than 10 pixels and normalized by cell number.
For micromass culture, fixed micromasses were washed and permeabilized using TBS + 0.1% Triton X-100. Then, micromasses were incubated in blocking solution (TBS + 0.1% Triton X-100 + 10% FBS + 2% DMSO) for 30 min. Immunostaining was performed using anti-calnexin (Abcam), anti-Col2a1 (DSHB), and a TUNEL staining kit (Roche). Secondary antibodies were Alexa Fluor 488–, 555–, or 633–conjugated antibodies (Invitrogen), and all samples were counterstained with DAPI (Molecular Probes). Stained micromasses were mounted using ProLong Gold antifade mounting solution (Invitrogen). Imaging was performed using an LSM780 confocal microscope (Zeiss). COL2A1 intensity was measured by ImageJ software (NIH) and normalized by micromass area.
Proinflammatory cytokine treatment, siRNA transfection, and adenovirus infection
Adenoviruses encoding mouse Sel1L shRNA (Ad-Sel1L shRNA) and control adenoviruses (Ad-C) were purchased from Vector Biolabs. Chondrocytes and FLS cells were cultured for 2 days, infected with adenovirus for 2 hours at the indicated multiplicity of infection (MOI), or treated with recombinant IL-1β, TNF-α, or IL-17 (all from GenScript) for 24 hours. For cartilage ex vivo culture, cartilage explants were obtained from mouse knee joints, cultured in DMEM (Gibco-BRL), and infected with Ad-Sel1L shRNA or Ad-C (800 MOI) for 36 hours. Sulfated proteoglycan accumulation was assessed by Alcian blue staining (1 volume of 0.3% Alcian blue 8GX in 70% ethanol, 1 volume of 100% acetic acid, and 18 volumes of 100% ethanol), as described previously (33).
In situ hybridization, RT-PCR, and qRT-PCR
For in situ hybridization, probes were synthesized using the indicated primer containing the SP6 promoter sequence (summarized in table S2). Whole-mount in situ hybridization was performed as previously described (35). To extract human chondrocyte RNA, finely chopped articular cartilage of human patient samples was directly reacted with TRIzol without additional digestion and vortexed.
Total RNA was isolated from articular chondrocytes or micromasses using TRIzol Reagent (Molecular Research Center). cDNA was obtained by reverse transcription using ImProm-II Reverse Transcriptase (Promega).
PCR primers and experimental conditions are summarized in table S3. PCR amplification was carried out using SYBR premix Ex Taq (TaKaRa Bio). For each target gene, transcript levels were normalized to glyceraldehyde-3-phosphate dehydrogenase and are expressed as fold change relative to the indicated controls, as previously described (33).
Immunoprecipitation samples were separated using 12% polyacrylamide gels and transferred to polyvinylfluoride (PVDF) membranes (Millipore). Membranes were blocked using blocking solution (TBS + 0.05% Tween-20 + 5% skim milk) for 30 min. Proteins were detected using anti-DDDDK (Abcam) and anti-HA (Santa Cruz Biotechnology). Visualization was performed using horseradish peroxidase (HRP)–conjugated antibodies (Invitrogen) with enhanced chemiluminescence (ECL) solution (Invitrogen). Images were captured using ChemiDoc MP (Bio-Rad).
Immunoprecipitation and Western blot analysis
Transfected HEK293T cells were homogenized using lysis buffer (TBS + 10% glycerol +1% Triton X-100) with protease inhibitors. Proteins were captured using anti-HA affinity gel (Biotool) at 4°C for 3 hours. Beads were washed several times using lysis buffer, and proteins were eluted using sodium dodecyl sulfate sample buffer.
Immunoprecipitation samples were separated using 12% polyacrylamide gels and transferred to the PVDF membranes (Millipore). Membranes were blocked using blocking solution (TBS + 0.05% Tween-20 + 5% skim milk) for 30 min. Proteins were detected using anti-DDDDK (Abcam) and anti-HA (Santa Cruz Biotechnology). Visualization was performed using HRP-conjugated antibodies (Invitrogen) with ECL solution (Invitrogen). Images were captured using ChemiDoc MP (Bio-Rad).
IA injection
Adenoviruses Ad-C (1060), control shRNA (1122N), and Ad-Sel1L shRNA (ADV-252021) were purchased from Vector Biolabs. C57BL/6 mice received twice weekly IA injections of adenovirus (1 × 109 plaque-forming units in a total volume of 10 μl) in the knee joint. Mice were sacrificed 3 weeks after the first adenoviral injection. Adenovirus-infected tissues were analyzed as described previously (36). All animal experiments were approved by the University of Ajou Animal Care and Use Committee and complied with all relevant ethical regulations.
Histology and immunohistochemical analyses
Human OA cartilage was sectioned (10 μm) and fixed in 4% PFA. Cartilage sections were stained with Alcian blue to detect sulfated proteoglycans. Mouse knee joints were excised, fixed in 4% PFA, decalcified for 2 weeks in 0.5 M EDTA, and embedded in paraffin. Paraffin blocks were serially sectioned (5 μm) at 40-μm intervals as previously described (36). Sel1L, MMP3, MMP13, and Cox2 were detected by immunohistochemical staining of human and mouse cartilage sections with the following antibodies: anti-MMP3 (ab52915, Abcam), anti-MMP13 (ab51072, Abcam), and anti-Sel1L. The signal of IHC was quantified using the ImageJ software v1.60.
OA, synovial inflammation, and osteophyte maturity scoring
To assess OA cartilage histopathology, stained images of each cartilage section were scored by four student observers under blinded condition according to the Osteoarthritis Research Society International (OARSI) scoring system (grade 0 = normal, 0.5 = loss if safranin-D without loss of cartilage, 1 = small fibrillations without loss of cartilage, 2 = vertical clefts down to the later immediately below the superficial layer and some of surface lamina, 3 = vertical clefts/erosion to the calcified cartilage extending to <25% of the articular surface, 4 = vertical clefts/erosion to the calcified cartilage extending to 25 to 50% of the articular surface, 5 = vertical clefts/erosion to the calcified cartilage extending to 50 to 75% of the articular surface, and 6 = vertical clefts/erosion to the calcified cartilage extending to >75% of the articular surface) (33, 37, 38).
Synovial histological changes were evaluated on a scale from 0 to 3 depending on infiltration of inflammatory cells into the synovial membrane (0 = no inflammation; 1 = mild inflammation; 2 = moderate inflammation; and 3 = severe inflammation) as described previously (37, 39). Osteophyte maturity was scored on images of the identical part of the anterior-medial tibia in each cartilage (0 = none, 1 = predominantly cartilaginous, 2 = mixed cartilage and bone with active vascular invasion and endochondral ossification, and 3 = predominantly bone) (40).
Statistical analysis
All experiments were performed independently at least three times. Statistical comparisons of two independent groups were performed using the Shapiro-Wilk test for normality, Levene’s test for homogeneity of variance, and a two-tailed independent t test. Multiple comparisons were conducted using the Shapiro-Wilk test, Levene’s test, and one-way analysis of variance (ANOVA) with a post hoc Bonferroni test. Data based on ordinal grading systems were analyzed using a nonparametric Mann-Whitney U test. A P value <0.05 was considered significant.
Ethics approval
The Institutional Review Board of the Catholic University of Korea (UC14CNSI0150) approved the use of human material. All animal experiments were approved by the University of Ajou Animal Care and Use Committee and UNIST Institutional Animal Care and Use Committee (UNISTIACUC-19-22).
Acknowledgments
We thank M. Ko and S. H. Park for critical discussions.
Funding: This work was supported by Korea National Research Foundation (2021R1A2B5B02002285, 2019M3E5D5067273, 2019R1I1A1A01060664, 2020R1A6A3A0109857311, SRC-2017R1A5A1014560, and NRF- 2021M3E5E7023855). This work was also supported by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (HI16C0992). This work was partially supported by the Institute for Basic Science (IBS-R022-D1).
Author contributions: H.J.S. and C.C. conducted most of the in vitro and in vivo experiments and analyzed the data. H.E.K. and J.Y.H. conducted Xenopus in situ hybridization analysis. E.K.S. and D.G.J. constructed Sel1L plasmids and synthesized antisense probes and mRNAs. K.Y.K. performed microinjection. S.-J.K. obtained and analyzed the data from the human OA patients. C.L., H.-S.L, T.K., S.Y., and T.J.P. conceived the study, designed and conducted the research, analyzed the data, wrote the manuscript, and directed the project.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1 to S6
REFERENCES AND NOTES
- 1.Walter P., Ron D., The unfolded protein response: From stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Chiti F., Dobson C. M., Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Guerriero C. J., Brodsky J. L., The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol. Rev. 92, 537–576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hwang J., Qi L., Quality control in the endoplasmic reticulum: Crosstalk between ERAD and UPR pathways. Trends Biochem. Sci. 43, 593–605 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christianson J. C., Ye Y., Cleaning up in the endoplasmic reticulum: Ubiquitin in charge. Nat. Struct. Mol. Biol. 21, 325–335 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olzmann J. A., Kopito R. R., Christianson J. C., The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb. Perspect. Biol. 5, a013185 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu C., Ng D. T. W., Glycosylation-directed quality control of protein folding. Nat. Rev. Mol. Cell Biol. 16, 742–752 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Qi L., Tsai B., Arvan P., New insights into the physiological role of endoplasmic reticulum-associated degradation. Trends Cell Biol. 27, 430–440 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiani C., Chen L., Wu Y. J., Yee A. J., Yang B. B., Structure and function of aggrecan. Cell Res. 12, 19–32 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Kvist A. J., Nyström A., Hultenby K., Sasaki T., Talts J. F., Aspberg A., The major basement membrane components localize to the chondrocyte pericellular matrix—A cartilage basement membrane equivalent? Matrix Biol. 27, 22–33 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Bateman J. F., Boot-Handford R. P., Lamandé S. R., Genetic diseases of connective tissues: Cellular and extracellular effects of ECM mutations. Nat. Rev. Genet. 10, 173–183 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Schwartz N. B., Domowicz M., Chondrodysplasias due to proteoglycan defects. Glycobiology 12, 57R–68R (2002). [DOI] [PubMed] [Google Scholar]
- 13.He Y., Yao W., Zhang M., Zhang Y., Zhang D., Jiang Z., Ma T., Sun J., Shao M., Chen J., Changes in osteogenic gene expression in hypertrophic chondrocytes induced by SIN-1. Exp. Ther. Med. 16, 609–618 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiebiger E., Hirsch C., Vyas J. M., Gordon E., Ploegh H. L., Tortorella D., Dissection of the dislocation pathway for type I membrane proteins with a new small molecule inhibitor, eeyarestatin. Mol. Biol. Cell 15, 1635–1646 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Healy C., Uwanogho D., Sharpe P. T., Regulation and role of Sox9 in cartilage formation. Dev. Dyn. 215, 69–78 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Akiyama H., Control of chondrogenesis by the transcription factor Sox9. Mod. Rheumatol. 18, 213–219 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Zhao Q., Eberspaecher H., Lefebvre V., de Crombrugghe B., Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev. Dyn. 209, 377–386 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Byun H., Gou Y., Zook A., Lozano M. M., Dudley J. P., ERAD and how viruses exploit it. Front. Microbiol. 5, 330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delom F., Emadali A., Cocolakis E., Lebrun J. J., Nantel A., Chevet E., Calnexin-dependent regulation of tunicamycin-induced apoptosis in breast carcinoma MCF-7 cells. Cell Death Differ. 14, 586–596 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Klein S. L., Strausberg R. L., Wagner L., Pontius J., Clifton S. W., Richardson P., Genetic and genomic tools for Xenopus research: The NIH Xenopus initiative. Dev. Dyn. 225, 384–391 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Sargent T. D., Jamrich M., Dawid I. B., Cell interactions and the control of gene activity during early development of Xenopus laevis. Dev. Biol. 114, 238–246 (1986). [DOI] [PubMed] [Google Scholar]
- 22.Song E. K., Jeon J., Jang D. G., Kim H. E., Sim H. J., Kwon K. Y., Medina-Ruiz S., Jang H. J., Lee A. R., Rho J. G., Lee H. S., Kim S. J., Park C. Y., Myung K., Kim W., Kwon T., Yang S., Park T. J., ITGBL1 modulates integrin activity to promote cartilage formation and protect against arthritis. Sci. Transl. Med. 10, eaam7486 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Jeong H., Sim H. J., Song E. K., Lee H., Ha S. C., Jun Y., Park T. J., Lee C., Crystal structure of SEL1L: Insight into the roles of SLR motifs in ERAD pathway. Sci. Rep. 6, 20261 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldring M. B., Osteoarthritis and cartilage: The role of cytokines. Curr. Rheumatol. Rep. 2, 459–465 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Kapoor M., Martel-Pelletier J., Lajeunesse D., Pelletier J. P., Fahmi H., Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 7, 33–42 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Scanzello C. R., Goldring S. R., The role of synovitis in osteoarthritis pathogenesis. Bone 51, 249–257 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchs S., Skwara A., Bloch M., Dankbar B., Differential induction and regulation of matrix metalloproteinases in osteoarthritic tissue and fluid synovial fibroblasts. Osteoarthr. Cartil. 12, 409–418 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Bernasconi R., Galli C., Calanca V., Nakajima T., Molinari M., Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J. Cell Biol. 188, 223–235 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R., OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1–SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 10, 272–282 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mueller B., Lilley B. N., Ploegh H. L., SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J. Cell Biol. 175, 261–270 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes A., Oxford A. E., Tawara K., Jorcyk C. L., Oxford J. T., Endoplasmic reticulum stress and unfolded protein response in cartilage pathophysiology; contributing factors to apoptosis and osteoarthritis. Int. J. Mol. Sci. 18, 665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhattacharya A., Qi L., ER-associated degradation in health and disease—From substrate to organism. J. Cell Sci. 132, jcs232850 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J. H., Jeon J., Shin M., Won Y., Lee M., Kwak J. S., Lee G., Rhee J., Ryu J. H., Chun C. H., Chun J. S., Regulation of the catabolic cascade in osteoarthritis by the zinc-ZIP8-MTF1 axis. Cell 156, 730–743 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Kang L.-J., Kwon E.-S., Lee K. M., Cho C., Lee J.-I., Ryu Y. B., Youm T. H., Jeon J., Cho M. R., Jeong S.-Y., Lee S.-R., Kim W., Yang S., 3′-Sialyllactose as an inhibitor of p65 phosphorylation ameliorates the progression of experimental rheumatoid arthritis. Br. J. Pharmacol. 175, 4295–4309 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harland R. M., Appendix G: In situ hybridization: An improved whole-mount method for Xenopus embryos. Methods Cell Biol. 36, 685–695 (1991). [DOI] [PubMed] [Google Scholar]
- 36.Bonnans C., Chou J., Werb Z., Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 15, 786–801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang S., Kim J., Ryu J. H., Oh H., Chun C. H., Kim B. J., Min B. H., Chun J. S., Hypoxia-inducible factor-2alpha is a catabolic regulator of osteoarthritic cartilage destruction. Nat. Med. 16, 687–693 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Yang S., Ryu J. H., Oh H., Jeon J., Kwak J. S., Kim J. H., Kim H. A., Chun C. H., Chun J. S., NAMPT (visfatin), a direct target of hypoxia-inducible factor-2α, is an essential catabolic regulator of osteoarthritis. Ann. Rheum. Dis. 74, 595–602 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Midwood K., Sacre S., Piccinini A. M., Inglis J., Trebaul A., Chan E., Drexler S., Sofat N., Kashiwagi M., Orend G., Brennan F., Foxwell B., Tenascin-C is an endogenous activator of Toll-like receptor 4 that is essential for maintaining inflammation in arthritic joint disease. Nat. Med. 15, 774–780 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Little C. B., Barai A., Burkhardt D., Smith S. M., Fosang A. J., Werb Z., Shah M., Thompson E. W., Matrix metalloproteinase 13-deficient mice are resistant to osteoarthritic cartilage erosion but not chondrocyte hypertrophy or osteophyte development. Arthritis Rheum. 60, 3723–3733 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 to S6