Abstract
Objectives
To develop a simple DNA sequencing test for simultaneous identification and antimicrobial resistance (AMR) detection of multiple sexually transmitted infections (STIs).
Methods
Real-time PCR (qPCR) was initially performed to identify Neisseria gonorrhoeae (NG), Chlamydia trachomatis (CT), Mycoplasma genitalium (MG) and Trichomonas vaginalis (TV) infections among a total of 200 vulvo-vaginal swab samples from female sex workers in Ecuador. qPCR positive samples plus qPCR negative controls for these STIs were subjected to single gene targeted PCR MinION-nanopore sequencing using the smartphone operated MinIT.
Results
Among 200 vulvo-vaginal swab samples 43 were qPCR positive for at least one of the STIs. Single gene targeted nanopore sequencing generally yielded higher pathogen specific read counts in qPCR positive samples than qPCR negative controls. Of the 26 CT, NG or MG infections identified by qPCR, 25 were clearly distinguishable from qPCR negative controls by read count. Discrimination of TV qPCR positives from qPCR negative controls was poorer as many had low pathogen loads (qPCR cycle threshold >35) which produced few specific reads. Real-time AMR profiling revealed that 3/3 NG samples identified had gyrA mutations associated with fluoroquinolone resistance, 2/10 of TV had mutations related to metronidazole resistance, while none of the MG samples possessed 23S rRNA gene mutations contributing to macrolide resistance.
Conclusions
Single gene targeted nanopore sequencing for diagnosing and simultaneously identifying key antimicrobial resistance markers for four common genital STIs shows promise. Further work to optimise accuracy, reduce costs and improve speed may allow sustainable approaches for managing STIs and emerging AMR in resource poor and laboratory limited settings.
Introduction
Sexually transmitted infections (STIs) remain a major public health problem worldwide, with an estimated 357 million new cases of Chlamydia trachomatis (CT), Neisseria gonorrhoeae (NG), Treponema pallidum (TP) and Trichomonas vaginalis (TV) per year [1]. High rates of the STI, Mycoplasma genitalium (MG), have also been reported worldwide [2] and is associated with genital discharge syndrome in men and reproductive sequelae in women [3]. All these STIs are generally curable with existing, effective single-dose antibiotic regimens but, if left undiagnosed and/or untreated, can result in serious long-term reproductive health sequelae, particularly for women. Antimicrobial resistance (AMR) among some STIs to multiple classes of antibiotics has spread rapidly in recent years. For NG, loss of extended spectrum cephalosporins as first line empirical treatment is a major concern [4] as circulating multi- and extensive-drug resistance clones have been detected internationally [5]. For MG, macrolide resistance is now widely but not universally reported with increasing rates of resistance to fluoroquinolones also detected [6]. These developments have made treatment, and particularly empirical treatment challenging.
New World Health Organization (WHO) guidelines reinforce the need to treat these STIs with the right antibiotic, at the right dose, and the right time to reduce spread and improve sexual and reproductive health. Laboratory based nucleic-acid amplification tests (NAAT) for detection of NG, CT, TV and MG are well established using various gene targets, for example, GenoQuick® CT (Hain Lifescience, Germany) which specifically and simultaneously detects both the gene encoding outer membrane protein 1 (omp-1) and the cryptic plasmid. For both NG and MG, accurate and rapid diagnostics which also predict antibiotic susceptibility are likely to be needed to achieve this. Laboratory based NAAT for detection of NG, CT, TV and MG are the current gold standard for detection, providing high sensitivity and specificity. NAAT are widely used in high-income countries but are often unavailable in resource-poor settings. Culture-based antimicrobial susceptibility testing (AST) for NG remains gold standard for phenotypically predicting AMR and commonly takes usually two to five days to obtain a result in UK microbiology laboratories (personal communication, Sadiq). This in practical terms may be too late to initiate targeted antibiotic therapy, particularly for hard to reach vulnerable populations. MG is difficult to cultivate and requires cell culture, not usually feasible in most clinical settings.
Advances in understanding AMR in NG, MG and TV have allowed for development of NAAT- based AMR detection. For example, absence of gyrA mutations at amino acid positions S91 and D95 of NG accurately predicts fluoroquinolone susceptibility [7, 8], presence of mutations at positions 2058 and 2059 (Escherichia coli numbering) in region V of the 23S rRNA gene in MG is associated with failure of treatment with azithromycin [9–11], and a single nucleotide polymorphism (SNP) in ntr6 gene (ntr6 A238T) of TV is associated with metronidazole resistance, which may have diagnostic value for metronidazole resistance [12]. Use of NAAT-based AMR tests, however, may have limitations due to continually changing mutations and novel mechanisms of resistance evolving under ongoing treatment selection pressures. Whole genome sequencing (WGS) using high throughput sequencing platforms enables detection of multiple genes/pathogens in a single run, which may address this challenge to some degree and also give added value in identifying phylogenetic relationships in identified infections [13, 14]. However, for diagnostic purposes WGS itself can be constrained by sample preparation, cost etc., making it unsuitable for near patient applications.
Oxford Nanopore Technologies’ (ONT) portable MinION DNA sequencer, together with its recent MinIT hand-held processor, may offer advantages as an accurate diagnostic in resource-limited settings. Herein we report early phase study among female sex workers (FSWs) in Ecuador, of using the MinION, controlled by a smartphone-operated MinIT to detect the four common STIs (NG, MG, TV and CT) and profile AMR to ciprofloxacin in NG, azithromycin in MG and metronidazole in TV utilising a single gene targeted approach.
Materials and methods
Ethics statement
Written and verbal informed consent was obtained from FSW participants all of whom were 18 years or older and none of whom received any compensation for their participation. The study was conducted according to the Declaration of Helsinki and approved by the Ethical Committee of Universidad Internacional del Ecuador (02-02-17).
Clinical sample collection and DNA preparation
A cross-sectional study of STIs was conducted among FSWs in a primary health centre in Quito, Ecuador, during the last quarter of 2017 [15]. Vulvo-vaginal swab samples were collected by clinicians using Xpert® CT/NG Patient-Collected Vaginal Swab Specimen Collection Kit (Cepheid). DNA from swab samples was prepared using PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions and quantified using a NanoDrop Spectrometer. STI identification of swab samples was performed by singleplex qPCR for individual STI pathogen. The DNA samples were stored at -20 °C till the present study.
STI identification of swab samples by qPCR
Swab samples were initially screened by singleplex qPCR to identify NG, TV, MG and CT infections, with well-studied gene targets and primers, as shown in Table 1. qPCR was performed using Applied Biosystems 7500 Fast Real-Time PCR System in a volume of 10 μl containing 5 μl of TaqMan™ Fast Universal PCR Master Mix, 1 μl of 10x Exogenous Internal Positive Control (IPC) Mix, 0.2 μl of 50x IPC DNA, 250 nM each of the primers, 100 nM each of the probes and 50 ng of template DNA. Cycling parameters were 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 1 min. Positive STI samples and some negatives identified by qPCR were used for this study.
Table 1. Gene targets, primers and probes used in this study.
| a) Initial real-time PCR (qPCR) for STI testing on samples |
| Trichomonas vaginalis: T. vaginalis specific repeat DNA fragment (amplicon 92 bp) [16] |
| TV_F: AAAGATGGGTGTTTTAAGCTAGATAAGG |
| TV_R: TCTGTGCCGTCTTCAAGTATGC |
| TV_P: [6FAM]AGTTCATGTCCTCTCCAAGCGTAAGT[BHQ1] |
| Mycoplasma genitalium: MgPa gene (amplicon 78 bp) [17] |
| MG_F: GAGAAATACCTTGATGGTCAGCAA |
| MG_R: GTTAATATCATATAAAGCTCTACCGTTGTTATC |
| MG_P: [6FAM]ACTTTGCAATCAGAAGGT[MGBNFQ] |
| Chlamydia trachomatis: The cryptic plasmid (amplicon 71 bp) [18] |
| CT_F: CATGAAAACTCGTTCCGAAATAGAA |
| CT_R: TCAGAGCTTTACCTAACAACGCATA |
| CT_P: [6FAM]TCGCATGCAAGATATCGA[MGBNFQ] |
| Neisseria gonorrhoeae: opa (amplicon 90 bp) [19] |
| NG_F: TTGAAACACCGCCCGGAA |
| NG_R: TTTCGGCTCCTTATTCGGTTTGA |
| NG_P: [6FAM]CCGATATAATCCGCCCTTCAACATCAG[BHQ1] |
| b) Targeted PCR (tPCR) for nanopore sequencing * |
| Trichomonas vaginalis: Nitroreductase family protein (ntr6; amplicon 746 bp) |
| TV_NTR6_BC_F: |
| 5’TTTCTGTTGGTGCTGATATTGCCTTCATTGAATTTATTCGTTCAAAATT |
| TV_NTR6_BC_R: |
| 5’ACTTGCCTGTCGCTCTATCTTCTTATTCAATGTATGTAACCTTTCTAA |
| Mycoplasma genitalium: 23S ribosomal RNA (amplicon 688 bp) |
| Mg 23S_BC_1992F: |
| 5’ TTTCTGTTGGTGCTGATATTGCCCATCTCTTGACTGTCTCGG |
| Mg 23S_BC_2679R: |
| 5’ ACTTGCCTGTCGCTCTATCTTCTCCTCTCGTACTAGAAGCAAAG |
| Chlamydia trachomatis: Major outer membrane protein 1 (omp1; amplicon 1060 bp) |
| CT_OMP1_BC_F: |
| 5’ TTTCTGTTGGTGCTGATATTGCTTTGCCGCTTTGAGTTCTGCT |
| CT_OMP1_BC_R: |
| 5’ ACTTGCCTGTCGCTCTATCTTCCAATACCGCAAGATTTTCTAGATTTC |
| Neisseria gonorrhoeae: gyrA (amplicon 1181 bp) |
| NG_gyrA_BC_F: |
| 5’ TTTCTGTTGGTGCTGATATTGCATCCGCCACGACCACAAATT |
| NG_gyrA_BC_R: |
| 5’ ACTTGCCTGTCGCTCTATCTTCATATTGGACAGTGCGACGGC |
*developed in this study.
Targeted PCR (tPCR) amplification and barcoding
Both qPCR positive and negative samples for NG, MG, TV and CT plus an NG positive control were amplified by tPCR, targeting genes gyrA (NG), 23S rRNA (MG), ntr6 (TV) and omp1 (CT) with the primers listed in Table 1. tPCR reactions for each target were performed in a 50 μl volume consisting of 25 μl LongAmp® Taq 2X Master Mix (NEB), 200 nM each of tPCR primers and 100 ng DNA template as follows: 95 °C for 3 min followed by 35 cycles of 95 °C for 30 sec, 58 °C for 30 sec, 65 °C for 1 min 30 sec, and one cycle of 65 °C for 5 min. tPCR amplicons were purified and subjected to a second PCR for barcoding using PCR Barcoding Expansion 1–96 (EXP-PBC096, ONT) according to the manufacturer’s instructions. In this study, we used a sequence-based method for pathogen detection by mapping a ‘unique’ gene sequence of pathogens of interest to a reference database, and therefore specificity of detection lies within the sequence itself. This is different from conventional nucleic acid amplification methods, in which primer specificity is crucial to detection accuracy. The AMR-associated gene targets we used have a unique nucleotide sequence specific to pathogen of interest and can differentiate itself from other species. As a general practice we have searched nucleotide databases by BLAST when designing tPCR primers in order to discriminate as much as possible the target from other closely related bacterial species. Furthermore, to enhance the read counts of pathogen of interest we selectively designed the tPCR primers with some mismatch base(s) to other closely related bacterial species at the 3’ end for preferential target amplification.
Nanopore sequencing and data analysis
A DNA sequencing library was constructed from a pool of barcoded tPCR amplicons of different clinical samples using Ligation Sequencing Kit (SQK-LSK108) and sequenced using FLO-MIN106 R9 Version Flow Cell MK I Spot-ON on portable DNA sequencer MinION MK I which was controlled by a smartphone operated MinIT (ONT) according to the manufacturer’s instructions. Sequencing data were uploaded onto Metrichor Epi2ME (ONT) and analyzed using the workflow Fastq Antimicrobial Resistance r3.3.2 which included three components: QC and Barcoding [rev. 3.10.4], WIMP [rev. 3.4.0] and ARMA CARD [rev. 1.1.6]. QC and Barcoding component contains quality score (qscore) and barcode filter, cutting off reads with a qscore below the threshold (min qscore:7) which were uploaded for analysis, and demultiplexing barcodes if the sequenced samples were barcoded. WIMP (what’s In My Pot) allows for species identification by classifying read sequences against the standard Centrifuge database (including RefSeq complete genomes for bacteria). ARMA (antimicrobial resistance mapping application) performs antimicrobial resistance identification by aligning input reads with minmap2 against all reference sequences available in the CARD database (CARD version 1.1.3). TV, an anaerobic, flagellated protozoan parasite, was not included in RefSeq for bacteria and therefore a TV G3 nitroreductase family protein (TVAG_354010, ntr6) reference was uploaded using Fasta Reference Upload r3.2.2 and TV identification was performed using Fastq Custom Alignment r3.2.2 against the ntr6 reference sequence. Read counts were log-transformed and compared between qPCR positive and negative samples by t-test.
BLAST search
DNA sequences of NG, MG and TV reads were extracted from read FASTQ files and BLAST-aligned against the reference sequences: Neisseria gonorrhoeae FA 1090 gyrA reference (NC_002946.2:c621189-618439), Mycoplasma genitalium strain G-37 23S rRNA gene (NR_077054.1) and Trichomonas vaginalis G3 nitroreductase family protein reference (TVAG_354010) respectively. Part of the aligned sequences flanking AMR-associated mutations were used to construct a consensus sequence using MultAlin [20]. AMR-associated mutations were confirmed by manually comparing the consensus sequence against the reference.
Results
STI identification of swab samples by qPCR
A total of 200 vulvo-vaginal swab samples from FSWs were initially screened by qPCR, and 43 were found positive, including 37 single infections (11 MG, 19 TV and 7 CT) and six co-infections (two CT/TV and one each of CT/NG, NG/TV, CT/NG/TV and CT/TV/MG), as shown in Table 2.
Table 2. STI identification of swab samples by qPCR and nanopore sequencing.
| Sample ID | NG | CT | TV | MG | Ct value | Read counts | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Analysed | NG | CT | TV | MG | ||||||
| 60 | - | - | - | + | 32.505 | 784 | 1 | 0 | 1 | 91 |
| 166 | - | - | - | + | 30.436 | 25188 | 2 | 0 | 9 | 19968 |
| 85 | - | - | - | + | 35.403 | 1931 | 1 | 3 | 4 | 477 |
| 31 | - | - | - | + | 34.976 | 7988 | 0 | 2 | 4 | 1416 |
| 230 | - | - | - | + | 28.800 | 23263 | 0 | 2 | 5 | 18676 |
| 103 | - | - | - | + | 33.375 | 18383 | 3 | 0 | 5 | 7262 |
| 13 | - | - | - | + | 32.980 | 26039 | 0 | 0 | 9 | 20902 |
| 109 | - | - | - | + | 29.870 | 39248 | 2 | 0 | 7 | 39248 |
| 239 | - | - | - | + | 36.210 | 3133 | 1 | 0 | 5 | 250 |
| 69 | - | - | - | + | 35.810 | 3412 | 0 | 0 | 9 | 737 |
| 263 | - | - | - | + | 29.592 | 2446 | 1 | 0 | 2 | 1457 |
| 260a | - | + | 36.672 | 633 | 1 | 1 | 4 | 1 | ||
| 51 | - | - | - | - | 2952 | 2 | 1 | 1 | 4 | |
| 75 | - | - | - | - | 589 | 0 | 2 | 4 | 2 | |
| 77 | - | - | - | - | 252 | 2 | 0 | 0 | 1 | |
| 24 | - | - | + | - | 20.607 | 20815 | 3 | 0 | 10368 | 2 |
| 58 | - | - | + | - | 25.250 | 23714 | 3 | 0 | 17712 | 2 |
| 235 | - | - | + | - | 36.170 | 29942 | 1 | 3 | 5 | 4 |
| 256 | - | - | + | - | 39.995 | 25061 | 2 | 1 | 8 | 2 |
| 146 | - | - | + | - | 39.695 | 68032 | 3 | 0 | 8 | 4 |
| 144 | - | - | + | - | 24.409 | 22359 | 1 | 2 | 6026 | 0 |
| 145 | - | - | + | - | 38.037 | 32349 | 2 | 0 | 10 | 4 |
| 261 | - | - | + | - | 38.596 | 37283 | 2 | 0 | 8 | 6 |
| 90 | - | - | + | - | 23.581 | 17230 | 2 | 1 | 9824 | 4 |
| 106 | - | - | + | - | 39.839 | 18291 | 0 | 2 | 8 | 2 |
| 92 | - | - | + | - | 40.00 | 19009 | 1 | 0 | 1 | 0 |
| 72 | - | - | + | - | 36.897 | 20163 | 2 | 0 | 11 | 2 |
| 140 | - | - | + | - | 35.249 | 22397 | 1 | 0 | 6 | 6 |
| 253 | - | - | + | - | 37.778 | 30217 | 1 | 0 | 5 | 5 |
| 41 | - | - | + | - | 38.867 | 10297 | 2 | 2 | 7 | 11 |
| 124 | - | - | + | - | 27.540 | 23714 | 2 | 1 | 17882 | 7 |
| 71 | - | - | + | - | 27.565 | 4949 | 2 | 1 | 2101 | 5 |
| 258 | - | - | + | - | 38.636 | 14925 | 10 | 17 | 5 | |
| 105 | - | - | + | - | 30.327 | 9029 | 4 | 2 | 1937 | 8 |
| 221b | - | + | - | 38.171 | 2513 | 1 | 1 | 6 | 2 | |
| 89b | - | + | - | 35.079 | 30401 | 2 | 0 | 7 | 5 | |
| 91c | - | + | - | 21.186 | 29511 | 1 | 1 | 16907 | 2 | |
| 39d | + | - | 21.381 | 41735 | 1 | 0 | 41486 | 0 | ||
| 260a | - | + | 27.556 | 35209 | 0 | 1 | 15706 | 0 | ||
| 79 | - | - | - | - | 28262 | 1 | 0 | 12 | 1 | |
| 84 | - | - | - | - | 6617 | 3 | 0 | 8 | 4 | |
| 98 | - | - | - | - | 21575 | 3 | 0 | 7 | 4 | |
| 260a | - | + | 35.905 | 17328 | 5 | 82 | 3 | 6 | ||
| 94 | - | + | - | - | 29.812 | 12745 | 3 | 530 | 4 | 3 |
| 259 | - | + | - | - | 30.331 | 8309 | 2 | 672 | 8 | 6 |
| 242 | - | + | - | - | 31.946 | 8443 | 1 | 1840 | 10 | 3 |
| 236 | - | + | - | - | 35.936 | 10335 | 3 | 65 | 7 | 8 |
| 157 | - | + | - | - | 30.126 | 10277 | 1 | 25 | 9 | 4 |
| 114 | - | + | - | - | 30.464 | 5086 | 1 | 2317 | 6 | 1 |
| 6 | - | + | - | - | 31.232 | 7582 | 1 | 127 | 18 | 24 |
| 221b | - | + | - | 35.968 | 2978 | 2 | 72 | 4 | 3 | |
| 89b | - | + | - | 29.932 | 6565 | 0 | 220 | 10 | 3 | |
| 30e | + | - | - | 30.886 | 14878 | 2 | 13796 | 3 | 1 | |
| 39d | + | - | 30.536 | 999 | 1 | 797 | 3 | 1 | ||
| 102 | - | - | - | - | 3095 | 0 | 7 | 5 | 1 | |
| 107 | - | - | - | - | 4664 | 2 | 1 | 7 | 2 | |
| 125 | - | - | - | - | 101 | 1 | 0 | 9 | 2 | |
| 30e | + | - | - | 19.033 | 27210 | 21087 | 0 | 1 | 0 | |
| 91c | + | - | - | 20.035 | 39223 | 36590 | 1 | 5 | 0 | |
| 39d | + | - | 21.504 | 28317 | 12325 | 3 | 3 | 0 | ||
| 126 | - | - | - | - | 14121 | 2 | 0 | 2 | 1 | |
| 189 | - | - | - | - | 9168 | 3 | 1 | 1 | 5 | |
| 193 | - | - | - | - | 11334 | 1 | 0 | 3 | 2 | |
| NG positive control | 7937 | 7659 | 0 | 1 | 0 | |||||
a CT, TV and MG mixed infection.
b CT and TV mixed infection.
c NG and TV mixed infection.
d NG, CT and TV mixed infection.
e Ng and CT mixed infection.
STI identification and AMR detection by single gene tPCR nanopore sequencing
All 43 qPCR positives plus 12 negatives (3 for each pathogen) and an NG positive control were subjected to single gene tPCR nanopore sequencing in one barcoded sequencing library. A sequencing run of 10 hours produced 1,499,872 reads of which 1,127,703 (76%) reads passed QC, with total yield of 1.2 gigabases, average qscore 8.34 (S1 Fig) and average sequence length 816 bases. To identify MG, NG and CT, all reads (1,127,703) which passed QC were analyzed by WIMP, resulting in 810,683 (72%) classified reads of which 43,491 (5.4%) were non-barcoded. For TV identification, Fastq Custom Alignment workflow produced 148,458/1,127,703 (13%) read alignments with TV ntr6 sequence of which 66,500 (5.9%) were non-barcoded.
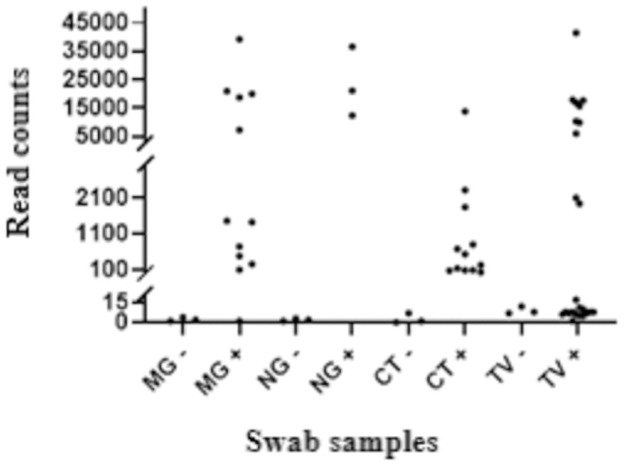
Targeted amplicon sequencing from clinical samples, as expected, still non-specifically produced some reads from other microbial and/or host DNA in the sample. Among 810,683 classified reads of the whole sequencing library, 560,601 (69%) mapped to the human genome. Details on the bacterial species that was mapped are shown in S1 Table. The ranges of classified reads mapped to the pathogens of interest are 0.5–95% (median 9.6%) for the CT positive samples, 0–86.6% (median 48.5%) for the MG positive samples and 92.3–95% (median 94.7%) for the NG positive samples, while the range of analysed reads mapped to TV in the TV positive samples is 0–99.4% (median 0.1%). However, for STI pathogens, qPCR positive samples generally yielded higher specific read counts than qPCR negative controls, all of the latter of which had fewer than 15 reads (Fig 1). Mean log10 read counts were higher in qPCR positive NG, MG and CT samples compared to negative by t-test (S2 Table) but not significantly for TV as 14/24 TV read counts in TV qPCR positive samples were very low (<20 reads). NG, MG and CT read counts clearly separated qPCR positive and negative samples except for one MG qPCR positive sample that had an absolute read count of 1 (Fig 1). The samples having <20 reads generally had a low load of pathogen, as evidenced by qPCR cycle threshold (Ct) being >35 (Table 2; S2 Fig).
Fig 1. Pathogen read counts in the qPCR positive and negative STI samples analyzed by single gene targeted PCR nanopore sequencing.
The ARMA workflow assigned 86,430/810,683 (11%) classified reads to antibiotic resistance genes in the CARD database, >99.99% of which were identified to be fluoroquinolone resistant gyrA gene in NG, using protein variant model. This analysis indicated that 3/3 NG clinical strains identified were fluoroquinolone-resistant, while none of the MG strains were resistant to macrolide antibiotics. The NG positive control, phenotypically susceptible to fluoroquinolones, and genotypically wild-type by BLAST (see below), was mis-identified to be fluoroquinolone-resistant. TV antimicrobial resistance was not profiled by ARMA due to the absence of TV ntr6 entries in the databases. Instead, it was analyzed by manual BLAST search against TV G3 ntr6 reference sequence (see below).
AMR confirmation by manual BLAST search
A manual BLAST search was performed to ascertain the accuracy of the pathogen AMR identified by the ARMA workflow. The BLAST confirmed, as shown in Table 3, that all (3/3) of NG had fluoroquinolone-resistant mutations, 2/10 (20%) of TV had mutations related to metronidazole resistance, and none of MG had macrolide resistance-associated mutations. It also showed that the gyrA of NG positive control did not possess fluoroquinolone-resistant mutations.
Table 3. BLAST confirmation of antimicrobial resistance associated mutations identified by single gene targeted PCR nanopore sequencing*.
| Bacterial gene | Sample | Antimicrobial resistance region: nucleotide sequence (NS) and amino acid (AA) change |
|---|---|---|
| Neisseria gonorrhoeae: gyrA | Wild type | NS: -TCCGCAGTTTACGAC- |
| AA: S91 D95 | ||
| All NG samples | NS -TTCGCAGTTTACGCC- | |
| AA change: S91F D95A | ||
| Positive control | NS: -TCCGCAGTTTACGAC- | |
| AA: S91 D95 | ||
| Mycoplasma genitalium: 23S rRNA gene | Wild type | NS: -CGGGACGGAAAGACC- |
| A2058, A2059 (Escherichia coli numbering) | ||
| All MG samples | NS: -CGGGACGGAAAGACC- | |
| A2058, A2059 (Escherichia coli numbering) | ||
| Trichomonas vaginalis: ntr6 | Wild type | NS: -AATGCAAAAGCAGAC- |
| AA: K80 | ||
| 2 TV samples | NS: -AATGCAATAGCAGAC- | |
| AA change: K80STOP | ||
| 8 TV samples | NS: -AATGCAAAAGCAGAC- | |
| AA: K80 |
*One or more changes of the bold bases in the wild type cause antimicrobial resistance of strains.
Discussion
We demonstrated the potential to test four common genital STIs and simultaneously detect AMR in three of them among vulnerable FSWs in Quito, Ecuador using the portable MinION DNA sequencer, controlled by smartphone, through a single gene targeted approach.
Single gene approaches for detection of infection, such as 16S rRNA sequencing have been increasingly described [21, 22], particularly with the advent of long read sequencing such as PacBio and ONT [23], which has greater likelihood of taxonomically identifying organisms at species level. We intentionally evaluated the accuracy of sequencing of a single AMR gene, as opposed to a two-stage gene approach [24], to both diagnose infection and predict AMR simultaneously in order to evaluate potential for use in resource poor field settings where simplicity and cost become increasingly important factors to consider.
Our approach appeared to have some advantages over NAAT. We demonstrated initially that genes responsible for AMR have potential to be as useful as 16S rRNA genes for diagnosis and thus may serve as a ‘two-in-one’ target. We chose for both pathogen and AMR detection the NG gyrA, MG 23S rRNA and TV ntr6, all gene mutations in which have been shown to be predictive of AMR [7–12, 25, 26], and CT omp-1 for CT detection [27]. As sequencing allows for identification of evolving mutations in the same genes without changing the test, our approach will therefore have value as it is adapted to other gene targets undergoing continuous evolution under selection pressure such as penA in which changes may give rise to penicillin and extended spectrum cephalosporin resistance [28]. The approach can also allow for sequencing multiple genes simultaneously alongside vigilance over any changing nature of phenotypic to genotypic association for antibiotics where AMR involves multiple mechanisms.
Clinical STI samples contain a wide range of pathogen loads being drawn frequently from largely asymptomatic patients as in the case of the FSW cohort in this study. We found that except for TV, diagnosis of NG, CT and MG by the single gene targeted nanopore sequencing appeared correct as almost NG, CT and MG positive samples had high pathogen loads. Due to the small sample size used in this study we did not determine a cut-off value of read counts for a positive STI test, but even so, our single gene approach correctly identified the majority of STI infections in the samples which had qPCR Ct<35 (higher pathogen loads) by their distinctive read counts. In the group with lower pathogen loads (Ct>35), tPCR amplification appeared not to enhance the absolute read counts of pathogen targets, possibly due to low tPCR efficiency caused by the presence of a large quantity of background DNA and non-optimal tPCR conditions, resulting in the ‘false-negatives’ observed for TV. Further optimization of tPCR conditions, for example enrichment of pathogen DNA during sample DNA preparation, may maximize the diagnostic sensitivity for this group of samples.
Regarding the Fastq Antimicrobial Resistance workflow used in this study, the component WIMP correctly identified each pathogen of interest, but we found the component ARMA CARD appeared to mis-profile the AMR of the NG control strain. This might be partially because the ARMA CARD used individual reads for matching AMR associated mutations, supported with the result obtained for the NG control strain by the manual BLAST search using the consensus sequence. A mixed AMR profile may exist in the community of pathogens of clinical samples, and therefore targeted amplification of the AMR associated genes can lead to a mixture of different AMR alleles. Therefore, any further development of ARMA, or a separate workflow, which contains a method to detect and quantify different AMR alleles, would be more appropriate for accurate prediction of AMR.
This study had some limitations. Firstly, there was a small sample size, which impacted particularly on measures of specificity. Secondly no further confirmatory sequencing was performed to confirm the mutations present, largely due to the local regulations which prevented transporting material out of Ecuador. Thus, this study should be regarded as a proof of concept study. Future work is required on larger sample sets with composite reference standards in a formal diagnostic evaluation. Additionally, we used older generation of reagents LSK-108 and tools (MinIt) in this study, which potentially impacted the output and speed of the workflow. Optimisation of amplification and target multiplexing will also be important.
Nanopore sequencing with MinION requires substantially lower infrastructure and startup cost compared to other high throughput sequencing platforms. Although arguably consumables are not optimally priced for resource poor settings, the promise of combining automated library preparations and use of disposable flow-cells has potential to use nanopore sequencing for field diagnostics. We recently demonstrated that even in developed settings implementation of rapid ciprofloxacin NAAT resistance tests for NG still requires net investment [29]. More recently ONT has developed a tablet version, MinION™ Mk1C which enables full sequencing and analysis to be performed in the lab and field without need for internet connection, and released rapid barcoding kits and flongle for sequencing. This development could reduce cost and improve speed further to use nanopore sequencing as a diagnostic tool. However, this will be formally evaluated in a large-scale clinical study.
In conclusion, this study demonstrated that single gene targeted nanopore sequencing for diagnosing and simultaneously identifying key antimicrobial resistance markers for four common STIs shows promise. Further work to optimise accuracy, reduce costs and improve speed may allow sustainable approaches for managing STIs and emerging AMR in resource poor and laboratory limited settings.
Supporting information
(TIF)
(TIF)
(XLSX)
(DOCX)
Acknowledgments
The authors would like to thank the FSWs associations for supporting the development of this study.
Data Availability
All relevant data are within the paper and its Supporting information files.
Funding Statement
1) Wellcome Trust 204809/Z/16/Z to St. George’s, University of London, Syed Tariq Sadiq 2) UIDE UIDE-EDM04-2016-2017 Natalia Romero Sandoval.
References
- 1.Rowley J, Vander Hoorn S, Korenromp E et al. Chlamydia, gonorrhoea, trichomoniasis and syphilis: global prevalence and incidence estimates, 2016. Bull World Health Organ. 2019;97(8):548–562P. doi: 10.2471/BLT.18.228486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golden MR, Workowski KA, Bolan G. Developing a Public Health Response to Mycoplasma genitalium. J Infect Dis. 2017;216(suppl_2):S420–6. doi: 10.1093/infdis/jix200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soni S, Horner P, Rayment M, Pinto-Sander N, Naous N, Parkhouse A, et al. British Association for Sexual Health and HIV national guideline for the management of infection with Mycoplasma genitalium (2018). Int J STD AIDS. 2019. Sep;30(10):938–950. doi: 10.1177/0956462419825948 Epub 2019 Jul 7. Erratum in: Int J STD AIDS. 2019 Aug 12;:956462419870463. . [DOI] [PubMed] [Google Scholar]
- 4.Wi T, Lahra MM, Ndowa F et al. Antimicrobial resistance in Neisseria gonorrhoeae: Global surveillance and a call for international collaborative action. PLoS Med. 2017;14(7):e1002344. Published 2017 Jul 7. doi: 10.1371/journal.pmed.1002344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ndowa F, Lusti-Narasimhan M, Unemo M. The serious threat of multidrug-resistant and untreatable gonorrhoea: the pressing need for global action to control the spread of antimicrobial resistance, and mitigate the impact on sexual and reproductive health. Sex Transm Infect. 2012;88(5):317–318. doi: 10.1136/sextrans-2012-050674 [DOI] [PubMed] [Google Scholar]
- 6.Muller EE, Mahlangu MP, Lewis DA et al. Macrolide and fluoroquinolone resistance-associated mutations in Mycoplasma genitalium in Johannesburg, South Africa, 2007–2014. BMC Infect Dis. 2019;19(1):148. Published 2019 Feb 13. doi: 10.1186/s12879-019-3797-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donà V, Low N, Golparian D et al. Recent advances in the development and use of molecular tests to predict antimicrobial resistance in Neisseria gonorrhoeae. Expert Rev Mol Diagn. 2017;17(9):845–859. doi: 10.1080/14737159.2017.1360137 [DOI] [PubMed] [Google Scholar]
- 8.Pond MJ, Hall CL, Miari VF et al. Accurate detection of Neisseria gonorrhoeae ciprofloxacin susceptibility directly from genital and extragenital clinical samples: towards genotype-guided antimicrobial therapy. J Antimicrob Chemother. 2016;71(4):897–902. doi: 10.1093/jac/dkv432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen JS, Bradshaw CS, Tabrizi SN et al. Azithromycin treatment failure in Mycoplasma genitalium-positive patients with nongonococcal urethritis is associated with induced macrolide resistance. Clin Infect Dis. 2008;47(12):1546–1553. doi: 10.1086/593188 [DOI] [PubMed] [Google Scholar]
- 10.Yew HS, Anderson T, Coughlan E et al. Induced macrolide resistance in Mycoplasma genitalium isolates from patients with recurrent nongonococcal urethritis. J Clin Microbiol. 2011;49(4):1695–1696. doi: 10.1128/JCM.02475-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito S, Shimada Y, Yamaguchi Y et al. Selection of Mycoplasma genitalium strains harbouring macrolide resistance-associated 23S rRNA mutations by treatment with a single 1 g dose of azithromycin. Sex Transm Infect. 2011;87(5):412–414. doi: 10.1136/sextrans-2011-050035 [DOI] [PubMed] [Google Scholar]
- 12.Paulish-Miller TE, Augostini P, Schuyler JA et al. Trichomonas vaginalis metronidazole resistance is associated with single nucleotide polymorphisms in the nitroreductase genes ntr4Tv and ntr6Tv. Antimicrobial Agents and Chemotherapy. 2014. May;58(5):2938–2943. doi: 10.1128/AAC.02370-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Köser CU, Ellington MJ, Peacock SJ. Whole-genome sequencing to control antimicrobial resistance. Trends Genet. 2014. Sep;30(9):401–7. doi: 10.1016/j.tig.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schürch AC, van Schaik W. Challenges and opportunities for whole-genome sequencing-based surveillance of antibiotic resistance. Ann N Y Acad Sci. 2017;1388(1):108–120. doi: 10.1111/nyas.13310 [DOI] [PubMed] [Google Scholar]
- 15.Llangarí-Arizo LM, Sadiq ST, Márquez C, Cooper P, Furegato M, Zhou L, et al. Sexually transmitted infections and factors associated with risky sexual practices among female sex workers: A cross sectional study in a large Andean city. PLoS One. 2021. May 6;16(5):e0250117. doi: 10.1371/journal.pone.0250117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pillay A, Radebe F, Fehler G et al. Comparison of a TaqMan-based real-time polymerase chain reaction with conventional tests for the detection of Trichomonas vaginalis. Sex Transm Infect. 2007;83(2):126–129. doi: 10.1136/sti.2006.022376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen JS, Björnelius E, Dohn B et al. Use of TaqMan 5’ nuclease real-time PCR for quantitative detection of Mycoplasma genitalium DNA in males with and without urethritis who were attendees at a sexually transmitted disease clinic. J Clin Microbiol. 2004;42(2):683–692. doi: 10.1128/JCM.42.2.683-692.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaton K, Bille J, Greub G. A novel real-time PCR to detect Chlamydia trachomatis in first-void urine or genital swabs. J Med Microbiol. 2006;55(Pt 12):1667–1674. doi: 10.1099/jmm.0.46675-0 [DOI] [PubMed] [Google Scholar]
- 19.Tabrizi SN, Chen S, Tapsall J et al. Evaluation of opa-based real-time PCR for detection of Neisseria gonorrhoeae. Sex Transm Dis. 2005;32(3):199–202. doi: 10.1097/01.olq.0000154495.24519.bf [DOI] [PubMed] [Google Scholar]
- 20.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881–10890. doi: 10.1093/nar/16.22.10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007;45(9):2761–2764. doi: 10.1128/JCM.01228-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo PC, Lau SK, Teng JL et al. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect. 2008;14(10):908–934. doi: 10.1111/j.1469-0691.2008.02070.x [DOI] [PubMed] [Google Scholar]
- 23.Johnson JS, Spakowicz DJ, Hong BY et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10(1):5029. Published 2019 Nov 6. doi: 10.1038/s41467-019-13036-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham RM, Doyle CJ, Jennison AV. Epidemiological typing of Neisseria gonorrhoeae and detection of markers associated with antimicrobial resistance directly from urine samples using next generation sequencing. Sex Transm Infect. 2017;93(1):65–67. doi: 10.1136/sextrans-2015-052422 [DOI] [PubMed] [Google Scholar]
- 25.Hemarajata P, Yang S, Soge OO et al. Performance and Verification of a Real-Time PCR Assay Targeting the gyrA Gene for Prediction of Ciprofloxacin Resistance in Neisseria gonorrhoeae. J Clin Microbiol. 2016;54(3):805–808. doi: 10.1128/JCM.03032-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Korne-Elenbaas J, Pol A, Vet J et al. Simultaneous Detection of Neisseria gonorrhoeae and Fluoroquinolone Resistance Mutations to Enable Rapid Prescription of Oral Antibiotics. Sex Transm Dis. 2020;47(4):238–242. doi: 10.1097/OLQ.0000000000001141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molano M, Meijer CJ, Morré SA et al. Combination of PCR targeting the VD2 of omp1 and reverse line blot analysis for typing of urogenital Chlamydia trachomatis serovars in cervical scrape specimens. J Clin Microbiol. 2004;42(7):2935–2939. doi: 10.1128/JCM.42.7.2935-2939.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yahara K, Ma KC, Mortimer TD et al. Emergence and evolution of antimicrobial resistance genes and mutations in Neisseria gonorrhoeae. bioRxiv 2020.10.26.354993; 10.1101/2020.10.26.354993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harding-Esch EM, Huntington SE, Harvey MJ et al. Antimicrobial resistance point-of-care testing for gonorrhoea treatment regimens: cost-effectiveness and impact on ceftriaxone use of five hypothetical strategies compared with standard care in England sexual health clinics. Euro Surveill. 2020;25(43):pii = 1900402. 10.2807/1560-7917.ES.2020.25.43.1900402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting information files.