Abstract
During the winter of 1995–1996, eight of nine bone marrow transplantation (BMT) unit patients were infected with the same strain of respiratory syncytial virus (RSV). This RSV strain was not detected in 20 hospitalized patients from the community, suggesting that the BMT unit infections did not occur by independent incidents of transmission from the community.
A number of studies highlight respiratory syncytial virus (RSV) as an important cause of mortality in immunocompromised transplant patient groups (6, 8) such as bone marrow transplant (BMT) recipients. The BMT unit in Bristol, United Kingdom, is a specialist referral center for unrelated-donor BMTs and practices standard precautions for all patients undergoing transplantation (10). A recent retrospective analysis of RSV infection in patients at this unit over a 5-year period by McCarthy and colleagues (10) reported an RSV infection incidence of 6.3%, with a rate of mortality of 19.2% directly attributable to RSV. Using restriction enzyme analysis to genotype RSV, they identified a number of patients infected with strains of the same genotype during a single winter epidemic season. However, that type of genetic analysis is too crude to provide firm conclusions about the origin of infection. The study described here further investigates this outbreak of RSV by analyzing viral nucleotide sequences obtained from both BMT unit patients and hospitalized patients in the community.
RNA was extracted from stored clinical material, cDNA was synthesized (10), and the virus was genotyped (3). The nucleotide sequence of the second variable region (7) of the glycoprotein gene (nucleotides [nt] 649 to 918 for group A viruses; nt 652 to 921 for group B viruses) was amplified and sequenced (11). This region of the virus genome has been shown to serve as a reliable proxy for variability over the entire glycoprotein gene (11).
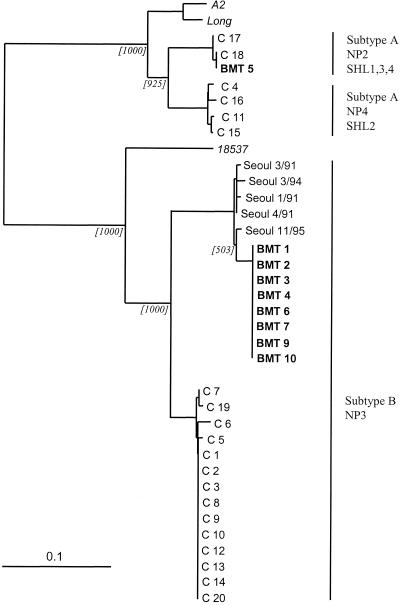
Twenty specimens from pediatric patients hospitalized during the winter of 1995–1996 were randomly selected from a collection of RSV-positive nasopharyngeal aspirate samples obtained from the Bristol area. Genotyping by restriction enzyme analysis revealed that the majority (14 of 20) of the infections were caused by RSV strains of the group B NP3 genotype. The remainder were classified as group A viruses, either genotype NP2 SHL1,3,4 (2 of 20 isolates) or genotype NP4 SHL2 (4 of 20 isolates). Phylogenetic analysis of the nucleotide sequences confirmed these genotyping results, with viruses classified as the same genotype clustering together (Fig. 1). There was, however, a low level of sequence variation between isolates within each genotype. Sequences classified as genotype NP4 SHL2, NP2 SHL1,3,4, and NP3 showed 98.1, 99.6, and 96.7% nucleotide identities, respectively.
FIG. 1.
Phylogenetic relationship of RSV nucleotide sequences obtained from both hospitalized community (C) and BMT unit (BMT) patient specimens. The genotypes assigned by restriction enzyme analysis are indicated. The numbers in brackets refer to the number of times that the node appeared in 1,000 bootstrap replicates. RSV sequences from the GenBank database include those from strains isolated in Seoul, South Korea (indicated on the diagram as Seoul followed by the month and year of isolation), and RSV type strains Long, A2, and 18537.
During the winter of 1995–1996, a total of 10 patients in the BMT unit were diagnosed with RSV infection by virus culture or direct immunofluorescence staining of nasopharyngeal aspirate or bronchoalveolar lavage specimens. RSV genetic material could not be amplified from the RSV-positive specimen obtained from one patient (Fig. 2, BMT 8), and this patient was not considered further. McCarthy and colleagues (10) had previously shown that of the remaining nine patients within the BMT unit whose isolates could be genotyped, the infection in one was caused by a group A RSV (genotype NP2 SHL1,3,4), while the infections in the remaining eight patients were caused by group B RSVs of identical genotypes (genotype NP3). Phylogenetic analysis of the RSV nucleotide sequences revealed that all eight of the genotype NP3 RSV isolates obtained from BMT patients had identical nucleotide sequences (henceforth referred to as the BMT strain of RSV).
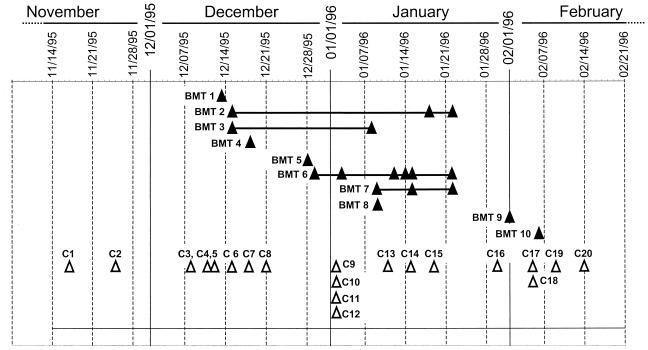
FIG. 2.
Temporal distribution of diagnoses of RSV infection for BMT and community patients. The sampling dates of the specimens BMT (closed triangles) or hospitalized community (open triangles) patients specimens are shown. Four of the BMT patients had multiple specimens that were RSV positive by immunofluorescence or virus culture. These are indicated by horizontal lines.
It was striking that the sequence of the BMT strain of RSV was distinct from the sequences of all of isolates obtained from the hospitalized community patient specimens, even including those sequences that were classified as belonging to the same NP3 genotype (range of sequence identity, 87.0 to 88.8%). The absence of the BMT strain of RSV from a total of 20 RSV specimens obtained from hospitalized community patients would suggest that its prevalence within the community during that epidemic season was low. As the BMT strain accounted for eight of the nine BMT unit infections, it appears unlikely that the infections within the BMT unit could have occurred by independent incidents of transmission from the community. The evidence strongly supports the fact that, once it was introduced into the BMT unit, the BMT strain of RSV infected all but one of these patients by nosocomial transmission. The isolation of this group of patients from each other would appear to rule out direct patient-to-patient spread. Therefore, transmission must have involved either illness in unit staff or mechanical transmission from environmental contamination. However, the absence of reported illness from staff members prevents a definitive description of the mechanism of transmission from being given. We could not investigate environmental contamination and mechanical transmission in this retrospective analysis.
RSV infections within the BMT unit, as determined by the date of the first diagnosis of RSV infection, could be grouped into three distinct clusters (Fig. 2). The cases in the first cluster (BMT 1 to BMT 4) were diagnosed within a 5-day period (13 to 18 December), the cases in the second cluster (BMT 5 to BMT 8) were diagnosed within a 12-day period (28 December to 9 January), and the cases in the third cluster (BMT 9 and BMT 10) were diagnosed within a 5-day period (1 to 6 February). Repeat RSV-positive nasopharyngeal aspirates were obtained from four patients (BMT 2, 3, 6, and 7), indicating that these patients were infected with RSV for prolonged periods of time. Prolonged viral shedding from these patients may provide a mechanism by which an identical strain of virus could be transmitted to temporally separated clusters of patients.
Comparison of the nucleotide sequence of the BMT strain of RSV with those RSV sequences held in the GenBank database that provided information on the location and date of isolation returned a number of sequences that clustered with the BMT strain of RSV (Fig. 1). These were isolated in Seoul, South Korea, between 1991 and 1995 (E. H. Choi and H. J. Lee, unpublished data) and Birmingham, Ala., during the winter of 1994 (data not shown) (4) (sequence identity range, 97.8 to 98.5%). RSV strains very similar to the strain isolated from the BMT unit patients were therefore circulating globally prior to and during the winter of 1995–1996. Multiple studies examining the transmission of RSV at the global level (1, 2, 5) have simultaneously isolated RSV strains with virtually identical G-gene sequences from widely separated geographical locations. Therefore, it is not inconceivable that this strain was present, albeit at a low prevalence, in the Bristol area during the winter of 1995–1996.
Mazzulli and colleagues (9) have recently studied RSV infection within an adult leukemia-lymphoma ward. Three strains, each with its own identical nucleotide sequence, were responsible for seven cases of RSV, suggesting that instead of a single nosocomial outbreak, multiple introductions of virus were transmitted nosocomially. Our data suggest that, once it was introduced into the BMT unit, the BMT strain of RSV infected eight patients by nosocomial transmission.
Acknowledgments
We are grateful to the medical staff of the Bristol bone marrow transplantation center for submitting clinical samples.
This work was supported by the Public Health Laboratory Service's small scientific initiative fund.
REFERENCES
- 1.Cane P A, Pringle C R. Evolution of subgroup A respiratory syncytial virus: evidence for progressive accumulation of amino acid changes in the attachment protein. J Virol. 1995;69:2918–2925. doi: 10.1128/jvi.69.5.2918-2925.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cane P A, Pringle C R. Molecular epidemiology of human respiratory syncytial virus. Semin Virol. 1995;6:371–378. [Google Scholar]
- 3.Cane P A, Pringle C R. Molecular epidemiology of respiratory syncytial virus: rapid identification of subgroup A lineages. J Virol Methods. 1992;40:297–306. doi: 10.1016/0166-0934(92)90088-u. [DOI] [PubMed] [Google Scholar]
- 4.Coggins W B, Lefkowitz E J, Sullender W M. Genetic variability among group A and group B respiratory syncytial viruses in a children's hospital. J Clin Microbiol. 1998;36:3552–3557. doi: 10.1128/jcm.36.12.3552-3557.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia O, Martin M, Dopazo J, Arbiza J, Rabasile S, Russi J, Hortal M, Perez-Brena P, Martinez I, Garcia-Barreno B, Melero J A. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J Virol. 1994;68:5448–5459. doi: 10.1128/jvi.68.9.5448-5459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hertz M I, England J A, Snover D, Bitterman P B, McGlave P B. Respiratory syncytial virus-induced acute lung injury in adult patients with bone marrow transplants: a clinical approach and review of the literature. Medicine. 1989;68:269–281. doi: 10.1097/00005792-198909000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Johnson P R, Spriggs M K, Olmsted R A, Collins P L. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc Natl Acad Sci USA. 1987;84:5625–5629. doi: 10.1073/pnas.84.16.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krinzman S, Basgoz N, Kradin R, Shepard J A, Flieder D B, Wright C D, Wain J C, Ginns L C. Respiratory syncytial virus-associated infections in adult recipients of solid organ transplants. J Heart Lung Transplant. 1998;17:202–210. [PubMed] [Google Scholar]
- 9.Mazzulli T, Peret T C, McGeer A, Cann D, MacDonald K S, Chua R, Erdman D D, Anderson L J. Molecular characterization of a nosocomial outbreak of human respiratory syncytial virus on an adult leukemia/lymphoma ward. J Infect Dis. 1999;180:1686–1689. doi: 10.1086/315085. [DOI] [PubMed] [Google Scholar]
- 10.McCarthy A J, Kingman H M, Kelly C, Taylor G S, Caul E O, Grier D, Moppett J, Foot A B, Cornish J M, Oakhill A, Steward C G, Pamphilon D H, Marks D I. The outcome of 26 patients with respiratory syncytial virus infection following allogeneic stem cell transplantation. Bone Marrow Transplant. 1999;24:1315–1322. doi: 10.1038/sj.bmt.1702078. [DOI] [PubMed] [Google Scholar]
- 11.Peret T C, Hall C B, Schnabel K C, Golub J A, Anderson L J. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J Gen Virol. 1998;79(Pt. 9):2221–2229. doi: 10.1099/0022-1317-79-9-2221. [DOI] [PubMed] [Google Scholar]