Summary
Background
The emergence of new SARS-CoV-2 variants of concern B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma) and B.1.617.2 (Delta) that harbor mutations in the viral S protein raised concern about activity of current vaccines and therapeutic antibodies. Independent studies have shown that mutant variants are partially or completely resistant against some of the therapeutic antibodies authorized for emergency use.
Methods
We employed hybridoma technology, ELISA-based and cell-based S-ACE2 interaction assays combined with authentic virus neutralization assays to develop second-generation antibodies, which were specifically selected for their ability to neutralize the new variants of SARS-CoV-2.
Findings
AX290 and AX677, two monoclonal antibodies with non-overlapping epitopes, exhibit subnanomolar or nanomolar affinities to the receptor binding domain of the viral Spike protein carrying amino acid substitutions N501Y, N439K, E484K, K417N, and a combination N501Y/E484K/K417N found in the circulating virus variants. The antibodies showed excellent neutralization of an authentic SARS-CoV-2 virus representing strains circulating in Europe in spring 2020 and also the variants of concern B.1.1.7 (Alpha), B.1.351 (Beta) and B.1.617.2 (Delta). In addition, AX677 is able to bind Omicron Spike protein just like the wild type Spike. The combination of the two antibodies prevented the appearance of escape mutations of the authentic SARS-CoV-2 virus. Prophylactic administration of AX290 and AX677, either individually or in combination, effectively reduced viral burden and inflammation in the lungs, and prevented disease in a mouse model of SARS-CoV-2 infection.
Interpretation
The virus-neutralizing properties were fully reproduced in chimeric mouse-human versions of the antibodies, which may represent a promising tool for COVID-19 therapy.
Funding
The study was funded by AXON Neuroscience SE and AXON COVIDAX a.s.
Keywords: SARS-CoV-2, COVID-19, Neutralizing antibodies, Escape mutation, Variants of concern
Abbreviations: S, SARS-CoV-2 spike protein; RBD, receptor-binding domain; mAbs, monoclonal antibodies; NAbs, neutralizing antibodies; CPE, cytopathic effect; PRNT, plaque reduction neutralization test; MLV, mouse leukemia virus
Research in context.
Evidence before this study
We have reviewed the knowledge on the appearance of SARS-CoV-2 mutations and variants in the human population and their effect on the neutralization by monoclonal antibodies and sera using PubMed (August10, 2021) with the search term combination “COVID AND ((monoclonal antibody) OR sera) AND (variants OR mutations)”.The published evidence showed that the mutations in the Spike protein of the virus can reduce the efficiency of both vaccines and monoclonal antibodies developed against the virus circulating in the population in the spring and summer of 2020. The data also showed that overwhelming majority of the monoclonal antibodies with neutralization potencies were identified and isolated from human subjects recovering from the SARS-CoV-2 infection.
Added value of this study
We have isolated mouse monoclonal antibodies that target two main epitopes on the Spike receptor binding domain. Two most active antibodies neutralize current SARS-CoV-2 variants of concern Alpha, Beta and Delta, and prevent systemic disease in an ACE2-humanized mouse model.
Implications of all the available evidence
Identification of monoclonal antibodies with various epitope properties will provide source of therapeutics that could prove essential for the treatment of breakthrough virus variants in the future. The combined functional and structural information on the collection of such antibodies will also help to expand our knowledge on the functional and immunogenic properties of both the Spike protein and the SARS-CoV-2 virus.
Alt-text: Unlabelled box
Introduction
The recent outbreak of COVID-19 showed us how inadequately prepared we are against newly emerging (zoonotic) viruses. Vaccines are the best strategy to suppress the COVID-19 pandemic. However, their waning protection and a large pool of unvaccinated individuals will provide ground for spread of the virus and generation of novel variants, posing a constant risk of infection for vulnerable people with inadequate immune responses. For those who become infected efficient treatment will need to be developed. Neutralizing antibodies (NAbs) against SARS-CoV-2 are being evaluated for prophylaxis and as therapeutic agents for COVID-19 patients.1 Human antibodies isolated from COVID-19 convalescent patients often target the receptor-binding domain (RBD) of the Spike (S) protein of SARS-CoV-2, and recognize distinct, sometimes non-overlapping epitopes.2, 3, 4, 5, 6, 7, 8 Only a small subset of these antibodies can block viral entry, usually by interfering with the binding of the viral S protein to the cellular receptor ACE2.2,4,5 Clinical trials suggest that antibody treatments can prevent deaths and hospitalizations among people with mild or moderate COVID‑19. Some of the therapeutic antibodies against COVID-19 were already authorized for emergency use9, however they were not initially selected for the neutralization activity against emerging variants of SARS-CoV-2.
The majority of NAbs against SARS-CoV-2 in the clinical development was derived from convalescent patients who recovered from COVID-19.10, 11, 12, 13 Antibodies derived from human B cells do not require an extensive humanization process and can be fast-forwarded to manufacture.14 However, several studies demonstrated that neutralizing monoclonal antibodies obtained from individuals during the early convalescence period may exhibit low levels of maturation associated with viral suppression of germinal center development.15, 16, 17 Optimal immunization procedure and exploration of mouse immunoglobulin repertoire may overcome these limitations and survey an orthogonal conformation space of antibody specificities.18
The emergence of new SARS-CoV-2 variants of concern (VOCs) B.1.1.7 (Alpha),19,20 B.1.351 (Beta),21 P.1 (Gamma)22,23 and B.1.617.2 (Delta)24 that harbor mutations in the viral S protein raised alarm whether current vaccines and therapeutic antibodies were sufficiently effective. Indeed, several independent studies have shown that mutant variants are partially or completely resistant against therapeutic antibodies that were authorized for emergency use.24, 25, 26, 27, 28, 29, 30, 31
Here we present two mouse-human chimeric monoclonal antibodies against the S protein RBD with non-overlapping epitopes and excellent neutralization potency not only against a live authentic SARS-CoV-2 virus isolate that circulated in Europe in spring 2020, but also variants of concern B.1.1.7 (Alpha), B.1.351 (Beta) and B.1.617.2 (Delta) currently spreading in the population. The combination of the two antibodies prevents generation of escape mutations of the live authentic SARS-CoV-2 virus, making the mixture a promising candidate for a therapeutic application.
Methods
Cell lines
Human embryonic kidney HEK293T/17-hACE2 cells with stable expression of human angiotensin-converting enzyme ACE2 (AXON Neuroscience SE) were prepared by stable transfection of pDUO2-hACE2-TMPRSS2a (InvivoGen) into HEK293T/17 cells (ATCC Cat# ACS-4500, RRID:CVCL_4V93) with transfection reagent Lipofectamine 3000 (L3000001, Thermo Fisher Scientific), according the to the manufacturer's recommendations. The hygromycin resistant colonies of stable transfectants were isolated and screened for the expression of the ACE2 protein by Western blotting (Reagent Validation file) and immunohistochemistry (Figure S1). The expression of the serine protease TMPRSS2 was not evaluated. Stable cell line HEK293T/17-hACE2 were maintained in selection medium Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% heat-inactivated foetal bovine serum (FBS), 2 mM L-glutamine (GIBCO), 5 µg/ml gentamicin (SIGMA) and 100µg/ml hygromycin B (Invitrogen) at 37°C in 5%CO2. In PRNT, Vero E6 cells (ECACC Cat# 85020206, RRID:CVCL_0574) were cultured in Eagle's minimal essential medium (EMEM, Lonza) supplemented with 5% FBS (GIBCO), Penicillin-Streptomycin-Amphotericin B Solution (10ml/l, Lonza, Switzerland). Expi293F™ (A14527, Thermo Fisher Scientific, RRID:CVCL_D615) cells were used for the expression of recombinant Spike, RBD and ACE2 proteins. They were grown in a chemically defined, protein-free Expi293 Expression medium (Cat. No.: A1435101, Thermo Fisher Scientific) with no human or animal-origin components, in the polycarbonate Erlenmeyer flasks with ventilated cap (Product numbers: 431143, 431145, 431147, Corning® USA). The flasks were kept on a shaker platform at 120 RPM in a humidified CO2 incubator at 37°C (32°C during the protein expression stage) and 5% of CO2. The cells were sub-cultured regularly 2-3 times a week, keeping them at the exponential growth phase. No antibiotics, feed, or other supplements were added to the culture before or after the transient transfections. NS0 mouse myeloma cells (ECACC Cat# 85110503, RRID:CVCL_3940) are routinely used for hybridoma preparation. All cell lines were purchased from certified suppliers, upon delivery they were thawed, amplified and frozen in working aliquots according to the suggestion of the suppliers. Upon thawing they are verified for consistency in morphology. The HEK293T/17 cell line was recently genetically authenticated. All mammalian and hybridoma cell lines are regularly tested for mycoplasma using the PCR Mycoplasma Test Kit I/C (PromoCell GmbH, Heidelberg, Germany).
Proteins
DNAs coding for the prefusion-stabilized SARS-CoV-2 S protein ectodomain32 (residues 1−1208 from GenBank: MN908947), with proline substitutions at residues 986 and 987, a “GSAS” substitution at the furin cleavage site residues 682–685, a C-terminal T4 fibritin trimerization motif, an HRV3C protease cleavage site, a TwinStrepTag and an 8XHisTag), the RBD fragment of the S protein (amino acids 319-591), containing the His-tag and Twin-Strep-tag, and modified human ACE2 (angiotensin-converting enzyme 2), containing tags for purification were synthesized at BioCat Co. (order no. 200408, BioCat GmbH, Germany). The SARS-CoV-2 Spike trimer protein of variant B.1.1.529 (Omicron) was purchased from ACROBiosystems Inc. (Newark, DE, USA, #SPN-C52Hz). It is the ectodomain of SARS-CoV-2 spike protein expressed in HEK293 cells, which contains amino acids Val16 - Pro1213 of Spike (GenBank: QHD43416.1) with T4 fibritin trimerization motif, a polyhistidine tag at the C-terminus, proline substitutions (F817P, A892P, A899P, A942P, K986P, V987P) and alanine substitutions (R683A and R685A) to stabilize the trimeric pre-fusion conformation, and mutations identified in the Omicron variant (A67V, HV69-70del, T95I, G142D, VYY143-145del, N211del, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F).
Viruses
Three cell culture isolates of SARS-CoV-2 were used in the in vitro characterization and validation of the anti-Spike antibodies. They were all isolated from clinical samples of COVID-19 patients in Slovakia. All three isolates were deposited in the European virus archive GLOBAL. The strain Slovakia/SK-BMC5/2020 (available at https://www.european-virus-archive.com/virus/sars-cov-2-strain-slovakiask-bmc52020) represents strains circulating in Europe in spring 2020 and carries the Spike D614G mutation (lineage B.1). The strain Slovakia/SK-BMC-P1A/2021 (available at https://www.european-virus-archive.com/virus/sars-cov-2-strain-slovakiask-bmc-p1a2021-b117-variant-voc-20201201) was isolated in January 2021 and belongs to the B.1.1.7 lineage (VOC 202012/01, Alpha). The strain Slovakia/SK-BMC-BA11/2021 (available at https://www.european-virus-archive.com/virus/sars-cov-2-strain-slovakiask-bmc-ba112021-b1351-variant-aka-20h501yv2-or-south-african-variant) was isolated in March 2021 and belongs to the B.1.351 lineage (20H/501Y.V2, Beta). The strain Slovakia/SK-BMC-BA15/2021 (available at https://www.european-virus-archive.com/virus/sars-cov-2-strain-slovakiask-bmc-ba152021-b16172-variant-aka-delta-voc-or-indian-variant) was isolated in June 2021 and belongs to the lineage B.1.617.2 (Delta). The complete genome sequences of all four viruses were deposited in the GISAID.org database under the accession IDs EPI_ISL_417879, EPI_ISL_77965.1, EPI_ISL_1234458 and EPI_ISL_2657324, respectively.
The strain SARS-CoV-2/human/Czech Republic/951/2020, kindly provided by Dr. J. Weber (Institute of Organic Chemistry and Biochemistry, Prague, Czech Republic), was used for infection of ACE2 humanized mice in the course of the in vivo testing of the antibodies. The virus was isolated from a clinical sample at the National Institute of Public Health, Prague, Czech Republic, and passaged in Vero E6 cells five times before its use in this study.
Preparation of plasmids coding for S, S-RBD and ACE2 proteins
For protein production, the synthetic genes (BioCat) coding for the spike protein (S, order No.200408 from BioCat), receptor binding domain (RBD, order No.200408 from BioCat) or human ACE2 (puc57 ACE2_order No.200408 from BioCat) were cloned under the CMV promoter in a pCMV-based mammalian expression vector immediately after a signal peptide accomplishing secretion of recombinant proteins from the cells into medium. The plasmids were amplified in DH5α bacterial cells. The cells carrying the plasmid were inoculated into 5 ml LB medium with kanamycin for 8 hrs, transferred into 250 ml cultures and incubated overnight with shaking at 37°C. The plasmid DNA was isolated using PureLink® HiPure Plasmid Filter Maxiprep Kit (#K210017, Invitrogen) with the last ethanol washes done in a sterile laminar-flow hood. After overnight incubation in sterile water at 4°C, DNA concentration was measured using NanoPhotometer and the DNA was aliquoted and stored at -20°C. Insertion of the desired point mutations in S-RBD was done using QuickChange II site directed mutagenesis kit (#200523, Agilent Technologies). Primers for mutagenesis were designed using The QuickChange® Primer Design Program accessible at the manufacturer's web site. The following HPLC-purified primer pairs (synthesized by Generi Biotech, Czech Republic) were used: N501Y, GGCTTTCAGCCGACCTATGGCGTGGGCTATCAG, and CTGATAGCCCACGCCATAGGTCGGCTGAAAGCC; N439K, GATTGCGTGGAACAGC AAGAACCTGGATAGCAAAG and CTTTGCTATCCAGGTTCTTGCTGTTCCACGCAATC; E484K, CCG TGCAACGGCGTGAAAGGCTTTAACTGCT and AGC AGTTAAAGCCTTTCACGCCGTTGCACGG; 417N, CGGGCCAGACCGGCAATATTGCGGATTATAACTA and TAGTTATAATCCGCAATATTGCCGGTCTGGCC CG; T345N, CGAAGTGTTTAACGCGAACCGCTTTGCGAGCG and CGCTCGCAAAGCGGTTCGCGTTAAACACTTCG; S477R, CGAAATTTATCAGGCG GGCCGCACCCCGTGCAACG and CGTTGCAC GGGGTGCGGCCCGCCTGATAAATTTCG. Prepared mutations were transformed into DH5α bacterial cells and individual clones verified by DNA sequencing. The positive clones were further amplified in DH5α for mammalian cell transfections.
Production of the proteins in human cell line Expi293
Proteins were produced in human cell line Expi293FTM (Expi293™ Expression System Kit A14635, Thermo Fisher Scientific, Expi293 cells A14527, RRID:CVCL_D615). Plasmids that drive protein expression from the CMV promoter were either transfected with polyethyleneimine (PEI) or by electroporation into Expi293 cells that were in exponential growth phase (2·5-3·0 × 106 cells/ml) in Expi293 expression medium (A14331-1, Thermo Fisher Scientific). For PEI transfections, 3·75ml PEI (PEI MAX®, 24765-1, Polysciences Europe GmbH, Germany) and 1·25mg DNA was added to a suspension of 109 cells in a total volume of 50 ml, shaken at 37°C for 3 hrs and diluted into working concentrations. For electroporation, 100 µg/ml DNA was electroporated in a suspension of 2·108 cells/ml in an electroporation buffer EPB1 using MaxCyte STX (both from MaxCyte, MD, USA), the cells were let to rest for 30 min and then diluted into the final concentration of 3·106 cells/ml in Expi293 Expression medium. The cultures were incubated (polycarbonate Erlenmayer flasks/vented cap, #431144, Corning) with shaking (120 rpm) in 8% CO2 at 37°C for 24 hrs, transferred to 32°C and further cultured until cell viability decreased to <50% (approx. 4-6 days), at which point the medium was harvested by centrifugation at 300xg for 12 min and stored at -20°C until purification.
Protein purification
The proteins were purified from cell media using Åkta FPLC (GE Healthcare) as follows. The culture medium was clarified by centrifugation at 20,000xg, NaCl was added to a final concentration of 0·5 M and the solution was filtered through a 0·2 μm membrane filter. A 5 ml His-Trap affinity column (#17524802, Cytiva) charged with nickel ions was equilibrated in a 20 mM sodium phosphate buffer, pH 7·4, 0·5 M NaCl and the culture medium was loaded onto the column. Subsequently, the column was washed with 20 ml of the 20 mM sodium phosphate buffer, pH 7·4, and His-tagged protein was eluted by 0·5 M imidazole in 20 mM sodium phosphate buffer, pH 7·4. The fractions containing the target protein were pooled and further purified on the Strep-Tactin® media (#2-1206-025, IBA GmbH, Germany) as described by the manufacturer, using 10 mM desthiobiotin (#2-1000-005, IBA GmbH, Germany) as an elution reagent. Purified proteins were concentrated by ultrafiltration and buffer-exchanged into PBS on a 5 ml HiTrap Desalting column (GE Healthcare). The concentration was determined from UV absorbance at 280 nm. The proteins were sterile-filtered and stored at -20°C. Extracellular part of the human ACE2 protein fused to human IgG Fc fragment, preceded by a HRV3C protease cleavage site was purified by His-Trap and Strep-Tactin® media as above. The Fc fragment was cleaved out by an overnight incubation with HRV3C protease (#71493-3, Merck) at +6°C. ACE2 was afterwards polished by size-exclusion chromatography on a Superdex 75 16/60 column (GE Healthcare), concentrated by ultrafiltration with a Amicon Ultra centrifugal filter Ultracel with 10 kDa MW cut off (#UFC901096, Millipore), sterile-filtered and stored at +6°C.
Preparation of hybridoma cell lines producing monoclonal antibodies specific to SARS-CoV-2 spike protein and RBD
Six-week-old BALB/cAnNCrl mice (RRID:IMSR_CRL:028, Charles River Laboratories, delivered by Velaz s.r.o., Czech Republic) were primed subcutaneously either with 30 μg of recombinant SARS-Cov-2 Spike protein (S) or spike RBD in the complete Freund's adjuvant (#F 5881, Sigma-Aldrich) and boosted three times at three-week intervals with 20 μg of the same antigen in the incomplete Freund`s adjuvant (F5506, Sigma-Aldrich). Three days before the spleen extraction, mice were injected intraperitoneally with 20 μg of the same antigens in PBS. Spleen cells from immunized mice were fused with NS0 myeloma cells according to the method described in Kontsekova et al.43 Splenocytes were mixed with NS0 myeloma cells (in a ratio 5:1, ECACC Cat# 85110503, RRID:CVCL_3940) and fused for 1 minute in 1 ml of 50% polyethylene glycol (PEG) 1550 (Serva) in serum free Dulbecco's modified Eagle's medium (DMEM, #11960-044, ThermoFisher Scientific) supplemented with 10% dimethyl sulfoxide (DMSO). The fused cells were resuspended in DMEM containing 20% horse serum, L-glutamine (2 mM), HAT media supplement Hybri-Max™ and gentamycin (40 U/ml) (both from Sigma-Aldrich), at a density of 2·5 × 105 spleen cells per well in 96-well plates and incubated for 10 days at 37°C.
ELISA for screening of monoclonal antibodies
The growing hybridomas were screened for the production of monoclonal antibodies specific to the S protein and RBD by an enzyme-linked immunosorbent assay (ELISA). Microtiter plates were coated overnight with one of the proteins (2 μg/ml, 50 μl/well) at 37°C in PBS. After blocking with PBS-0·1% Tween 20 to reduce nonspecific binding, the plates were washed with PBS-0·05% Tween 20 and incubated with 50 μl/well of hybridoma culture supernatant for 1 hr at 37°C. Bound monoclonal antibodies were detected with goat anti-mouse immunoglobulin (Ig) conjugated with horse radish peroxidase (P0447, Dako, Denmark). If chimeric mouse-human versions of the tested antibodies were used they were detected with Goat anti-Human IgG, IgM, IgA (H+L) Secondary Antibody, HRP (#31418, Thermo Fisher Scientific) The reaction was developed with TMB one (4380A, Kementec Solutions A/S, Demark) as a peroxidase substrate and stopped with 50 μl of 0·25 M H2SO4. Absorbance at 450 nm was measured using a PowerWave HT (Bio-Tek). Readouts with an absorbance value of at least twice the value of the negative controls (PBS) were considered positive. ELISA positive hybridoma cultures were further subcloned in soft agar according to the procedure described previously.33 After purified hybridoma clones were obtained, the antibodies isotypes were determined by ELISA using a mouse Ig isotyping kit (ISO-2, SIGMA).
Determination of kinetic characteristics of the antibody-RBD interaction by surface plasmon resonance (SPR)
Kinetic characteristics and affinities of mouse and chimeric antibodies to RBD was measured by SPR. A BIACORE3000 instrument with a CM5 sensor chip (Cytiva, USA) was used for the SPR assays. Amine-coupling reagents (EDC, NHS, ethanolamine pH 8·5), were obtained from Cytiva. Typically, 10 000 RU (response units) of polyclonal anti-mouse antibody (No. Z 0420; Dako, Denmark) or polyclonal anti-human antibody (I2136, Merck, Germany) was coupled simultaneously in four flow cells. One of them was used as a reference cell in kinetic measurements. The experiments were done at 25°C in PBS (pH 7·4) supplemented with 0·005% of Tween 20 (PBS-P). In each analysis cycle, three different antibodies were captured in three analytical flow cells to reach an immobilization level of 100-150 RU. Subsequently, four serial dilutions of RBD protein ranging from 3·125-25 nM or the running buffer (PBS-P) as a control were injected in duplicates at a flow rate of 100 µl/min over the sensor chip. Regeneration of the chip surface was accomplished by a 6 s injection of 100 mM HCl. Kinetic binding data were double referenced by subtracting the reference cell signal and the PBS-P buffer response and fitted by BIA evaluation software 4.1 (Biacore AB) to a 1:1 interaction model as described previously.34 On- and off-rates were fitted locally, maximal response was fitted globally and the bulk response was set to zero. The equilibrium dissociation constants KD of antibody-RBD complexes were calculated from the averaged association and dissociation rate constants. The final error of affinities was propagated from errors of the experimental rate constants used for KD calculation.
Epitope binning by competitive ELISA
For analysis of cross-competition between RBD/S specific mAbs, competitive ELISA was developed. Fab fragment of anti-mouse IgG (0·5 µg/ml, 50 μl/well in PBS) was immobilized overnight on microtiter ELISA plate at 37°C. After blocking with PBS-0·1% Tween 20 to reduce nonspecific binding, the plates were washed with PBS-0·05% Tween 20 and incubated with 50 μl/well of monoclonal antibody 1 (0·6 µg/ml) for 1hr at 37°C. Horseradish peroxidase (HRP)-conjugated RBD (#4120, Kementec Solutions A/S, Denmark), at a concentration of 20 ng/ml, was pre-incubated with monoclonal antibody 2 (at concentrations of 0·25 – 2 µg/ml) for 1 h at room temperature, then the mixture was added to the ELISA plate and incubated at 37°C for 1 h. Binding of HRP-conjugated RBD to immobilized antibody 1 was detected with TMB one substrate (4380A, Kementec Solutions A/S, Demark) and stopped with 50 μl of 0·25 M H2SO4. Absorbance at 450 nm was measured using a PowerWave HT (Bio-Tek) plate reader.
RBD-ACE2 binding inhibition assay
With the aim to select the antibodies blocking the interaction of receptor binding domain (RBD) with angiotensin-converting enzyme 2 (ACE2) receptor, ELISA-based inhibition test was used. 96-well microtitre plates (Nunc MaxiSORP, # 44-2404-21, Thermo Fisher Scientific) were coated with human ACE2 recombinant protein at 350 ng per well in 50 μl of phosphate-buffered saline (PBS) overnight at 4°C, followed by blocking with PBS supplemented with 0·1% Tween-20 (PBS-T). Horseradish peroxidase (HRP)-conjugated RBD at a concentration of 0·4 μg/ml was pre-incubated with tested hybridoma culture supernatant producing mAbs (diluted 1:6 with PBS-T) for 1 h at room temperature (final volume of 50 μl), followed by addition into the ELISA plates coated with ACE2 and incubated for 1 h at 37°C. Unbound HRP-conjugated RBD were removed by five washes with PBS-T. A colorimetric signal was developed by the enzymatic reaction of HRP with a chromogenic substrate TMB one (4380A, Kementec Solutions A/S, Denmark). An equal volume of stop solution (0·25 M H2SO4) was added to stop the reaction, and the absorbance of the product at 450 nm was measured using a PowerWave HT microplate reader (BioTek). Inhibition activity of the antibody was determined as follows:
(1 – (absorbance of antibody/absorbance of positive control) × 100.
S-ACE2 binding inhibition assay
HEK 293T/17 cells (ATCC Cat# ACS-4500, RRID:CVCL_4V93) stably expressing human ACE2 protein (HEK 293T/17-hACE2, described above) were seeded at 60-70% plating density in 48-well plates and cultivated O/N at 37°C, 5% CO2 in a humidified incubator in DMEM (11960-044, Thermo Fisher Scientific) supplemented with 10% (v/v) foetal calf serum, 2 mM L-glutamine, 100 units/mL penicillin/streptomycin (all from Life Technologies Invitrogen, Carlsbad, CA, USA) and 100 µg/ml hygromycin (Thermo Fisher Scientific). Recombinant S protein was labelled with Alexa Fluor™ 546 (A-20002, Thermo Fisher Scientific) according to the manufacturer's recommendations. Labelled S protein (40 ng/ml) was preincubated with tested hybridoma culture supernatants producing mAbs (diluted 1:50 in DMEM) for 30 min at 37°C. Then the preincubation mixtures were added to HEK 293T/17-hACE2 cells and incubated for 2 hrs at 37°C, 5% CO2 in a humidified incubator. Subsequently, cells were gently re-suspended in 500 µl PBS, transferred into flow cytometry tubes and immediately evaluated for the S protein internalization by flow cytometry (BD LSRFortessa II, Becton Dickinson). Cells were gated using forward and side scatter to exclude cellular debris. The FSC-H/FSC-A gating approach was used to eliminate doublets. Measurements were recorded as mean fluorescent intensity of Alexa Fluor 546. Approx. 10 000 cells were analysed for each microtiter plate well. Fluorescence data were normalized to cells treated with the labelled S protein pre-incubated with a hybridoma culture supernatant from a clone producing an irrelevant antibody. IC50 values for particular experiments were calculated by nonlinear regression analysis using GraphPad Prism (GraphPad Software Inc.).
Pseudovirus SARS-CoV-2 neutralization assay
For testing the neutralization ability of monoclonal antibodies, the procedure adapted according to Nie et al., (2020) was used. Briefly, the recipient cells HEK293/17-hACE2 were seeded in 96-well plate and cultured overnight in a humidified CO2 incubator (at 5% CO2 and 37°C) in 100 µL of normal growth medium (DMEM, 11960-044, Thermo Fisher Scientific) with 10% of FCS. In parallel, the optimal S-typed pseudoviral particles (S-PVPs) dilution was determined. “SARS-CoV-2 Pseudoviral Particles Spike” from MyBiosource (San Diego, CA, USA) were used in the assay (Cat. No. MBS434275, Lot # C932EL). PVPs concentration of the material was ≥ 1 × 107 (determined from a TEM scan by the manufacturer). PVPs were aliquoted and stored at -80˚C.
Optimal dilution of the original PVPs stock for testing of the antibody neutralization efficiency was set in the range from 1:100 to 1:200, since this dose results in 50% of recipient cells infected with S-PVPs. The neutralization assay was performed as follows: the serially diluted selected monoclonal antibodies were mixed in DMEM with a defined amount of S-PVPs. The mix was incubated at 37°C in a CO2 incubator for 1 hr, then added to the cells in a 96 well–plate (50 μl of mix per well) in triplicates. After 48 hrs of incubation of the recipient cells with the S-PVP mix the luciferase activity was measured using a luminometer (Fluoroskan Ascent® FL, Labsystems) and half maximal effective concentration was calculated for the measured monoclonal antibodies. The measurement of relative luminescent units (RLU) and comparison to the positive control (a control antibody not recognizing S protein) and a negative control (cell extracts with no S-PVPs or extracts from the cells infected with S-deficient PVPs) was performed in all experiments.
SARS-CoV-2 plaque reduction neutralization assay
The plaque reduction neutralization test (PRNT) with the live SARS-CoV-2 virus (performed in a containment laboratory of Biosafety level 3 by the Department of Virus Ecology at the Biomedical Research Center of the Slovak Academy of Sciences, Slovak Republic) was used as a test for determination of neutralization capacity for the coronavirus of mAbs elicited by immunization with the Spike protein or its RBD. Serial dilutions of supernatants from the hybridoma clones producing the selected mAbs were incubated with 100 plaque-forming units of SARS-CoV-2 at 37°C for 2 hrs. MAb–virus mixtures were then added to Vero E6 cell (ECACC Cat# 85020206, RRID:CVCL_0574) monolayer in 24-well plates and incubated at 37°C for additional 1 h. After incubation, cells were overlaid with 2% (w/v) carboxymethylcellulose (9004-32-4, Sigma Aldrich) in Dulbecco's modified essential medium (DMEM Low Glucose w/ Stable Glutamine w/ Sodium Pyruvate; LM-D1102/500; Biosera) supplemented with 5% foetal bovine serum (SUPERIOR Supplemented FBS, EU approved, S0615-500ML, Sigma Aldrich). Plates were incubated at 37°C for 72 hrs. Then, the cells were fixed for 30 min with 4% formaldehyde in a phosphate-buffered saline (PBS). After incubation the SARS-CoV-2 plaques were visualized by staining with 0·5% crystal violet (1.01408.0025, Merck) at room temperature for 10 min. After washing the wells with water, the number of plaques was counted. The antibody neutralization activity was determined as the reciprocal of the highest dilution resulting in an infection reduction of 50% (PRNT50). Data were fitted to a logistic 4-parameter sigmoidal dose response curve using GraphPad Prism (GraphPad Software Inc.), the goodness of fit was R2>0.9, except for AX266 on B1.1.7 (R2=0·7888), AX96 on B.1.1.7 (R2=0·8766), AX290ch on B1.1.7 (R2=0·8730) and AX677ch on B.1.351 (R2=0·8598).
Virus escape from neutralizing antibodies
For the first round of the escape mutant selection, SARS-CoV-2 virus at a high multiplicity of infection (MOI) of 0·5 was added to each dilution of mAbs (5-fold serial dilution starting at 50 µg/mL) and incubated at 37°C for 1hr. The mixture was then transferred to 12-well plates containing sub-confluent Vero E6 cell monolayer. Plates were incubated three to four days at 37°C and 5% CO2 and examined for the presence of a cytopathic effect (CPE). Supernatants from the first wells in the mAb dilution series with the evident CPE (approx. 30% cells affected) were collected. For the second passage, these supernatants were diluted 1:5 in DMEM (LM-D1102/500; Biosera) and pre-incubated for 1hr at 37°C in the presence of serially 5-fold diluted mAbs starting at 100 µg/mL. After incubation at 37°C for 1hr, the mixture was transferred to 12-well plates with Vero E6 cells and further incubated three to four days at 37°C. Total RNA, including the viral RNA, was extracted from the cells in the first wells of the mAb dilution series that show CPE and used for the sequencing.
To evaluate the impact of the escape mutations on the viral fitness, a co-infection study was performed where the isolated mutant and parental viruses were mixed in approx. 1:1 ratio and used for infection of the Vero E6 cells in the absence of any antibody selection pressure. For this purpose, the viral mutants isolated in the virus escape experiment, labelled A1 (containing the mutation Spike S477R), B11 (containing the mutation Spike T345N), and C25 (containing the insertion E 13IV->IACLV) were mixed with the parental SARS-CoV-2 strain Slovakia/SK-BMC5/2020 in order to obtain equal amounts of infectious viruses. To verify the obtained mutant:parent ratio, part of the prepared mixtures was subjected to the RNA extraction and nanopore sequencing. The mixed viral stocks were further diluted in DMEM and used for infection of Vero E6 cells with the MOI of 0·001 (100 pfu per 100,000 cells). The experiment was performed in 24-well plates in triplicates. The virus inoculum of 100 µl was incubated with the cells (100,000 cells per well) for 60 minutes at 37°C. The inoculum was then removed and 1mL/well of fresh cultivation medium was added to the cells. After three days of incubation at 37°C, the cell culture supernatants were harvested. From each replicate, 140 µl of the supernatant was used for RNA extraction with the Viral RNA Minikit (#52904, QIAGEN) according to manufacturer's instructions. The isolated RNA was then subjected to the nanopore sequencing analysis as described below. Changes in the proportion of the mutant virus after three days of cultivation in the absence of the monoclonal antibodies were considered as a read-out indicator of improved or reduced viral fitness of the mutant viruses in comparison to the parental virus.
Nanopore sequencing of viral RNA
The sequencing libraries were constructed essentially as described in the Eco PCR tiling of SARS-CoV-2 virus with native barcoding protocol (Oxford Nanopore Technologies), except the amplicons spanning the SARS-CoV-2 genome sequence were generated using the ∼2·5-kbp primer panel35 in which the rightmost primer pair was replaced by corresponding pair from the ∼·0-kbp panel.36 For analysis of the escape variants, the amplicons were generated using the ∼2·0-kbp panel, in which the primers P2_hCoV_F6_2kb and P1_hCoV_F13_2kb were replaced by the oligonucleotides 5‘-CGATATTACGCACAACTAATGGTGACT-3‘ and 5‘-AAAGGAGTTGCACCAGGTACAG-3‘, respectively. The primers were custom synthesized by Microsynth (Microsynth Austria GmbH). Briefly, purified RNA (8 µl) was converted into cDNA using a LunaScript RT SuperMix Kit (E3010, New England Biolabs) and used as a template in two separate amplification reactions generating odd- and even-numbered tiled amplicons. The PCR was performed using a Q5 Hot Start High-Fidelity 2X Master Mix (M0494, New England Biolabs) and the cycling conditions were: 30 sec at 98°C (initial denaturation), followed by 30 cycles of 15 sec at 98°C (denaturation) and 5 min at 65°C (combined annealing and polymerization). The overlapping amplicons were pooled and purified using 0·5 volume of AMPure XP magnetic beads (A63881, Beckman Coulter). The ends of amplicons were treated with a NEBNext Ultra II End repair / dA-tailing Module (E7442L, New England Biolabs) and barcoded using a Native Barcoding Expansion 96 kit (EXP-NBD196, Oxford Nanopore Technologies) and a Blunt/TA Ligation Master Mix (M0367L, New England Biolabs). Barcoded samples (96) were pooled and purified using 0·5 volume of AMPure XP magnetic beads. The AMII sequencing adapter (EXP-AMII001, Oxford Nanopore Technologies) was ligated to about 300 ng of barcoded pools using Quick T4 DNA ligase (M2200, New England Biolabs) and the library was purified using 0·5 volume of AMPure XP magnetic beads. About 90 ng of the library was loaded on an R9.4.1 flow cell and the sequencing was performed using a MinION Mk-1b device (Oxford Nanopore Technologies).
Variant calling
Raw nanopore data was base called and demultiplexed using Guppy v.4.4.1. Barcodes at both ends were required in demultiplexing. Variants were called using the ARTIC pipeline (https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html), which internally filters reads based on quality and length, aligns them to the reference (Wuhan-Hu-1 isolate, NC_045512) using minimap2 (https://github.com/lh3/minimap2), trims primers, and calls variants using Nanopolish (https://github.com/jts/nanopolish). All variants reported in Table S1 were determined based on read coverage of at least 380. All new nonsynonymous mutations were supported by at least 90% of the analyzed reads overlapping a particular site. Note that due to higher error rates of nanopore sequencing, even fixed variants typically do not achieve 100% support.
Immunocytochemistry
HEK293T/17-hACE2 cells were plated on cover glass pre-coated with rat-tail collagen, type I (C3867, Sigma-Aldrich) and cultivated for 24 h in DMEM with 10% FCS. Cells were fixed with 4% paraformaldehyde and labelled with anti-human ACE2 antibody at 10 µg/ml (cat. # 600-401-X59; Rockland Immunochemicals), followed by goat anti-rabbit IgG secondary antibody at 1/500 dilution (Alexa Fluor 488; #A-11008; Invitrogen). The samples were mounted in Fluoroshield™ medium with DAPI (Sigma-Aldrich). Images were captured by LSM 710 confocal microscope (Zeiss, Jena, Germany).
Hydrogen Deuterium Exchange Mass Spectrometry (HDX-MS)
RBD and AX290 or AX677 were dissolved in non‐deuterated PBS buffer (Phosphate Buffered Saline, pH 7·4) and digested individually with flow through an immobilized nepenthesin-2 column, 2·1 mm X 20 mm (Affipro, Praha, Czech Republic) for peptide mapping. Prior to the HDX experiments, RBD and the AX290 or AX677 were incubated for 1 h to allow complex formation. To initiate HDX, proteins were diluted with D2O buffer (10 mM phosphate buffer, D2O, pD 7·0). Different aliquots were submitted to HDX for three times: 0·3, 2, 20, and 120 min. The reaction mixture was then quenched by adding quenching buffer (6M urea, 2M thiourea and 0·4 M TCEP, pH 2·3). The deuterated mixture was then digested online by using the same nepenthesin-2 column and the resultant peptides were desalted with flow through on a trap column (Phenomenex UHPLC fully Porous Polar C18, 2·1mm, Torrance, CA, USA) with 0·4% formic acid in water. The eluted peptides were further separated on an analytical column (Luna® Omega 1·6 μm Polar C18 100 Å, 100 × 1·0 mm, Torrance, CA, USA), by using a gradient of acetonitrile and water with 0·1% formic acid. Peptides were introduced to a timsTOF (Bruker, Brno, Czech Republic) mass spectrometer for mass analysis. The peptide‐level deuterium uptakes were calculated by using MS Tools software.37
In vivo antibody testing in ACE2 humanized mouse model
This study was carried out in a strict accordance with the Czech national laws and guidelines on the use of experimental animals and protection of animals against cruelty (Animal Welfare Act No. 246/1992 Coll.). The protocol was approved by the Committee on the Ethics of Animal Experiments of the Institute of Parasitology, Institute of Molecular Genetics of the Czech Academy of Sciences, and the Departmental Expert Committee for the Approval of Projects of Experiments on Animals of the Academy of Sciences of the Czech Republic (permits 82/2020 and 101/2020).
Humanization of mice by application of AAV-hACE2 viral particles
The preparation and susceptibility to SARS-CoV-2 infection of the ACE2 humanized (AVV-hACE2) mouse model using pHelper (Cat. No 320202, Cell BioLabs), pAAV2/9n (RRID:Addgene_112865) and AAV-hACE2 were described previously.38 4 × 109 viral genome copies in 40 ul of PBS were applied to 11-week-old C57BL/6N female mice by forced inhalation. Mice were anaesthetized by intraperitoneal injection of ketamine and xylazine (0·1 mg per g body weight (Biopharm) and 0·01 mg per g (Bioveta), respectively).
Testing of antibodies on ACE2 humanized mice
The AAV humanized mice were moved to BSL3 animal housing facility on day eight after transduction with AAV-hACE2, and after the necessary acclimatization (three days) the treatment study was started. The mice were randomly assigned to cages and the cages were then randomized into groups. AX290 and AX677 antibodies, their mixture and an isotype antibody control (DC8E8 antibody against pathological neuronal protein tau39) were administered subcutaneously (high-dose experiment; 1·25 mg/0·2 ml PBS, 10 animals per group) or intraperitoneally (low-dose experiment; 0·5 mg/0·2 ml PBS, 10 animals per group) twelve days after applying the AAV-hACE2 virus.
24 hrs later the mice were infected intranasally with SARS-CoV-2 (1 × 104 plaque-forming units, strain SARS-CoV-2/human/Czech Republic/951/2020, isolated from a clinical sample at the National Institute of Health, Prague, Czech Republic, and passaged in Vero E6 cells) in a total volume of 50 μl DMEM under isoflurane anesthesia as described previously.38 This day was marked as the Day zero and used as a reference day for all analyses of the treatment efficacy. Mice were monitored for health status and weighed daily for the remainder of the experiment. It has been noted, that during the intraperitoneal application of the antibody mix (AX677+AX290) in the low-dose group one animal urinated, suggesting that the urinary bladder was accidentally injected and antibody was incorrectly applied. For this reason, this animal was excluded from the experiment resulting in four animals being monitored in this group.
Terminal necropsy
On the day three and seven post-infection selected groups of mice (five animals per group per collection day) were sacrificed and lung tissues collected for analyses. One lobe was homogenized in sterile PBS using 30 Hz sonication for 3 min, centrifuged at 10,000 xg for 10 min at 4°C, and resulting suspension used for viral plaque titration analysis (day three post infection) and for viral RNA analysis (days three and seven post infection; high-dose experiment only). The other lung lobe was fixed in 4% paraformaldehyde in PBS and stored in a fridge at 2-8°C till further histopathological processing (high-dose experiment only).
Virus titer plaque assay
Plaque assays were performed in Vero E6 cells (ATCC CRL-1586; mycoplasma-free) grown at 37 °C and 5% CO2 in DMEM supplemented with 10% FBS, and 100 U/ml penicillin, 100 μg/ml streptomycin, and 1% L-glutamine using a modified version of a previously published protocol.40 In brief, serial dilutions of virus were prepared in 24-well tissue culture plates and cells were added to each well (0·6 × 105–1·5 × 105 cells per well). After 4 h, the suspension was overlaid with 1·5 % (w/v) carboxymethylcellulose in DMEM. Following a 5-day incubation at 37 °C and 5% CO2, plates were washed with phosphate-buffered saline and the cell monolayers were stained with naphthol blue black. The virus titer was expressed as plaque-forming units per gram of tissue with the limit of detection of 2·14 log pfu/g of tissue.
Viral RNA analysis
RNA was isolated from tissue homogenates (collected from the high-dose experiment) using the QIAmp Viral RNA mini kit (Cat. no. 52906, Qiagen) following manufacturer's instructions. RNA was eluted in 50 µl of the elution buffer. Viral RNA was quantified using EliGene COVID19 Basic a RT (#90077-RT-A, Elisabeth Pharmacon) according to the manufacturer's instructions. A calibration curve was constructed from four dilutions of a sample quantified using Quanty COVID-19 kit (#RT-25; Clonit) according to the recommendations from the manufacturer. All real-time PCR reactions were performed using a LightCycler 480 (Roche).
Histopathological processing
The fixed lung tissues (collected from the high-dose experiment) were processed on Leica ASP6025 automatic vacuum tissue processor and embedded in Leica EG1150 H+C embedding station. For sectioning, Leica RM2255 rotary microtome (2 µm sections) was used. Samples were stained with Leica ST5020 + CV5030 stainer and coverslipper. Hematoxylin (Cat.no. HHS32-1L, Sigma-Aldrich)/eosin (Cat.no. X883.2, ROTH) staining was used as a standard descriptive histopathological staining according to an internal SOP. To assess the presence of macrophages a rabbit anti-mouse F4/80 monoclonal antibody (Cat.no.70076, Cell Signalling Technology) was used as primary antibody. This is a well-established marker for mouse macrophages. The lung sections (thickness 4-5 µm) were deparafinized in Multistainer Leica ST5020 (Leica Biosystems). Antigens were retrieved by heating the slides in a citrate buffer pH 6 (Cat.no. ZUC028-500, Zytomed – Systems, Germany). Endogenous peroxidase was neutralised with 3% H2O2 (diluted from 30% H2O2, Cat.no. 23980-11000, with methanol, Cat.no. 21210-20005, Penta Chemicals, Czech Republic). Sections were incubated for 1 hour at RT with a 1:800 dilution of the primary antibody. After washing they were incubated with anti-rabbit secondary antibody conjugated with HRP (Cat.no. ZUC032-100, Zytomed – Systems, Germany). Staining of the sections was developed with a diaminobenzidine substrate kit (Cat.no. K3468, DAKO - Agilent) and sections were counterstained with Harris Hematoxylin (Cat.no. HHS32-1L, Sigma Aldrich – Merck) in Multistainer Leica.
Statistical analysis
Statistical methods used for the evaluation of the data are described in the figure legends. All evaluations were performed using GraphPad Prism Version 6.07 (GraphPad Software Inc.), if other software was not explicitly mentioned.
Role of funding source
The funders approved the study design and data collection, participated in data analysis, interpretation of the data, and writing the report. The corresponding authors had full access to all the data and had final responsibility for submission of the report for publication.
Results
Identification of monoclonal antibodies neutralizing SARS-CoV-2
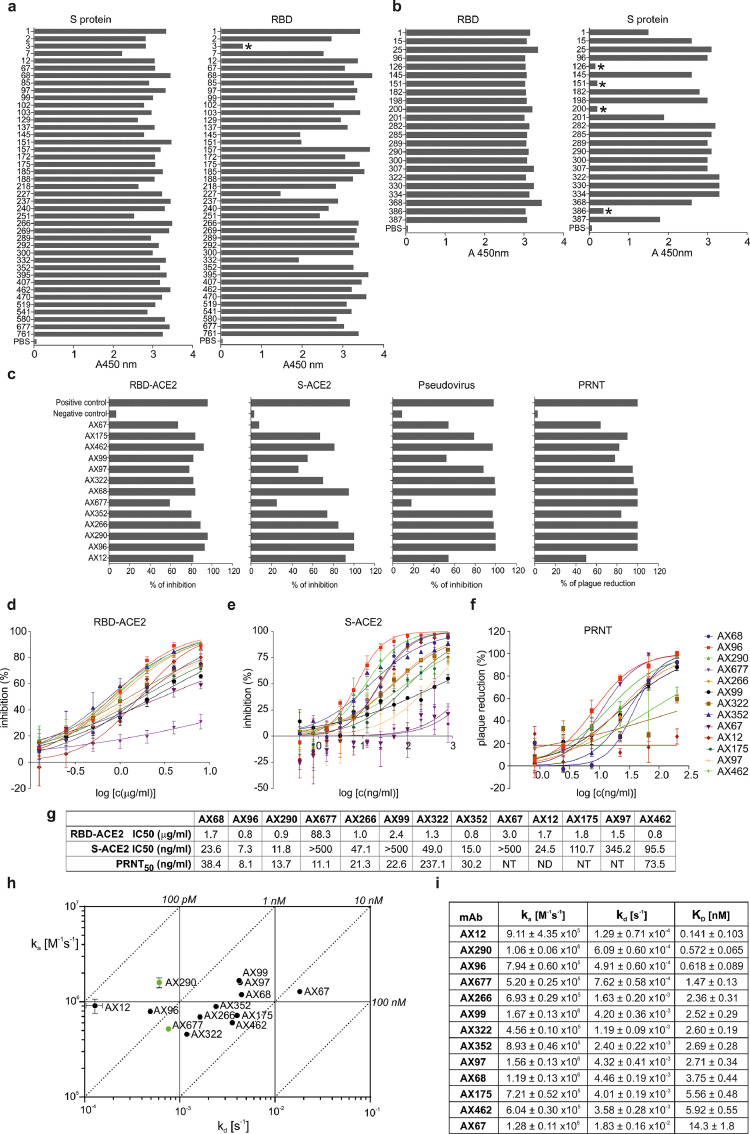
To isolate fully matured antibodies with high neutralizing potency against live SARS-CoV-2 virus, we immunized mice either with the recombinant S protein or its RBD. Screening of the antibody-expressing mouse hybridoma clones generated from the S-immunized mice revealed that all except one (97·6%) target RBD of the SARS-CoV-2 S protein (Figure 1a). Nineteen out of 23 hybridoma clones from RBD-immunized mice produced antibodies recognizing the S protein (Figure 1b). Out of the 65 RBD- and S-positive hybridoma clone supernatants, thirteen inhibited the interaction between RBD and ACE2 in the competition ELISA, twelve inhibited the interaction between the S protein and ACE2 expressed on the surface of HEK293T/17-hACE2 cells (the assay mimics the first step in the infection of the human cells by SARS-CoV-2), twelve inhibited cell entry of replication-deficient mouse leukemia virus (MLV) pseudotyped with the SARS-CoV-2 spike protein, and thirteen neutralized the live authentic virus (Slovakia/SK-BMC5/2020 isolate, B.1 lineage) in PRNT (Figure 1c).
Figure 1.
Binding characteristics and inhibition activities of monoclonal antibodies specific to SARS-CoV-2 S/RBD.
(a) Immunoreactivity of hybridoma clones culture supernatants derived from mice immunized with the spike (S) protein of SARS-CoV-2 and with RBD of Spike (b) in ELISA against the pre-fusion stabilized and trimerized S protein ectodomain (“S protein”) or RBD (“RBD”). Asterisks mark non-binders.
(c) Blocking of interaction between RBD and ACE2 in a competition assay based on ELISA (RBD-ACE2), inhibition of the S protein internalization by cells expressing ACE2 (S-ACE2), inhibition of cell infection by MLV pseudotyped with SARS-CoV-2 spike protein (Pseudovirus) and plaque reduction neutralization test performed with an authentic SARS-CoV-2 virus isolate Slovakia/SK-BMC5/2020 (PRNT). Experiments were done with hybridoma culture supernatants adjusted with fresh cell culture medium to the same mAb concentration and then diluted for the assays as follows: RBD-ACE2 competitive ELISA 1:6 in PBS-T; S-ACE2 cell assay 1:50 in DMEM; pseudoviral assay 1:25 in DMEM; live virus PRNT 1:50 in EMEM. Positive control: serum of a mouse immunized with the S protein, diluted 1:200; Negative control: irrelevant mAb. All experiments are an average of at least two measurements, pseudoviral tests were all done in tetraplicates.
(d) The purified monoclonal antibodies were tested for blocking the RBD-ACE2 interaction in competitive ELISA, the inhibition of ACE2-mediated S protein internalization by HEK293/17-hACE2 cells (e), and plaque reduction neutralization test (f) performed with strain Slovakia/SK-BMC5/2020. The curves are calculated from two replicates using Prism 6 for Windows (GraphPad Software). (g) Summary of assays from d-f. (h, i) Kinetic characteristics of the interactions of RBD with selected neutralizing antibodies. On-rate (ka), off-rate (kd) constants and equilibrium dissociation constant (KD) of individual antibody-RBD complexes (±SD). Green dots mark antibodies AX290 and AX677 selected for further development.
Quantitative analyses of purified 13 best-performing monoclonal antibodies (mAbs) revealed that the majority of them inhibited the interaction of the RBD and ACE2 proteins in competitive ELISA with IC50 of 0·8 – 3·0 µg/ml (Figure 1d, g). MAbs AX96, AX290 and AX352 had the strongest inhibitory effect on the binding and internalization of the S protein by HEK293T/17-hACE2 cells (Figure 1e, g, Figure S1, S2), with IC50 values of 7·3 ng/ml, 11·8 ng/ml and 15·0 ng/ml, respectively. Five antibodies displayed high neutralization potency of the live SARS-CoV-2 virus in PRNT (Figure 1f, g). Importantly, AX677 showed the second highest live virus neutralization activity of all tested antibodies (PRNT50=11·1 ng/ml), although it had relatively limited ability to inhibit S-ACE2 interaction in in vitro assays and poorly inhibited the S-typed MLV pseudovirus.
The neutralizing antibodies bind RBD with nanomolar or sub-nanomolar affinities (Figure 1h). Kinetic SPR experiments revealed that antibodies exhibited a large distribution of dissociation kinetic rates, spanning two orders of magnitude (1·3 × 10−4 – 1·8 × 10−2 s−1; Figure 1h, i; Figure S3). The association kinetics also varied, but to a lesser extent (4·6 × 105 – 1·7 × 106 M−1s−1). Four antibodies with the highest (picomolar) affinities, AX12, AX96, AX290 and AX677, had very slow dissociation rates below 10-3 s-1. Six antibodies showed nearly identical affinities of 2 to 3 nM (AX68, AX97, AX99, AX266, AX322 and AX352), but with contrasting kinetic behaviour. In general, the antibodies exhibit RBD binding affinities similar to or better than antibodies isolated from patient sera.12,25,26
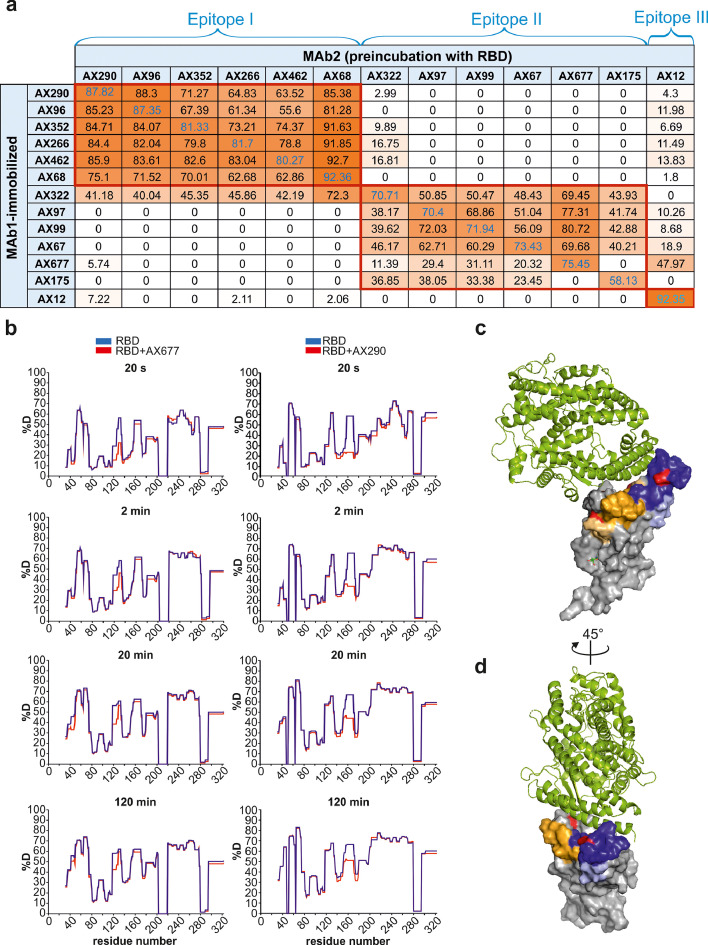
Neutralizing antibodies recognize two immunodominant epitopes on RBD
Mutual competition of the selected antibodies for binding to RBD revealed three non-overlapping antibody epitopes (Figure 2a, Epitopes I, II, III). Epitope I was recognized by the group of six antibodies, which appeared among the best neutralizers of the authentic live SARS-CoV-2 virus. Within this group, the virus neutralization activities highly correlated with their affinities to RBD (R2>0·9, Figure S4). The Epitope II group comprised six antibodies, where AX677 and AX175 did not compete with each other. One antibody (AX12) recognised a non-overlapping independent region (Epitope III).
Figure 2.
The panel of selected neutralization antibodies target three non-overlapping epitopes on RBD.
(a) Competitive ELISA was used for determination of the epitopes on RBD by NAbs. Signal reduction for more than 30 % was considered a positive competition. The experiments were performed in duplicates.
(b) Difference plots depicting the changes in deuterium uptake in the RBD peptides upon binding to AX677 (left panels) and AX290 (right panels). The HDX reaction times are indicated in seconds. Numbers on x-axis represent amino acid positions of the recombinant RBD protein (adding 316 aligns them with the S protein numbering). Each plot represents an average of three technical replicates.
(c, d) The positions of the peptides bound by the antibodies and identified in the HDX experiments are highlighted in the structure of SARS-CoV-2 spike receptor-binding domain bound to ACE2 reproduced from PDB 6M0J[41] . The AX290 binding site is represented by shades of blue, AX677 by orange and yellow. Darker colours represent peptides that exhibit reduction in deuterium exchange (%D) ≥20%, light colours represent peptides with reduction in %D between 5-20%. ACE2 is shown in a green cartoon model, RBD as a grey surface model, mutations N439K (within AX677 binding site) and E484K (within AX290 binding site) are shown in red. The model was rendered using PyMOL Molecular Graphics System, Version 2.5.0a0 Open-Source, Schrödinger, LLC.
AX290 and AX677 showed the highest binding affinities to RBD of all members of the Epitope I and II groups (Fig. 1h, i), respectively, and showed the highest neutralizing activities against the live authentic virus. We applied hydrogen-deuterium exchange coupled to mass spectrometry (HDX) to identify their binding sites on RBD. Deuterium uptake was monitored on 288 unique peptides, covering 89% of the RBD sequence (Figure S5). The peptides protected by AX677 antibody included amino acids 42-48 and 118-139 (RBD numbering) (Figure 2b), which correspond to amino acids 358-364 and 434-455, respectively, of the S protein. The RBD peptides protected by the AX290 antibody encompassed amino acids 151-179 (Figure 2b), which correspond to peptides 467-495, respectively, of the S protein. We used the published model of ACE2-bound RBD from PDB 6M0J41 and highlighted the peptides protected in HDX experiments (Figure 2c, d). The epitope of AX290 is located in the region of RBD that comprises some of the contacts to ACE2, is often targeted by NAbs,2 and overlaps with epitopes of REGN10933 and LY-CoV555.12,42 The epitope of AX677 is located outside of the ACE2-binding interface, which explains why it poorly inhibits ACE2-RBD interaction in the competition ELISA and cellular inhibition assays. The deuterium uptake data suggest that the binding of AX677 partially overlaps with that of REGN10987 and COV2-2130.12,43 The AX677 mode of neutralization might be allosteric or via interference with attachment to other cell surface molecules.
Selection of antibodies that neutralize SARS-CoV-2 variants of concern B.1.1.7 and B.1.351
In order to test the ability of the selected mAbs to bind RBD with the mutations that appear in the current SARS-CoV-2 VOCs, B.1.1.7 (N501Y) and B.1.351 (K417N/E484K/N501Y), we screened the mutated RBDs by ELISA (Figure 3a, b). None of the mutations completely prevented recognition by the antibodies. Individual mutations N501Y, K417N and E484K, and N439K mutation (widespread in Europe), had only a marginal impact on the antibody binding. The combination K417N/E484K/N501Y exhibited either no or only minor influence on the binding of the antibodies of the Epitope I group. The mutations showed practically no influence on antibodies AX99, AX677 and AX97 of the Epitope II group. E484K, however, reduced binding of AX322, AX175 and partially also AX67 to RBD. E484K and N501Y made the highest impact on the binding of these mAbs to RBD containing the triple mutation N501Y/E484K/K417N present in the variant B.1.351.
Figure 3.
Effect of RBD mutations on immunoreactivity of monoclonal antibodies in ELISA.
(a, b) Binding of the antibodies to the RBD carrying the individual mutations (N501Y, N439K, E484K, K417N) and RBD with triple mutation N501Y/E484K/K417N were analysed. Effectivity of binding for each antibody was expressed by EC50 values. All experiments were done at least in technical duplicates.
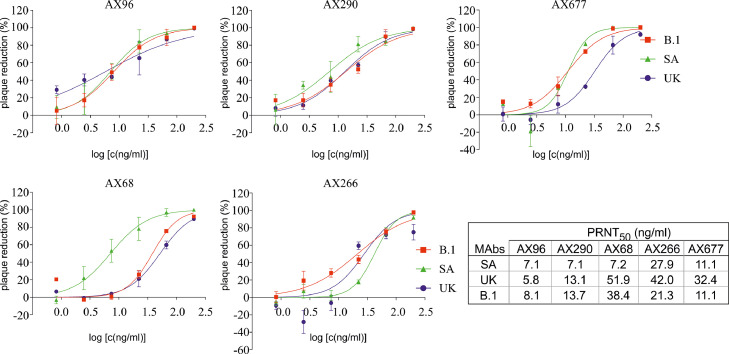
Selected mAbs resistant against RBD mutations were tested for their neutralization activities in the plaque reduction neutralization test using the live authentic SARS-CoV-2 virus B.1 and its VOCs B.1.1.7 and B.1.351 (Figure 4). AX96 and AX290 effectively neutralized all three virus variants. AX677 was the third best NAb, highly efficient against the B.1 and B.1.351 viruses, with slightly reduced activity against B.1.1.7. Interestingly, AX68 showed relatively high activity against the B.1.351 variant with triple mutation in RBD, but 5- to 7-fold lower activity against the B.1 and B.1.1.7 variants, respectively.
Figure 4.
Selected mouse monoclonal antibodies neutralize SARS-CoV-2 variants of concern. Live authentic SARS-CoV-2 virus variants: Slovakia/SK-BMC5/2020 isolate (B.1), B.1.1.7 (UK) and B.1.351 (SA), were pre-incubated with serial dilutions of Abs (250 ng/ml – 2 ng/ml) and then added to Vero E6 cells. Plaque reductions (%) relative to negative control were calculated 72 h post infection. Data are plotted as the mean from two wells of one experiment. PRNT50 mAb concentrations are shown in the table. The experiments were done at least in triplicates.
Combination of antibodies with non-overlapping epitopes prevents rapid mutational escape of the authentic SARS-CoV-2 virus
SARS-CoV-2 virus has accumulated mutations resulting in compromised activities of some vaccines and therapeutic antibodies.42,44 We, therefore, examined the ability of the authentic virus to generate escape mutations under the selection pressure of AX290 and AX677, the best NAbs with non-overlapping epitopes, alone and in combination (Figure 5a, b).
Figure 5.
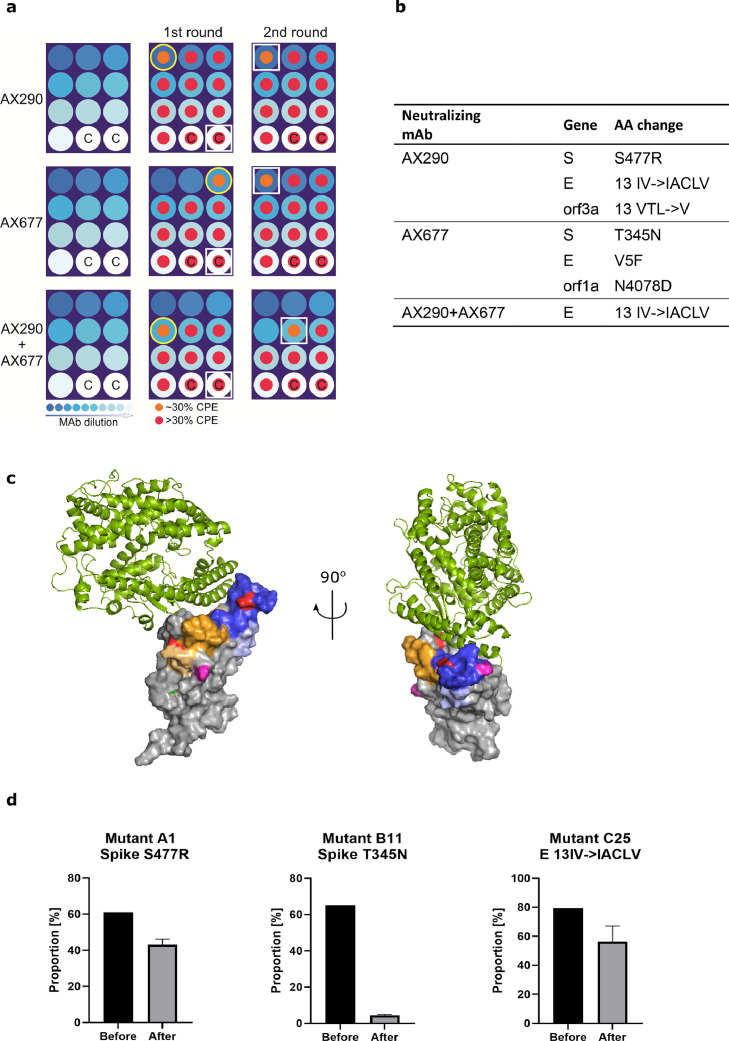
A combination of AX290 and AX677 prevents rapid mutational escape of authentic live SARS-CoV-2 virus.
(a) A schematic diagram of microplate well allocations in the SARS-CoV-2 escape mutants experiment. MAbs were serially five-fold diluted starting with 50 µg/mL (left panels), pre-incubated with live SARS-CoV-2 (Slovakia/SK-BMC5/2020) virus (diluted to MOI=0·5) and added to Vero E6 cells (1st passage, middle panels). Culture medium from the first wells of the mAb dilution series where CPE appeared (indicated with the yellow circles) were used for the 2nd passage (right panels). White squares highlight wells with CPE where virus genomes were sequenced. C - control wells with virus without mAbs were sequenced to monitor possible tissue culture adaptations.
(b) The table shows amino acid changes resulting from nonsynonymous mutations in the virus genome that appeared under selection pressure of mAbs. Combination of the two antibodies prevented appearance of escape mutations in the Spike protein.
(c) The positions of the amino acids that were mutated in the viruses that escaped from the neutralization by the antibodies are highlighted in pink in the structure of SARS-CoV-2 Spike receptor-binding domain bound to ACE2 (reproduced from PDB 6M0J, see also Figure 2c, d). T345 is located close to the AX677 binding site (orange and yellow, the structure on the left), S477 is located in the binding site of AX290 (blue, structure on the right). (d) Changes in the proportion of the mutant viruses in the mixture with the parental virus before and after three days of co-cultivation with Vero E6 cells in the absence of the monoclonal antibodies determined by nanopore sequencing. The mutant viruses were arbitrarily named A1, B11 and C25 for the escape viruses from AX290, AX677 and AX290+AX677 mix, respectively. Mean and SD are shown from three separate passages of the viral mixtures.
It has been shown that S-typed recombinant viruses might not fully recapitulate the complex behaviour of the authentic SARS-CoV-2 as measured by neutralizing activities of tested antibodies.45 We have also observed that our highly neutralizing antibodies behave differently on S-pseudotyped MLV, AX677 did not neutralize it while AX290 did (Fig. 1c). Therefore, we have used live authentic virus isolate Slovakia/SK-BMC5/2020, that represents strains circulating in Europe in spring 2020, for the mutational escape. The virus particles, preincubated with serially diluted antibodies (individual or their 1:1 mixture), were used to infect Vero E6 cells. When the cytopathic effect of the escaping viruses became evident, the cell culture media with the escaping viruses (wells with approx. 30% cells affected) were collected, again preincubated with the respective serially diluted antibodies and added to fresh Vero E6 cells for another round of mutant virus amplification (Figure 5a). Sequence analysis of the viruses that escaped from the neutralization by the individual antibodies revealed only one mutation in the S protein per antibody: S477R substitution for AX290, and T345N substitution for AX677 (Figure 5b). Additional mutations were detected in the envelope protein, orf3a and orf1ab, but their contribution to the viral escape from the antibodies needs further evaluation (Tab. S1).
The S477R substitution that allowed escape of the virus from AX290 is located in the region of RBD that was identified by HDX as the contact site of the antibody (Figure 5c). It was found in 0.025% of the sequenced SARS-CoV-2 isolates so far (https://www.gisaid.org/hcov19-mutation-dashboard/, database version as of Dec 21, 2021, 6055957 sequences analysed). The alternative substitution S477N that is more widespread in circulating viruses (1.482%) was not found in the sequenced viral genomes.
The mutation T345N present in the virus mutant that escaped from AX677 is positioned close to the peptides protected by the antibody in the HDX-MS experiment, but not directly within their sequence (Figure 5c). It is a very rare mutation, only one T-N substitution at the position 345 of spike has been found in circulating viruses, and only 582 substitutions of T345 were detected in the GISAID database (130 if the conservative T-S substitutions are excluded). It is possible that this region of Spike RBD is less targeted by human immune repertoire resulting in a low selection pressure, or mutations there compromise the viral fitness. The latter is supported by data from the deep mutational scanning of the S protein, which showed that amino acid exchange T-N at the position 345 reduced expression levels of the protein.46
The impact of the escape mutations on the viral fitness was directly tested in a co-infection study where the isolated mutant viruses were mixed with the parental virus isolate Slovakia/SK-BMC5/2020 at the 1:1 ratio and used for infection of the Vero E6 cells in the absence of antibody selection pressure. After a single passage of cultivation of the cells for three days, the cell culture medium was harvested and proportion of the two viruses was analysed by nanopore sequencing. The analyses showed that T345N greatly reduced replication capacity of the virus and was almost eliminated from the pool (Figure 5d, Table S2, Mutant B11). The AX290 escape virus (Mutant A1, containing the S477R mutation in the Spike protein) showed only slight decline in viral fitness indicated by a slight reduction in the proportion of the mutant in the mixture after the cultivation. A similar drop in the fitness was observed for the virus passaged in the presence of both AX677 and AX290 (Figure 5d, Mutant C25). These two escape viruses (A1 and C25) share the insertion 13IACLV in the E protein, which might also account for the observed reduction in their fitness.
The T345N escape mutation has been identified previously using an S-typed VSV pseudovirus for antibody 2H04,47 which also did not compete with ACE2 for binding to the S protein.
Importantly, no escape mutation in the Spike protein was detected with the antibody mixture. The cytopathic effect appeared simply due to the dilution of the antibody concentration below the threshold required to neutralize the virus (625-fold higher dilution than any single mAb in the second passage). This shows that the combined therapy with both AX290 and AX677 would provide a potent barrier impeding viral replication and would protect against the rapid mutational escape.
Mouse-human chimeric versions of AX290 and AX677 maintain their potent neutralizing activities against authentic SARS-CoV-2 VOCs
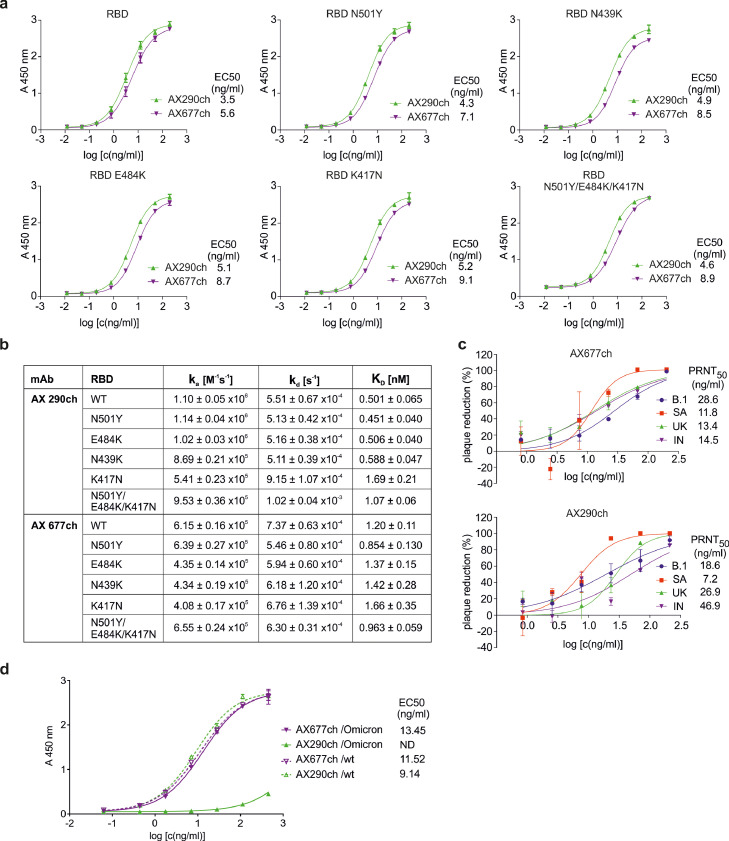
In order to prepare AX290 and AX677 for clinical testing, their variable regions were fused to human kappa and IgG1 constant regions. The chimeric mAbs AX290ch and AX677ch exhibited similar binding and functional properties as parental mouse versions. Both chimeric antibodies bound various mutant forms of RBD in ELISA with similar activities (Figure 6a).
Figure 6.
Functional characteristics of chimeric monoclonal antibodies.
(a) Binding of the chimeric antibodies AX290ch and AX677ch to the Spike protein RBD and RBD carrying the individual and triple mutations were analysed by ELISA and binding to each antibody was expressed by EC50 value. (b) Neutralization activity of the two chimeric antibodies against live authentic SARS-CoV-2 virus variants: Slovakia/SK-BMC5/2020 isolate (B.1), B.1.1.7 (UK), B.1.351 (SA) and B.1.617.2 (IN) determined in plaque reduction neutralization test (%). Data are plotted as the mean from two replicates within one experiment. (c) Kinetic characteristics of the interactions of WT and mutant RBD with selected neutralizing chimeric antibodies. (d) Recombinant ectodomain of wild type S protein and S containing mutations of the lineage B.1.1.529 Omicron (#SPN-C52Hz, ACROBiosystems Inc., Newark, DE, USA) were immobilized on ELISA plate, blocked and incubated with serially diluted chimeric antibodies AX677ch and AX290ch. The bound antibodies were visualized by HRP-labelled anti-human antibodies. The experiment was performed in technical duplicates and data were evaluated by fitting to a sigmoidal four-parameter logistic curve using GraphPad Prism Version 6.07 (GraphPad Software Inc.). ND, could not be determined.
The chimeric antibodies recognised all tested mutants with nanomolar or sub-nanomolar affinities, which guarantees their unmodified neutralizing capacity (Figure 6b, Figure S6). AX290ch maintained its picomolar affinity to all mutants except those containing K417N mutation alone or in combination N501Y/E484K/K417N (3-fold and 2-fold decrease in the affinity, respectively). AX677ch binds N501Y mutation, with 1·4-fold higher affinity than wild type RBD, reaching picomolar value (Figure 6b, Figure S6). Furthermore, this positive effect of the N501Y mutation is propagated into the triple mutant, which combines mutations N501, E484K, and K417N, and binds AX677ch with higher affinity than either of the single mutants E484K and K417N, and even wild type RBD (1·4-, 1·7- and 1·2-fold, respectively).
Most importantly, the chimeras efficiently neutralized live authentic SARS-CoV-2 virus, Slovakia/SK-BMC5/2020 isolate (B.1 lineage), and three variants of concern circulating in the population B.1.1.7 (Alpha), B.1.351 (Beta) and B.1.617.2 (Delta), in PRNT (Figure 6c). The AX677 chimera showed very consistent, slightly higher activities (approx. 2-fold) against all three VOCs compared to the original (B.1) virus. AX290 chimera showed exceptionally high activity against the variant B.1.351, and slightly reduced (two- and three-fold) activities against the original virus B.1 and B.1.1.7. Its neutralization efficiency was reduced (approx. 6-fold) against the B.1.617.2 variant, most likely due to the T478K mutation in the RBD domain of Spike. This mutation lies next to serine 477, where escape mutation against AX290 appeared, and suggests that this region of RBD provides important contacts for the antibody. Nevertheless, these tests confirmed that the neutralizing activity was preserved in the chimeric versions of the antibodies.
The currently emerged variant B.1.1.529 (Omicron) harbours unusually high number of mutations and poses a serious threat due to its high potential to evade both, therapeutic antibodies and immune protection acquired through vaccination or infection.49 We have performed a test of binding of chimeric antibodies AX677ch and AX290ch to the ectodomain trimer of the Omicron S protein (carrying mutations A67V, HV69-70del, T95I, G142D, VYY143-145del, N211del, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F) in ELISA (Figure 6d). Binding of AX677ch was not affected at all by the mutations and might preserve the full neutralization potency against this virus variant. Binding of AX290ch was reduced, most likely due to the mutation S477N.
AX290, AX677 and their combination protect ACE2 humanized mice against SARS-CoV-2
Potential clinical efficacy of the AX290 and AX677 mAbs was tested in an ACE2 humanized mouse model that expresses human ACE2 (hACE2) in the cells of the upper and lower respiratory tract following inhalation of a modified adeno-associated virus (AAV-hACE2).38 These ACE2 humanized mice, upon infection with SARS-CoV-2, suffer from respiratory pathology and disease, progressive weight loss and require culling on day eight post infection.
The ACE2 humanized mice were injected subcutaneously with 1·25 mg of either antibody alone (approx. 50 mg/kg), their equimolar combination, or an isotype control antibody, 24 hours before infection with SARS-CoV-2. After intranasal inoculation of 104 pfu of SARS-CoV-2, mice were weighed daily, and terminal lung tissue sampling was done three and seven days post infection (dpi) to assess viral load, viral RNA and tissue histopathology.
Mice treated with the isotype control antibody experienced deterioration of their overall health status reflected in weight loss starting at day three post-infection (dpi) (Figure 7b). By seven dpi, most mice treated with the isotype control had lost approximately 25-30% of their body weight. In contrast, prophylactic administration of AX290 and AX677, either individually or in a combination, neutralized the effect of the virus and fully prevented the weight loss in all mice, which appeared indistinguishable from the control non-infected mice.
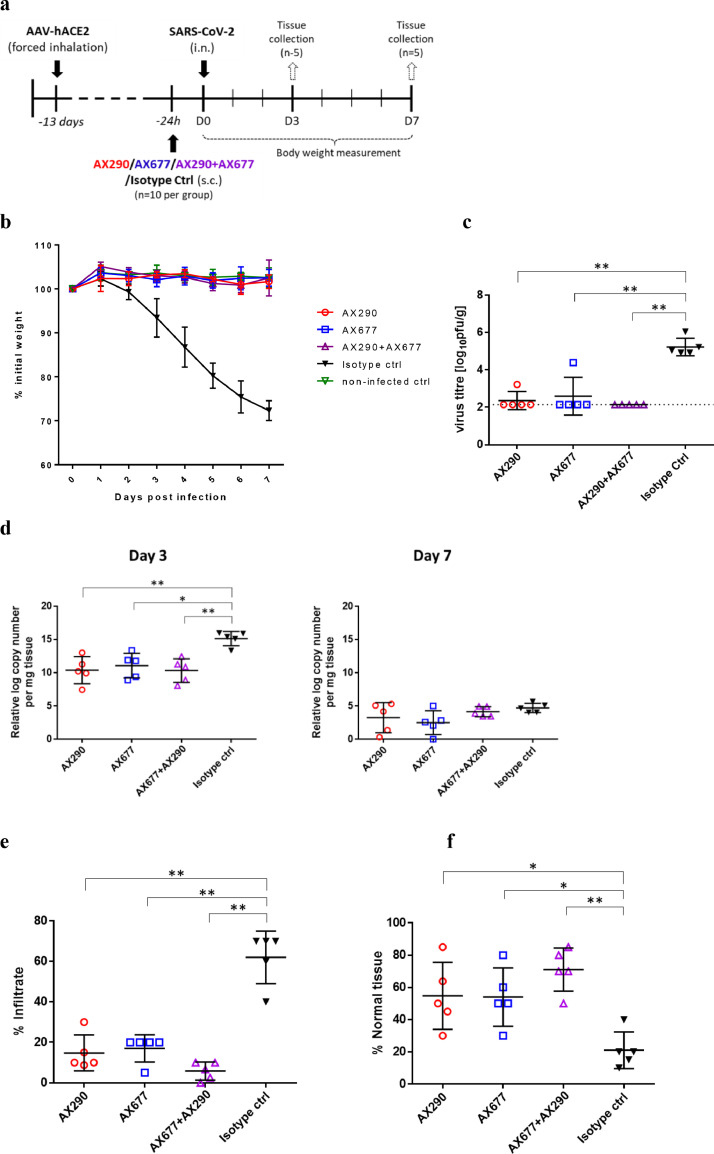
Figure 7.
Antibody prophylaxis against SARS-CoV-2 infection in a mouse model.
(a) Schematic of the in vivo experimental procedures. Wild-type mice were transduced with AAV-hACE2 by forced inhalation. After 11 days, the mice were inoculated subcutaneously (s.c.) with 1·25 mg of the indicated mAbs antibodies one day (-24 h, n=10 animals per group) before being infected intranasally (i.n.) with SARS-CoV-2 (1 × 104 pfu). Health characteristics and body weight were monitored daily for the duration of the experiment. 5 animals per group were sacrificed 3 and 7 days after infection and lung tissue analyzed for viral load (on day 3 only), viral RNA and histopathological changes. (b) Body weight was monitored daily for 7 days (n=5 mice per group). Mean with s.d. is shown. (c) Viral burden in the lungs of mice infected with SARS-CoV-2 (n = 5 per group) as measured 3 dpi by plaque assays. Mean with s.d. is shown. Dashed line indicates the limit of detection (2·14 log pfu/g of tissue). All three treated groups were compared to the Isotype Ctrl group, p values were calculated using non-parametric Mann-Whitney test, in all comparisons p=0·0079 (exact p values were calculated). (d) The analysis of viral RNA copy numbers in the lungs shows clear reduction in all animals treated with anti-Spike antibodies compared to isotype control on day 3 (p=0·0079 for AX290, p=0·0159 for AX677 and p=0·0079 for AX290+AX677; evaluated by non-parametric Mann-Whitney test). On day 7 the copy numbers were greatly reduced in all animals, probably due to the elimination of the replicating viruses. The bars represent mean with s.d. Histological analysis of hematoxylin/eosin-stained lung sections collected 7 days after infection showed accumulation of the interstitial inflammatory infiltrates (e) and reduction of normal tissue (f) in animals treated with isotype antibody control. These histopathological changes were reduced by the anti-Spike antibodies AX290 and AX677 applied either alone or in combination. Data were evaluated by Mann-Whitney test (** p=0·0079, * p=0·0159).
Analysis of the viral titer in the lungs of the animals three days post-infection by plaque assay revealed that the disease-free status of the animals pre-treated with the anti-Spike antibodies was due to the greatly reduced viral burden (Figure 7c). While mice that received the isotype control antibody had high virus titers in the lungs, the mice treated with AX290 or AX677 had virus titers mostly below the level of detection with only one mouse per group having detectable, but greatly reduced, virus titers. No detectable virus titer was found in the lungs of mice treated with the mixture of the antibodies. Both single antibody treatment groups (AX290 and AX677) and their combination showed strongly reduced levels of viral RNA in the autopsy lung tissues on day three post-infection (Figure 7d). The effect is diminished on day seven post-infection, where amounts of viral RNA were substantially reduced in all animal groups.
The same prophylactic effect could also be seen and quantified using histopathology assessment of lung tissues. The lung tissues of ACE2 humanized mice infected with SARS-CoV-2 showed pathological changes typical for inflammation characterised by the presence of inflammatory infiltrate, mostly represented by activated macrophages, which filled and structurally damaged alveoli38 (see also Figure S7). The lung alveoli were beside macrophages also accompanied by lymphocytes, neutrophils and fibroblasts, from which we could assume significant cytokine production and proinflammatory action leading to lung fibrosis.
In the treated animals the extend of the lung tissue inflammatory damage and macrophage infiltration was clearly much smaller than in the control group (Figure 7e and Figure S7), although some regions with thickening of alveolar septa and inflammatory infiltrates with increased presence of F4/80 positive macrophages were noticeable. Importantly, the combination of AX290 and AX677 provided enhanced protective effect, which was clearly visible on the 7th day after infection. Inflammatory infiltrate creates less than 10% of the evaluated tissue in all samples from animals treated with the antibody combination, in comparison with the untreated group, where infiltrate occupies 40-70% of the evaluated tissue (Figure 7e). The presence of macrophages was also reduced and, especially, spreading of macrophages towards so far healthy tissue was minimal in all three groups of animals treated with anti-S antibodies, with additive effect when the antibodies were in combination. Such findings suggest that the antibody treatment is not fully protecting lungs from viral induced damage, however, based on weight curve and health status monitoring, each single antibody treatment provided protection from severe (or lethal) COVID-19 symptoms, and the combination of both antibodies further enhanced the lung tissue protection on histological level.
The in vivo protection experiment was repeated with a lower dose of antibodies, 0·5 mg in 0·2 ml per animal (approx. 20 mg/kg) applied intraperitoneally (Figure S8). The protection was demonstrated by strongly reduced viral loads in the lungs of the animals treated with the two anti-S antibodies on day three post-infection compared to the viral titers measured in the lungs of animals injected with the isotype control (Figure S8a). AX677 and the combination AX677+AX290 reduced the viral titers below the limit of detection (<2·14 log10 pfu/g); in case of AX290 the measured titer was lowered to 2·4-3·4 log10 pfu/g. In contrast, lungs from the control animals exhibited virus titer of 5·5-6·3 log10 pfu/g (Figure S8a). None of the SARS-CoV-2-infected animals treated with AX677, AX290, or a combination AX677+AX290 showed any weight loss during the whole experiment, while SARS-CoV-2-infected mice treated with the isotype control showed a weight loss of about 20-25% by day seven post-infection (Figure S8b). Consistent with the stable weight findings, gross lung samples collected on day seven post-infection from SARS-CoV-2-infected mice treated with AX677, AX290, or a combination AX677+AX290, showed no or minimal pathological lesions, while lungs from mice treated with the isotype control were massively affected (Figure S8c).
Discussion
Monoclonal antibodies targeting the viral protein S have enormous potential to prevent SARS-CoV-2 infection and treat patients with mild to moderate COVID-19.35 Several antibodies targeting RBD of the Spike protein have already been authorised for emergency use, including REGN10933/REGN10987, LY-CoV555 and JS016/LyCoV016 and VIR-7831[1] with others in the pipeline (TY027, CT-P59, BRII-196 and BRII-198, SCTA01 etc.). The emergence of new SARS-CoV-2 variants of concern B.1.1.7, B.1.351, P.1, and B.1.1.529 with mutations in Spike protein and increased human-human transmissibility,20, 21, 22, 23, 49 raised concern about the efficacy of vaccines and therapeutic antibodies.27,42,44
It was indeed shown that SARS-CoV-2 variants B.1.351 and P.1 were partially or completely resistant against antibodies used for COVID-19 treatment.42 This is in concordance with a study showing that the interaction between the mutant RBD containing three mutations N417/K484/Y501 and LY-CoV555/Bamlanivimab was completely abolished.44 To avoid this therapeutic limitation, we screened and selected the antibodies showing neutralization activity against several variants of SARS-CoV-2.
We took advantage of the mouse immunoglobulin repertoire18 through the hybridoma technology to isolate monoclonal antibodies neutralizing live authentic SARS-CoV-2 virus. The in vitro and live virus screening assays allowed us to identify two NAbs, AX290 and AX677, each of them neutralizing live authentic SARS-CoV-2 virus circulating in Europe in the spring of 2020 and three fast-spreading authentic variants of concern B.1.1.7 (Alpha), B.1.351 (Beta) and B.1.617.2 (Delta). The antibodies were oblivious to the E484K mutation, which has been recently shown to endow the S-typed VSV pseudovirus with resistance to several human convalescent sera.47 The recently emerged Omicron variant (Pango lineage B.1.1.529) proves highly infectious and has been shown to evade most of the therapeutic antibodies currently approved for use in humans.49 AX677ch might be another antibody effectively neutralizing the Omicron variant, since its binding to Spike is not at all influenced by the Omicron mutations. The neutralizing activity needs to be verified with the authentic Omicron SARS-CoV-2 virus in the PRNT assay.
The use of a live authentic SARS-CoV-2 virus to identify the escape mutations, as opposed to other studies that employed an S-typed pseudovirus,10,30,47,50, 51, 52 allowed to eliminate a potential bias from an inadequate infection and replication machinery of a model virus. It has been noted that only some of the spike escape mutations, identified in SARS-CoV-2 S-typed infectious vesicular stomatitis virus, were found so far in the context of authentic SARS-CoV-2 virus isolates, it is possible that those not identified might compromise the fitness of the genuine virus.47
Our authentic virus approach identified antibody-specific escape mutations in RBD of the Spike protein, S477R and T345N, that directly affect the recognition by the antibodies. It also allowed us to evaluate the cost the mutations pose on the fitness of the authentic virus. We found that the AX677 escape virus carrying the T345N mutation in Spike had greatly reduced fitness in comparison with the parental virus under antibody-free conditions (Figure 5d). This observation is supported by an extremely rare occurrence of mutations altering T345 in SARS-CoV-2 isolates and indicates an advantage of using AX677 for prophylaxis and treatment. In addition, the use of the authentic SARS-CoV-2 virus also allowed to identify mutations in other regions of the viral genome, the envelope (E) protein, Orf3a and Orf1a, that might help the SARS-CoV-2 virus to adapt to and escape from the selection pressure imposed by the S protein-binding antibodies. The AX290 selection pressure led to an insertion of three amino acids in the transmembrane region of the viroporin E protein, 13IV to 13IACLV. Remarkably, exactly the same insertion was also found in the virus isolated from the cells ‘protected’ by the cocktail of AX290+AX677, but not in the virus that escaped the AX677 antibody. The AX677 virus contained different mutation in the E protein, the V5F substitution. Both E mutations are in the N-terminal part of the protein that is predicted to be on (V5F) or close to (13IACLV) the surface of the virus and the luminal side of endoplasmic reticulum (ER)-Golgi intermediate compartment.53 In the viral fitness experiment we found that the insertion 13IACLV slightly reduced the fitness of the virus. Recently, several large in-frame deletions were identified in the E protein in various SARS-CoV-2 viral lineages in India,54 however all were located in its C-terminal half oriented on the cytoplasmic side of the ER membrane.
The escape mutant virus from AX290 contained another mutation, deletion of two amino acids 13VTL to 13V in orf3a, another ion channel protein encoded by the SARS-CoV-2 virus. 55 The E and orf3a ion channels can potentially transport cations from the ER and thus activate the inflammasome and downregulate the type-I interferon, but mostly are implicated in the later stages of the virus cycle – assembly and release.53,55 Thus, it is important to evaluate how can mutations in these proteins contribute to the viral fitness and potential antibody resistance, since it might have implications for conceivable treatment options.
The combination of the two antibodies exhibits a strong neutralizing effect against the authentic live SARS-CoV-2 virus observable at >600-fold lower concentration than each antibody alone. More importantly, the antibody combination also prevented emergence of escape mutations of the authentic SARS-CoV-2 virus. This is important for potential therapeutic application, where an antibody monotherapy might lead to the emergence of escape mutations in humans, as it was shown for bamlanivimab.56
Most importantly, the antibodies neutralized the virus in vivo in a novel mouse model in which human ACE2 (hACE2) is expressed by both upper and lower respiratory tract cells upon inhalation of Adeno Associated Virus (AAV-hACE2).38 Infection of these mice with SARS-CoV-2 results in progressive weight loss and respiratory pathology requiring euthanasia on day 8.38 Both antibodies AX290 and AX677, applied either individually or in combination, practically eliminated live viral loads, greatly reduced viral RNA and inflammation in the lungs and completely prevented systemic disease.
Appearance of novel variants of SARS-CoV-2 warrants development of novel types of vaccines and therapeutic approaches. Monoclonal antibodies provide safe, highly selective and efficient ways of viral neutralization; however, their high selectivity with relatively slow transition to the public use is also a weakness against the ever-mutating virus. It is, therefore, necessary to identify antibodies and their combinations that would prove impervious to the viral mutants. We have identified a pair of monoclonal antibodies from the mouse B-cell repertoire. Mouse immunoglobulins map slightly different structural space than human antibodies,18 and thus these antibodies might prove more resistant to future mutations emerging against human immune system than human monoclonal antibodies.
The antibodies AX290 and AX677 recognize non-overlapping epitopes on functionally different portions of the S protein RBD and apparently different mechanisms of how the they neutralize the virus. They protect hACE2-sensitized mice against lethal infection of SARS-CoV-2, efficiently neutralize the currently spreading authentic SARS-CoV-2 variants of concern, B.1.1.7 (Alpha), B.1.351 (Beta) and B.1.617.2 (Delta), in cell assays, and AX677 might even retain its full potency against B.1.1.529 (Omicron). The cocktail of the two antibodies might thus provide an effective COVID-19 treatment and protection against antibody resistance. The dose-sparing opportunity provided by topical administration57,58 would even allow a combination of more than two well-selected antibodies and provide further benefits preventing viral resistance.
Limitations of the study
We recognize that the resistance of the antibody cocktail against the virus escape mutations was not confirmed in PRNT. The mechanism of virus neutralization by AX677 has not yet been fully understood. Since we used prophylactic approach in our in vivo studies, we were not able to properly evaluate the contribution of the effector function to the therapeutic effect of the candidate antibodies. Based on previous reports, the neutralizing activity of AX677 might be further enhanced by its effector functions, since it efficiently neutralizes SARS-CoV-2 variants of concern without interfering with the Spike-ACE2 interaction.12,48 The effector function of antibodies was found indispensable for their post-infection therapeutic activities, but seems less important when antibodies are applied prophylactically or topically (e.g. by inhalation).59, 60, 61, 62 Hence, the effector functions of the antibodies need to be addressed during further development for therapeutic use.
Declaration of interests
All authors affiliated with AXON COVIDAX a.s., AXON Neuroscience SE, AXON Neuroscience R&D Services SE (BKo, LF, PF, RSk, MZ, NP-I, AK, DP, GPR, KT, NTC, KM, PM, VP, KS, NB, JH, MPr, OC, MC, JP, MF, MN, NZ, EK) receive a salary from or were employed by the respective companies. Biomedical Research Center, the employer of MS and BKl, received reimbursement from AXON Neuroscience SE for neutralization assays performed according to the research contract. KB, VC, BB, TVi, JN, LE, VH, MPa, DR, TVy, PS, BM, DZ, GK, VN, JP and RSe have no conflict of interest.
Acknowledgments
Contributors
BKo, NZ, MN, MF, EK conceived and designed the experiments and supervised the study. EK, LF generated hybridoma. LF, MZ, NPI, NTC, MPr, DP, KM performed primary hybridoma screening and carried out in vitro Ab assays. BKo, RSk, OC, KT performed purifications of Abs and recombinant antigens and affinity measurements. PF, MC, MS, GPR, KS, NB, JP expressed and purified recombinant proteins and chimeric antibodies, performed pseudovirus experiments. BKl, MS performed plaque reduction neutralization assay and virus escape experiments. KB, VC, BB, TVi, JN sequenced viral RNAs and performed bioinformatics analysis. AK, PM, VP, JP did proteomic analyses and HDX data evaluation. LE, VH, DR, MPa, TVy, and PS carried out the in vivo antibody testing on live SARS-CoV-2 virus in ACE2 humanized mice. BM, DZ, GK, VN, JP and RSe prepared the ACE2 humanized mouse model and histological evaluation of the animal samples. RSk, BKl, BB, DR, RSe, BKo, EK and NZ supervised the acquisition, and analysed and verified the data. BKo, EK and NZ verified the underlying data and prepared the manuscript with substantial input from all co-authors. All authors read and approved the final version of the manuscript and decided to proceed with publication.
Acknowledgements
The study was funded by AXON Neuroscience SE and AXON COVIDAX a.s.
Data sharing statement
Further information and requests for data should be directed to and will be fulfilled by the lead contact, Branislav Kovacech (kovacech@axon-neuroscience.eu). Sequences of the escape viruses A1, B11 and C25 will be available from European Nucleotide Archive (ENA, project PRJEB49042), and data from the in vivo experiments can be provided on request. Such requests should be accompanied by well-defined research questions.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2022.103818.
Contributor Information
Branislav Kovacech, Email: kovacech@axon-neuroscience.eu.
Norbert Zilka, Email: zilka@axon-neuroscience.eu.
Appendix. Supplementary materials
References
- 1.Yang L, Liu W, Yu X, Wu M, Reichert JM, Ho M. COVID-19 antibody therapeutics tracker: a global online database of antibody therapeutics for the prevention and treatment of COVID-19. Antib Ther. 2020;3(3):205–212. doi: 10.1093/abt/tbaa020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes CO, Jette CA, Abernathy ME, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brouwer PJM, Caniels TG, van der Straten K, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369(6504):643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao Y, Su B, Guo X, et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients' B Cells. Cell. 2020;182(1):73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584(7819):115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 6.Liu L, Wang P, Nair MS, et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584(7821):450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 7.Rogers TF, Zhao F, Huang D, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369(6506):956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zost SJ, Gilchuk P, Case JB, et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584(7821):443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplon H, Reichert JM. Antibodies to watch in 2021. MAbs. 2021;13(1) doi: 10.1080/19420862.2020.1860476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baum A, Fulton BO, Wloga E, et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369(6506):1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen P, Nirula A, Heller B, et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N Engl J Med. 2021;384(3):229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen J, Baum A, Pascal KE, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369(6506):1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinreich DM, Sivapalasingam S, Norton T, et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N Engl J Med. 2021;384(3):238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiaojie S, Yu L, Lei Y, Guang Y, Min Q. Neutralizing antibodies targeting SARS-CoV-2 spike protein. Stem Cell Res. 2020;50 doi: 10.1016/j.scr.2020.102125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaneko N, Kuo HH, Boucau J, et al. Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell. 2020;183(1):143–157.e13. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreer C, Zehner M, Weber T, et al. Longitudinal Isolation of Potent Near-Germline SARS-CoV-2-Neutralizing Antibodies from COVID-19 Patients. Cell. 2020;182(6):1663–1673. doi: 10.1016/j.cell.2020.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreye J, Reincke SM, Kornau HC, et al. A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model. Cell. 2020;183(4):1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins AM, Jackson KJL. On being the right size: antibody repertoire formation in the mouse and human. Immunogenetics. 2018;70(3):143–158. doi: 10.1007/s00251-017-1049-8. [DOI] [PubMed] [Google Scholar]
- 19.Rambaut A, Loman N, Pybus O, et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations 2020. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (Accessed 9 December 2020).
- 20.Zhao S, Lou J, Cao L, et al. Quantifying the transmission advantage associated with N501Y substitution of SARS-CoV-2 in the UK: an early data-driven analysis. J Travel Med. 2021;28(2) doi: 10.1093/jtm/taab011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kidd M, Richter A, Best A, et al. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral loads in samples tested by ThermoFisher TaqPath RT-qPCR. J Infect Dis. 2021 doi: 10.1093/infdis/jiab082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francisco RDS, Jr., Benites LF, Lamarca AP, et al. Pervasive transmission of E484K and emergence of VUI-NP13L with evidence of SARS-CoV-2 co-infection events by two different lineages in Rio Grande do Sul, Brazil. Virus Res. 2021;296 doi: 10.1016/j.virusres.2021.198345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mwenda M, Saasa N, Sinyange N, et al. Detection of B.1.351 SARS-CoV-2 Variant Strain - Zambia, December 2020. MMWR Morb Mortal Wkly Rep. 2021;70(8):280–282. doi: 10.15585/mmwr.mm7008e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mlcochova P, Kemp S, Dhar MS, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021 doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones BE, Brown-Augsburger PL, Corbett KS, et al. LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection. bioRxiv. 2020 [Google Scholar]
- 26.Kim C, Ryu DK, Lee J, et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat Commun. 2021;12(1):288. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tada T, Dcosta BM, Zhou H, Vaill A, Kazmierski W, Landau NR. Decreased neutralization of SARS-CoV-2 global variants by therapeutic anti-spike protein monoclonal antibodies. bioRxiv. 2021 [Google Scholar]
- 28.Thomson EC, Rosen LE, Shepherd JG, et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184(5):1171–1187.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P, Casner RG, Nair MS, et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29(5):747–751.e4. doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen RE, Zhang X, Case JB, et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med. 2021;27(4):717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferreira I, Kemp S, Datir R, et al. SARS-CoV-2 B.1.617 mutations L452 and E484Q are not synergistic for antibody evasion. J Infect Dis. 2021 doi: 10.1093/infdis/jiab368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kontsekova E, Novak M, Macikova I, Kontsek P. One-step method for establishing 8-azaguanine-resistant hybridomas suitable for the preparation of triomas. J Immunol Methods. 1991;145(1-2):247–250. doi: 10.1016/0022-1759(91)90333-b. [DOI] [PubMed] [Google Scholar]
- 34.Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12(5):279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Eden JS, Sim E. SARS-CoV-2 Genome Sequencing Using Long Pooled Amplicons on Illumina Platforms 2020. https://www.protocols.io/view/sars-cov-2-genome-sequencing-using-long-pooled-amp-befyjbpw (Accessed 4 April 2020).
- 36.Resende PC, Motta FC, Roy S, et al. SARS-CoV-2 genomes recovered by long amplicon tiling multiplex approach using nanopore sequencing and applicable to other sequencing platforms. bioRxiv. 2020 2020.04.30.069039. [Google Scholar]
- 37.Kavan D, Man P. MSTools—Web based application for visualization and presentation of HXMS data. International Journal of Mass Spectrometry. 2011;302:53–58. [Google Scholar]
- 38.De Gasparo R, Pedotti M, Simonelli L, et al. Bispecific IgG neutralizes SARS-CoV-2 variants and prevents escape in mice. Nature. 2021;593(7859):424–428. doi: 10.1038/s41586-021-03461-y. [DOI] [PubMed] [Google Scholar]
- 39.Kontsekova E, Zilka N, Kovacech B, Skrabana R, Novak M. Identification of structural determinants on tau protein essential for its pathological function: novel therapeutic target for tau immunotherapy in Alzheimer's disease. Alzheimer's research & therapy. 2014;6(4):45. doi: 10.1186/alzrt277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Madrid AT, Porterfield JS. A simple micro-culture method for the study of group B arboviruses. Bull World Health Organ. 1969;40(1):113–121. [PMC free article] [PubMed] [Google Scholar]
- 41.Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 42.Hoffmann M, Arora P, Gross R, et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184(9):2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong J, Zost S, Greaney A, et al. Genetic and structural basis for recognition of SARS-CoV-2 spike protein by a two-antibody cocktail. bioRxiv. 2021 [Google Scholar]
- 44.Liu H, Wei P, Zhang Q, et al. 501Y.V2 and 501Y.V3 variants of SARS-CoV-2 lose binding to Bamlanivimab in vitro. bioRxiv. 2021 doi: 10.1080/19420862.2021.1919285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asarnow D, Wang B, Lee WH, et al. Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia. Cell. 2021 doi: 10.1016/j.cell.2021.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Starr TN, Greaney AJ, Hilton SK, et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182(5):1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Z, VanBlargan LA, Bloyet LM, et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29(3):477–488.e4. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med. 2014;20(2):143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoffmann M, Krüger N, Schulz S, et al. The Omicron variant is highly resistant against antibody-mediated neutralization – implications for control of the COVID-19 pandemic. bioRxiv. 2021 doi: 10.1016/j.cell.2021.12.032. 2021.12.12.472286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu H, Yuan M, Huang D, et al. A combination of cross-neutralizing antibodies synergizes to prevent SARS-CoV-2 and SARS-CoV pseudovirus infection. Cell Host Microbe. 2021;29(5):806–818.e6. doi: 10.1016/j.chom.2021.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffmann M, Arora P, Gross R, et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021 doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koenig PA, Das H, Liu H, et al. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371(6530) doi: 10.1126/science.abe6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mandala VS, McKay MJ, Shcherbakov AA, Dregni AJ, Kolocouris A, Hong M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat Struct Mol Biol. 2020;27(12):1202–1208. doi: 10.1038/s41594-020-00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar BK, Rohit A, Prithvisagar KS, Rai P, Karunasagar I, Karunasagar I. Deletion in the C-terminal region of the envelope glycoprotein in some of the Indian SARS-CoV-2 genome. Virus Res. 2021;291 doi: 10.1016/j.virusres.2020.198222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barrantes FJ. Structural biology of coronavirus ion channels. Acta Crystallogr D Struct Biol. 2021;77(Pt 4):391–402. doi: 10.1107/S2059798321001431. [DOI] [PubMed] [Google Scholar]
- 56.Peiffer-Smadja N, Bridier-Nahmias A, Ferre VM, et al. Emergence of E484K Mutation Following Bamlanivimab Monotherapy among High-Risk Patients Infected with the Alpha Variant of SARS-CoV-2. Viruses. 2021;13(8) doi: 10.3390/v13081642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Halwe S, Kupke A, Vanshylla K, et al. Intranasal Administration of a Monoclonal Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. Viruses. 2021;13(8) doi: 10.3390/v13081498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nambulli S, Xiang Y, Tilston-Lunel NL, et al. Inhalable Nanobody (PiN-21) prevents and treats SARS-CoV-2 infections in Syrian hamsters at ultra-low doses. Sci Adv. 2021;7(22) doi: 10.1126/sciadv.abh0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamin R, Jones AT, Hoffmann HH, et al. Fc-engineered antibody therapeutics with improved anti-SARS-CoV-2 efficacy. Nature. 2021;599(7885):465–470. doi: 10.1038/s41586-021-04017-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gorman MJ, Patel N, Guebre-Xabier M, et al. Fab and Fc contribute to maximal protection against SARS-CoV-2 following NVX-CoV2373 subunit vaccine with Matrix-M vaccination. Cell Rep Med. 2021;2(9) doi: 10.1016/j.xcrm.2021.100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winkler ES, Gilchuk P, Yu J, et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184(7):1804–1820.e16. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vigil A, Frias-Staheli N, Carabeo T, Wittekind M. Airway Delivery of Anti-influenza Monoclonal Antibodies Results in Enhanced Antiviral Activities and Enables Broad-Coverage Combination Therapies. J Virol. 2020;94(22) doi: 10.1128/JVI.00052-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.