Abstract
Although tobacco smoking is a risk factor for lung adenocarcinoma (LUAD), the mechanisms by which tobacco smoking induces LUAD development remain elusive. Histone methylation levels in human bronchial epithelial cells have been reported to increase after exposure to cigarettes. In this study, we explored the mechanisms regulating histone methylation in LUAD in response to smoking. We found that the histone H3K9 methylation reader CBX3 was upregulated in current smokers with LUAD, and that CBX3 overexpression promoted LUAD progression. Functional enrichment analyses revealed that CBX3 regulated the activation of Rho GTPases in LUAD. We also found that by forming a complex with TRIM28, TRIM24, and RBBP4, CBX3 repressed the expression of ARHGAP24 and increased the amount of active Rac1 in LUAD cells. Collectively, these results suggest that smoking associated upregulation of CBX3 promotes LUAD progression by activating the ARHGAP24/Rac1 pathway. Hence, the CBX3/ARHGAP24/Rac1 axis may represent a promising therapeutic target in smoking-induced LUAD.
Subject terms: Non-small-cell lung cancer, Oncogenes
Introduction
Lung cancer is one of the most common malignancies and the leading cause of cancer-related death worldwide, with approximately 2,207,000 new cases and 1,796,000 deaths in 2020 [1]. Lung adenocarcinoma (LUAD) is the most common histological subtype of lung cancer, accounting for approximately 40% of lung cancer cases. The incidence of LUAD has significantly increased in recent years and continues to rise globally. Tobacco smoking has been identified as a strong risk factor for lung cancer. Smoking has been shown to affect the epigenetic modification of genes in lung tissues and the blood [2, 3]. In addition, exposure to cigarettes has been linked with increased levels of whole-genome histone methylation in human bronchial epithelial cells; this was particularly evident for mono- and dimethylation of histone H3 and H4 residues [4]. The expression of histone methyltransferase (HMT), an enzyme catalyzing histone methylation, has also been found to be regulated by smoking [5, 6]. Therefore, understanding the role of smoking in histone methylation and transcriptional regulation may facilitate the development of novel therapies for smoking-induced LUAD.
Chromobox homolog 3 (CBX3) belonging to the heterochromatin-associated protein 1 (HP1) family, binds to chromatin and trimethyl lysine 9 of histone H3 (H3K9me3) to promote the formation of heterochromatin [7]. By suppressing the expression of various genes, CBX3 regulates numerous cellular processes, including cell growth, cell differentiation, and DNA damage [8–10]. Aberrant CBX3 expression has been implicated in the development of various cancer types [11]. However, the role of CBX3 in LUAD remains understudied [12]. In addition, little is known about the relationship between smoking, CBX3, and LUAD progression.
Here, through in silico analyses of publicly available datasets of genes involved in histone methylation, we found that the expression of the histone H3K9 methylation reader CBX3 was upregulated in current smokers with LUAD. We also found that CBX3 overexpression was associated with LUAD progression. Functional enrichment analyses revealed that CBX3 regulates numerous signaling pathways, including Rho GTPase signaling. We also found that CBX3 represses the expression of ARHGAP24, a negative regulator of the Rho GTPase pathway [13], thereby increasing the amount of active Rac1 in LUAD cells. Collectively, these data unveiled a previously unknown relationship between smoking, CBX3, and Rac1 in LUAD.
Material and methods
Data mining and bioinformatics analysis
The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), Clinical Proteomic Tumor Analysis Consortium (CPTAC), CancerSEA, and ChIP-Atlas were used for data mining and bioinformatics analysis (see Supplementary Methods for details).
Cell lines
Lung adenocarcinoma cell lines including A549 (CM-0016) and H1299 (CM-0165) were purchased from Procell (Wuhan, China). Both cell lines were authenticated by short tandem repeat (STR) profiling. A549 cells were cultured in Ham’s F-12K medium (PM150910, Procell) supplemented with 10% FBS (164210-500, Procell) and 1% P/S (PB180120, Procell). H1299 cells were cultured in RPMI-1640 medium (PM150110, Procell) plus 10% FBS (164210-500, Procell) and 1% P/S(PB180120, Procell). All cell lines were routinely maintained at 37 °C in a 5% CO2 incubator.
Western blot analysis and coimmunoprecipitation (Co-IP)
Cells were harvested and lysed with lysis buffer containing 1% protease and phosphatase inhibitors for 15 min on ice as described previously [14, 15]. A protein assay kit (Pierce Biotechnology, USA) was used to determine the protein concentration. Equal amounts of proteins were separated in SDS-PAGE gels and detected in PVDF membranes after incubation with primary antibodies and subsequent secondary antibodies. Antibodies against the following proteins were used: CBX3 (cat. no. 11650-2-AP, Proteintech; 1:1000 dilution); ARHGAP24 (cat. no. 18834-1-AP, Proteintech; 1:2500 dilution); Rac1 (cat. no. 66122-1-Ig, Proteintech; 1:2500 dilution); GAPDH (cat. no. 60004-1-Ig, Proteintech; 1:5000 dilution); TRIM24 (cat. no. 14208-1-AP. Proteintech; 1:1000 dilution), TRIM28 (cat. no. 15202-1-AP; Proteintech; 1:2000 dilution), H3K9me3 (cat. no. ab8898; Abcam; 1:1000 dilution) and RBBP4 (cat. no. ab79416; Abcam; 1:1000 dilution).
For immunoprecipitation, the cell lysate was incubated with Pierce Protein G Agarose (Thermo Fisher Scientific, USA) and primary antibody or IgG. Then the precipitates were analyzed by immunoblotting. To detect active Rac1, we followed the manufacturer’s instruction for an Active Rac1 Detection Kit (cat. no. 8815, Cell Signaling Technology).
Quantitative real-time PCR (RT-qPCR), chromatin immunoprecipitation (ChIP) and ChIP-qPCR
The RT-qPCR procedure was described previously. A PrimeScript™ RT reagent kit (cat. no. RR037A) and TB Green™ Fast qPCR Mix kit (cat. no. RR430A) purchased from Takara Bio Inc. (Shiga, Japan) were used to construct cDNA and perform RT-qPCR analysis. The sequences of primers are provided in Table S1.
A chromatin Extraction Kit (Abcam, ab117152, USA) and ChIP Kit Magnetic - One Step (Abcam, ab156907, USA) were used to preform ChIP as described previously. The detailed antibodies as follows: CBX3 (Proteintech; 11650-2-AP; 1:100), TRIM24 (Proteintech; 14208-1-AP; 1:500), TRIM28 (Proteintech; 15202-1-AP; 1:200), H3K9me3 ((Abcam; ab8898; 1:500) and RBBP4 (Abcam; ab79416; 1:100). The sequences of primers are provided in Table S2.
Tissue microarray and immunohistochemistry (IHC)
A tissue microarray (cat. no. D881001, Bioaitech, CN) and IHC were employed to assess the levels of CBX3 (cat. no. 11650-2-AP, Proteintech; 1:2500 dilution) and ARHGAP24 (cat. no. 18834-1-AP, Proteintech; 1:2500 dilution) in lung adenocarcinoma. The IHC score was evaluated as previously reported.
Xenografts assay
Ethical approval was obtained from the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology for all animal procedures. BALB/c-nude mice (4–5 weeks old, 18–20 g) were obtained from Vitalriver (Beijing, China). Power analysis was used to calculate the sample size required for animal experiments and animals were randomized into the different groups. A459 cells were transduced with different lentiviral particles. After puromycin selection for 72 h, cells (1 × 107 per mouse) were subcutaneously injected into the backs of mice. The xenografts assay procedure was described previously [16]. At the study endpoint, the volume and mass of xenografts were measured. Nicotine (cat. no. MED24104) was purchased from Medbio (CN).
Statistical analysis
All data are presented as the means ± standard deviation (SD). The sample size (n) for each statistical analysis is provided in the figure legends. Statistical significance was determined with Student’s t test, and one-way or two-way ANOVA using GraphPad Prism 5 software. P < 0.05 were considered statistically significant.
Other methods, the sequenced of gene-specific shRNAs (Table S3), and demographic information of the clinical data (Table S4, S5) are provided in the Supplementary Information.
Results
CBX3 is upregulated in current smokers with LUAD, and CBX3 overexpression predicts worse survival in patients with LUAD
Genes (n = 347) involved in histone methylation-related biological processes and signaling pathways were identified from the Molecular Signatures Database (MSigDB) (Fig. 1a). Through univariate Cox regression analysis, 22 genes were identified to be associated with the recurrence-free survival (RFS) of LUAD patients in the TCGA-LUAD dataset (P < 0.05). Lasso-Cox regression analysis with 1000 replications further showed that CBX3 and TAF9 were two key genes (both 1000 times) associated with RFS in patients with LUAD (Fig. 1b). Then, we combined the transcriptome data of the TCGA-LUAD and TCGA-LUSC datasets, and found that CBX3 but not TAF9 was expressed at significantly higher levels in the current smoking group than in the former smoking group in both datasets (all P < 0.05; Fig. 1c). The low expression level of CBX3 was mainly concentrated in former smokers and never smokers in the TCGA-LUAD dataset (Fig. 1d). Kaplan–Meier survival analyses showed that high CBX3 expression levels were associated with poor RFS and overall survival (OS) in both the TCGA-LUAD and GSE68465 datasets (all P < 0.05; Fig. 1e–h). Additionally, CBX3 was significantly upregulated in tumor tissues compared with tumor-adjacent tissues in the TCGA-LUAD and CPTAC-LUAD datasets (all P < 0.001, Fig. 1i, j). Similarly, through IHC staining, we detected CBX3 expression in a tissue microarray of lung adenocarcinoma specimens (nontumor lung tissues (n = 8) and lung adenocarcinoma tissues (n = 59), cat. no. R881001, Bioaitech, China). The IHC images stained with CBX3 are shown in Supplementary figure 1a. We found that CBX3 was downregulated in the nontumor lung tissue compared to the lung adenocarcinoma (P = 0.0031) (Supplementary Fig. 1a, b and Table S5).
Fig. 1. CBX3 is upregulated in current smokers with LUAD, and CBX3 overexpression predicts worse survival in patients with LUAD.
a The flow chart of how to identify CBX3 associated with survival and smoking in LUAD. b 10-fold cross-validation with 1000 replications for selection of genes associated with LUAD RFS in the LASSO-COX model by minimum criteria (the 1-SE criteria). c Venn diagrams showing numbers of upregulated genes involved in the histone methylation in current smokers compared to former smokers in TCGA-LUAD and TCGA-LUSC datasets. d Heatmap of top 10 averagely highly expressed genes in TCGA-LUAD samples out of the 46 genes showed in Venn diagram. Red indicates overexpression and blue underexpression. The columns correspond to the LUAD samples. The real status of the samples is given in a color scale: blue for never smokers, red for former smokers and green for current smokers. e, f Kaplan–Meier analysis with two-sided log-rank test was conducted to evaluate the differences in RFS (e) and OS (f) between the patients with high and low expression of CBX3 in TCGA-LUAD dataset. Median expression level of CBX3 was used as cutoff. g, h Kaplan–Meier analysis with two-sided log-rank test was conducted to evaluate the differences in RFS (g) and OS (h) between the patients with high and low expression of CBX3 in GSE68465 dataset. Median expression level of CBX3 was used as cutoff. i, j Differential expression analyses of CBX3 between tumor and normal tissues in TCGA-LUAD (i) and CPTAC-LUAD (j) datasets.
CBX3 promotes LUAD cell growth and invasion
Since the single-cell sequence of LUAD indicated that CBX3 may regulate cell cycle, proliferation, invasion, and metastasis (Fig. 2a and Supplementary Fig. 2a), we further explored the protumorigenic role of CBX3 in LUAD cells. To this end, we silenced CBX3 expression in A549 and H1299 cells using two different short-hairpin RNAs (shRNAs) (Fig. 2b, c). MTS assays and colony formation assays showed that CBX3 silencing decreased the proliferation ability of LUAD cells (Fig. 2d–f). Similarly, CBX3 knockdown suppressed cell invasion in A549 and H1299 cells (Fig. 2g). Conversely, CBX3 overexpression enhanced cell proliferation and invasion in LUAD cells (Fig. 2h–k, Supplementary Fig. 2b). Moreover, rescuing CBX3 expression in A549 cells with CBX3 knockdown reinforced the proliferation of cells in vitro and in vivo (Fig. 2l–p, Supplementary Fig. 2c). Together, these data suggest that CBX3 acts as an oncoprotein in LUAD. In addition, we found that there is a close relationship between CBX3 and smoking in LUAD. Next, we investigated the role of CBX3 in smoking associated LUAD. A549 cells were infected with shControl or shCBX3 for 72 h. After puromycin selection, the cells were subcutaneously injected into nude mice. The silencing effect of CBX3 was evaluated via Western blotting and RT-qPCR analysis (Supplementary Fig. 3a, b). Then, each group of mice was randomly divided into three subgroups that were treated with or without cigarette smoke extract (CSE) (0.3 ml/20 g, i.p.) or nicotine (0.5 mg/kg, i.p.) (Supplementary Fig. 3c) [17]. We demonstrated that CSE and nicotine treatment promoted tumor growth in the shControl group (Supplementary Fig. 3d–f). However, knockdown of CBX3 diminished the tumor growth-promoting effect induced by CSE or nicotine treatment in vivo (Supplementary Fig. 3d–f). Furthermore, knockdown of CBX3 by infection with shRNAs or treatment with CSE or nicotine had no effect on the liver and renal function of nude mice according to serum analysis of aminoleucine transferase (ALT), aspartate transaminase (AST), creatinine (CRE), or blood urea nitrogen (BUN) in each group of nude mice (Supplementary Fig. 3g–j). Together, these data suggest that CBX3 plays a role in modulating smoking-associated lung adenocarcinoma cell growth.
Fig. 2. CBX3 promotes LUAD cell growth and invasion.
a Analysis the potential cancer-related function of CBX3 in lung adenocarcinoma by the single cell sequencing dataset. b–g A549 and H1299 cells were infected with shControl, shCBX3 #1, or shCBX3 #2. Seventy two hours post infection, cells were collected for Western blotting analysis (b), RT-qPCR analysis (c), MTS assay (d), colony formation assay (e and f) and transwell assay (g). Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test. Data presented as Mean ± SD with three replicates (n = 3). ***P < 0.001. h–k A549 and H1299 were transfected with pcDNA3.1 (as empty vector) or Flag-CBX3 for 24 h. Cells were collected for RT-qPCR analysis (h), MTS assay (i) and transwell assay (j, k). Statistical significance was determined by two-side student t test. Data presented as Mean ± SD with three replicates (n = 3). ***P < 0.001. l, m A549 cells were infected with Tsin-EV + shControl, Tsin-EV + shCBX3, or Tsin-CBX3 + shCBX3 for 72 h. Cells were harvested for Western blotting analysis (l) and MTS assay (m). Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test. Data presented as Mean ± SD with three replicates (n = 3). ***P < 0.001. n–p A549 cells were infected with Tsin-EV + shControl, Tsin-EV + shCBX3, or Tsin-CBX3 + shCBX3 as indicated. After 72 h puromycine selection, cells were harvested and subcutaneously injected into nude mice for xenografts assay. The image of tumor was shown in panel (n). The tumor growth curve was indicated in panel (o). The tumor mass was demonstrated in panel (p). Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test. Data presented as Mean ± SD with five replicates. NS not significant; ***P < 0.001.
CBX3 increases the amount of active Rac1 in LUAD
To explore the mechanisms underlying the oncogenic effects of CBX3 in LUAD, we performed functional enrichment analyses of transcriptomics data of bulk tissues and single cells. Gene set enrichment analysis (GSEA) showed that CBX3 activated Rho GTPase signaling in bulk tissues (TCGA-LUAD and GSE68465) and A459 and LC-2/ad single cells (all P < 0.001; Fig. 3a–d). RNA-seq analysis of A549 cells after CBX3 knockdown (GSE173858) confirmed a role of CBX3 in the activation of Rho GTPase signaling (Fig. 3e, f) and indicated that CBX3 downregulated several negative regulators of Rho GTPase signaling in A549 cells (Fig. 3g). Consistently, CBX3 silencing decreased the levels of activated Rac1 in LUAD cells (Fig. 3h, i); Rac1 is considered an indicator of Rho GTPase signaling activation [18]. Conversely, CBX3 overexpression increased Rac1 levels in A549 and H1299 cells (Fig. 3j, k). Taken together, these data suggest that CBX3 plays a critical role in upregulating the amount of active Rac1 in LUAD cells.
Fig. 3. CBX3 increases the amount of active Rac1 in LUAD.
a, b GSEA for CBX3 in TCGA-LUAD (a) and GSE68465 (b) datasets. Samples were first divided into two groups according to the median expression level of the CBX3. Then, differential expression analysis was applied between the high and low expression groups. Input genes for GSEA were sorted by their logFC values. Signaling pathways activated or suppressed by CBX3 were decided by the NES value derived from GSEA. c, d Rho GTPase signaling pathway was activated by the over expression of CBX3 in single cells of A459 (c) and LC-2/ad (d). Median expression level of CBX3 was used as cutoff. e, f Knockdown of CBX3 in A549 cell line leaded to the suppression of Rho GTPases signaling pathway. g Key genes involved in the regulation of Rho GTPases signaling pathway by the knockdown of CBX3 in A459 cell line. h, i A459 and H1299 cells were infected with shConrtol or shCBX3 for 72 h. Cells were divided in to two equal parts. The first part of cells was collected for Western blotting analysis and detected by the CBX3, Rac1 and Beta-actin antibodies. The second part of cells were lysed and subjected to GST-pull down following the protocol of Active Rac1 Detection Kit, the active Rac1 was detected by Western blotting analysis. Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test. Data presented as Mean ± SD with two replicates (n = 2). *P < 0.05. j, k A459 and H1299 cells were transfected with indicated constructs for 24 h. Cells were divided in to two equal parts. The first part of cells was collected for Western blotting analysis and detected by the CBX3, Rac1 and Beta-actin antibodies. The second part of cells were lysed and subjected to GST-pull down following the protocol of Active Rac1 Detection Kit, the active Rac1 was detected by Western blotting analysis. Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test. Data presented as Mean ± SD with two replicates (n = 2). *P < 0.05; **P < 0.01; ***P < 0.001.
CBX3 negatively regulates ARHGAP24 expression in LUAD
Next, we explored the molecular mechanisms by which CBX3 regulates the Rac1 pathway. We evaluated the correlation between the levels of CBX3 and regulators of Rho GTPase signaling. ARHGAP24 was the only Rho GTPase regulator negatively associated with CBX3 levels in the TCGA-LUAD, GSE68465, and CPTAC-LUAD datasets (Fig. 4a). In addition, ARHGAP24 mRNA and protein levels were lower in LUAD specimens than in nonmalignant tissues (TCGA-LUAD dataset and CPTAC-LUAD dataset) (Fig. 4b, c). Interestingly, low ARHGAP24 mRNA levels were associated with poor disease-free survival and OS in patients with LUAD (Fig. 4d, e).
Fig. 4. CBX3 negatively regulates ARHGAP24 expression in LUAD.
a Venn diagram showed number of genes negatively correlated with CBX3 in TCGA-LUAD, GSE68465 and CPTAC-LUAD datasets. b, c Differential expression analyses of ARHGAP24 between tumor and normal tissues in TCGA-LUAD (b) and CPTAC-LUAD (c) datasets. d, e Kaplan–Meier analysis with two-sided log-rank test was conducted to evaluate the differences in RFS (d) and OS (e) between the patients with high and low expression of ARHGAP24 in TCGA-LUAD dataset. The dashed lines in panel (d) and (e) represented the 95% confidence intervals of the KM curves. f Volcano diagram showed differential expressed genes caused by the knockdown of CBX3 in A549 cell line. g–i ARHGAP24 was negatively correlated with CBX3 in TCGA-LUAD (g), GSE68465 (h) and CPTAC-LUAD (i) datasets. j, k The tissue microarray of lung adenocarcinoma was stained with CBX3 and ARHGAP24, respectively (n = 59). The typical IHC images stained with CBX3 and ARHGAP24 were shown in panel (j). The size of the scale bar on microscopy images as indicated in the figure. The correlation of these two proteins was shown in panel (k). Spearman correlation was used to determine statistical significance, P = 0.0427.
RNA-seq analysis indicated that ARHGAP24 was upregulated in A549 cells after CBX3 silencing (Fig. 4f). Moreover, CBX3 expression levels were negatively correlated with those of ARHGAP24 in different LUAD datasets (TCGA, GSE68465 and CPTAC; all P < 0.001; Fig. 4g–i). A microarray of LUAD tissues (including tissues of 59 patients with LUAD; cat. no. R881001, Bioaitech, China) was employed to evaluate the protein levels of CBX3 and ARHGAP24 (Fig. 4j). In line with the transcriptomics data, CBX3 protein levels were negatively correlated with ARHGAP24 protein levels in LUAD tissues (Spearman r = −0.2648, P = 0.0427; Fig. 4k). These results suggest that CBX3 negatively regulates ARHGAP24 expression in LUAD.
CBX3 indirectly increases the amount of active Rac1 by repressing ARHGAP24 expression in LUAD cells
As ARHGAP24 was reported to inhibit the Rac1 pathway [19], we next assessed whether CBX3 upregulates the levels of active Rac1 due to a reduction in the rate of deactivation via suppression of ARHGAP24 expression. CBX3 knockdown increased the protein and mRNA levels of ARHGAP24 in A549 and H1299 cells (Fig. 5a, b). Conversely, CBX3 overexpression reduced ARHGAP24 mRNA and protein levels in A549 and H1299 cells (Fig. 5c, d). Analyses using ChIP-Atlas revealed a CBX3 binding peak in the promoter of ARHGAP24 (Fig. 5e). CBX3 represses the transcription of target genes by recognizing histone H3K9 trimethylation (H3K9me3) [20], and H3K9me3-binding sites overlap with the CBX3-binding site in the promoter region of ARHGAP24 (Fig. 5f). ChIP-qPCR analysis revealed the presence of CBX3 and H3K9me3 in the promoter of ARHGAP24 (Fig. 5g, h), suggesting that CBX3 regulates ARHGAP24 expression by recognizing methylation of the ARHGAP24 promoter. In addition, overexpression of CBX3 in ARHGAP24 ablation cells did not further increase the levels of active Rac1 compared to knockdown of ARHGAP24 alone in A549 cells (Fig. 5i). Coknockdown of ARHGAP24 and CBX3 diminished the CBX3 silencing ability to indirectly reduce Rac1 deactivation (Fig. 5J) and to inhibit the in vitro and in vivo growth of LUAD cells (Fig. 5k–n). These data suggest that CBX3 downregulates ARHGAP24 to increase the amount of active Rac1 and promote tumor growth in LUAD.
Fig. 5. CBX3 indirectly increases the amount of active Rac1 by repressing ARHGAP24 expression in LUAD cells.
a, b A549 and H1299 cells were infected with shControl, shCBX3 #1, or shCBX3 #2 for 72 h. Cells were collected for Western blotting analysis (a) and RT-qPCR analysis (b). Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test. Data presented as Mean ± SD with three replicates. **P < 0.01. c, d A549 and H1299 cells were transfected with indicated plasmids for 24 h. Cells were collected for Western blotting analysis (c) and RT-qPCR analysis (d). Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test. Data presented as Mean ± SD with three replicates. **P < 0.01; ***P < 0.001. e, f The ChIP-seq of CBX3 on the promoter region of ARHGAP24. g The ChIP-qPCR of CBX3 on the promoter region of ARHGAP24 in A549 and H1299 cells. Statistical significance was determined by two-side Student t test. Data presented as Mean ± SD with three replicates. NS not significant; **P < 0.01; ***P < 0.001. h The ChIP-qPCR of H3K9me3 on the promoter region of ARHGAP24 in A549 and H1299 cells. Statistical significance was determined by two-side Student t test. Data presented as Mean ± SD with three replicates. NS not significant; *P < 0.05; ***P < 0.001. i A549 cells were infected with shControl or shARHGAP24 for 48 h. Then, cells were transfected with pcDNA3.1 or Flag-CBX3 as indicated. After 24 h, cells were harvested for Western blotting analysis. Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test. For quantification of active Rac1, data presented as Mean ± SD with two replicates. NS not significant; *P < 0.05; **P < 0.01. j, k A549 cells were infected with indicated shRNAs for 72 h. Cells were collected for Western blotting analysis (j) and MTS assay (k). Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test. For quantification of active Rac1, data presented as Mean ± SD with two replicates. For MTS assay, data presented as Mean ± SD with three replicates. NS not significant; *P < 0.05; **P < 0.01; ***P < 0.001. l–n A549 cells were infected with indicated shRNAs. After 72 h puromycin selection, cells were harvested and subcutaneously injected into nude mice for xenografts assay. The image of tumor was shown in panel (l). The tumor growth curve was indicated in panel (m). The tumor mass was demonstrated in panel (n). Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test. Data presented as Mean ± SD with five replicates. NS not significant; ***P < 0.001.
CBX3 binds to TRIM28, TRIM24, or RBBP4 to regulate ARHGAP24 expression in LUAD cells
Protein–protein interaction (PPI) network analyses revealed that CBX3 interacts with various proteins to regulate numerous cellular processes (Fig. 6a). Among these proteins, TRIM28, TRIM24, and RBBP4 exhibited the closest relationship with CBX3 (Fig. 6a). Intriguingly, a co-IP assay showed that CBX3 interacted with TRIM28, TRIM24, or RBBP4 in A549 and H1299 cells (Fig. 6b–e; Supplementary Fig. 4a). In addition, we performed a proximity ligation assay (PLA) to confirm the interaction between CBX3 and TRIM28, TRIM24, or RBBP4 in A549 cells (Supplementary Fig. 4b). Moreover, an immunofluorescence assay showed that CBX3 colocalized with TRIM28, TRIM24 or RBBP4 in A549 cells (Supplementary Fig. 4c). TRIM28 and TRIM24 belong to the same TRIM subfamily of proteins, which contain bromodomains [21]. We found that a TRIM28 mutant lacking the bromodomain could not bind to CBX3 in A549 cells (Supplementary Fig. 5a), suggesting that CBX3 may interact with TRIM28 through its bromodomain. As bromodomains recognize di-acetylated proteins [22, 23], we analyzed the amino acid sequence of CBX3 and identified a consensus FBP1 or TWIST1 motif for acetylation [22, 23] (Supplementary Fig. 5b). Expression of CBX3 K81R/K84R (KR) mutants mimicking deacetylated CBX3 decreased the interaction between CBX3 and TRIM28 (Supplementary Fig. 5c) and further downregulated ARHGAP24 compared with wild-type CBX3 (Supplementary Fig. 5d–f). On the other hand, knockdown of TRIM28, TRIM24, and RBBP4 increased ARHGAP24 expression (Fig. 6f–k; Supplementary Fig. 5g–i). Furthermore, we identified a negative correlation between the levels of ARHGAP24 and TRIM28, TRIM24, or RBBP4 in LUAD specimens (Supplementary Fig. 5j–l). Expression of the TRIM28 mutant lacking the bromodomain did not affect the expression levels of ARHGAP24 in A549 cells (Supplementary Fig. 5m). RBBP4, together with the PRC component, catalyzes the trimethylation of histone H3 [24]. Thus, we evaluated whether CBX3 could bind TRIM28, TRIM24, and RBBP4 to regulate ARHGAP24 expression. Analysis of ChIP-seq data revealed the presence of a common binding region for TRIM28, RBBP4, TRIM24, CBX3, and H3K9me3 in the promoter of ARHGAP24 (Supplementary Fig. 6a). ChIP-qPCR analyses confirmed the binding of TRIM28, TRIM24, and RBBP4 to the ARHGAP24 promoter in A549 and H1299 cells (Fig. 6l–n; Supplementary Fig. 6b–d). In addition, knockdown of TRIM28, TRIM24, or RBBP4 reduced the binding of CBX3 to the promoter of ARHGAP24 in A549 cells (Fig. 6o–q). Conversely, overexpression of wild-type TRIM28 but not TRIM28 lacking the bromodomain enhanced CBX3 binding to the ARHGAP24 promoter (Supplementary Fig. 6e). Furthermore, the reduction in ARHGAP24 expression levels induced by overexpression or TRIM28 and the upregulation of ARHGAP24 expression levels induced by knockdown of TRIM28, TRIM24, or RBBP4 were not obvious after coknockdown of CBX3 in A549 cells (Fig. 6r–t; Supplementary Fig. 6f–j). In addtiton, we also found that knockdown of TRIM24 or RBBP4 combined with TRIM28 silencing could not further obviously enhance ARHGAP24 expression in A549 cells (Supplementary Fig. 6k–p), which indicated that TRIM28 is the most important factor in modulating the expression of ARHGAP24. Taken together, these data demonstrate that CBX3 forms a complex with TRIM28, TRIM24, or RBBP4 to regulate ARHGAP24 expression in LUAD.
Fig. 6. CBX3 binds to TRIM28, TRIM24, or RBBP4 to regulate ARHGAP24 expression in LUAD cells.
a PPI network between CBX3 and proteins correlated with CBX3. b The whole cell lysates (WCL) of A549 were collected to undergo immunoprecipitation by using the IgG and CBX3 antibodies. Western blotting analysis was using used to detect the RBBP4, TRIM24, TRIM28 and CBX3. c–e The whole cell lysates (WCL) of A549 were collected to undergo immunoprecipitation by using the IgG and TRIM28, TRIM2, or RBBP4 antibodies, respectively. Western blotting analysis was using used to detect the RBBP4, TRIM24, TRIM28 and CBX3. f, g A549 cells were infected with shControl, shTRIM28 #1, or shTRIM28 #2 for 72 h. Cells were collected for Western blotting analysis (f) and RT-qPCR analysis (g). Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test. Data presented as Mean ± SD with three replicates. *P < 0.05; **P < 0.01; ***P < 0.001. h, i A549 cells were infected with shControl, shTRIM24 #1, or shTRIM24 #2 for 72 h. Cells were collected for Western blotting analysis (h) and RT-qPCR analysis (i). Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test. Data presented as Mean ± SD with three replicates. *P < 0.05; **P < 0.01; ***P < 0.001. j, k A549 cells were infected with shControl, shRBBP4 #1, or shRBBP4 #2 for 72 h. Cells were collected for Western blotting analysis (j) and RT-qPCR analysis (k). Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test. Data presented as Mean ± SD with three replicates. *P < 0.05; **P < 0.01; ***P < 0.001. l The ChIP-qPCR of TRIM28 on the promoter region of ARHGAP24 in A549 cells. Statistical significance was determined by two-side Student t test. Data presented as Mean ± SD with three replicates (n = 3). NS not significant; ***P < 0.001. Primer I indicated the pair of primer located in the common binding peak of RBBP4, TRIM24, TRIM28, CBX3 and H3K9me3; Primer O indicated the the pair of primer located outside the common binding peak of RBBP4, TRIM24, TRIM28, CBX3 and H3K9me3. m The ChIP-qPCR of TRIM24 on the promoter region of ARHGAP24 in A549 cells. Statistical significance was determined by two-side Student t test. Data presented as Mean ± SD with three replicates (n = 3). NS not significant; ***P < 0.001. n The ChIP-qPCR of RBBP4 on the promoter region of ARHGAP24 in A549 cells. Statistical significance was determined by two-side Student t test. Data presented as Mean ± SD with three replicates (n = 3). NS not significant; **P < 0.01. o A549 cells were infected with shControl and shTRIM28 for 72 h. Cells were collected for the ChIP-qPCR of CBX3 on the promoter region of ARHGAP24 in A549 cells. Statistical significance was determined by two-side Student t test. Data presented as Mean ± SD with three replicates (n = 3). *P < 0.05; **P < 0.01. p A549 cells were infected with shControl and shTRIM24 for 72 h. Cells were collected for the ChIP-qPCR of CBX3 on the promoter region of ARHGAP24 in A549 cells. Statistical significance was determined by two-side Student t test. Data presented as Mean ± SD with three replicates. ***P < 0.001. q A549 cells were infected with shControl and shRBBP4 for 72 h. Cells were collected for the ChIP-qPCR of CBX3 on the promoter region of ARHGAP24 in A549 cells. Statistical significance was determined by two-side Student t test. Data presented as Mean ± SD with three replicates. *P < 0.05; **P < 0.01; ***P < 0.001. r–t A549 cells were infected with indicated shRNAs for 72 h. Cells were harvested for Western blotting analysis.
Discussion
Accumulating evidence shows that smoking regulates gene expression by modulating DNA methylation. For example, the methylation status of AHRR, P16, F2RL3, and DAPK has been reported as a biomarker for evaluating the progression of smoking-related diseases, such as lung cancer [25, 26]. However, unlike DNA methylation, histone methylation induced by smoking remains understudied [26]. In this study, through in silico analyses of genes involved in histone methylation, we found that the expression of the histone H3K9 methylation reader CBX3 was upregulated in current smokers with LUAD. We also identified CBX3 as a critical mediator of LUAD progression. However, the relationship between smoking and CBX3 overexpression in the lungs remains to be determined.
CBX3 is upregulated in various types of cancer, including lung cancer, osteosarcoma, liver cancer, and colorectal cancer [11, 27, 28]. However, the clinicopathological and prognostic value of CBX3 in cancer remains controversial. A previous meta-analysis demonstrated that high CBX3 expression was associated with poor prognosis in patients with lung cancer, tongue squamous cell carcinoma, digestive cancer, and urinary cancer [11]. CBX3 has been shown to interact with H3K9me3 to inhibit the expression of downstream target genes [29]. Recently, CBX3 was found to sustain the pluripotency of embryonic stem cells in an H3K26me3-dependent manner by recognizing H3K9me3 [20]. CBX3 plays a pivotal role in tumorigenesis. By inhibiting the P53/P21 pathway, CBX3 maintains the self-renewal ability of stem cells and enhances tumor growth in esophageal squamous cell carcinoma [30]. CBX3 has also been shown to regulate the c-Met/AKT/mTOR signaling pathway in glioma [31]. In prostate cancer, CBX3 has been demonstrated to regulate androgen receptor signaling and upregulate c-Myc to promote tumor progression [32, 33]. Furthermore, CBX3 has been found to be critical for the proliferation and migration of LUAD cells by inhibiting the expression of NCOR2 and ZBTB7A [34]. Our bioinformatic analyses of publicly available datasets and our RNA-seq data from LUAD cells indicated that CBX3 regulates the Rho GTPase signaling pathway in LUAD cells. We also found that CBX3, in collaboration with RBBP4, TRIM28, and TRIM24, represses the expression of ARHGAP24 to upregulate the amount of active Rac1. Nonetheless, the specific mechanisms underlying how these three proteins in complex with CBX3 regulate LUAD progression remain unknown.
Rho family GTPases belong to the Ras superfamily [35]. They have been shown to promote tumor growth and cancer cell invasion and migration and to modulate the tumor microenvironment [36–38]. Typical Rho family members include RhoA, Rac1, and Cdc42 [39]. Rho-associated protein kinases (ROCK1 and ROCK2) are important downstream effectors of Rho GTPases [40]. Rho-ROCK signaling has been extensively studied as a therapeutic target in cancer [38]. The cycle of Rho GTPases is regulated by three types of proteins: (1) guanine nucleotide exchange factors (GEFs), which activate Rho GTPases by exchanging GTP with GDP; [41] (2) GTPase-activating proteins, which promote GTP hydrolysis and inactivate Rho GTPases; [42] and (3) guanine nucleotide ionization inhibitors (GDIs), which maintain the inactive state of Rho proteins in the cytoplasm, thereby terminating the signaling pathway [43]. Rho GTPases are also regulated by posttranslational modifications, including lipid modification, phosphorylation, ubiquitination, and SUMOylation [44]. As a histone H3K9 methylation reader, CBX3 indirectly increases the amount of active Rac1 by inhibiting the GTPase-activating protein ARHGAP24. Because of the role of Rac1 in tumor progression [45], our findings provide a theoretical basis for the development of drugs targeting CBX3 to treat LUAD.
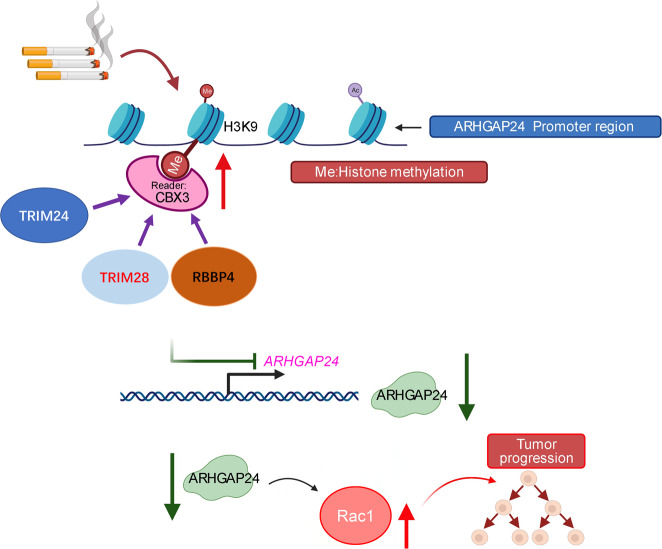
In conclusion, our results suggest that CBX3 is upregulated in current smokers with LUAD. CBX3 overexpression promotes LUAD cell proliferation and invasion and contributes to poor survival outcomes in patients with LUAD. This oncogenic role of CBX3 is mediated via the ARHGAP24/Rac1 pathway. Our data also suggest that TRIM28, TRIM24, and RBBP4 regulate the CBX3/ARHGAP24 axis in LUAD (Fig. 7). These findings strongly suggest that the CBX3/ARHGAP24/Rac1 axis plays a key role in smoking-induced LUAD.
Fig. 7. A model depicting that smoking induced CBX3 up-regulating in the lung adenocarcinoma.
The overexpressed CBX3 coupled with TRIM28, TRIM24 or RBBP4 to repress ARHGAP24 and indirectly increase the amount of active Rac1, which finally promotes the progression of lung adenocarcinoma.
Supplementary information
Acknowledgements
This work was supported by grants from the Chinese National Natural Science Foundation Grant No. 82073321 (XJ).
Author contributions
XJ: methodology, writing - original rraft, project administration; BZ: conceptualization, formal analysis, methodology; HZ: investigation, project administration; HY: methodology, investigation, project administration.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding authors (haixin.y@outlook.com) on reasonable request.
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
The study was conducted in accordance with the principles of the Declaration of Helsinki principles. It was approved by the Animal Use and Care Committees at Tongji Medical College, Huazhong University of Science and Technology.
Consent for publication
All subjects have written informed consent.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xin Jin, Bin Zhang.
Contributor Information
Hao Zhang, Email: zhanghao7106@csu.edu.cn.
Haixin Yu, Email: haixin.y@outlook.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41388-021-02114-8.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Stueve TR, Li WQ, Shi J, Marconett CN, Zhang T, Yang C, et al. Epigenome-wide analysis of DNA methylation in lung tissue shows concordance with blood studies and identifies tobacco smoke-inducible enhancers. Hum Mol Genet. 2017;26:3014–27. doi: 10.1093/hmg/ddx188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu H, Raut JR, Schottker B, Holleczek B, Zhang Y, Brenner H. Individual and joint contributions of genetic and methylation risk scores for enhancing lung cancer risk stratification: data from a population-based cohort in Germany. Clin Epigenetics. 2020;12:89. doi: 10.1186/s13148-020-00872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sundar IK, Nevid MZ, Friedman AE, Rahman I. Cigarette smoke induces distinct histone modifications in lung cells: implications for the pathogenesis of COPD and lung cancer. J Proteome Res. 2014;13:982–96. doi: 10.1021/pr400998n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang N, Chen P, Chen Y, Zeng H, He X, Zhu Y. PRMT6 mediates CSE induced inflammation and apoptosis. Int Immunopharmacol. 2015;24:95–101. doi: 10.1016/j.intimp.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 6.He X, Li T, Kang N, Zeng H, Ren S, Zong D, et al. The protective effect of PRMT6 overexpression on cigarette smoke extract-induced murine emphysema model. Int J Chron Obstruct Pulmon Dis. 2017;12:3245–54. doi: 10.2147/COPD.S144881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oyama K, El-Nachef D, Fang C, Kajimoto H, Brown JP, Singh PB, et al. Deletion of HP1gamma in cardiac myocytes affects H4K20me3 levels but does not impact cardiac growth. Epigenetics Chromatin. 2018;11:18. doi: 10.1186/s13072-018-0187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abe K, Naruse C, Kato T, Nishiuchi T, Saitou M, Asano M. Loss of heterochromatin protein 1 gamma reduces the number of primordial germ cells via impaired cell cycle progression in mice. Biol Reprod. 2011;85:1013–24. doi: 10.1095/biolreprod.111.091512. [DOI] [PubMed] [Google Scholar]
- 9.Bot C, Pfeiffer A, Giordano F, Manjeera DE, Dantuma NP, Strom L. Independent mechanisms recruit the cohesin loader protein NIPBL to sites of DNA damage. J Cell Sci. 2017;130:1134–46. doi: 10.1242/jcs.197236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang C, Su T, Xue Y, Cheng C, Lay FD, McKee RA, et al. Cbx3 maintains lineage specificity during neural differentiation. Genes Dev. 2017;31:241–6. doi: 10.1101/gad.292169.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin H, Zhao X, Xia L, Lian J, You J. Clinicopathological and prognostic significance of CBX3 expression in human cancer: a systematic review and meta-analysis. Dis Markers. 2020;2020:2412741. doi: 10.1155/2020/2412741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maeda T, Hiraki M, Jin C, Rajabi H, Tagde A, Alam M, et al. MUC1-C induces PD-L1 and immune evasion in triple-negative breast cancer. Cancer Res. 2018;78:205–15. doi: 10.1158/0008-5472.CAN-17-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura F. FilGAP and its close relatives: a mediator of Rho-Rac antagonism that regulates cell morphology and migration. Biochem J. 2013;453:17–25. doi: 10.1042/BJ20130290. [DOI] [PubMed] [Google Scholar]
- 14.Jin X, Yang C, Fan P, Xiao J, Zhang W, Zhan S, et al. CDK5/FBW7-dependent ubiquitination and degradation of EZH2 inhibits pancreatic cancer cell migration and invasion. J Biol Chem. 2017;292:6269–80. doi: 10.1074/jbc.M116.764407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin X, Pan Y, Wang L, Ma T, Zhang L, Tang AH, et al. Fructose-1,6-bisphosphatase Inhibits ERK activation and bypasses gemcitabine resistance in pancreatic cancer by blocking IQGAP1-MAPK interaction. Cancer Res. 2017;77:4328–41. doi: 10.1158/0008-5472.CAN-16-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang B, Cheng X, Zhan S, Jin X, Liu T. MIB1 upregulates IQGAP1 and promotes pancreatic cancer progression by inducing ST7 degradation. Mol Oncol. 2021;15:3062–3075. doi: 10.1002/1878-0261.12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Wang S, Takayama K, Harada T, Okamoto I, Iwama E, et al. Nicotine induces resistance to erlotinib via cross-talk between alpha 1 nAChR and EGFR in the non-small cell lung cancer xenograft model. Lung Cancer. 2015;88:1–8. doi: 10.1016/j.lungcan.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 18.Bid HK, Roberts RD, Manchanda PK, Houghton PJ. RAC1: an emerging therapeutic option for targeting cancer angiogenesis and metastasis. Mol Cancer Ther. 2013;12:1925–34. doi: 10.1158/1535-7163.MCT-13-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H, Wang W, Shen W, Wang L, Zuo Y. ARHGAP24 ameliorates inflammatory response through inactivating Rac1/Akt/NF-kappaB pathway in acute pneumonia model of rat. Ann Transl Med. 2020;8:1289. doi: 10.21037/atm-20-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaidan NZ, Sridharan R. HP1gamma regulates H3K36 methylation and pluripotency in embryonic stem cells. Nucleic Acids Res. 2020;48:12660–74. doi: 10.1093/nar/gkaa1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fong KW, Zhao JC, Song B, Zheng B, Yu J. TRIM28 protects TRIM24 from SPOP-mediated degradation and promotes prostate cancer progression. Nat Commun. 2018;9:5007. doi: 10.1038/s41467-018-07475-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210–25. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang C, Zhu S, Yang H, Fan P, Meng Z, Zhao J, et al. FBP1 binds to the bromodomain of BRD4 to inhibit pancreatic cancer progression. Am J Cancer Res. 2020;10:523–35. [PMC free article] [PubMed] [Google Scholar]
- 24.Glancy E, Ciferri C, Bracken AP. Structural basis for PRC2 engagement with chromatin. Curr Opin Struct Biol. 2021;67:135–44. doi: 10.1016/j.sbi.2020.10.017. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Elgizouli M, Schottker B, Holleczek B, Nieters A, Brenner H. Smoking-associated DNA methylation markers predict lung cancer incidence. Clin Epigenetics. 2016;8:127. doi: 10.1186/s13148-016-0292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaur G, Begum R, Thota S, Batra S. A systematic review of smoking-related epigenetic alterations. Arch Toxicol. 2019;93:2715–40. doi: 10.1007/s00204-019-02562-y. [DOI] [PubMed] [Google Scholar]
- 27.Zhong X, Kan A, Zhang W, Zhou J, Zhang H, Chen J, et al. CBX3/HP1gamma promotes tumor proliferation and predicts poor survival in hepatocellular carcinoma. Aging (Albany NY) 2019;11:5483–97. doi: 10.18632/aging.102132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saini V, Hose CD, Monks A, Nagashima K, Han B, Newton DL, et al. Identification of CBX3 and ABCA5 as putative biomarkers for tumor stem cells in osteosarcoma. PLoS ONE. 2012;7:e41401. doi: 10.1371/journal.pone.0041401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh PB. HP1 proteins—what is the essential interaction? Genetika. 2010;46:1424–9. [PubMed] [Google Scholar]
- 30.He Z, Chen J, Chen X, Wang H, Tang L, Han C. microRNA-377 acts as a suppressor in esophageal squamous cell carcinoma through CBX3-dependent P53/P21 pathway. J Cell Physiol. 2021;236:107–20. doi: 10.1002/jcp.29631. [DOI] [PubMed] [Google Scholar]
- 31.Cai H, Yu Y, Ni X, Li C, Hu Y, Wang J, et al. LncRNA LINC00998 inhibits the malignant glioma phenotype via the CBX3-mediated c-Met/Akt/mTOR axis. Cell Death Dis. 2020;11:1032. doi: 10.1038/s41419-020-03247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang C, Liu J, He W, Qu M, Huang X, Deng Y, et al. A regulatory circuit HP1gamma/miR-451a/c-Myc promotes prostate cancer progression. Oncogene. 2018;37:415–26. doi: 10.1038/onc.2017.332. [DOI] [PubMed] [Google Scholar]
- 33.Itsumi M, Shiota M, Yokomizo A, Kashiwagi E, Takeuchi A, Tatsugami K, et al. Human heterochromatin protein 1 isoforms regulate androgen receptor signaling in prostate cancer. J Mol Endocrinol. 2013;50:401–9. doi: 10.1530/JME-13-0024. [DOI] [PubMed] [Google Scholar]
- 34.Alam H, Li N, Dhar SS, Wu SJ, Lv J, Chen K, et al. HP1gamma promotes lung adenocarcinoma by downregulating the transcription-repressive regulators NCOR2 and ZBTB7A. Cancer Res. 2018;78:3834–48. doi: 10.1158/0008-5472.CAN-17-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray JL, von Delft F, Brennan PE. Targeting the small GTPase superfamily through their regulatory. Proteins Angew Chem Int Ed Engl. 2020;59:6342–66. doi: 10.1002/anie.201900585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porazinski S, Parkin A, Pajic M. Rho-ROCK signaling in normal physiology and as a key player in shaping the tumor microenvironment. Adv Exp Med Biol. 2020;1223:99–127. doi: 10.1007/978-3-030-35582-1_6. [DOI] [PubMed] [Google Scholar]
- 37.Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582:2093–101. doi: 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 38.Haga RB, Ridley AJ. Rho GTPases: regulation and roles in cancer cell biology. Small GTPases. 2016;7:207–21. doi: 10.1080/21541248.2016.1232583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svensmark JH, Brakebusch C. Rho GTPases in cancer: friend or foe? Oncogene. 2019;38:7447–56. doi: 10.1038/s41388-019-0963-7. [DOI] [PubMed] [Google Scholar]
- 40.Johan MZ, Samuel MS. Rho-ROCK signaling regulates tumor-microenvironment interactions. Biochem Soc Trans. 2019;47:101–8. doi: 10.1042/BST20180334. [DOI] [PubMed] [Google Scholar]
- 41.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–80. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 42.Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- 43.Cho HJ, Kim JT, Baek KE, Kim BY, Lee HG. Regulation of Rho GTPases by RhoGDIs in Human Cancers. Cells. 2019;8:1037. doi: 10.3390/cells8091037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hodge RG, Ridley AJ. Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol. 2016;17:496–510. doi: 10.1038/nrm.2016.67. [DOI] [PubMed] [Google Scholar]
- 45.Yang J, Qiu Q, Qian X, Yi J, Jiao Y, Yu M, et al. Long noncoding RNA LCAT1 functions as a ceRNA to regulate RAC1 function by sponging miR-4715-5p in lung cancer. Mol Cancer. 2019;18:171. doi: 10.1186/s12943-019-1107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding authors (haixin.y@outlook.com) on reasonable request.