Abstract
Haematological traits are linked to cardiovascular, metabolic, infectious and immune disorders, as well as cancer. Here, we examine the role of genetic variation in shaping haematological traits in two isolated Mediterranean populations. Using whole-genome sequencing data at 22× depth for 1457 individuals from Crete (MANOLIS) and 1617 from the Pomak villages in Greece, we carry out a genome-wide association scan for haematological traits using linear mixed models. We discover novel associations (p < 5 × 10–9) of five rare non-coding variants with alleles conferring effects of 1.44–2.63 units of standard deviation on red and white blood cell count, platelet and red cell distribution width. Moreover, 10.0% of individuals in the Pomak population and 6.8% in MANOLIS carry a pathogenic mutation in the Haemoglobin Subunit Beta (HBB) gene. The mutational spectrum is highly diverse (10 different mutations). The most frequent mutation in MANOLIS is the common Mediterranean variant IVS-I-110 (G>A) (rs35004220). In the Pomak population, c.364C>A (“HbO-Arab”, rs33946267) is most frequent (4.4% allele frequency). We demonstrate effects on haematological and other traits, including bilirubin, cholesterol, and, in MANOLIS, height and gestation age. We find less severe effects on red blood cell traits for HbS, HbO, and IVS-I-6 (T>C) compared to other b+ mutations. Overall, we uncover allelic diversity of HBB in Greek isolated populations and find an important role for additional rare variants outside of HBB.
Subject terms: DNA sequencing, Next-generation sequencing, Genetic association study, Medical genomics, Quantitative trait, Biomarkers
Introduction
The number, size and shape of blood cells vary widely between individuals. The genetic architecture of these haematological traits is complex1. Genome-wide association studies (GWAS) have identified more than 100 common variants with relatively small effects1–7. Moreover, autosomal recessive mutations in the Haemoglobin Subunit Alpha and Beta genes can cause haemoglobin disorders8. These mutations lead to either reduced levels or abnormal structure of the haemoglobin subunits. The most common disorders are sickle cell disease, alpha and beta thalassemia9. Inherited anaemia is particularly prevalent in parts of Africa, the Mediterranean region, the Middle East, the Indian subcontinent, and Southeast Asia9. The geographic distribution overlaps regions with a history of long-standing endemic malaria10 and there is strong evidence that heterozygous carrier status provides a protective effect against malaria9.
Haemoglobin disorders represent the most common type of monogenic disorder globally11. With almost 500 thalassemia mutations in the HbVar database12 and substantial heterogeneity in disease manifestations13, the genetic architecture of thalassemia in particular is complex. Knowledge of the full mutational spectrum and carrier frequencies is still limited for many populations9. Heterozygous mutations can lead to thalassemia trait, a milder form of haemoglobin-related abnormalities. A comprehensive assessment of the physiological effects in a large sample of heterozygotes and comparisons of individual mutations is lacking for most variants. Little is known about the role of genetic variation outside of the haemoglobin genes in areas with high prevalence of haemoglobin disorders as the vast majority of previous studies has assessed common variation only and used data from individuals of European ancestries outside historic malaria regions1–7. Studying isolated populations offers power gains in detecting associations involving rare and low-frequency variants14.
We carried out whole-genome sequencing in 3,074 individuals from two Mediterranean populations, the Hellenic Isolated Cohorts (HELIC). The HELIC cohorts (https://www.helmholtz-muenchen.de/index.php?id=53481)15–17 consist of MANOLIS which includes individuals from the mountainous Mylopotamos villages on Crete. The Pomak cohort includes individuals who were recruited at the Pomak villages, a set of mountainous villages in the North of Greece. Genetic isolatedness has been demonstrated for both cohorts16. We assess the role of common and rare variation across the genome for haematological traits. We characterise the mutational spectrum for haemoglobin genes and determine the impact of mutations across the phenome in non-clinically ascertained samples.
Results
Novel genetic associations with haematological traits
Following quality control, data were available for 1,457 individuals for MANOLIS and 1,617 individuals for the Pomak population. Five rare, previously unreported loci were associated with haematological traits in either cohort after adjusting for multiple testing (p < 5 × 10–9) (Table 1).
Table 1.
Association results of the lead SNPs of novel genome-wide significant loci with haematological traits.
| Trait | Chr | Gene | Rs-id | Position | Type | A1* | A0* | AF* | AF Topmed | Beta | SE | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pomak | ||||||||||||
| White blood cell count | 2 | LOC105374834 | rs551751343 | 81,484,692 | Intron | T | C | 0.004 | 0.0001 | 1.72 | 0.29 | 4.1 × 10–9 |
| 2 | – | 81,745,516 | – | A | G | 0.004 | – | 1.72 | 0.29 | 4.1 × 10–9 | ||
| 2 | LOC102724542 | rs1041582657 | 81,759,339 | Intron | A | G | 0.004 | 0.00001 | 1.72 | 0.29 | 4.1 × 10–9 | |
| 2 | rs556280089 | 81,954,850 | Intergenic | T | C | 0.004 | 0.00001 | 1.72 | 0.29 | 4.1 × 10–9 | ||
| 2 | rs989421555 | 82,175,919 | Intergenic | G | C | 0.004 | 0.00009 | 1.72 | 0.29 | 4.1 × 10–9 | ||
| 2 | – | 82,474,946 | – | G | A | 0.004 | – | 1.72 | 0.29 | 4.1 × 10–9 | ||
| 2 | rs576414992 | 82,654,697 | Intergenic | G | C | 0.004 | 0.00006 | 1.72 | 0.29 | 4.1 × 10–9 | ||
| 2 | LOC105374831 | rs73941786 | 82,835,859 | Intron | C | A | 0.004 | 0.00833 | 1.72 | 0.29 | 4.1 × 10–9 | |
| 2 | LOC105374834 | rs190806297 | 83,537,814 | Intron | A | G | 0.004 | 0.00004 | 1.72 | 0.29 | 4.1 × 10–9 | |
| 2 | rs140340075 | 83,662,388 | Intergenic | C | A | 0.004 | – | 1.72 | 0.29 | 4.1 × 10–9 | ||
| 2 | rs188113595 | 83,930,989 | Intergenic | T | C | 0.004 | 0.00144 | 1.72 | 0.29 | 4.1 × 10–9 | ||
| 2 | rs573444805 | 84,150,305 | Intergenic | G | C | 0.004 | 0.00004 | 1.72 | 0.29 | 4.1 × 10–9 | ||
| 2 | rs548781149 | 84,159,599 | Intergenic | T | C | 0.004 | 0.00004 | 1.72 | 0.29 | 4.1 × 10–9 | ||
| 2 | – | 84,341,726 | – | C | C | 0.004 | – | 1.72 | 0.29 | 4.1 × 10–9 | ||
| 2 | – | 84,464,745 | – | T | G | 0.004 | – | 1.72 | 0.29 | 4.1 × 10–9 | ||
| 2 | – | 84,535,416 | – | C | C | 0.004 | – | 1.72 | 0.29 | 4.1 × 10–9 | ||
| Red cell distribution width | 9 | SVEP1 | rs189173017 |
110,549,951 110,453,573 |
Intron | G | A | 0.004 | 0.00294 | − 1.90 | 0.30 | 8.4 × 10–10 |
| Red cell distribution width | 9 | LPAR1 | rs145221983 | 110,885,694 | Intron | G | C | 0.002 | 0.00178 | − 2.64 | 0.39 | 3.9 × 10–11 |
| platelet distribution width | 20 | APCDD1L | rs73183273 | 58,478,356 | Intron | A | C | 0.007 | 0.00483 | 1.44 | 0.23 | 4.1 × 10–9 |
| MANOLIS | ||||||||||||
| Red blood cell count | 15 | rs1320751535 | 101,913,651 | Intergenic | G | A | 0.009 | 0.00002 | 1.52 | 0.23 | 6.2 × 10–10 | |
Betas are reported in units of standard deviation of the traits. The lead SNP for the locus on chromosome 2 is represented by 16 variants in perfect linkage disequilibrium covering an area of 3 Mb.*A0 is the reference allele and A1 is the effect allele for which the allele frequency (AF) is reported in the current sample and in TopMed.
The G-allele (minor allele frequency (MAF) = 0.009) of rs1320751535 at 15q26 was associated with an increase in red blood cell count by 1.52 units of standard deviation error (SE = 0.23, p = 6.2 × 10–10) in MANOLIS. This single nucleotide variant (SNV) is extremely rare in reference samples. It is not observed in the 1000 Genomes Project data and is carried by 2 out of 125,568 individuals in TopMed18. Of note, the credible set in this region, a group of variants that is likely to contain the causal one, included only two markers, neither of which has been reported in previous GWAS for blood traits.
In the Pomak population, we identified four novel loci, including an association at 2p11.2 with increased white blood cell count (WBC) (beta = 1.72, SE = 0.29, p value = 4.1 × 10–9). The locus contains 16 highly associated variants in perfect linkage disequilibrium that span a region of 3 Mb (Supplementary Material, Supplementary Table 2). One of the seven variants in the credible set was associated with WBC at nominal significance in a large cosmopolitan GWAS1 (Supplementary Material, Supplementary Table 3). There was also a novel association with platelet distribution width (PDW) at 20q13.32. Variant rs73183273 (beta = 1.44, SE = 0.23, p value = 4.1 × 10–9) is located within an intron of the APC down-regulated 1 like (APCDD1L) gene. We also identified an association of rs189173017 with red cell distribution width (RDW) (beta = - 1.90, SE = 0.30, p value = 8.4 × 10–10). This variant is located in an intron of sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 (SVEP1) at 9q31.3 and has a Regulome Database (RDB) score of 5, indicating regulatory function (Supplementary Material, Supplementary Table 2). Variants in the credible set, including rs201343203 intronic to FSD1L and rs186868542 intronic to TMEM38B, overlap with enhancer sites that are active in blood. In a previous study, an association with platelet distribution width has been reported for another variant in SVEP1: missense mutation rs61751931, but not with RDW. There was another novel association on chromosome 9. The rare G-allele of a variant at 9q31.3, rs145221983, had a large effect on RDW with a beta of − 2.62 (SE = 0.39, p value = 3.9 × 10–11) in the Pomak population. It is intronic to lysophosphatidic acid receptor 1 (LPAR1). The encoded membrane protein belongs to a group known as endothelial differentiation gene receptors which mediate platelet aggregation. Common variants in LPAR1 have previously been linked to haematological traits1.
We replicated several previously-reported associations1 at genome-wide significance, mostly with platelet traits. Variant rs11553699 at 12q24.31 was associated with mean platelet volume (beta = 0.39, SE = 0.06, p value = 4.9 × 10–11) and platelet distribution width (beta = 0.39, SE = 0.06, p value = 2.3 × 10–10) in MANOLIS, and rs1354034 at 3p14.3 and rs342293 at 7q22.3 with large platelet distribution ratio (beta = 0.23, SE = 0.037, p value = 7.0 × 10–10 and beta = 0.22, SE = 0.04, p value = 1.4 × 10–9, respectively), mean platelet volume (beta = 0.24, SE = 0.04, p value = 1.4 × 10–10 and beta = 0.23, SE = 0.04, p value = 3.8 × 10–10, respectively) and platelet distribution width (beta = 0.23, SE = 0.04, p value = 2.1 × 10–9 and beta = 0.22, SE = 0.04, p value = 3.9 × 10–9, respectively) in the Pomak population (Supplementary Table 4).
HBB mutations
Across the entire genome, a region on chromosome 11 displayed the strongest association with all measured red blood cell traits, except haematocrit (HCT) in Pomak and mean corpuscular haemoglobin concentration (MCHC) in MANOLIS (Supplementary Material, Supplementary Fig. 1). Conditional analyses demonstrated that the chr11 peak in the Pomak population could be explained by the three most frequent mutations in the HBB gene, c.364C>A, IVS-II-745 and IVS-I-6 (Fig. 1). The same was true for MANOLIS, again with three independent signals from HBB mutations, IVS-I-110, CD8/9+G and CD39C>T. We identified all mutations in the HBB gene that have previously been classified as pathogenic, as described in the methods section. In both groups, we observed high proportions of carriers. A total of 99 individuals in MANOLIS (6.8% of the individuals) carried a pathogenic HBB mutation and 162 individuals in the Pomak population (10.0% of the individuals). Different mutation spectra were observed in the two populations (Table 2). There were ten mutations overall, six observed in MANOLIS and six in the Pomak population. Consequences included missense, splice site, splice donor, stop gained and frameshift mutations.
Figure 1.
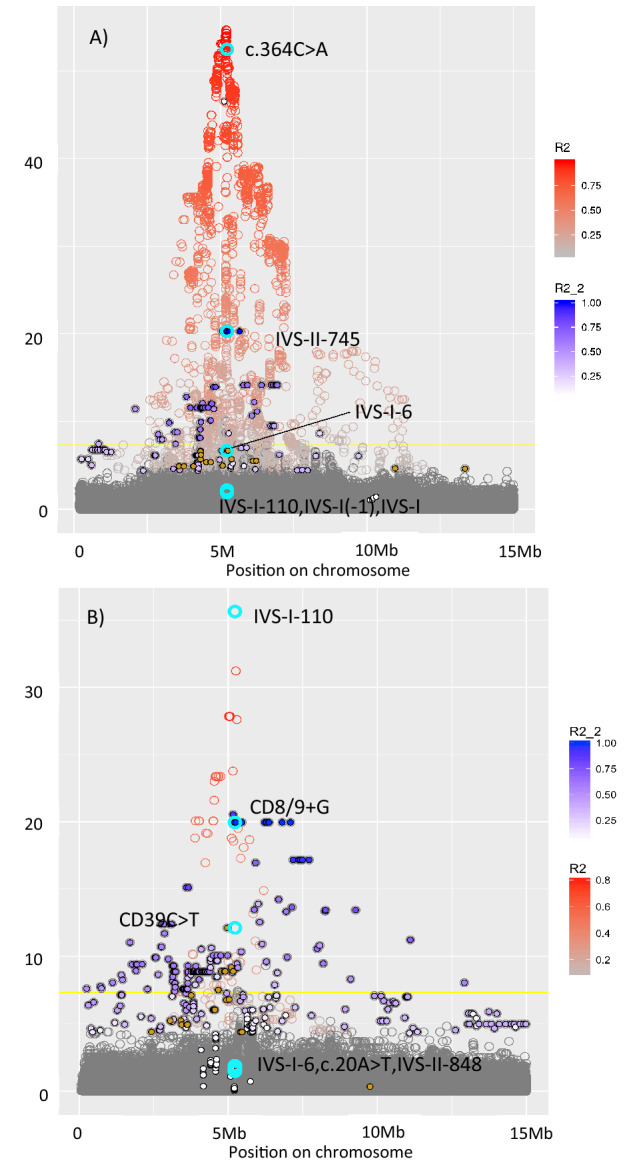
Regional association plot for variants located between 0 and 15 Mb on chromosome 11. Each circle represents a genetic variant. They are arranged on the x-axis by their location. The y-axis shows the p-value for their association with red cell distribution width in (A) Pomak and (B) MANOLIS. Pathogenic HBB mutations are highlighted in turquoise and labelled. The colouring of the circle (R2) indicates the strength of linkage disequilibrium (LD) with the most strongly associated HBB mutation, c.364C>A in MANOLIS and IVS-I-110 in Pomak. The blue filling of points (R2_2) indicates the strength of LD with the second most strongly associated HBB mutation, IVS-II-745 in MANOLIS and CD8/9+G in Pomak. The brown filling of the circle indicates variants in LD with the third most strongly associated HBB mutation, IVS-I-6 in MANOLIS and CD39C>T in Pomak.
Table 2.
Pathogenic HBB mutations in Pomak and MANOLIS and their associations with red cell distribution width.
| Mutation | rs-id | Consequence | type | Position | Allele frequency | N carriers | Beta* | SE | P-value* |
|---|---|---|---|---|---|---|---|---|---|
| Pomak (N = 1617) | |||||||||
| HbO-Arab c.364G>A (p.Glu122Lys) | rs33946267 | Missense | HbO | 5,225,678 | 0.044 | 139 | − 1.35 | 0.085 | 1.3 × 10–51 |
| IVS-II-745c.316-106C> G | rs34690599 | Splice site | β + | 5,225,832 | 0.004 | 14 | − 2.79 | 0.291 | 4.7 × 10–21 |
| IVS-I-110 c.93-21G>A | rs35004220 | Splice site | β + | 5,226,820 | 0.0003 | 1 | − 2.59 | 0.949 | 0.006 |
| IVS-I-6 c.92 + 6T>C | rs35724775 | Splice site | β + | 5,226,924 | 0.002 | 7 | − 1.99 | 0.383 | 2.3 × 10–7 |
| IVS-I-1 c.92+1G>A | rs33971440 | Splice donor | β0 | 5,226,929 | 0.0003 | 1 | − 2.39 | 0.968 | 0.014 |
| IVS-I (-1) c.92G>A (p.Arg31Lys) | rs33960103 | Missense | β0 | 5,226,930 | 0.0003 | 1 | − 2.40 | 0.908 | 0.008 |
| MANOLIS (N = 1457) | |||||||||
| IVS-II-848 c.316-3C>A | rs33913413 | Splice site | β + | 5,225,729 | 0.0003 | 1 | − 2.12 | 0.991 | 0.033 |
| CD39 c.118C>T (p.Gln40Ter) | rs11549407 | Stop gained | β0 | 5,226,774 | 0.005 | 13 | − 2.06 | 0.285 | 7.4 × 10–13 |
| IVS-I-110 c.93-21G>A | rs35004220 | splice site | β + | 5,226,820 | 0.015 | 44 | − 2.16 | 0.166 | 3.2 × 10–36 |
| IVS-I-6 c.92 + 6T>C | rs35724775 | splice site | β + | 5,226,924 | 0.001 | 3 | − 1.40 | 0.562 | 0.013 |
| CD8/9 + G c.27dupG (p.Ser10Valfs*14) | rs35699606 | frameshift | β0 | 5,226,995 | 0.009 | 23 | − 2.35 | 0.247 | 1.1 × 10–20 |
| c.20A>T (p.Glu7Val) | rs334 | missense | HbS | 5,227,002 | 0.004 | 15 | − 0.79 | 0.342 | 0.021 |
* Regression coefficient beta and p value for the association of the variant with red cell distribution width in units of standard deviation.
Mutation spectrum in the Pomak population
The missense variant c.364C>A (rs33946267), also known as HbO-Arab19, was the most common pathogenic mutation in HBB with 139 carriers and an allele frequency of 4.4%. While no carriers of this mutation were observed in MANOLIS, 85% of HBB carriers in the Pomak group had the c.364C>A mutation. Variants in linkage disequilibrium (LD) with c.364C>A in the Pomak population spanned a range of almost 10 Mb from approximately 2,500,000 to 11,000,000 (Fig. 1).
Mutation spectrum in MANOLIS
IVS-I-110 (G>A) was also present in MANOLIS, where it was the most common HBB mutation with 44 carriers of this mutation which represent 44% of all carriers in MANOLIS. Additional mutations in MANOLIS included (Table 2): CD39 (C>T) (rs11549407), CD8/9+G (rs35699606) and the sickle cell HbS mutation c.20A>T (rs334). CD8/9+G in MANOLIS was located on a haplotype ranging from 0 to 15,000,000.
Characterisation of phenotypic effects of HBB mutations
To assess the effects on haematological traits, we grouped HBB mutations into those that either reduce (b+) or abolish (b0) expression of beta-globin (see Table 2 for list of mutations) and analysed c.364C>A (HbO-Arab) in Pomak and sickle cell HbS c.20A>T (rs334) in MANOLIS separately. Carriers of thalassemia variants were characterised by microcytosis (decreased mean corpuscular volume), hypochromia (decreased mean corpuscular haemoglobin), mild anaemia (decreased haemoglobin) as well as decreased mean corpuscular haemoglobin concentration, haematocrit, red cell distribution width and increased red blood cell count (Table 3). We also observed increased platelet distribution width and large platelet distribution ratio in Hb0 carriers in MANOLIS. In line with previous reports20, c.364C>A (HbO-Arab) led to increased mean corpuscular haemoglobin concentration while thalassemia variants were linked to decreased values. The effects of the HbO-Arab mutation extended beyond red blood cell traits: for 12 out of 15 measured haematological traits, we observed significant differences after Bonferroni correction (p < 0.0013). These included associations with platelet traits: increased platelet distribution width (beta = 0.62, SE = 0.18, p value = 4.2 × 10–4), mean platelet volume (beta = 0.34, SE = 0.09, p value = 1.6 × 10–4) and large platelet distribution ratio (beta = 2.77, SE = 0.70, p value = 7.5 × 10–5). We also observed an increased white blood cell count (beta = 1.25, SE = 0.17, p value = 1.6 × 10–13) for HbO-Arab carriers and consistent effects on lymphocyte (beta = 0.36, SE = 0.06, p value = 1.3 × 10–8) and neutrophil count (beta = 0.77, SE = 0.14, p value = 9.8 × 10–8).
Table 3.
Differences in haematological, cardiometabolic and anthropometrics traits between carriers and non-carriers of HBB mutations. width RDW-SD in Pomak, RDW-a in Manolis.
| Trait | Unit | Pomak | MANOLIS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Type | Beta | SE | P value | Type | Beta | SE | P value | ||
| Red cell distribution | fl | b+ | − 10.45 | 0.74 | < 2e−16 | b+ | − 16.78 | 0.90 | < 2e−16 |
| b0 | − 12.06 | 2.28 | 1.4e−07 | b0 | − 17.28 | 0.97 | < 2e−16 | ||
| HbO | − 6.42 | 0.30 | < 2e−16 | HbS | − 6.92 | 1.81 | 1.4E−04 | ||
| Red blood cell count | 10^12/l | b+ | 1.02 | 0.08 | < 2e−16 | b+ | 0.88 | 0.08 | < 2e−16 |
| b0 | 0.82 | 0.26 | 1.3E−03 | b0 | 1.03 | 0.08 | < 2e−16 | ||
| HbO | 0.30 | 0.03 | < 2e−16 | HbS | 0.15 | 0.14 | 0.30 | ||
| Haemoglobin | g/l | b+ | − 1.92 | 0.28 | 4.5e−12 | b+ | − 1.85 | 0.20 | < 2e−16 |
| b0 | − 2.70 | 0.84 | 1.4e−03 | b0 | − 2.15 | 0.21 | < 2e−16 | ||
| HbO | 0.20 | 0.11 | 0.07 | HbS | − 0.68 | 0.37 | 0.07 | ||
| Haematocrit | % | b+ | − 2.82 | 0.71 | 7.4e−05 | b+ | − 4.70 | 0.59 | 3.7E−15 |
| b0 | − 5.71 | 2.17 | 8.7e−03 | b0 | − 5.50 | 0.62 | < 2e−16 | ||
| HbO | − 1.09 | 0.29 | 1.3e−04 | HbS | − 2.39 | 1.09 | 0.03 | ||
| Mean corpuscular volume | fl | b+ | − 20.35 | 1.06 | < 2e−16 | b+ | − 21.42 | 0.83 | < 2e−16 |
| b0 | − 24.49 | 3.27 | 1.1e−13 | b0 | − 24.20 | 0.87 | < 2e−16 | ||
| HbO | − 7.43 | 0.43 | < 2e−16 | HbS | − 7.21 | 1.54 | 3.2E−06 | ||
| Mean corpuscular haemoglobin | pg | b+ | − 8.47 | 0.46 | < 2e−16 | b+ | − 7.79 | 0.31 | < 2e−16 |
| b0 | − 9.67 | 1.42 | 1.6e−11 | b0 | − 8.77 | 0.33 | < 2e−16 | ||
| HbO | − 1.34 | 0.19 | 1.3e−12 | HbS | − 2.18 | 0.58 | 1.9E−04 | ||
| Mean corpuscular haemoglobin concentration | g/dl | b+ | − 2.51 | 0.25 | < 2e−16 | b+ | − 0.64 | 0.15 | 2.2E−05 |
| b0 | − 2.40 | 0.77 | 1.8e−03 | b0 | − 0.67 | 0.16 | 2.9E−05 | ||
| HbO | 1.39 | 0.10 | < 2e−16 | HbS | 0.41 | 0.28 | 0.14 | ||
| Platelet count | 10^9/l | b+ | 26.81 | 12.64 | 0.03 | b+ | 6.38 | 8.73 | 0.46 |
| b0 | − 7.31 | 38.76 | 0.85 | b0 | − 8.64 | 9.19 | 0.35 | ||
| HbO | 5.53 | 5.11 | 0.28 | HbS | − 41.88 | 16.18 | 9.8E−03 | ||
| Platelet distribution width | fl | b+ | 0.21 | 0.46 | 0.64 | b+ | 0.42 | 0.19 | 0.03 |
| b0 | 0.33 | 1.87 | 0.86 | b0 | 0.87 | 0.21 | 5.4E−05 | ||
| HbO | 0.62 | 0.18 | 4.2e−04 | HbS | − 0.56 | 0.40 | 0.16 | ||
| Mean platelet volume | fl | b+ | − 0.22 | 0.24 | 0.35 | b+ | 0.18 | 0.14 | 0.21 |
| b0 | − 0.71 | 0.97 | 0.46 | b0 | 0.44 | 0.15 | 3.1E−03 | ||
| HbO | 0.34 | 0.09 | 1.6e−04 | HbS | − 0.36 | 0.26 | 0.16 | ||
| Plateletcrit | % | b+ | b+ | 0.01 | 0.01 | 0.08 | |||
| b0 | b0 | 0.01 | 0.01 | 0.31 | |||||
| HbO | HbS | − 0.05 | 0.01 | 5.8E−04 | |||||
| Large platelet distribution ratio | % | b+ | − 0.45 | 1.82 | 0.80 | b+ | 2.24 | 0.92 | 0.02 |
| b0 | − 3.30 | 7.45 | 0.66 | b0 | 4.70 | 1.00 | 2.9E−06 | ||
| HbO | 2.77 | 0.70 | 7.5e−05 | HbS | − 2.74 | 1.86 | 0.14 | ||
| Granulocyte count | 10^9/l | b+ | b+ | 0.38 | 0.25 | 0.12 | |||
| b0 | b0 | 0.79 | 0.27 | 3.4E−03 | |||||
| HbO | HbS | − 0.35 | 0.50 | 0.49 | |||||
| White blood cell count | 10^9/l | b+ | 0.53 | 0.42 | 0.21 | b+ | 0.67 | 0.30 | 0.03 |
| b0 | 0.70 | 1.28 | 0.58 | b0 | 0.87 | 0.32 | 6.5E−03 | ||
| HbO | 1.25 | 0.17 | 1.6e−13 | HbS | − 0.76 | 0.56 | 0.18 | ||
| Lymphocyte count | 10^9/l | b+ | 0.38 | 0.16 | 0.01 | b+ | 0.22 | 0.12 | 0.05 |
| b0 | 1.29 | 0.48 | 7.2e−03 | b0 | − 0.03 | 0.12 | 0.78 | ||
| HbO | 0.36 | 0.06 | 1.3e−08 | HbS | − 0.38 | 0.22 | 0.08 | ||
| Neutrophil count | 10^3/L | b+ | − 0.27 | 0.39 | 0.48 | b+ | |||
| b0 | − 0.54 | 1.03 | 0.60 | b0 | |||||
| HbO | 0.77 | 0.14 | 9.8e−08 | HbS | |||||
| Mixed cell count | 10^3/L | b+ | 0.05 | 0.07 | 0.47 | b+ | 0.02 | 0.02 | 0.34 |
| b0 | − 0.05 | 0.18 | 0.76 | b0 | 0.02 | 0.03 | 0.49 | ||
| HbO | 0.03 | 0.02 | 0.19 | HbS | − 0.04 | 0.05 | 0.44 | ||
| C-reactive protein | nmol/L | b+ | − 1.32 | 14.12 | 0.93 | b+ | 3.08 | 16.76 | 0.85 |
| b0 | − 8.13 | 40.97 | 0.84 | b0 | 3.83 | 19.69 | 0.85 | ||
| HbO | 0.10 | 5.85 | 0.99 | HbS | − 2.16 | 28.97 | 0.94 | ||
| Ferritin | pmol/L | b+ | 45.05 | 41.42 | 0.28 | b+ | 12.46 | 41.02 | 0.76 |
| b0 | 302.18 | 127.04 | 0.02 | b0 | 91.95 | 46.69 | 0.05 | ||
| HbO | 24.78 | 17.07 | 0.15 | HbS | − 66.81 | 71.68 | 0.35 | ||
| Iron | mmol/L | b+ | − 1.14 | 1.59 | 0.47 | b+ | 0.52 | 0.86 | 0.54 |
| b0 | 3.86 | 4.86 | 0.43 | b0 | 1.59 | 0.98 | 0.10 | ||
| HbO | 1.57 | 0.65 | 0.02 | HbS | − 2.70 | 1.50 | 0.07 | ||
| Glucose | mmol/l | b+ | 0.01 | 0.40 | 0.99 | b+ | − 0.31 | 0.28 | 0.26 |
| b0 | − 1.61 | 1.21 | 0.19 | b0 | 0.38 | 0.31 | 0.23 | ||
| HbO | − 0.07 | 0.16 | 0.66 | HbS | 0.98 | 0.48 | 0.04 | ||
| Insulin | pmol/L | b+ | − 8.46 | 28.31 | 0.77 | b+ | − 11.73 | 22.27 | 0.60 |
| b0 | − 38.26 | 86.83 | 0.66 | b0 | 92.40 | 25.09 | 0.00 | ||
| HbO | − 11.76 | 11.67 | 0.31 | HbS | 37.28 | 38.51 | 0.33 | ||
| High-density lipoprotein | mmol/L | b+ | − 0.21 | 0.07 | 3.2e−03 | b+ | − 0.04 | 0.05 | 0.47 |
| b0 | − 0.25 | 0.22 | 0.25 | b0 | − 0.10 | 0.06 | 0.09 | ||
| HbO | − 0.05 | 0.03 | 0.12 | HbS | − 0.12 | 0.09 | 0.15 | ||
| Low-density lipoprotein | mmol/L | b+ | − 0.14 | 0.21 | 0.50 | b+ | − 0.43 | 0.14 | 2.5E−03 |
| b0 | 1.21 | 0.64 | 0.06 | b0 | − 0.44 | 0.16 | 6.4E−03 | ||
| HbO | 0.01 | 0.09 | 0.90 | HbS | 0.02 | 0.25 | 0.93 | ||
| Triglycerides | mmol/L | b+ | − 0.06 | 0.21 | 0.79 | b+ | − 0.16 | 0.17 | 0.35 |
| b0 | 0.02 | 0.63 | 0.98 | b0 | − 0.25 | 0.19 | 0.19 | ||
| HbO | 0.01 | 0.08 | 0.89 | HbS | 0.25 | 0.30 | 0.41 | ||
| Total cholesterol | mmol/L | b+ | − 0.38 | 0.23 | 0.11 | b+ | − 0.53 | 0.16 | 9.6E−04 |
| b0 | 0.96 | 0.72 | 0.18 | b0 | − 0.65 | 0.18 | 4.3E−04 | ||
| HbO | − 0.03 | 0.10 | 0.77 | HbS | 0.01 | 0.28 | 0.97 | ||
| Thyroid stimulating hormone | uIU/ml | b+ | − 0.03 | 0.87 | 0.97 | b+ | − 0.23 | 0.49 | 0.64 |
| b0 | 0.75 | 3.36 | 0.82 | b0 | − 0.54 | 0.58 | 0.35 | ||
| HbO | − 0.19 | 0.32 | 0.56 | HbS | − 0.20 | 0.87 | 0.82 | ||
| Free thyroxine | ng/dl | b+ | − 0.12 | 0.05 | 0.01 | b+ | 0.07 | 0.03 | 0.03 |
| b0 | − 0.11 | 0.19 | 0.56 | b0 | 0.00 | 0.04 | 0.99 | ||
| HbO | 0.05 | 0.02 | 0.01 | HbS | 0.01 | 0.06 | 0.85 | ||
| Osteocalcin | ng/ml | b+ | 2.18 | 3.20 | 0.50 | b+ | 2.19 | 1.34 | 0.10 |
| b0 | 9.15 | 10.56 | 0.39 | b0 | − 1.31 | 1.59 | 0.41 | ||
| HbO | 1.07 | 1.18 | 0.36 | HbS | − 2.39 | 2.31 | 0.30 | ||
| Bilirubin | mg/dl | b+ | 0.08 | 0.02 | 1.2e−04 | b+ | 0.04 | 0.01 | 2.2E−03 |
| b0 | 0.07 | 0.08 | 0.39 | b0 | 0.05 | 0.01 | 8.8E−04 | ||
| HbO | 0.04 | 0.01 | 2.8e−07 | HbS | − 0.02 | 0.02 | 0.39 | ||
| Alanine aminotransferase | iu/l | b+ | − 1.15 | 1.84 | 0.53 | b+ | − 5.42 | 1.79 | 2.5E−03 |
| b0 | − 4.95 | 7.10 | 0.49 | b0 | − 3.82 | 2.13 | 0.07 | ||
| HbO | 0.62 | 0.67 | 0.36 | HbS | − 4.37 | 3.10 | 0.16 | ||
| Gamma-glutamyl transferase | iu/l | b+ | 4.49 | 3.95 | 0.26 | b+ | − 5.76 | 3.24 | 0.08 |
| b0 | − 6.96 | 15.22 | 0.65 | b0 | − 0.80 | 3.86 | 0.84 | ||
| HbO | 2.53 | 1.44 | 0.08 | HbS | − 6.24 | 5.79 | 0.28 | ||
| Leptin | ng/ml | b+ | b+ | − 1.79 | 3.51 | 0.61 | |||
| b0 | b0 | − 7.42 | 4.52 | 0.10 | |||||
| HbO | HbS | 13.75 | 6.35 | 0.03 | |||||
| Adiponectin | ug/ml | b+ | b+ | − 0.54 | 0.49 | 0.27 | |||
| b0 | b0 | − 0.77 | 0.54 | 0.15 | |||||
| HbO | HbS | − 2.31 | 0.95 | 0.01 | |||||
| Weight | kg | b+ | 3.94 | 3.29 | 0.23 | b+ | − 0.71 | 2.22 | 0.75 |
| b0 | − 4.97 | 10.09 | 0.62 | b0 | 1.34 | 2.50 | 0.59 | ||
| HbO | 0.12 | 1.34 | 0.93 | HbS | 6.91 | 3.67 | 0.06 | ||
| Height | cm | b+ | 0.66 | 1.54 | 0.67 | b+ | − 0.41 | 0.09 | 7.8E−06 |
| b0 | 5.30 | 4.60 | 0.25 | b0 | − 2.01 | 1.12 | 0.07 | ||
| HbO | − 0.53 | 0.61 | 0.39 | HbS | 5.98 | 1.69 | 4.0E−04 | ||
| Waist-hip ratio | b+ | 0.03 | 0.02 | 0.11 | b+ | − 0.01 | 0.01 | 0.52 | |
| b0 | 0.02 | 0.06 | 0.70 | b0 | 0.02 | 0.01 | 0.13 | ||
| HbO | 0.01 | 0.01 | 0.08 | HbS | 0.00 | 0.02 | 0.85 | ||
| Body-mass-index | b+ | 1.65 | 1.21 | 0.18 | b+ | − 0.38 | 0.82 | 0.65 | |
| b0 | − 3.75 | 3.62 | 0.30 | b0 | 1.25 | 0.92 | 0.17 | ||
| HbO | 0.27 | 0.48 | 0.57 | HbS | 0.61 | 1.37 | 0.66 | ||
| Gestation age | Months | b+ | b+ | − 0.41 | 0.09 | 7.8E−06 | |||
| b0 | b0 | 0.03 | 0.08 | 0.66 | |||||
| HbO | HbS | 0.03 | 0.10 | 0.72 | |||||
We grouped HBB mutations into those that either reduce (b +) or abolish (b0) expression of beta-globin (see Table 2). We also separated c.364C>A (HbO-Arab) in Pomak and sickle cell HbS in MANOLIS. Associations significant after Bonferroni correction (p value < 0.0013) were bolded.
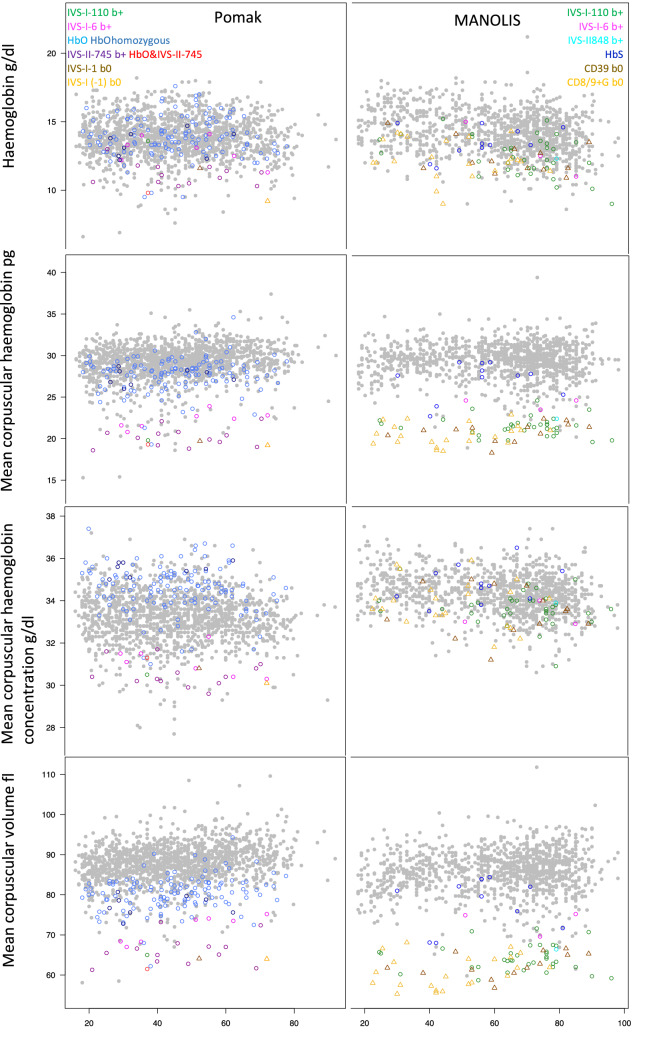
We tested whether the effects on red cell traits differ between specific HBB mutations. Across the mutational spectrum, there was a clear separation between carriers and non-carriers (Fig. 2), and each mutation was individually associated with red cell traits, such as RDW levels (Table 2). A case-only analysis demonstrated differences between the structural variants and thalassemia mutations (Supplementary Material, Supplementary Table 5). Moreover, in both Pomak and MANOLIS, IVS-I-6 had significantly less severe effects on red cell distribution width, mean corpuscular volume and mean corpuscular haemoglobin compared to the most common b+ mutation in each group.
Figure 2.
Values for different red cell traits (y-axis) by age (x-axis) for carriers of different HBB mutations in Pomak and MANOLIS. Individuals without a detected HBB mutation are shown as grey points. Values for carriers of particular mutations are shown using different colours as indicated on the plot.
We also observed effects of HBB mutations on several cardiometabolic-related blood biomarkers. Carriers of all HBB mutations except HbO had increased bilirubin levels in both populations. In MANOLIS, thalassemia mutation carriers had decreased total cholesterol (beta = − 0.53 and beta = − 0.65 for b+ and b0, respectively) which coincided with decreased values of low-density lipoprotein (LDL) cholesterol (beta = − 0.43 and beta = − 0.44, respectively). Finally, there were differences in other traits in MANOLIS. Carriers of HbS were significantly taller (beta = 5.98, SE = 1.69, p-value = 4.0 × 10–4). Earlier gestation age was seen in carriers of b+ mutations (beta = − 0.41, SE = 0.09, p value = 7.8 × 10–6).
Discussion
We present a first detailed characterisation of the mutational spectrum of haemoglobin in two isolated Mediterranean populations, HELIC MANOLIS (Crete) and the HELIC Pomak cohort. A large proportion of the variation in the haematological traits can be explained by HBB mutations in these populations. We provide the first effect estimates for two large non-clinically ascertained samples. We also demonstrate an important role for rare non-coding variation. Finally, we replicate associations of common variants with smaller effects that have previously been reported for cosmopolitan populations with European ancestry.
We discover novel associations of variants at 15q26, 2p11.2, 20q13.32, and 9q31.3 with red and white blood cell count, platelet and red cell distribution width, respectively. All of these variants are rare and located outside of coding regions, making it difficult to understand the exact mechanisms through which these variants affect blood traits. Several genes in the 9q31.3 region have been associated with abnormal lipid profiles and coronary artery disease, including SVEP121 and ATP Binding Cassette Subfamily A Member 1 gene (ABCA1)22,23. Links between haematological traits and lipid profiles have been previously demonstrated24. Another variant at the locus with likely regulatory function, rs201343203, is intronic to Fibronectin Type III and SPRY Domain Containing 1 Like (FSD1L), a gene that has been previously linked to red cell distribution width25.
For the PDW-associated variants at 20q13.32, results from chromatin interaction experiments26 further implicate APCDD1L as a likely target gene. An important paralog of this gene (APCDD1) is a negative regulator of the Wnt signaling pathway that is involved in the regulation of platelet function27. Chromatin immunoprecipitation sequencing experiments have also shown that the Histone-lysin N-methyltransferase SETDB1 protein binds in the lead SNP in this region. Furthermore, female heterozygous mutant mice have abnormal peripheral blood lymphocytes data28. Further investigation is required to fully elucidate the underlying mechanisms of these novel associations.
In each of the two populations studied here, moderate to large effects are observed, with alleles conferring effects of 1.44–2.63 units of standard deviation. This contrasts with other studies. For example, a study based on whole-genome sequencing of 3,781 individuals from a cosmopolitan European population did not discover any novel associations with blood traits29. We hypothesize that the observed enrichment of novel haematological association of rare variants is a consequence of population history30. Genetic drift due to the founder event in these isolated groups may have resulted in increased allele frequencies for the associated variants, which provides better statistical power for discovery of rare variants with large impact16. In fact, most of the lead variants at the novel loci have risen in frequency compared to large reference populations. For example, the rare allele of rs1320751535 at chromosome 15 is only carried by two individuals in TopMed (MAF = 0.000016) but has a frequency of almost 1% in MANOLIS. Limitations of genetic association studies include the possibility of false positive associations. Relative to some array-based genotyping efforts, our sample size was smaller. To fully confirm these novel associations, replication studies would be warranted. However, the low frequency of these variants in other populations and the need for large-scale sequencing to detect them has prevented replication testing using data from other available studies.
Previous reports of high levels of mutational diversity were limited to cosmopolitan populations. We found six different pathogenic HBB mutations in MANOLIS and six in the Pomak group, two of which were seen in both populations. We compared the mutational spectrum observed in the two Greek isolated populations to published data from 3,796 individuals, who were referred to genetic counselling at the NTC from all over Greece31. The two most common variants seen in the Greek NTC samples, IVS-I-110G>A (42.1% of carriers) and CD39C>T (18.8% of carriers), were both relatively common in MANOLIS (44% and 13% of carriers, respectively)). However, some of the mutations from the NTC sample, such as IVS-I-1 (G>A) (12.8% of carriers in NTC) and IVS-II-745C>G (6.3%), were not present in MANOLIS. Conversely, the second most frequent variant in MANOLIS, CD8/9+G, was rare in the NTC sample (0.1% of carriers). It should be noted, however, that recruitment for NTC was for symptomatic cases and relatives which may affect the mutation spectrum. Therefore, the carrier frequencies may not be comparable. There were marked differences in frequency patterns between the Pomak population and NTC sample with the top three mutations from NTC sample, IVS-I-110G>A, CD39C>T, and IVS-I-1, either not present or carried by only one individual in the Pomak population. The most common mutation in Pomak, c.364C>A (HbO), was not observed in the NTC sample. It has been previously postulated that this variant originated in the Pomak population19.
We provide detailed additional information to characterise the effects of this variant based on data of heterozygous and homozygous carriers. Firstly, we confirm a high allele frequency of 4.4% in the Pomak sample. In line with previous reports, we observe decreased levels of mean corpuscular volume but increases in mean corpuscular haemoglobin concentration20,32. This can be explained by the strong positive charge of the HbO molecule which results in their accumulation below the inner surface of the negatively charged erythrocyte membrane, leading to more space that can be filled with haemoglobin as well as denser, more spherical cells33. However, in discordance with previous research20,32, we do not find statistically significant differences between heterozygotes and homozygotes for these traits. Moreover, we observe novel effects of HbO on white blood cell traits and platelets, including increased platelet distribution width, mean platelet volume, large platelet distribution ratio, as well as white blood cell, lymphocyte and neutrophil count.
The isolated Cretan population has low levels of cardiometabolic complications despite exposure to risk factors such as obesity15. In line with previous reports34–36, we found decreased levels of total and LDL-cholesterol in carriers of thalassemia variants in MANOLIS. This has previously been linked to a decreased risk of atherosclerotic cardiovascular disease37–42. In MANOLIS we also found an association between the sickle cell HbS mutation with increased height. However, it cannot be ruled out that carrier status tags a specific ancestral group with taller stature. There was also a novel association of b+ mutations with increased risk of being born pre-term.
We show that c.364C>A (HbO-Arab), IVS-II-745 (C>G), CD8/9 + G and IVS-I-110 (G>A) are located on a very large haplotype each, extending over up to 15 Mb, a range that has previously only been reported for the major histocompatibility complex (MHC) region. This pattern likely represents a trace of the natural history of haemoglobin disorders. Positive natural selection can lead to increased linkage disequilibrium43. There is strong evidence that heterozygous carrier status of certain haemoglobin mutations provides a protective effect against malaria9 and Greece is one of the regions with a history of long-standing endemic malaria10.
In conclusion, whole-genome sequencing enabled a detailed characterisation of the spectrum of mutations, providing important insights into the allelic architecture of medically-relevant haematological traits. This can provide important guidance for mutation screening in these regions. Future research should extend this work to other populations with a high prevalence of haemoglobin disorders.
Materials and methods
Samples
The HELIC cohorts (https://www.helmholtz-muenchen.de/itg/projects-and-cohorts/helic/index.html) have previously been described in detail15–17. Briefly, MANOLIS includes individuals from the mountainous Mylopotamos villages on Crete. Individuals for the Pomak cohort were recruited at the Pomak villages, a set of mountainous villages in the North of Greece. Genetic isolatedness has been demonstrated for both cohorts16. A wide range of phenotypic information was collected including anthropometric and biometric measurements, biochemical and haematological blood measures, medical history, demographic, socioeconomic and lifestyle information. All participants provided written informed consent. Ethical approval was obtained from the Harokopio University Bioethics Committee. All methods were performed in accordance with the relevant guidelines and regulations.
Phenotype data
The distribution of each haematological trait was assessed. For those not sufficiently approximating a normal distribution, log- or rank-based inverse normal transformation was applied to phenotype measures and outliers were excluded (Supplementary Material, Supplementary Table 1). Values were adjusted for sex, age and squared age if any of these were significantly (p < 0.05) associated with the trait in a linear regression analysis. Standardised residuals from these analyses were used as outcome for the genome-wide association analysis. For the characterisation of pathogenic mutations in haemoglobin genes, we used un-transformed values as outcomes in the regression analyses to retain interpretability in original units. These analyses were conducted in R v3.444.
Sequencing
Whole-genome sequencing was carried out for 1,482 samples from MANOLIS and 1,642 samples from Pomak using Illumina’s HiSeqX platform with a target depth of 30x. Processing followed the GATK best practice guideline and has been described in detail elsewhere45. Alignment was carried out using BWA mem 0.7.8 using hg38 as the reference. Picard was used to mark duplicates. HaplotypeCaller v.3.5 was used to call genotypes. VQSR was used to for variant quality control (QC) using a tranche threshold of 99.4% for single nucleotide polymorphisms (SNP). For indels, we used the recommended threshold of 1%. We also filtered out 14% of variants with call rates < 99%.
We excluded 25 samples from MANOLIS that failed one or more of the following checks: four samples failed sex checks, eight had low concordance with previous genotyping efforts17, eleven were duplicates, twelve samples contaminated. For Pomak a total of 25 samples failed QC: three were duplicates, thirteen were heterozygosity outliers, eight were sex check failures and one was a depth outlier.
Genome-wide association analyses
The association of genetic variants with each haematological trait was evaluated using a linear mixed model implemented in GEMMA46. This approach accounts for relatives in the sample as well as any population substructure. GEMMA was used to estimate the genetic relatedness matrix after filtering for minor allele frequency (MAF) < 0.05, missingness < 1% and linkage disequilibrium (LD)-based pruning. We considered associations of variants with minor allele count of at least 10. To determine the multiple-testing burden, we estimated the effective number of traits by carrying out a principal component analysis for the correlation matrix of traits. The first 10 principal components explained 99% of the variants, therefore the effective number of traits was estimated to be 10. We also accounted for number of variants which resulted in an adjusted p-value threshold of 5 × 10–9. Measures of linkage disequilibrium, D’ and r2, were calculated using plink47.
For each of the previously unreported variants significantly associated with haematological traits, we considered all variants in a ± 500 Kb distance. In order to identify potentially causal variants, we excluded SNPs with a likelihood of being causal of less than 1:100, by comparing the likelihood of each SNP from the association analysis with the one of the most strongly associated SNP48. The remaining variants at each locus, henceforth called credible set, were annotated used FUMA49, Ensembl including VEP50, HaploReg51 and Open Targets Genetics52 to characterise their putative functional impact.
HBB mutations
We identified all mutations in the Haemoglobin Subunit Beta (HBB) gene that are classified as pathogenic with a review status of at least one star in the ClinVar data base53. These were referenced against the HbVar data bank12. The HELIC sequence data were queried for these 83 variants.
In addition to single variant association analyses as described above, we also carried out conditional analyses where we included either the most strongly associated variant or the pathogenic HBB variants as covariates in the model.
We used burden testing to evaluate the combined effect of variants in HBB and linked regulatory elements on blood traits. We followed the approach outlined in45. Briefly, we used an extended SKAT-O model54 to account for relatedness or population structure as implemented in MONSTER55. Boundaries for HBB were extracted from GENCODE v25. We applied eleven different conditions: regions of interest (coding regions only, coding and regulatory regions and regulatory regions only), variant filters (inclusion criteria based on severity of predicted consequence) and weighting schemes.
Ethics approval and consent to participate
All participants provided written informed consent. Ethical approval was obtained from the Harokopio University Bioethics Committee.
Supplementary Information
Acknowledgements
We would like to thank Professor Dallas Swallow, UCL, for her advice on methods and interpretation from an evolutionary genetics perspective. The MANOLIS study is dedicated to the memory of Manolis Giannakakis, 1978-2010. We thank the residents of the Pomak villages and of the Mylopotamos villages for taking part. This work was supported by the Wellcome Trust (WT098051) and the European Research Council (ERC-2011-StG 280559-SEPI).
Abbreviations
- APCDD1L
APC down-regulated 1 like
- GWAS
Genome-Wide Association Study
- HBB
Haemoglobin subunit beta
- HELIC
Hellenic isolated cohorts
- LD
Linkage disequilibrium
- LDL
Low-density lipoprotein
- LPAR1
Lysophosphatidic acid receptor 1
- MAF
Minor allele frequency
- PDW
Platelet distribution width
- QC
Quality control
- RDW
Red cell distribution width
- RDW
Regulome database
- SNP
Single nucleotide polymorphism
- SNV
Single nucleotide variant
- SE
Standard error
- SVEP1
Sushi, Von Willebrand Factor Type A, EGF and pentraxin domain containing 1
- WBC
White blood cell count
Author contributions
K.K. conceptualised the research question, carried out the analyses and wrote the manuscript. E.Z. designed the study and reviewed the manuscript. A.G. and D.S. carried out quality control of the data and carried out the genome-wide association analyses. L.S. and B.K. prepared the phenotype data. O.G. annotated the novel associations. E.T., M.K. and A.-E.F. collected data. G.D. and E.Z. are the PIs of the HELIC studies. M.S. is one of the PIs of the GPC Uganda study and D.G. analysed the Uganda data. J.D., I.B. are PIs of the INTERVAL study. A.B. and K.K. analysed the INTERVAL data.
Data availability
The data generated and/or analysed during the current study (i.e. the HELIC genotype and WGS datasets) are availableon the the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/home): EGAD00010000518; EGAD00010000522; EGAD00010000610; EGAD00001001636, EGAD00001001637.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-04436-9.
References
- 1.Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanai M, Akiyama M, Takahashi A, Matoba N, Momozawa Y, Ikeda M, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018;50:390–400. doi: 10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
- 3.Keller MF, Reiner AP, Okada Y, van Rooij FJA, Johnson AD, Chen M-H, et al. Trans-ethnic meta-analysis of white blood cell phenotypes. Hum. Mol. Genet. 2014;23:6944–6960. doi: 10.1093/hmg/ddu401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiner AP, Lettre G, Nalls MA, Ganesh SK, Mathias R, Austin MA, et al. Genome-wide association study of white blood cell count in 16,388 African Americans: the continental origins and genetic epidemiology network (COGENT) PLoS Genet. 2011;7:e1002108. doi: 10.1371/journal.pgen.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain D, Hodonsky CJ, Schick UM, Morrison JV, Minnerath S, Brown L, et al. Genome-wide association of white blood cell counts in Hispanic/Latino Americans: the Hispanic Community Health Study/Study of Latinos. Hum. Mol. Genet. 2017;26:1193–1204. doi: 10.1093/hmg/ddx024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CHARGE Consortium Hematology Working Group Meta-analysis of rare and common exome chip variants identifies S1PR4 and other loci influencing blood cell traits. Nat. Genet. 2016;48:867–876. doi: 10.1038/ng.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nalls MA, Couper DJ, Tanaka T, van Rooij FJA, Chen M-H, Smith AV, et al. Multiple loci are associated with white blood cell phenotypes. PLoS Genet. 2011;7:e1002113. doi: 10.1371/journal.pgen.1002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weatherall DJ. The inherited diseases of hemoglobin are an emerging global health burden. Blood. 2010;115:4331–4336. doi: 10.1182/blood-2010-01-251348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams TN, Weatherall DJ. World Distribution, Population Genetics, and Health Burden of the Hemoglobinopathies. Cold Spring Harb. Perspect. Med. 2012 doi: 10.1101/cshperspect.a011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flint J, Harding RM, Boyce AJ, Clegg JB. The population genetics of the haemoglobinopathies. Baillieres Clin. Haematol. 1998;11:1–51. doi: 10.1016/S0950-3536(98)80069-3. [DOI] [PubMed] [Google Scholar]
- 11.Higgs DR, Engel JD, Stamatoyannopoulos G. Thalassaemia. Lancet. 2012;379:373–383. doi: 10.1016/S0140-6736(11)60283-3. [DOI] [PubMed] [Google Scholar]
- 12.Patrinos GP, Giardine B, Riemer C, Miller W, Chui DHK, Anagnou NP, et al. Improvements in the HbVar database of human hemoglobin variants and thalassemia mutations for population and sequence variation studies. Nucleic Acids Res. 2004;32:D537–541. doi: 10.1093/nar/gkh006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rund D, Rachmilewitz E. β-Thalassemia. N. Engl. J. Med. 2005;353:1135–1146. doi: 10.1056/NEJMra050436. [DOI] [PubMed] [Google Scholar]
- 14.Hatzikotoulas K, Gilly A, Zeggini E. Using population isolates in genetic association studies. Brief Funct. Genomics. 2014;13:371–377. doi: 10.1093/bfgp/elu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farmaki A-E, Rayner NW, Matchan A, Spiliopoulou P, Gilly A, Kariakli V, et al. The mountainous Cretan dietary patterns and their relationship with cardiovascular risk factors: the Hellenic Isolated Cohorts MANOLIS study. Public Health Nutr. 2017;20:1063–1074. doi: 10.1017/S1368980016003207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panoutsopoulou K, Hatzikotoulas K, Xifara DK, Colonna V, Farmaki A-E, Ritchie GRS, et al. Genetic characterization of Greek population isolates reveals strong genetic drift at missense and trait-associated variants. Nat. Commun. 2014;5:5345. doi: 10.1038/ncomms6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Southam L, Gilly A, Süveges D, Farmaki A-E, Schwartzentruber J, Tachmazidou I, et al. Whole genome sequencing and imputation in isolated populations identify genetic associations with medically-relevant complex traits. Nat. Commun. 2017;8:15606. doi: 10.1038/ncomms15606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papadopoulos V, Dermitzakis E, Konstantinidou D, Petridis D, Xanthopoulidis G, Loukopoulos D. HbO-Arab mutation originated in the Pomak population of Greek Thrace. Haematologica. 2005;90:255–257. [PubMed] [Google Scholar]
- 20.Papadopoulos V, Vassiliadou D, Xanthopoulidis G, Petridis D, Agorasti A, Loukopoulos D. The implications of haemoglobin O-Arab mutation. Haema. 2003;6.
- 21.Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators, Stitziel NO, Stirrups KE, Masca NGD, Erdmann J, Ferrario PG, et al. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N Engl J Med. 2016;374: 1134–1144. 10.1056/NEJMoa1507652 [DOI] [PMC free article] [PubMed]
- 22.Sałacka A, Bińczak-Kuleta A, Kaczmarczyk M, Hornowska I, Safranow K, Clark JSC. Possible association of ABCB1:c.3435T>C polymorphism with high-density-lipoprotein-cholesterol response to statin treatment-a pilot study. Bosn. J. Basic Med. Sci. 2014;14:144–149. doi: 10.17305/bjbms.2014.3.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q, Jin Y-Z, Zhou S-B, Wang L-J, Lei M-M, Wang Y, et al. ABCB1 C3435T polymorphism and the risk of ischemic heart disease: a meta-analysis. Genet. Test Mol. Biomark. 2014;18:636–645. doi: 10.1089/gtmb.2014.0065. [DOI] [PubMed] [Google Scholar]
- 24.Antwi-Baffour S, Kyeremeh R, Boateng SO, Annison L, Seidu MA. Haematological parameters and lipid profile abnormalities among patients with Type-2 diabetes mellitus in Ghana. Lipids Health Dis. 2018;17:283. doi: 10.1186/s12944-018-0926-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kichaev G, Pasaniuc B. Leveraging functional-annotation data in trans-ethnic fine-mapping studies. Am. J. Hum. Genet. 2015;97:260–271. doi: 10.1016/j.ajhg.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung I, Schmitt A, Diao Y, Lee AJ, Liu T, Yang D, et al. A compendium of promoter-centered long-range chromatin interactions in the human genome. Nat. Genet. 2019;51:1442–1449. doi: 10.1038/s41588-019-0494-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steele BM, Harper MT, Macaulay IC, Morrell CN, Perez-Tamayo A, Foy M, et al. Canonical Wnt signaling negatively regulates platelet function. Proc. Natl. Acad. Sci. USA. 2009;106:19836–19841. doi: 10.1073/pnas.0906268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardin A, White J. The sanger mouse genetics programme: High throughput characterisation of knockout mice. Acta Ophthalmol. (Copenh.). 2011 doi: 10.1111/j.1755-3768.2011.4451.x. [DOI] [Google Scholar]
- 29.The UK10K Consortium. The UK10K project identifies rare variants in health and disease. Nature. 2015;526: 82–90. 10.1038/nature14962 [DOI] [PMC free article] [PubMed]
- 30.Kuchenbaecker K, Telkar N, Reiker T, Walters RG, Lin K, Eriksson A, et al. The transferability of lipid loci across African, Asian and European cohorts. Nat. Commun. 2019;10:4330. doi: 10.1038/s41467-019-12026-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boussiou M, Karababa P, Sinopoulou K, Tsaftaridis P, Plata E, Loutradi-Anagnostou A. The molecular heterogeneity of beta-thalassemia in Greece. Blood Cells Mol. Dis. 2008;40:317–319. doi: 10.1016/j.bcmd.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Vassiliadou D, Papadopoulos V, Konstantinidou C, Arnaoutoglou M. HbOThrace trait, HbOThrace hemoglobinopathy and HbOThrace/Hb beta zero hemoglobinopathy: a retrospective study of 118 cases. Folia. Med. (Plovdiv). 1998;40:46–50. [PubMed] [Google Scholar]
- 33.Ballas SK, Embi K, Goshar D, Burka ER. Binding of beta S, beta C and beta O Arab globins to the erythrocyte membrane. Hemoglobin. 1981;5:501–505. doi: 10.3109/03630268108991827. [DOI] [PubMed] [Google Scholar]
- 34.Triantafyllou AI, Vyssoulis GP, Karpanou EA, Karkalousos PL, Triantafyllou EA, Aessopos A, et al. Impact of β-thalassemia trait carrier state on cardiovascular risk factors and metabolic profile in patients with newly diagnosed hypertension. J. Hum. Hypertens. 2014;28:328–332. doi: 10.1038/jhh.2013.102. [DOI] [PubMed] [Google Scholar]
- 35.Fessas P, Stamatoyannopoulos G, Keys A. Serum-cholesterol and thalassemia trait. Lancet Lond Engl. 1963;1:1182–1183. doi: 10.1016/S0140-6736(63)92478-4. [DOI] [PubMed] [Google Scholar]
- 36.Maioli M, Pettinato S, Cherchi GM, Giraudi D, Pacifico A, Pupita G, et al. Plasma lipids in beta-thalassemia minor. Atherosclerosis. 1989;75:245–248. doi: 10.1016/0021-9150(89)90182-2. [DOI] [PubMed] [Google Scholar]
- 37.Gallerani M, Scapoli C, Cicognani I, Ricci A, Martinelli L, Cappato R, et al. Thalassaemia trait and myocardial infarction: low infarction incidence in male subjects confirmed. J. Intern. Med. 1991;230:109–111. doi: 10.1111/j.1365-2796.1991.tb00416.x. [DOI] [PubMed] [Google Scholar]
- 38.Crowley JP, Sheth S, Capone RJ, Schilling RF. A paucity of thalassemia trait in Italian men with myocardial infarction. Acta Haematol. 1987;78:249–251. doi: 10.1159/000205887. [DOI] [PubMed] [Google Scholar]
- 39.Wang CH, Schilling RF. Myocardial infarction and thalassemia trait: an example of heterozygote advantage. Am. J. Hematol. 1995;49:73–75. doi: 10.1002/ajh.2830490112. [DOI] [PubMed] [Google Scholar]
- 40.Tassiopoulos T, Stamatelos G, Zakopoulos N, Fessas P, Eliopoulos GD. Low incidence of acute myocardial infarction in beta-thalassaemia trait carriers. Haematologia (Budap). 1995;26:199–203. [PubMed] [Google Scholar]
- 41.Namazi MR. Minor thalassemia as a protective factor against cerebrovascular accidents. Med. Hypotheses. 2002;59:361–362. doi: 10.1016/S0306-9877(02)00264-5. [DOI] [PubMed] [Google Scholar]
- 42.Liu DJ, Peloso GM, Yu H, Butterworth AS, Wang X, Mahajan A, et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat. Genet. 2017;49:1758. doi: 10.1038/ng.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet. Res. 1974;23:23–35. doi: 10.1017/S0016672300014634. [DOI] [PubMed] [Google Scholar]
- 44.R Core Team. R: A language and environment for statistical computing. 2021. Available: https://www.R-project.org/
- 45.Gilly A, Suveges D, Kuchenbaecker K, Pollard M, Southam L, Hatzikotoulas K, et al. Cohort-wide deep whole genome sequencing and the allelic architecture of complex traits. Nat. Commun. 2018;9:4674. doi: 10.1038/s41467-018-07070-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Udler MS, Tyrer J, Easton DF. Evaluating the power to discriminate between highly correlated SNPs in genetic association studies. Genet. Epidemiol. 2010;34:463–468. doi: 10.1002/gepi.20504. [DOI] [PubMed] [Google Scholar]
- 49.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward LD, Kellis M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carvalho-Silva D, Pierleoni A, Pignatelli M, Ong C, Fumis L, Karamanis N, et al. Open Targets Platform: New developments and updates two years on. Nucleic Acids Res. 2019;47:D1056–D1065. doi: 10.1093/nar/gky1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am. J. Hum. Genet. 2012;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang D, McPeek MS. Robust rare variant association testing for quantitative traits in samples with related individuals. Genet Epidemiol. 2014;38:10–20. doi: 10.1002/gepi.21775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated and/or analysed during the current study (i.e. the HELIC genotype and WGS datasets) are availableon the the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/home): EGAD00010000518; EGAD00010000522; EGAD00010000610; EGAD00001001636, EGAD00001001637.