Abstract
Leptospirosis is a global zoonotic disease caused by pathogenic bacteria of the genus Leptospira. We sought to determine if rodents in U.S. Virgin Islands (USVI) are carriers of Leptospira. In total, 140 rodents were sampled, including 112 Mus musculus and 28 Rattus rattus. A positive carrier status was identified for 64/140 (45.7%); 49 (35.0%) were positive by dark-field microscopy, 60 (42.9%) by culture, 63 (45.0%) by fluorescent antibody testing, and 61 (43.6%) by real-time polymerase chain reaction (rtPCR). Molecular typing indicated that 48 isolates were L. borgpetersenii and 3 were L. kirschneri; the remaining nine comprised mixed species. In the single culture-negative sample that was rtPCR positive, genotyping directly from the kidney identified L. interrogans. Serotyping of L. borgpetersenii isolates identified serogroup Ballum and L. kirschneri isolates as serogroup Icterohaemorrhagiae. These results demonstrate that rodents are significant Leptospira carriers and adds to understanding the ecoepidemiology of leptospirosis in USVI.
Subject terms: Microbiology, Diseases
Introduction
Leptospirosis is a zoonosis of global distribution caused by pathogenic species in the genus Leptospira that infects people, wildlife, and domestic animals1. Rodent species act as reservoir hosts without clinical disease from Leptospira, which colonize renal tubules and are excreted through urine contaminating water and soil where it can survive for weeks2. Humans are incidental hosts and exposure occurs by direct contact with infected animals or indirectly through contact with contaminated water or soil. Human leptospirosis ranges in severity from a mild, self-limited febrile illness to a fulminant life-threatening illness3. Leptospirosis in domestic animals is characterized by similar acute clinical features, but persistent chronic infection in animals can occur4,5.
Acute infections are more common in low-resource, tropical and subtropical locations where outbreaks can occur after natural disasters, such as hurricanes and concurrent rainfall and flooding1. The U.S. Virgin Islands (USVI) is a U.S. territory located in the Caribbean Ocean with three main islands, St. Croix (STX), St. Thomas (STT), and St. John (STJ), totaling ~ 133 square miles6, with a population of 106,405 persons7. The climate is tropical, with average high temperature ranges of 82°F to 90°F, and average rainfall of 43 inches per year. USVI was directly struck by Category 5 Hurricanes Irma and Maria in September 2017, generating record-breaking rainfalls and flooding. Afterwards, the Virgin Islands Department of Health documented the first-known human cases of leptospirosis on the islands6. A follow-up seroprevalence study among residents reported evidence of exposure to Leptospira, with highest reacting titers to serogroups Icterohaemorrhagiae, Australis, Canicola, Pyrogenes, Tarassovi, Autumnalis, Bataviae, Djasiman, Ballum and Sejröe (Esther Ellis, USVI Department of Health, Personal Communication 2019). Assessment of animal leptospirosis in USVI has been limited to a single serologic study of small ruminants conducted in 1992, which showed reactivity to serogroups Autumnalis, Ballum, Bataviae, Australis, Canicola, Icterohaemorrhagiae, Sejröe and Pyrogenes8.
Rodents are a principal reservoir host for the transmission of leptospirosis9,10. Given our limited understanding of leptospirosis disease transmission in USVI, this investigation sought to determine the extent to which wild rodents act as carriers of leptospires and identify any associated species of Leptospira. Such information is critical for efficacious surveillance, control, and prevention strategies.
Materials and methods
Sample collection
Field activities and euthanasia procedures were in accordance with CDC IACUC Protocol Numbers 2879SALMULX-A4 and AVMA Guidelines for Euthanasia of Animals11, and in compliance with the ARRIVE guidelines. A pilot study was performed in STX at a single unique study site in September 2019 to assess logistics of field sampling and laboratory processing and shipment. In total, 39 Sherman Traps® (H.B. Sherman Traps, Tallahassee, Florida, USA) were deployed, and three Mus musculus were sampled. The cross-sectional field study was carried out during June 15–June 30, 2020 and employed single sample events at 20 different study sites from three islands as follows: eight in STX, six in STT and six in STJ (Table 1 and Fig. 1). Trapping consisted of ten 6 × 6 × 18-inch (15 × 15 × 46 cm) Tomahawk® live traps (Tomahawk Live Trap Co, Tomahawk, Wisconsin, USA) and 80 Sherman Traps®, 15 m apart at each study site. For bait, oat and peanut butter mixture were used in the Sherman Trap®; Vienna sausage was used in Tomahawk traps. Traps were placed in rural areas (e.g., farms, parks, or bush) throughout USVI in the evening and collected at dawn for a single sampling event per site.
Table 1.
Number of rodents captured and sampled, by island and field site as shown in Fig. 1.
| Island | Field site | Rodents captured | Rodents sampled* | ||||
|---|---|---|---|---|---|---|---|
| Mus muscularis captured | Rattus rattus captured | Total captured | Mus muscularis sampled | Rattus rattus sampled | Total sampled | ||
| St. Croix (STX) | Concordia* | 3 | 0 | 3 | 3 | 0 | 3 |
| Jolly Hill | 8 | 0 | 8 | 8 | 0 | 8 | |
| Sandy Point | 8 | 0 | 8 | 8 | 0 | 8 | |
| Haypenny Beach† | 31 | 2 | 33 | 18 | 2 | 20 | |
| Lower Love | 4 | 0 | 4 | 4 | 0 | 4 | |
| Cramer Park | 9 | 0 | 9 | 9 | 0 | 9 | |
| Prune Bay | 1 | 2 | 3 | 1 | 2 | 3 | |
| Recovery Hill | 13 | 3 | 16 | 7 | 3 | 10 | |
| Southgate | 7 | 1 | 8 | 7 | 1 | 8 | |
| St. Thomas (STT) | Stumpy Bay | 13 | 8 | 21 | 5 | 5 | 10 |
| Magens Bay | 6 | 0 | 6 | 6 | 0 | 6 | |
| STT Airport | 31 | 0 | 31 | 10 | 0 | 10 | |
| Tutu | 0 | 0 | 0 | 0 | 0 | 0 | |
| Vessup Beach | 0 | 0 | 0 | 0 | 0 | 0 | |
| Red Hook Point | 2 | 0 | 2 | 2 | 0 | 2 | |
| St. John (STJ) | Gifft Hill Landfill | 20 | 6 | 26 | 4 | 6 | 10 |
| Cinnamon Bay | 7 | 0 | 7 | 7 | 0 | 7 | |
| Lameshur Bay | 0 | 1 | 1 | 0 | 1 | 1 | |
| Salt Pond | 2 | 0 | 2 | 2 | 0 | 2 | |
| Brown Bay | 6 | 3 | 9 | 6 | 3 | 9 | |
| Annaberg Plantation | 13 | 5 | 18 | 5 | 5 | 10 | |
| Totals | 184 | 31 | 215 | 112 | 28 | 140 | |
*Pilot study site sampled on September 3, 2019; in total, 39 Sherman traps deployed.
†Because of laboratory resources and field safety, sampling was limited to 10 rodents per site after sampling at Haypenny Beach.
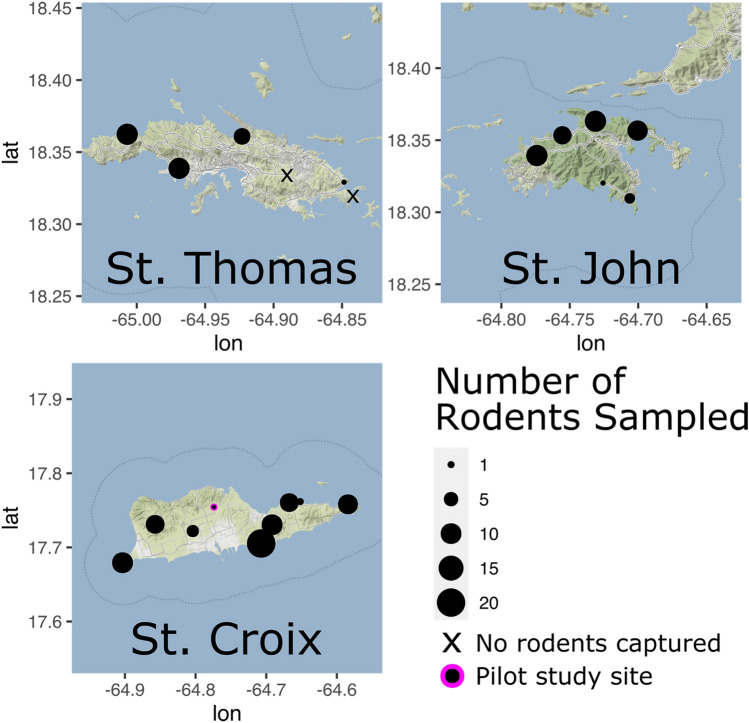
Figure 1.
Map of U.S. Virgin Islands showing the sampled areas among the three islands: St. Croix, St. Thomas, and St. John. Black dots identify each location, with the size correlating to the numbers of rodents sampled. Map base layer available under CC BY 3.0 license: http://maps.stamen.com/terrain/#10/18.0114/-64.7823.
Because of finite laboratory resources and field and animal safety in a tropical climate, sampling was limited to a maximum of 10 rodents per field site after capture of 33 rodents in Haypenny Beach during the second day of the study (Table 1). Thereafter, any rodents captured surplus to sampling needs were immediately released back into their environment. To avoid selection bias, if more than 10 rodents were captured at a sampling site, traps with captured rodents were randomly selected using a random number generator for sampling. Species was confirmed visually, and selection preferred a 50/50 split between Mus musculus and Rattus rattus, if possible.
Rodents were rapidly anesthetized with isoflurane until euthanasia by cervical dislocation. Sex, mass, and morphometrics (e.g., total body length, ear, hind foot, and tail lengths) were recorded. Blood samples were collected by cardiac puncture and stored on ice. Samples were centrifuged (15,000×g for 15 min) and serum collected and stored at −20 °C. Frozen serum was then transported to the National Veterinary Services Laboratories, APHIS, U.S. Department of Agriculture (USDA), Ames, Iowa. One kidney from each rodent was removed using aseptic technique by necropsy and immediately stored in Hornsby-Alt-Nally (HAN) media12, then transported by overnight delivery services at ambient temperature to the National Animal Disease Center, ARS, USDA, Ames, Iowa.
Microscopic agglutination test (MAT)
The microscopic agglutination test (MAT) was performed according to World Organization for Animal Health guidelines using a panel of 18 antigens representative of 15 serogroups (Supplementary Table 1)13. A titer was considered positive at ≥ 1:100.
Kidney sample processing
The kidney was macerated in 9 mL of HAN media in 710 mL Whirl–Pak® bags (Nasco).
Dark-field microscopy (DFM)
Ten microliters of the macerate were placed on a microscope slide and cover-slipped. Ten fields were examined by DFM (× 200 and × 400) for leptospires.
Culture
One mL of the kidney macerate was used to inoculate 9 mL HAN liquid medium and 200 µl of this dilution was inoculated into 5 mL of three different media: semisolid T80/40/LH14 that was incubated at 29℃, and 5 mL of liquid and semisolid HAN, which were incubated at 37℃ in 3% CO212. Semisolid cultures were observed using a lighted black background to examine for development of a Dinger’s zone (DZ), and if noted, were confirmed as positive by DFM, at days 3 and 5, weekly for one month, and monthly thereafter for six months. From the positive cultures, average time for a DZ to appear was noted. Inoculated tubes of liquid HAN were examined daily by DFM for leptospires from day 3 to day 8.
Fluorescent Antibody Testing (FAT)
A 10 µl aliquot of the kidney macerate was placed on a glass slide within a 7 mm well, in duplicate, and FAT performed as previously described15.
DNA extraction
DNA was extracted from 500 μL of kidney macerate using the Maxwell RSC Purefood Purification Pathogen kit (Promega Corporation, Madison, Wisconsin, USA), following manufacturer's instructions, but using 1 h of incubation with lysis buffer A and a 100μL elution volume. For cultures, DNA was extracted from 5 mL of each isolate in HAN media, which was harvested by centrifugation at 10,000×g for 15 min.
Real-time polymerase chain reaction (rtPCR)
After DNA extraction from 500 µl of kidney macerate, 5 µl was used for rtPCR. The lipL32 gene was amplified using a set of primers and protocol as described previously: LipL32-47Fd (5′-GCATTACMGCTTGTGGTG -3′) and LipL32-301Rd (5′-CCGATTTCGCCWGTTGG -3′), the probe LipL32-189P (6-carboxyfluorescein [FAM]-5′-AA AGC CAG GAC AAG CGC CG-3′-black hole quencher 1 [BHQ1]), using PerfeCTa qPCR ToughMix®, Low ROX™ (Quanta Biosciences, Gaithersburg, Maryland, USA)16,17. To control for rtPCR inhibitors, the TaqMan® (Thermo Fischer Scientific, Waltham, MA, USA) Exogenous Internal Positive Control (IPC) was added to the master mix to confirm DNA amplification and detect false negatives and to qualitatively detect presence of amplification inhibitory substances in a sample. If IPC negative samples indicated the sample contained rtPCR inhibitors, samples were diluted 1:10 and rtPCR repeated. If the diluted sample was still negative for IPC, a new DNA extraction was performed. All samples were assayed in triplicate and considered positive when duplicate or triplicates were positive with Ct value < 4016,17.
Molecular and serological typing of cultured Leptospira species
Concentration of reconstituted genomic DNA was determined by Qubit® (Qubit dsDNA BR assay, Qubit 3.0 fluorometer, Invitrogen, Carlsbad, CA, USA). Whole-genome sequence of all cultures was obtained (MiSeq Desktop Sequencer, 2 × 250 v2 paired-end chemistry and the Nextera XT DNA Library Preparation Kit, Ilumina, San Diego, California) per manufacturer’s instructions and draft assemblies of each genome were mined to retrieve full length secY coding regions which were analyzed with Geneious Prime 2020.2.2 (geneious.com). Consensus sequences of 522 bp were then compared with sequences in GenBank using BLAST (Basic Local Alignment Search Tool). secY sequences for USVI rodent isolates were deposited in the National Center for Biotechnology Information (NCBI) database, accession numbers MZ241244–MZ241294. A phylogenetic tree was made with Geneious Prime 2020.2.2 using the neighbor‐joining method, with the Tamura‐Nei nucleotide substitution model (https://www.geneious.com/).
Isolates were serotyped by MAT using a panel of polyclonal rabbit reference antisera representing 13 serogroups; Australis, Autumnalis, Ballum, Bataviae, Canicola, Grippotyphosa, Hebdomadis, Icterohaemorrhagiae, Mini, Pomona, Pyrogenes, Sejröe, and Tarassovi (National Veterinary Services Laboratories, APHIS, USDA, Ames, Iowa) (Supplementary Table 2). The serogroup for each isolate was assigned according to the antiserum that gave the highest agglutination titer18.
Genotyping of Leptospira directly from kidney samples
The secY housekeeping gene was amplified with the primers secY_F (5′ -ATGCCGATCATTTTTGCTTC-3′) and secY_R (5′-CCGTCCCTTAATTTTAGACTTCTTC-3′)19. PCR products were then purified and labeled using the Big Dye Terminator v3.1 cycle sequencing reagent (Applied Biosystems, Foster City, California, USA). Sequencing was performed using the ABI 3130XL Genetic Analyzer. Sequence data were analyzed with DNAStar’s Lasergene sequence analysis software. Consensus sequences were compared with available sequences in the GenBank database using BLAST. Phylogenetic analyses were performed as described previously. The secY sequence was deposited in NCBI, accession number MZ241295.
Evaluation of virulence
All animal experimentation was conducted in accordance with protocols as reviewed and approved by the Animal Care & Use Committee at the National Animal Disease Center (ARS-2018-745), and as approved by USDA institutional guidelines. Five representative rodent isolates of L. borgpetersenii (designated LR45, LR47, LR59, LR88, and LR131) were propagated in liquid HAN medium12 supplemented with 0.4% rabbit serum at 37 °C in 3% CO2 and evaluated for virulence by intraperitoneal injection into five groups of four golden Syrian hamsters (Mesocricetus auratus), as previously described15. One group of four animals was also inoculated through the conjunctival route with 108 of strain LR131 in 10 µl of HAN medium, which was applied to the conjunctival membrane of the left eye, as previously described20. Liver and kidney tissue were harvested for culture, FAT, and lipL32 rtPCR when hamsters met euthanasia criteria attributable to clinical signs of infection including weight loss, lethargy, bloody discharge from the nose or urogenital tract, and sudden death21.
Statistical analyses
Comparison of culture results in HAN and T80/40/LH media were assessed using the Students T-test. Analysis was conducted using SPSS statistical software (SPSS Inc., Chicago, Illinois, USA), and results were considered significant when p < 0.05.
Results
Rodent survey
In total, 140 rodents were sampled, including Mus musculus (n = 112) and Rattus rattus (n = 28): 73 in STX, 28 in STT and 39 in STJ (Table 1, Fig. 1). Overall trap success was 11.7% (215 rodents captured for 1,839 trap-nights deployed); rodents were sampled at 19/21 study sites (i.e., one pilot study site; 20 cross-sectional survey sites), and no rodents were captured at two study sites in St. Thomas. More female rodents (n = 82) were sampled than males (n = 58). Among sexually mature females, 3/80 (4%) were pregnant.
Seroprevalence
Of 112/140 (80.0%) sera tested by MAT, 60/112 (53.6%) were positive (titer ≥ 1:100). Twenty-eight (20.0%) sera samples were of inadequate volume to perform MAT. Of 60 positive sera samples, 51 had sufficient volumes remaining to determine titers against reacting serogroups (Table 2). The most frequent highest-reacting titer was to serogroup Ballum (30/51, 58.8%), followed by Icterohaemorrhagiae (5/51, 9.8%), Hebdomadis (5/51, 9.8%), Djasiman (2/51, 3.9%), Cynopteri (2/51, 3.9%) and Australis (1/51, 2.0%) (Table 2). Equivalent high titers were observed to more than one serogroup in six samples; three reacted with both Australis and Hebdomadis (LR119 with a titer of 1:100, LR138 and LR140 with a titer of 1:400), one (LR34) reacted to both Sejröe and Ballum (titer of 1:1600), one (LR71) reacted to both Autumnalis and Icterohaemorrhagiae (titer of 1:100) and one (LR113) with both Hebdomadis and Cynopteri (titer of 1:100).
Table 2.
Titers of rodent samples reactive with serogroups of Leptospira*.
| Titer Serogroup | 100 | 200 | 400 | 800 | 1600 | 3200 | 6400 | 12,800 | Total |
|---|---|---|---|---|---|---|---|---|---|
| Australis | – | 1 | – | – | – | – | – | – | 1 |
| Ballum | 3 | 3 | 3 | 2 | 10 | 7 | 2 | – | 30 |
| Cynopteri | 1 | 1 | – | – | – | – | – | – | 2 |
| Djasiman | 1 | 1 | – | – | – | – | – | – | 2 |
| Hebdomadis | 1 | 2 | – | – | 2 | – | – | – | 5 |
| Icterohaemorrhagiae | – | – | – | 1 | – | 2 | – | 2 | 5 |
| Total | 6 | 8 | 3 | 3 | 12 | 9 | 2 | 2 | 45 |
*Not included are those six samples most reactive to two different serogroups and as reported in Supplementary Table 3.
Detection of leptospires in rodent kidney
Forty-nine (35.0%; 49/140) kidney samples were positive for Leptospira by DFM (Table 3): 32/73 (43.8%) in STX, 5/28 (17.9%) in STT and 12/39 (30.8%) in STJ. By FAT, 63/140 (45.0%) were positive (Table 3): 39/73 (53.4%), 6/28 (21.4%) and 18/39 (46.2%) on STX, STT and STJ respectively. All samples positive by DFM were positive by FAT (Fig. 2). rtPCR for lipL32 detected 61 (43.6%) positive kidneys (Table 3): 38/73 (52.1%) in STX, 6/28 (21.4%) in STT and 17/39 (43.6%) in STJ. The average Ct value of positive samples was 24.8 ± 4.1 (95% CI).
Table 3.
Detection of Leptospira in different rodent species from different study sites by dark-field microscopy (DFM), fluorescent antibody test (FAT), real-time polymerase chain reaction (rtPCR), and culture.
| Species | DFM | FAT | rtPCR | Culture |
|---|---|---|---|---|
| Mus musculus | 47/112 (41.9%) | 56/112 (50%) | 52/112 (46.4%) | 51/112 (45.5%) |
| Rattus rattus | 2/28 (7.1%) | 7/28 (25%) | 9/28 (32.1%) | 9/28 (32.1%) |
| Total | 49/140 (35%) | 63/140 (45%) | 61/140 (43.7%) | 60/140 (42.8%) |
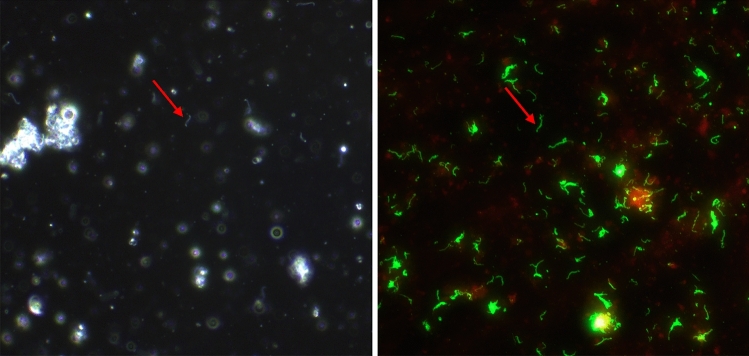
Figure 2.
Representative images of a rodent kidney sample that was positive for leptospires by (A) dark-field microscopy and (B) fluorescent antibody test (FAT). Red arrows indicate leptospires. Original magnification 400×.
Leptospira species were isolated by culture from 60 (42.9%) individual kidneys (Table 3): 37/73 (50.7%) in STX, 6/28 (21.4%) in STT and 17/39 (43.6%) in STJ. Fifty-seven isolates were recovered in all three media including liquid and semisolid HAN at 37℃ and T80/40/LH at 29℃ and three isolates were recovered in only liquid and semisolid HAN media. For positive cultures, the average time for a DZ to appear in semisolid HAN medium was 7.4 ± 3.3 (95% CI) days; whereas, the average number of days required for T80/40/LH medium was 17.2 ± 5.1 (95% CI). HAN media demonstrated a significant difference (p < 0.001) in the fewer days required from primary inoculation to development of a visible DZ. Positive cultures were detected in HAN liquid media at 5 ± 1 days postinoculation, as defined by detection of a single leptospire by dark-field microscopy.
A positive carrier status was identified for leptospires in 64 (45.7%) rodents as defined by a positive result in any of the assays used; 57 (89.1%) were positive by culture, FAT and rtPCR; three samples were positive by culture and rtPCR; three samples were positive only by FAT and one sample was positive only by rtPCR. Notably, two rodents identified as carriers were seronegative. All data is presented in Supplementary Table 3.
Molecular and serotyping of Leptospira isolates
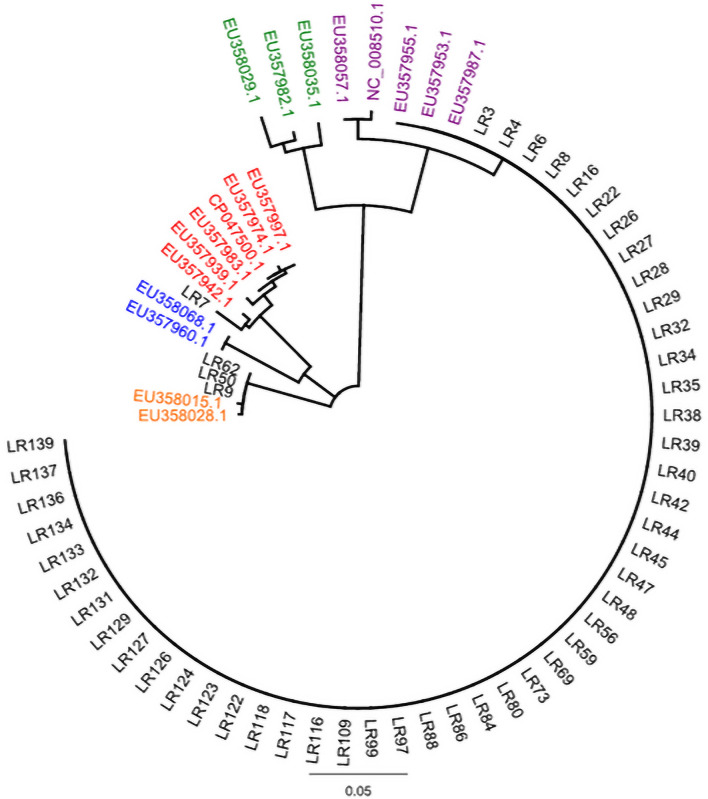
Molecular typing indicated that 48/60 isolates showed 100% secY identity with L. borgpetersenii and clustered with L. borgpetersenii serovar Polonica (EU357987.1), L. borgpetersenii serovar Ballum (EU357953.1) and L. borgpetersenii serovar Castellonis (EU357955.1); three additional isolates showed 99.8% identity with L. kirschneri (Fig. 3). The remaining 9/60 isolates were not readily identified to species level, and likely contain a mixed population of species of both L. borgpetersenii and L. kirschneri (data not shown). Serotyping of 48 isolates of L. borgpetersenii indicated that all belong to serogroup Ballum, and three isolates of L. kirschneri belong to serogroup Icterohaemorrhagiae. All rodent isolates obtained from STT and STJ were classified as L. borgpetersenii serogroup Ballum. However, cultures of Leptospira from rodent isolates in STX included those classified as L. borgpetersenii serogroup Ballum and L. kirschneri to serogroup Icterohaemorrhagiae, as well as cultures that were potentially mixed species.
Figure 3.
Phylogeny of Leptospira isolates based on secY gene sequence (522 bp) analysis using the neighbor-joining method. U.S. Virgin Islands isolates of Leptospira from rodents are annotated as LR and colored black, whereas accession numbers are provided for reference strains of L. borgpetersenii from different hosts (purple), L. santarosaii (green), L. kirschneri (orange), and L. noguchii (blue). Note sample LR7 which was genotyped directly from kidney and clades with reference strains of L. interrogans (red).
Genotyping of Leptospira directly from kidney
One kidney sample designated LR7 was positive only by rtPCR. Subsequent PCR amplification of partial sequence of secY followed by sequencing and BLAST/NCBI comparisons with GenBank indicated 100% identity with L. interrogans (Fig. 3).
Evaluation of virulence
Intraperitoneal inoculation of all hamsters with 108 leptospires of each of five strains resulted in acute disease requiring all to be euthanized at 5 days post-infection. Liver and kidney samples from each group were positive by culture, FAT and lipL32 rtPCR. A single group inoculated by the conjunctival route using isolate LR131 showed acute disease by 11 days post-infection; liver and kidney samples from this group also tested positive by culture, FAT and lipL32 rtPCR.
Discussion
Sampling of USVI rodents determined that 45.7% (64/140) were carriers of pathogenic Leptospira species (L. borgpetersenii, L. kirschneri, or L. interrogans) as defined by at least one positive FAT, rtPCR, or culture.
Seroprevalence for rodents in this study was relatively high (53.6%; 60/112), compared with previous findings from Central American countries including Barbados (32.6%)22, Grenada (24.5%)23, Guadeloupe (32%)22, and Trinidad (20.5%)24. Reactivity to serogroup Ballum was most frequently detected and differs from other studies with rodents in the same region, which reported reactivity primarily to serogroup Icterohaemorrhagiae9,22–24. Previous work demonstrated that goats in USVI were also reactive to serogroups Ballum and Icterohaemorrhagiae8. However, a positive serology in reservoir hosts of infection is indicative only of exposure and not active disease4.
Culture is the definitive diagnostic assay to detect shedding of leptospires, though it can have low sensitivity due to the fastidious nature of Leptospira4. The use of the newly described HAN media allowed for recovery of Leptospira species in a significantly shorter time frame at 37℃, compared with T80/40/LH at 29℃, and likely attributable to growth factors and conditions that more closely emulate the in vivo environment to support metabolic requirements of in vivo derived leptospires12. Recovery of isolates from rodent hosts allows for their more complete characterization at the genotypic and phenotypic level, and their use for enhanced diagnostic (i.e., animal and human) or bacterin-based vaccination strategies of animals to limit zoonotic transmission.
We attributed 80% of active rodent infections to L. borgpetersenii serogroup Ballum and 5% to L. kirschneri serogroup Icterohaemorrhagiae. One lipL32 rtPCR positive, but culture negative sample was genotyped as L. interrogans directly from kidney25. Among nine culture positive samples, species identification was not readily apparent and likely attributable to mixed populations of species. Further analysis of these mixed bacterial samples is underway to obtain clonal isolates, along with a comprehensive analysis of genomes of all recovered isolates. Notably, all mixed and L. kirschneri carriage in rodents were limited to those rodents trapped on St. Croix island. St. Croix rodent populations may have developed a unique carrier population of Leptospira because of its remote location and agricultural landscape.
Identification of L. borgpetersenii, L. kirschneri, and L. interrogans in kidneys of rodents in USVI is similar to recent findings in mongoose from USVI which were carriers of L. borgpetersenii, L. kirschneri and L. interrogans species26. However, and in contrast to USVI mongoose which were carriers of L. borgpetersenii serogroup Sejroe, USVI rodents were carriers of L. borgpetersenii serogroup Ballum. Seroprevalence investigations have also implicated serogroups Ballum in human leptospirosis in USVI (Esther Ellis, USVI Department of Health, personal communication, 2019) and livestock leptospirosis in USVI27 and across the Caribbean, including Puerto Rico28, Barbados29, Cuba30, Martinique25, Guadeloupe25,31 and Jamaica32; serogroup Ballum is also detected in other regions, such as New Caledonia33 and Australia34,35. Virulence of L. borgpetersenii serogroup Ballum from rodents in USVI was confirmed in the hamster model of leptospirosis, which emulates pathology associated with acute lethal forms of human leptospirosis and similar to that observed for serogroup Ballum isolates from Puerto Rico28 and New Caledonia36. Positive MAT titers against serogroup Ballum are indicative of human exposure, although its role in human disease has yet to be determined28. Rodents from rural and urban areas of Puerto Rico are carriers of L. interrogans and L. borgpetersenii28,37; rodents from Martinique and Guadeloupe are carriers of L. borgpetersenii, L. interrogans, and L. kirschneri, and rodents from urban areas of New Orleans are carriers of L. borgpetersenii, L. interrogans, and L. kirschneri38.
Two species of introduced (non-native) rodents were trapped; Mus musculus and Rattus rattus39. The rodent species diversity observed in rural areas in USVI was not as high as that observed in other rural settings in nearby countries40,41; a predominance of Rattus species are frequently noted within the Caribbean, including Grenada, Guadeloupe, Trinidad and Barbados9,22–24, which is in contrast to Puerto Rico37 and results reported here for USVI in which trapped rodents were predominantly M. musculus (80%).
Exposure to rodents is associated with an increased risk of leptospirosis42. Serogroup Ballum, the principal reservoir of which is mice36, is increasingly reported in human infections31,43 and our results highlight the need for rodent control to limit effects of leptospirosis36. Human leptospirosis infections usually reflect serogroups maintained by local animal populations highlighting the need for serovar-specific vaccine development in high-risk populations. Serogroup Ballum is not included in commercial bacterins for animals despite evidence of infection and its association with poor animal reproductive performance8,44.
USVI’s climate is typical of maritime tropical environments, with warm and stable temperatures and steady winds. Intense rainfall events generally occur in the form of tropical depressions, storms, or hurricanes. Occurrence of natural disasters and deficiencies in sanitary infrastructure can create a favorable environment for rodent proliferation and increase risk for infection45. The first documented case of human leptospirosis was identified in USVI after Hurricanes Irma and Maria, and associated with exposure to flood water and occupation of buildings with evidence of rodent infestation6. Research should be conducted in rural and urban USVI areas to identify region-specific risk factors for infection. Leptospirosis is a reemerging disease of public health importance with respect to morbidity and mortality both in humans and animals.
Limitations of this study include using a cross-sectional design that does provide a geographically encompassing assessment of rodents throughout USVI, but in a limited time frame (i.e., two weeks). Rodents can be transient carriers of Leptospira, but we did not control for seasonal variation.
In conclusion, this study confirms the presence of three species of pathogenic Leptospira (L. borgpetersenii, L. kirschneri, and L. interrogans) among USVI’s rodent populations. Local isolates of L. borgpetersenii serogroup Ballum and L. kirschneri serogroup Icterohaemorrhagiae should be included in MAT diagnostic panels for human and domestic animal samples from USVI, to increase capacity for disease detection in humans and animals. Control, public health surveillance, and prevention efforts need to be multidisciplinary and multisectoral, making it a prime candidate for the One Health approach.
Supplementary Information
Acknowledgements
This research was supported by funding Emergency Response Public Health Crisis Response 2017 Hurricane Recovery CDC-RFA-TP18-1802, in part by the U.S. Department of Agriculture, Agricultural Research Service and in part by an appointment to the Animal and Plant Health Inspection Service (APHIS) Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the U.S. Department of Agriculture (USDA). ORISE is managed by ORAU under DOE contract number DE-SC0014664. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official positions of USDA, CDC, CSTE, DOE, or ORAU/ORISE. USDA is an equal opportunity provider and employer. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Author contributions
Conceptualization: ASB, JEN, EME; Methodology: CH, ASB, LHW, RLH, KL, TA, TS, HMC, SKB, GB, DH, MLT, ME, NFA, JR, KB, JSS, IJS, BE, DPA, JEN; Formal analysis, CH, ASB, RLH, TS, JEN; Resources: SKB, GB, DH, ME, NFA, JR, KB, JSS, IJS, BE, DPA, LS, JEN, EME; Figures: CH, ASB, RLH, KL, TS; writing—original draft preparation, CH, ASB, RLH, JEN; writing—review and editing: All authors. All authors have read and agreed to the published version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-04846-3.
References
- 1.Costa F, et al. Global morbidity and mortality of leptospirosis: a systematic review. PLoS Negl Trop Dis. 2015;9(9):e0003898. doi: 10.1371/journal.pntd.0003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casanovas-Massana, A., et al. Quantification of Leptospira interrogans survival in soil and water microcosms. Appl Environ Microbiol. 84(13) (2018). [DOI] [PMC free article] [PubMed]
- 3.Haake DA, Levett PN. Leptospirosis in humans. Curr Top Microbiol Immunol. 2015;387:65–97. doi: 10.1007/978-3-662-45059-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellis WA. Animal leptospirosis. Curr Top Microbiol Immunol. 2015;387:99–137. doi: 10.1007/978-3-662-45059-8_6. [DOI] [PubMed] [Google Scholar]
- 5.Putz EJ, Nally JE. Investigating the immunological and biological equilibrium of reservoir hosts and pathogenic Leptospira: balancing the solution to an acute problem? Front. Microbiol. 2020;11:2005. doi: 10.3389/fmicb.2020.02005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marinova-Petkova, A., et al. First Reported Human Cases of Leptospirosis in the United States Virgin Islands in the Aftermath of Hurricanes Irma and Maria, September–November 2017. in Open Forum Infectious Diseases. 2019. Oxford University Press US. [DOI] [PMC free article] [PubMed]
- 7.U.S. Census Bureau, 2020. Understanding the Population of the U.S. Virgin Islands.https://www.census.gov/content/dam/Census/programs-surveys/sis/resources/2020/sis_2020map_usvi_k-12.pdf.
- 8.Ahl, A., D. Miller, and P. Bartlett, Leptospira serology in small ruminants on St. Croix, US Virgin Islands.Ann. N. Y. Acad. Sci.653(1): 168–171 (1992). [DOI] [PubMed]
- 9.Boey, K., K. Shiokawa, and S. Rajeev, Leptospira infection in rats: a literature review of global prevalence and distribution. PLoS Negl Trop Dis.13(8): e0007499 (2019). [DOI] [PMC free article] [PubMed]
- 10.Ido Y, et al. The rat as a carrier of Spirocheta icterohaemorrhagiae, the causative agent of Weil's disease (spirochaetosis icterohaemorrhagica. J. Exp. Med. 1917;26:341–353. doi: 10.1084/jem.26.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leary, S.L., et al. AVMA Guidelines for the Euthanasia of Animals: 2013 Edition. 2013. American Veterinary Medical Association Schaumburg, IL.
- 12.Hornsby RL, Alt DP, Nally JE. Isolation and propagation of leptospires at 37 degrees C directly from the mammalian host. Sci Rep. 2020;10(1):9620. doi: 10.1038/s41598-020-66526-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole JR, Sulzer CR, Pursell AR. Improved microtechnique for the leptospiral microscopic agglutination test. Appl. Microbiol. 1973;25(6):976–980. doi: 10.1128/am.25.6.976-980.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis W, Montgomery J, Cassells J. Dihydrostreptomycin treatment of bovine carriers of Leptospira interrogans serovar Hardjo. Res. Vet. Sci. 1985;39(3):292–295. doi: 10.1016/S0034-5288(18)31716-8. [DOI] [PubMed] [Google Scholar]
- 15.Nally JE, et al. Isolation and characterization of pathogenic leptospires associated with cattle. Vet. Microbiol. 2018;218:25–30. doi: 10.1016/j.vetmic.2018.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Stoddard RA, et al. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis. 2009;64(3):247–55. doi: 10.1016/j.diagmicrobio.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Galloway RL, Hoffmaster AR. Optimization of LipL32 PCR assay for increased sensitivity in diagnosing leptospirosis. Diagn. Microbiol. Infect. Dis. 2015;82(3):199–200. doi: 10.1016/j.diagmicrobio.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dikken H, Kmety E. Serological typing methods of leptospires. Methods Microbiol. 1978;11:259–307. doi: 10.1016/S0580-9517(08)70493-8. [DOI] [Google Scholar]
- 19.Ahmed N, et al. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann. Clin. Microbiol. Antimicrob. 2006;5(1):1–10. doi: 10.1186/1476-0711-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wunder EA, et al. Real-time PCR reveals rapid dissemination of Leptospira interrogans after intraperitoneal and conjunctival inoculation of hamsters. Infect. Immun. 2016;84(7):2105–2115. doi: 10.1128/IAI.00094-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Putz EJ, et al. Circulating foamy macrophages in the golden syrian hamster (Mesocricetus auratus) model of leptospirosis. J. Comput. Pathol. 2021;189:98–109. doi: 10.1016/j.jcpa.2021.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Desvars A, Cardinale E, Michault A. Animal leptospirosis in small tropical areas. Epidemiol. Infect. 2011;139(2):167–188. doi: 10.1017/S0950268810002074. [DOI] [PubMed] [Google Scholar]
- 23.Keenan J, et al. Seroprevalence of Leptospira in Rattus norvegicus in Grenada, West Indies. West Indian Med J. 2009;58(2):114–117. [PubMed] [Google Scholar]
- 24.Suepaul S, et al. Seroepidemiology of leptospirosis in dogs and rats in Trinidad. Trop Biomed. 2014;31(4):853–861. [PubMed] [Google Scholar]
- 25.Bourhy, P., et al. Serovar diversity of pathogenic Leptospira circulating in the French West Indies. PLoS Negl. Trop. Dis.7(3): e2114 (2013). [DOI] [PMC free article] [PubMed]
- 26.Cranford, H.M., et al. Mongooses (Urva auropunctata) as reservoir hosts of Leptospira species in the United States Virgin Islands, 2019–2020. PLoS Negl. Trop. Dis.15(11): e0009859 (2021). [DOI] [PMC free article] [PubMed]
- 27.Cranford HM, et al. Exposure and carriage of pathogenic leptospira in livestock in St. Croix, US Virgin Islands. Trop. Med. Infect. Dis. 2021;6(2):85. doi: 10.3390/tropicalmed6020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briskin EA, et al. Seroprevalence, risk factors, and rodent reservoirs of leptospirosis in an urban community of Puerto Rico, 2015. J. Infect. Dis. 2019;220(9):1489–1497. doi: 10.1093/infdis/jiz339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Everard CO, et al. The prevalence of severe leptospirosis among humans on Barbados. Trans. R. Soc. Trop. Med. Hyg. 1984;78(5):596–603. doi: 10.1016/0035-9203(84)90216-5. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez I, et al. Confirmación microbiológica de 2 brotes emergentes de leptospirosis humana en Cuba. Rev Cubana Med Trop. 2007;59(1):19–23. [PubMed] [Google Scholar]
- 31.Storck CH, et al. Changes in epidemiology of leptospirosis in 2003–2004, a two El Nino Southern Oscillation period, Guadeloupe archipelago, French West Indies. Epidemiol. Infect. 2008;136(10):1407–1415. doi: 10.1017/S0950268807000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGrowder D, Brown P. Clinical and laboratory findings in patients with leptospirosis at a tertiary teaching hospital in Jamaica. Res. Rep. Trop. Med. 2010;1:59–64. [Google Scholar]
- 33.Goarant C, et al. Outbreak of leptospirosis in New Caledonia: diagnosis issues and burden of disease. Trop. Med. Int. Health. 2009;14(8):926–929. doi: 10.1111/j.1365-3156.2009.02310.x. [DOI] [PubMed] [Google Scholar]
- 34.Lau CL, et al. The emergence of Leptospira borgpetersenii serovar Arborea in Queensland, Australia, 2001 to 2013. BMC Infect. Dis. 2015;15(1):1–11. doi: 10.1186/s12879-015-0982-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wynwood S, et al. The emergence of Leptospira borgpetersenii serovar Arborea as the dominant infecting serovar following the summer of natural disasters in Queensland, Australia 2011. Trop. Biomed. 2014;31(2):281–285. [PubMed] [Google Scholar]
- 36.Matsui M, et al. Experimental hamster infection with a strain of Leptospira borgpetersenii Ballum isolated from a reservoir mouse in New Caledonia. Am. J. Trop. Med. Hyg. 2015;92(5):982–985. doi: 10.4269/ajtmh.14-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benavidez KM, et al. The prevalence of Leptospira among invasive small mammals on Puerto Rican cattle farms. PLoS Negl. Trop. Dis. 2019;13(5):e0007236. doi: 10.1371/journal.pntd.0007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterson AC, et al. Amplification of pathogenic Leptospira infection with greater abundance and co-occurrence of rodent hosts across a counter-urbanizing landscape. Mol. Ecol. 2021;30(9):2145–2161. doi: 10.1111/mec.15710. [DOI] [PubMed] [Google Scholar]
- 39.Shiels, A.B., et al. Invasive rat establishment and changes in small mammal populations on Caribbean Islands following two hurricanes. Glob. Ecol. Conserv.22: e00986 (2020).
- 40.Torres-Castro M, et al. Detección molecular de leptospiras patógenas en roedores sinantrópicos y silvestres capturados en Yucatán, México. Biomedica. 2018;38:51–58. doi: 10.7705/biomedica.v38i3.3938. [DOI] [PubMed] [Google Scholar]
- 41.Ricardo, T., et al. Seroprevalence of leptospiral antibodies in rodents from riverside communities of Santa Fe, Argentina. PLoS Negl. Trop. Dis. 14(4): e0008222 (2020). [DOI] [PMC free article] [PubMed]
- 42.Costa, F., et al. Influence of household rat infestation on Leptospira transmission in the urban slum environment. PLoS Negl. Trop. Dis. 8(12): e3338 (2014). [DOI] [PMC free article] [PubMed]
- 43.Thornley C, et al. Changing epidemiology of human leptospirosis in New Zealand. Epidemiol. Infect. 2002;128(1):29–36. doi: 10.1017/S0950268801006392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brihuega, B., et al. First isolation of Leptospira borgpetersenii from Fetuses of Wild boars (Sus scrofa). (2017).
- 45.Gutierrez J. Effects of meteorological factors on human leptospirosis in Colombia. Int. J. Biometeorol. 2021;65(2):257–263. doi: 10.1007/s00484-020-02028-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.