Dear Editor,
Accumulation of misfolded and aggregation-prone proteins is the common hallmark of many neurodegenerative disorders, and lowering the levels of these proteins may provide promising strategies for the potential treatment of some of these diseases [1, 2]. Among them, Huntington’s disease (HD) is a monogenic disease caused by mutation of the HTT (huntingtin) gene [3], which encodes the mutant HTT protein (mHTT) with an expanded polyglutamine tract (polyQ). The monogenetic nature of HD provides high confidence for the causal relationship between mHTT and disease pathology, making HD suitable for testing the potential beneficial effects of reducing disease-causing proteins.
The gain of toxic function of mHTT is the major cause of HD, and lowering mHTT protein levels has been shown to effectively alleviate its toxicity. mHTT is known to be degraded by autophagy [4], and lowering its protein levels may ameliorate its downstream toxicity and treat HD [5]. Much evidence supports the role of apoptosis in HD; mHTT can induce apoptosis and then promote neuronal death [6], so the modulation of autophagy and apoptosis may be a potential means of reducing neuronal death. Turning off the transgene in a transgenic HD mouse model that expresses inducible mHTT N-terminal fragments reverses the neuropathology and motor deficits [7]. Other genetic strategies such as delivering short-hairpin RNAs, small interfering RNAs, antisense oligonucleotides [8], and CRISPR/Cas9-mediated genome editing [9] can attenuate the neuropathology in HD mouse models. We identified the kinase HIPK3 (homeodomain interacting protein kinase 3) as a novel modulator of mHTT protein levels from an unbiased genetic screen [10]. Knocking-down HIPK3 or loss of its kinase activity by mutagenesis lowers mHTT levels via autophagy [10]. Meanwhile, whether HIPK3 contributes to neurotoxicity and whether inhibiting its kinase function rescues HD-relevant phenotypes remained unknown.
In this study, we elucidated the potential role of HIPK3 in HD pathogenesis and the possible therapeutic effects of the small-molecule HIPK3 inhibitor AST487 in HD models, including HD mouse primary neurons, human induced pluripotent stem cell (iPSC)-derived neurons, and HD fly models.
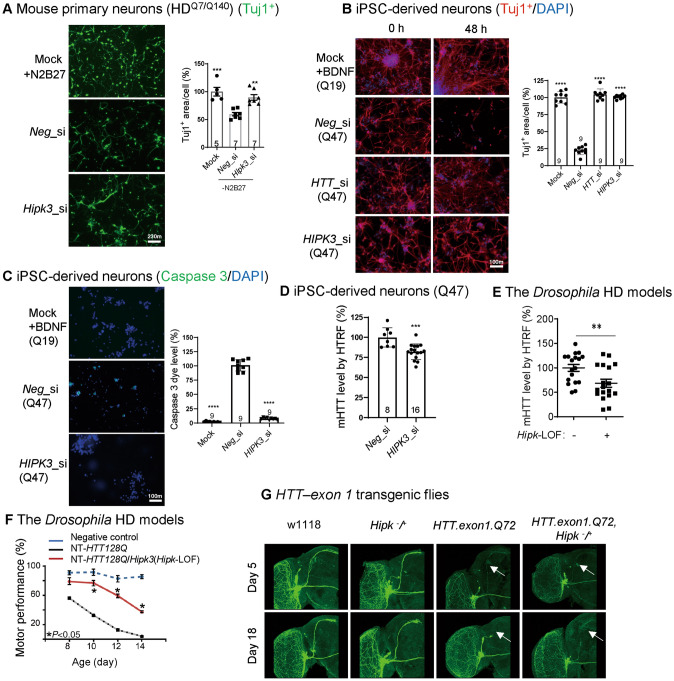
We first investigated the potential pathological role of HIPK3 in HD. mHTT-induced cytotoxicity under stressed culture conditions occurred in neurons from a knock-in mouse model (HDQ7/Q140, Q indicates polyglutamine) expressing endogenous mHTT protein from its original locus [11]. To induced apoptosis phenotypes, neurons were cultured in a medium without supplements N2 and B27, and their shrinkage was measured by the Tuj1+ (neuron-specific class III beta-tubulin) area in each neuron [12]. Knocking down HIPK3 rescued the apoptotic phenotype (Fig. 1A). To further confirm this in a human neuronal model, we cultured neurons derived from human iPSCs with or without brain-derived neurotrophic factor (BDNF). Knocking down HIPK3 rescued the neuronal shrinkage (Fig. 1B) and apoptotic phenotype (Fig. 1C), and decreased the mHTT level in the human iPSC-derived neurons (Q47) (Fig. 1D). Taken together, targeting the kinase HIPK3 lowered the mHTT level and ameliorated mHTT neurotoxicity and apoptosis. To further confirm the role of Hipk3 in vivo, we tested two HD Drosophila models. A loss-of-function (LOF) mutation of the Drosophila homolog of Hipk3 (Hipk-LOF) significantly decreased the mHTT level in the HD transgenic model expressing the N-terminal fragment of human mHTT with 128Q (NT-HTT-128Q, Fig. 1E). Consistent with this, the LOF mutation also rescued the climbing deficits in this model (Fig. 1F). We next investigated another HD fly model expressing the human mHTT exon 1 fragment (HTT-exon1-Q72) to validate the role of Hipk3 in vivo. In this model, the small ventral lateral clock neurons are labeled by mCD8GFP protein [13], so neurodegeneration in vivo is clearly indicated by the neuronal morphology, such as axon shrinkage and loss. Hipk-LOF rescued the mHTT-induced neuronal degeneration, while the same group of neurons in the wild-type flies were not influenced by Hipk-LOF (Fig. 1G), suggesting that the effects were HD-specific. Taken together, the effects of Hipk3 are likely to play a role in vivo and to be evolutionarily conserved.
Fig. 1.
The potential pathological role of HIPK3 in HD. A Representative images and quantification of the Tuj1+ area in mouse primary neurons (HDQ7/Q140). Data are normalized to controls with N2 and B27. Note that the mock group was the same for both the Fig. 1A and Fig. 2E quantification (scale bar, 10 µm; n, number of mice of each genotype). B Representative immunostaining results of the neuron-specific tubulin marker Tuj1 and DAPI showing the morphology of human iPSC-derived neurons (HD: polyQ = 47; WT: polyQ = 19). Note that the mock group was the same for both the Fig. 1B and Fig. 2F quantification [scale bar, 10 µm; data are normalized to the wild-type (WT)]. C Representative images and quantification of the caspase signal in human iPSC-derived neurons (HD: polyQ = 47; WT: polyQ = 19), note that the mock group was the same for both the Fig. 1C and Fig. 2G quantification, scale bar, 10 µm. D mHTT levels in HD iPSC-derived neurons (HD: polyQ = 47) treated with HIPK3 RNAi and controls assessed by the pair of antibodies 2B7/3B5H10 using HTRF (homogeneous time-resolved fluorescence). E mHTT levels in fly heads assessed by 2B7/3B5H10 using HTRF (fly model: NT-HTT128Q). F Hipk3 depletion rescues the climbing deficit in HD flies. G Neurodegeneration in axons of small ventral lateral clock neurons (sLNv) labeled by mCD8GFP protein using GMR61G12-GAL4 in a fly model. Mock, N2 and B27 were not removed from the medium for mouse primary neuron culture, BDNF was not removed from the medium for iPSC-derived neuron culture. Neg_si, siScramble; Hipk3_si, siHipk3. Data are presented as the mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ns, P > 0.05; one-way ANOVA and post hoc Dunnett’s tests (A–C, F), and unpaired t-test (D, E).
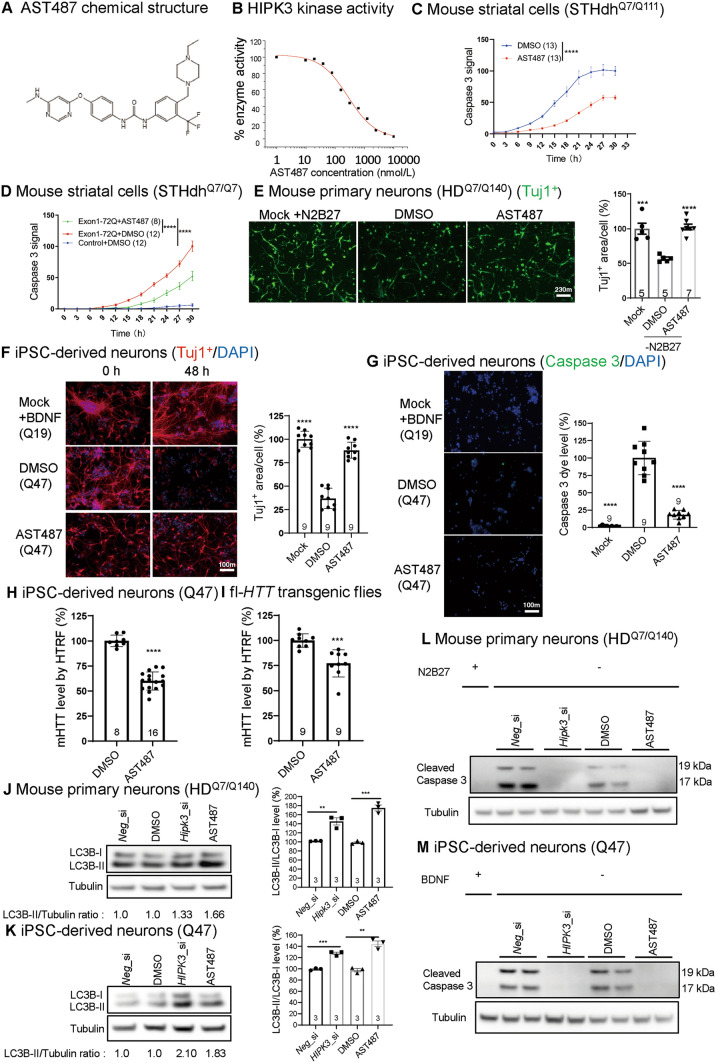
We then investigated the possibility of targeting HIPK3 using small molecular inhibitors, which may provide potential entry points to HD therapeutics. AST487 (Fig. 2A) has been reported to be a potent HIPK3 inhibitor based on a kinase screen [14]. We confirmed this by showing that AST487 blocked HIPK3 activity in vitro in a dose-dependent manner with an IC50 of 276.15 nmol/L (Fig. 2B). We then tested the effects of AST487 on mouse striatal HD cells (STHdhQ7/Q111), and found that mHTT toxicity was significantly rescued by AST487 at 2 µmol/L, as measured by an indicator dye for active caspase 3 (Fig. 2C). We further confirmed that mHTT toxicity was significantly rescued in an STHdhQ7/Q7-transfected model (Fig. 2D). AST487 was also sufficient to rescue the outgrowth deficits in mouse primary neurons (HDQ7/Q140) (Fig. 2E). Consistent with the results in mouse cells, AST487 rescued the disease-relevant phenotypes in the iPSC-derived neurons from HD patients after BDNF removal. Treatment with AST487 at 5 µmol/L rescued the neuronal shrinkage (Fig. 2F) and apoptosis (Fig. 2G), We further demonstrated that AST487 treatment decreased mHTT levels (Fig. 2H). Finally, to confirm the effects of AST487 in vivo, we examined the mHTT level in HD transgenic flies expressing full-length mHTT, and showed that AST487 significantly reduced the mHTT levels in fly transgenic models (Fig. 2I). Collectively, these results confirmed the significant role of HIPK3 and the rescue effect of inhibitors both in vitro and in vivo.
Fig. 2.
The potential pathological role of AST487 in HD. A Chemical structure of AST487. B AST487 regulates HIPK3 kinase activity. The plots are fitted with a logistic curve. C Apoptosis in STHdhQ7/Q111 cells at different time points with or without AST487 treatment (active caspase 3 was measured using NucView 488 dye). Data are presented as the mean ± SD. D As in C, but using STHdhQ7/Q7 cells transfected with exon 1 HTT-Q72 plasmid. E Representative images and quantification of primary neurons (HDQ7/Q140), note that the mock group was the same for both the Fig. 1A and Fig. 2E quantification, scale bar, 10 µm. F Representative immunostaining results of the neuron-specific tubulin marker Tuj1 and DAPI showing the morphology of human iPSC-derived neurons (HD: Q47; WT: Q19) [note that the mock group was the same for both the Fig. 1B and Fig. 2F quantification; scale bar, 10 µm; data are presented as the mean ± SD (normalized to the WT)]. G Representative images and quantification of the caspase 3 signal in human iPSC-derived neurons (HD: polyQ = 47; WT: polyQ = 19), note that the mock group was the same for both the Fig. 1C and Fig. 2G quantification, scale bar, 10 µm. H mHTT levels in human iPSC-derived neurons (HD: polyQ = 47; WT: polyQ = 19). I mHTT levels in fly heads assessed by 2B7/3B5H10 using HTRF (fly model: fl-HTT-Q128). J Representative images and quantification of LC3B Western blots for Hipk3 RNAi or Hipk3 inhibitor AST487 treatment in mouse primary neurons (HDQ7/Q140) (n = 3 replicates). K As in J, but in human iPSC-derived neurons (HD: polyQ = 47) (n = 3 replicates). L Representative caspase 3 Western blots for Hipk3 RNAi or Hipk3 inhibitor in mouse primary neurons (HDQ7/Q140). M As in L, but in human iPSC-derived neurons (HD: polyQ = 47). Mock, N2 and B27 were not removed from the medium for mouse primary neuron culture, BDNF was not removed from the medium for iPSC-derived neuron culture. Data are presented as the mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ns, P > 0.05; two-way ANOVA (C, D), one-way ANOVA and post hoc Dunnett’s tests (E–G), or the unpaired t-test (H–K).
We have demonstrated that HIPK3 modulates autophagy in a previously study. Here, we further showed that HIPK3 RNAi (RNA interference) or AST487 treatment enhanced the autophagosome marker protein LC3B-II levels in mouse primary neurons (Fig. 2J) and human iPSC-derived neurons (Fig. 2K), indicating that the autophagy level was up-regulated. HIPK3 RNAi decreased the caspase signal (Figs 1C and 2G) and significantly decreased the cleaved caspase 3 in mouse primary neurons (Fig. 2L) and human iPSC-derived neurons (Fig. 2M), suggesting that HIPK3 is involved in the pathogenesis in HD neurons. Collectively, inhibition of the HIPK3 kinase ameliorates mHTT-induced neurotoxicity and apoptosis.
Gain of toxic function of mHTT is the major cause of HD, so lowering mHTT protein levels may ameliorate its downstream toxic effects and treat HD. In neurodegenerative disorders, neuronal death occurs via one of two mechanisms: apoptosis or necrosis [15]. So far, there is no strong evidence to support that HD is correlated with apoptosis per se, however, the processes involved in initiating apoptosis happens before the onset of HD [15]. Autophagy and apoptosis are basic physiological processes, so the modulation of some regulators of autophagy and apoptosis may be an ideal way to treat neurodegenerative disorders.
The HTT protein is very large (348 kDa), its entire crystal structure and function are still unknown, so it is very difficult to develop therapeutic drugs that target it. So, protein kinases and receptors are ideal drug targets for HD and target-discovery approaches for similar diseases. The current study provides both genetic and chemical biology evidence demonstrating that HIPK3 is a potential therapeutic target of HD: knocking down HIPK3 or treatment with the HIPK3 inhibitor AST487 (Figs 1 and 2). Thus, targeting the kinase HIPK3 to lower the mHTT level would be a potential direction for developing HD drugs. Collectively, targeting HIPK3 using small-molecule inhibitors could be beneficial to HD patients, while further optimization of the compounds and further in vivo studies in the HD mouse models are needed.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Drs Lixiang Ma and Saiyin Hexige for sharing the HD iPSC lines. This work was supported by the National Natural Science Foundation of China (31970748, 92049301, and 81870990) and the Research Start-up Fund of Fudan University, China.
Conflict of interest
The authors claim no conflicts of interest.
References
- 1.Cheng HR, Li XY, Yu HL, Xu M, Zhang YB, Gan SR, et al. Correlation between CCG polymorphisms and CAG repeats during germline transmission in Chinese patients with Huntington's disease. Neurosci Bull. 2020;36:811–814. doi: 10.1007/s12264-020-00485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang WQ, Xie JM, Qiang Q, Li L, Lin X, Ren YQ, et al. Gedunin degrades aggregates of mutant huntingtin protein and intranuclear inclusions via the proteasomal pathway in neurons and fibroblasts from patients with Huntington's disease. Neurosci Bull. 2019;35:1024–1034. doi: 10.1007/s12264-019-00421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell 1993, 72: 971–983. [DOI] [PubMed]
- 4.Fu YH, Wu P, Pan YY, Sun XL, Yang HY, Difiglia M, et al. A toxic mutant huntingtin species is resistant to selective autophagy. Nat Chem Biol. 2017;13:1152–1154. doi: 10.1038/nchembio.2461. [DOI] [PubMed] [Google Scholar]
- 5.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo SQ, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 6.Vis JC, Schipper E, de Boer-van Huizen RT, Verbeek MM, de Waal RM, Wesseling P, et al. Expression pattern of apoptosis-related markers in Huntington's disease. Acta Neuropathol. 2005;109:321–328. doi: 10.1007/s00401-004-0957-5. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 8.Harper SQ, Staber PD, He XH, Eliason SL, Martins IH, Mao QW, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci U S A. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang S, Chang RB, Yang HM, Zhao T, Hong Y, Kong HE, et al. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington's disease. J Clin Invest. 2017;127:2719–2724. doi: 10.1172/JCI92087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu M, Fu YH, Liang YJ, Song HK, Yao Y, Wu P, et al. Suppression of MAPK11 or HIPK3 reduces mutant Huntingtin levels in Huntington's disease models. Cell Res. 2017;27:1441–1465. doi: 10.1038/cr.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington's disease with 140 CAG repeats. J Comp Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 12.Wang CC, Zhang YF, Guo SM, Zhao Q, Zeng YP, Xie ZC, et al. Correction to GPR52 antagonist reduces huntingtin levels and ameliorates Huntington's disease-related phenotypes. J Med Chem. 2021;64:1220–1221. doi: 10.1021/acs.jmedchem.0c01981. [DOI] [PubMed] [Google Scholar]
- 13.Wen X, An P, Li HX, Zhou ZJ, Sun YM, Wang J, et al. Tau accumulation via reduced autophagy mediates GGGGCC repeat expansion-induced neurodegeneration in Drosophila model of ALS. Neurosci Bull. 2020;36:1414–1428. doi: 10.1007/s12264-020-00518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 15.Ghavami S, Shojaei S, Yeganeh B, Ande SR, Jangamreddy JR, Mehrpour M, et al. Autophagy and apoptosis dysfunction in neurodegenerative disorders. Prog Neurobiol. 2014;112:24–49. doi: 10.1016/j.pneurobio.2013.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.