Abstract
Background & Aims
The I148M Patatin-like Phospholipase Domain-containing 3 (PNPLA3), the rs641738 in the Membrane bound O-acyltransferase domain containing 7-transmembrane channel-like 4 (MBOAT7-TMC4) locus, and the E167K Transmembrane 6 Superfamily Member 2 (TM6SF2) polymorphisms represent the main predisposing factors to nonalcoholic fatty liver disease (NAFLD) development and progression. We previously generated a full knockout of MBOAT7 in HepG2 cells (MBOAT7-/-), homozygous for I148M PNPLA3. Therefore, we aimed to investigate the synergic impact of the 3 at-risk variants on liver injury and hepatocellular carcinoma (HCC) in a large cohort of NAFLD patients, and create in vitro models of genetic NAFLD by silencing TM6SF2 in both HepG2 and MBOAT7-/- cells.
Methods
NAFLD patients (n = 1380), of whom 121 had HCC, were stratified with a semiquantitative score ranging from 0 to 3 according to the number of PNPLA3, TM6SF2, and MBOAT7 at-risk variants. TM6SF2 was silenced in HepG2 (TM6SF2-/-) and MBOAT7-/- (MBOAT7-/-TM6SF2-/-) through Clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9 (CRISPR/Cas9).
Results
In NAFLD patients, the additive weight of these mutations was associated with liver disease severity and an increased risk of developing HCC. In HepG2 cells, TM6SF2 silencing altered lipid composition and induced the accumulation of microvesicular lipid droplets (LDs), whereas the MBOAT7-/-TM6SF2-/- cells showed a mixed microvesicular/macrovesicular pattern of LDs. TM6SF2 deletion strongly affected endoplasmic reticulum and mitochondria ultrastructures, thus increasing endoplasmic reticulum/oxidative stress. The mitochondrial number was increased in both TM6SF2-/- and MBOAT7-/-TM6SF2-/- models, suggesting an unbalancing in mitochondrial dynamics, and the silencing of both MBOAT7 and TM6SF2 impaired mitochondrial activity with a shift toward anaerobic glycolysis. MBOAT7-/-TM6SF2-/- cells also showed the highest proliferation rate. Finally, the re-overexpression of MBOAT7 and/or TM6SF2 reversed the metabolic and tumorigenic features observed in the compound knockout model.
Conclusions
The co-presence of the 3 at-risk variants impacts the NAFLD course in both patients and experimental models, affecting LD accumulation, mitochondrial functionality, and metabolic reprogramming toward HCC.
Keywords: NAFLD, HCC, TM6SF2, ER Stress, Mitochondrial Dynamics
Abbreviations used in this paper: Akt, protein kinase B; ANOVA, analysis of variance; ApoB, apolipoprotein B-100; ATP, adenosine triphosphate; BMI, body mass index; BSA, bovine serum albumin; Cer, ceramide; CRISPR-Cas9, Clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9; DAG, diacylglycerol; DMEM, Dulbecco’s modified Eagle medium; DMSO, dimethyl sulfoxide; ER, endoplasmic reticulum; FBS, fetal bovine serum; GFP, green fluorescent protein; HCC, hepatocellular carcinoma; LD, lipid droplet; LDH, lactate dehydrogenase; lyso, lysophosphatidylinositol; mRNA, messenger RNA; mTOR, mammalian target of rapamycin; NADH, nicotinamide adenine dinucleotide; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; OR, odds ratio; ORF, open reading frame; ORO, Oil Red O; PC, phosphatidylcholine; PCA, principal component analysis; PCR, polymerase chain reaction; PGC1α, Peroxisome proliferator-activated receptor-γ (PPARγ) coactivator-1α; PI, phosphatidylinositol; MT-COX1, mitochondrially encoded cytochrome c oxidase subunit 1; SDHA, succinate dehydrogenase complex flavoprotein subunit A; sgRNA, small guide RNA; SNP, single-nucleotide polymorphism; TAG, triacylglycerol; TEM, transmission electron microscopy; T2D, type 2 diabetes; VLDL, very-low-density lipoprotein
Graphical abstract
Summary.
The co-presence of I148M Patatin-like Phospholipase Domain-containing 3 (PNPLA3), the rs641738 close to Membrane Bound O-acyltransferase Domain-containing 7 (MBOAT7) and the E167K Transmembrane 6 Superfamily Member 2 (TM6SF2) at-risk variants impacts on nonalcoholic fatty liver disease (NAFLD) course, in both patients and experimental models. They affect lipid droplets accumulation, mitochondrial functionality and metabolic reprogramming leading to hepatocarcinogenesis.
Nonalcoholic fatty liver disease (NAFLD) is a growing burden on global health care, and it is considered the most relevant liver disease of the 21st century, affecting both adults and children. It is predicted to become the leading cause of hepatocellular carcinoma (HCC) and the most common indication for liver transplantation by 2030.1 NAFLD encompasses a wide spectrum of hepatic conditions ranging from simple steatosis (hepatic fat, >5%) to nonalcoholic steatohepatitis (NASH), fibrosis, cirrhosis, and HCC.2 The pathogenesis of NAFLD is closely intertwined with increased adiposity, insulin resistance, and dyslipidemia.3 Besides environmental factors, the 50%–70% of hereditable traits contributed to NAFLD susceptibility and its interindividual phenotypic variability.4 Three main single-nucleotide polymorphisms (SNPs) have been identified in the PNPLA3, MBOAT7, and TM6SF2 genes through genome-wide association studies and have been associated with the NAFLD spectrum.5, 6, 7
Intracellular fat accumulation and aberrant lipid metabolism represent the earliest events occurring in NAFLD and genetics may participate to hasten steatosis development and its transition to NASH and eventually to HCC.8 The rs738409 C>G variant in the PNPLA3 gene, encoding isoleucine to methionine amino acidic substitution at position 148 (I148M), is the strongest genetic variant predisposing from fatty liver to HCC and its frequency ranges from 17% to 49% according to ethnicity and the geographic distribution of NAFLD.9 PNPLA3 localizes on the surface of lipid droplets (LDs) and functions as triacylglycerol (TAG) lipase. The I148M mutation leads to the accumulation of mutant PNPLA3 on LD surfaces, thus inhibiting TAG hydrolysis. The rs641738 C>T variant close to the MBOAT7 gene (∼35%–40% T-allele frequency) was associated with cirrhosis development in alcohol abusers,7 and, afterward, with NAFLD severity in European descendents.10 MBOAT7 enzymatically remodels acyl chains of phospholipids on cellular membranes by transferring polyunsaturated fatty acids to lysophosphatidylinositols. Both hyperinsulinemia and the presence of the rs641738 polymorphism reduced the expression of hepatic MBOAT7, determining changes in phosphatidylinositol (PI) species toward saturated ones, precursors of TAG synthesis, and favoring fat accumulation.11 Finally, Kozlitina et al5 identified the rs58542926 C>T genetic variant in the TM6SF2 gene, whose allele frequency hovers 7.2% in individuals of European ancestry, 4.7% in Hispanics, and 3.4% in African Americans, thereby showing a lower minor allele frequency in the general population compared with PNPLA3 and MBOAT7 mutations. TM6SF2 localizes in the endoplasmic reticulum (ER) and ER-Golgi compartments,12 and participates in TAG-rich lipoprotein lipidation and assembly in the ER cisternae.5,13 The rs58542926 C>T variant in TM6SF2 encoding lysine instead of glutamate at residue 167 (E167K) causes the retention of very-low-density lipoprotein (VLDL) in the liver and increases the intrahepatic TAG content, but it protects against cardiovascular complications.5,14 However, differently from I148M PNPLA3 and rs641738 MBOAT7 variants, the E167K TMS6F2 variant was associated with fatty liver, but its role in cell injury and carcinogenesis remains uncharted.
It is well established that the I148M PNPLA3, the rs641738 in MBOAT7, and the E167K TM6SF2 SNPs predispose to NAFLD and advanced liver injury. Recently, the opportunity has emerged to translate the genetics into clinics by aggregating these genetic variants in polygenic risk scores for the assessment of fatty liver development and progression.15 However, the additive weight of the 3 at-risk mutations on liver disease severity and the related mechanisms need further investigation.16 Therefore, we aimed to explore the synergic effects of the I148M PNPLA3, rs641738 MBOAT7, and E167K TM6SF2 variants on clinicopathologic features and liver disease severity in a large cohort of patients with NAFLD. Moreover, to reproduce in vitro a condition that parallels human genetic NAFLD in vitro, we silenced hepatoma cells in HepG2 homozygous for the I148M PNPLA3 variant, the TM6SF2 and MBOAT7 genes by exploiting Clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9 (CRISPR/Cas9) technology. We previously generated a full knockout of MBOAT7 (MBOAT7-/-) in HepG2 cells, which spontaneously developed LDs.11 In this study, we silenced TM6SF2 in both HepG2 (TM6SF2-/-) and MBOAT7-/- (MBOAT7-/-TM6SF2-/-) cells to elucidate whether TM6SF2 ablation, alone or in combination with that of MBOAT7, may induce pathologic features resembling human NAFLD.
Results
The I148M PNPLA3, rs641738 MBOAT7, and E167K TM6SF2 Genetic Variants Have a Synergic Effect on Liver Damage
In the overall cohort, 172 patients were wild type (12.46%); 574 were heterozygous or homozygous for the I148M PNPLA3, the rs641738 MBOAT7, or the E167K TM6SF2 (42.03%); 552 carried at least 2 different risk SNPs in variable combinations (40%); and 82 had all 3 at-risk variants (5.94%). In the NAFLD-HCC cohort the percentage of patients who carried 2 or 3 risk variants was higher compared with the Hepatology Service cohort (52% and 11.5% vs 38.84% and 5.4%, respectively; P = .004 and .006) (Table 1).
Table 1.
Demographic, Anthropometric, and Clinical Features of the Overall Cohort (n = 1380), Including the Hepatology Service Cohort (n = 1259) and the NAFLD-HCC Cohort (n = 121), Stratified for Enrollment Criteria
| Overall cohort (n = 1380) | Hepatology service cohort (n = 1259) | NAFLD-HCC cohort (n = 121) | P value∗a | |
|---|---|---|---|---|
| Sex, male | 739 (53.5) | 650 (51.6) | 89 (73.5) | .06 |
| Age, y | 50.3 ± 13.5 | 47.86 ± 12.6 | 67.64 ± 10.0 | <.0001 |
| BMI, kg/m2 | 34.3 ± 8.7 | 34.7 ± 8.8 | 28.7 ± 5.14 | .01 |
| IFG/T2D, yes | 369 (26.7) | 293 (23.3) | 76 (62.8) | .005 |
| HOMA-IR | 5.26 ± 7.96 | 5.01 ± 5.49 | 12.4 ± 31.3 | <.0001 |
| Insulin, IU/mL | 20.9 ± 24.0 | 20.3 ± 16.9 | 39.4 ± 91.6 | .0003 |
| Total cholesterol, mmol/L | 5.14 ± 1.07 | 5.18 ± 1.04 | 4.28 ±.1.18 | <.0001 |
| LDL cholesterol, mmol/L | 3.15 ± 0.97 | 3.19 ± 0.95 | 2.4 ± 0.99 | <.0001 |
| HDL cholesterol, mmol/L | 1.29 ± 0.38 | 1.29 ± 0.37 | 1.31 ± 0.51 | .44 |
| Triglycerides, mmol/L | 1.62 ± 1.00 | 1.63 ± 0.94 | 1.41 ± 1.81 | .03 |
| ALT, IU/L | 4.49 {2.99–4.04} | 3.46 {2.99–4.04} | 3.61 {3.29–3.98} | .77 |
| AST, IU/L | 3.21 {2.94–3.61} | 3.21{2.89–3.58} | 3.62 {3.22–4.04} | .0074 |
| Risk variants, n | P value∗b | |||
| 0 | 172 (12.46) | 164 (13.02) | 6 (4.9) | .009 |
| 1 | 574 (42.03) | 538 (42.73) | 38 (31.4) | .01 |
| 2 | 552 (40) | 489 (38.84) | 63 (52) | .004 |
| 3 | 82 (5.94) | 68 (5.4) | 14 (11.5) | .006 |
NOTE. Values are reported as means ± SD, number (%), or median {interquartile range}, as appropriate. Characteristics of participants were compared across class enrollment criteria using linear regression model (for continuous variables) or logistic regression model (for categoric characteristics).
ALT, alanine aminotransferase; AST, aspartate aminotransferase; HDL, high-density lipoprotein; HOMA-IR, homeostasis model assessment-estimated insulin resistance; IFG, impaired fasting glucose; LDL, low-density lipoprotein; T2D, type 2 diabetes.
Models were adjusted for sex, age, BMI, IFG/T2D, class enrollment, and number of 3 at-risk variants (I148M PNPLA3, E167K TM6SF2, and the rs641738 C>T MBOAT7). Bold P < .05 was considered statistically significant. ∗Hepatology service cohort vs the NAFLD-HCC cohort.
The frequencies for each risk variant subgroup were compared using the chi-squared test. Bold P < .05 was considered statistically significant. 0, absence of risk variants; 1–3, total number of risk variants carried. ∗Hepatology service cohort vs the NAFLD-HCC cohort.
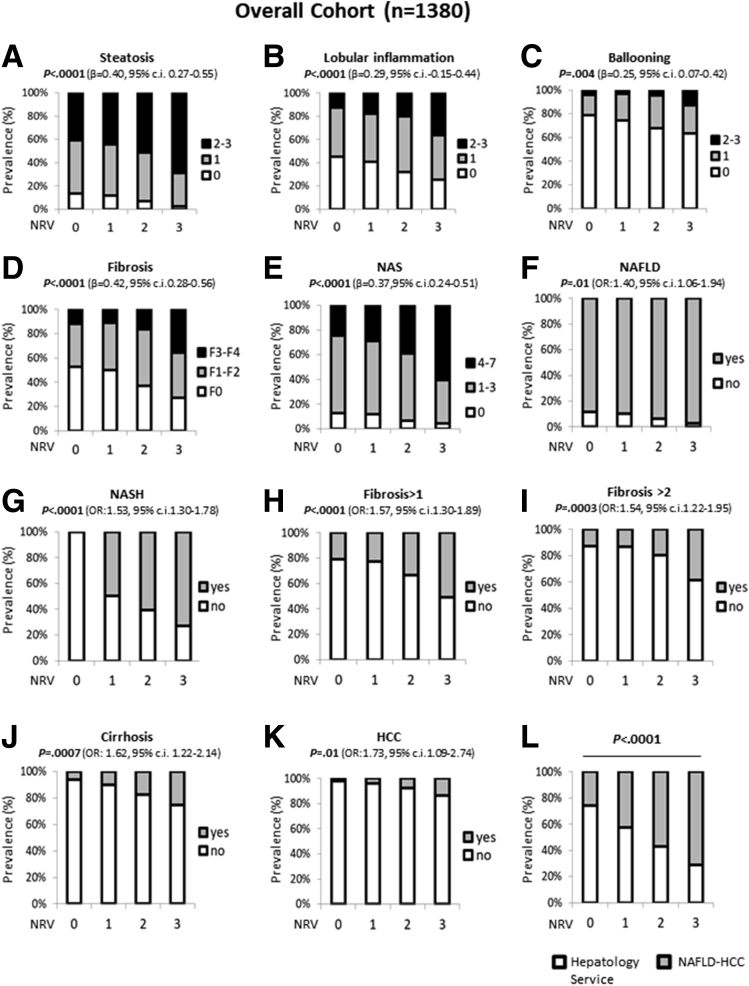
In a generalized linear model adjusted for age, sex, body mass index (BMI), and type 2 diabetes (T2D), the co-presence of the 3 risk variants in the overall cohort was associated with increased levels of markers of liver damage (P = .003, β = .07, 95% CI, -0.02 to 0.11; and P < .0001, β = .07, 95% CI, -0.04 to 0.11) (Table 2). At ordinal logistic regression analysis adjusted as described earlier, the co-presence of the 3 at-risk variants was associated with a higher grade of steatosis (P < .0001; β = .40; 95% CI, 0.27–0.55) (Figure 1A), lobular inflammation (P < .0001; β = .29; 95% CI, -0.15 to 0.44) (Figure 1B), ballooning (P = .004; β = .25; 95% CI, 0.07–0.42) (Figure 1C), fibrosis (P < .0001; β = .42; 95% CI, 0.28–0.56) (Figure 1D), and NAFLD activity score (P < .0001; β = .37; 95% CI, 0.24–0.51) (Figure 1E) in the overall cohort. At nominal logistic regression analysis adjusted for age, sex, BMI, and T2D, carriers of the 3 SNPs had an increased risk of developing NAFLD (odds ratio [OR], 1.40; 95% CI, 1.06–1.83; P = .01) (Figure 1F), NASH (OR, 1.53; 95% CI, 1.30–1.78; P < .0001) (Figure 1G), fibrosis greater than 1 (OR, 1.57; 95% CI, 1.30–1.89; P < .0001) (Figure 1H), fibrosis greater than 2 (OR, 1.54; 95% CI, 1.22–1.95; P = .0003) (Figure 1I), cirrhosis (OR, 1.62; 95% CI, 1.22–2.14; P = .0007) (Figure 1J), and an approximately 2-fold higher risk of developing HCC (OR, 1.73; 95% CI, 1.09–2.74; P = .01) (Figure 1K) even after the adjustment for the presence of fibrosis. In addition, at bivariate analysis we found that the prevalence of the 3 risk variants was approximately 2.5-fold enriched in patients in the NAFLD-HCC cohort compared with those in the Hepatology Service cohort (P < .0001) (Figure 1L).
Table 2.
Demographic, Anthropometric, and Clinical Features of the Overall Cohort (n = 1380) Stratified for Number of PNPLA3 I148M, MBOAT7 rs641738, and TM6SF2 E167K Risk Variants
| Number of risk variants |
P valuea | ||||
|---|---|---|---|---|---|
| 0 (n = 172) | 1 (n = 574) | 2 (n = 552) | 3 (n = 82) | ||
| Sex, male | 78 (45.9) | 304 (52.9) | 305 (55.3) | 51 (62.2) | .27 |
| Age, y | 49.02 ± 12.17 | 48.69 ± 13.17 | 49.71 ± 13.96 | 52.33 ± 14.70 | .80 |
| BMI, kg/m2 | 35.77 ± 8.37 | 34.15 ± 8.68 | 34.48 ± 8.85 | 31.71 ± 8.26 | .02 |
| IFG/T2D, yes | 42 (24.41) | 138 (24.04) | 159 (28.8) | 29 (35.36) | .10 |
| HOMA-IR | 4.34 ± 3.30 | 4.98 ± 3.82 | 5.81 ± 11.57 | 5.25 ± 3.92 | .16 |
| Insulin, IU/mL | 18.7 ± 13.03 | 20.08 ± 13.96 | 22.63 ± 34.01 | 20.50 ± 11.3 | .16 |
| Total cholesterol, mmol/L | 5.21 ± 1.09 | 5.24 ± 1.07 | 5.07 ± 1.08 | 4.77 ± 0.80 | .006 |
| LDL cholesterol, mmol/L | 3.23 ± 0.94 | 3.20 ± 0.98 | 3.13 ± 0.99 | 2.87 ± 0.70 | .06 |
| HDL cholesterol, mmol/L | 1.36 ± 0.37 | 1.33 ± 0.41 | 1.25 ± 0.34 | 1.22 ± 0.35 | .001 |
| Triglycerides, mmol/L | 1.51 ± 0.96 | 1.69 ± 1.07 | 1.59 ± 0.95 | 1.54 ± 1.02 | .57 |
| ALT, IU/L | 3.33 {2.91–3.97} | 3.40 {2.94–3.97} | 3.58 {3.07–4.13} | 3.67 {3.25–4.14} | .003 |
| AST, IU/L | 3.46 {3.17–3.79} | 3.17{2.89–3.55} | 3.29 {2.99–3.66} | 3.46 {3.17–3.79} | <.0001 |
NOTE. Values are reported as means ± SD, number (%), or median {interquartile range}, as appropriate. Characteristics of participants were compared across the increasing number of at-risk variants (I148M PNPLA3, E167K TM6SF2, and the rs641738 in TMC4/MBOAT7 locus) using linear regression model (for continuous variables) or logistic regression model (for categoric characteristics).
ALT, alanine aminotransferase; AST, aspartate aminotransferase; HDL, high-density lipoprotein; HOMA-IR, homeostasis model assessment-estimated insulin resistance; IFG, impaired fasting glucose; LDL, low-density lipoprotein; T2D, type 2 diabetes.
Models were adjusted for sex, age, BMI, IFG/T2D, and the number of 3 at-risk variants. 0, absence of risk variants; 1–3, total number of risk variants carried. Bold P < .05 was considered statistically significant.
Figure 1.
The co-presence of the PNPLA3 rs738409, MBOAT 7 rs641738, and TM6SF2 rs58542926 variants correlated with NAFLD severity and HCC risk. (A–E) At ordinal regression analysis adjusted for age, sex, BMI, and T2D, the co-presence of I148M PNPLA3, MBOAT7 rs641738, and E167K TM6SF2 SNPs was associated with steatosis, lobular inflammation, ballooning, fibrosis, and NAFLD activity score (NAS). (F–I) At nominal logistic regression analysis adjusted for age, sex, BMI, and T2D, the co-presence of 3 SNPs increased the risk of developing NAFLD, NASH, fibrosis score > 1, and fibrosis score > 2. (J and K) The increasing number of at-risk variants correlated with cirrhosis at nominal logistic regression analysis adjusted for age, sex, BMI, and T2D and with HCC after further adjustment for the presence of fibrosis. (L) The co-presence of the 3 SNPs was enriched significantly in the NAFLD-HCC cohort vs the Hepatology Service cohort (P < .0001). 0, indicates the absence of risk variants; 1–3 indicate the total number of risk variants (NRVs) carried.
CRISPR/Cas9-Mediated Gene Editing in Hepatocytes to Model NAFLD
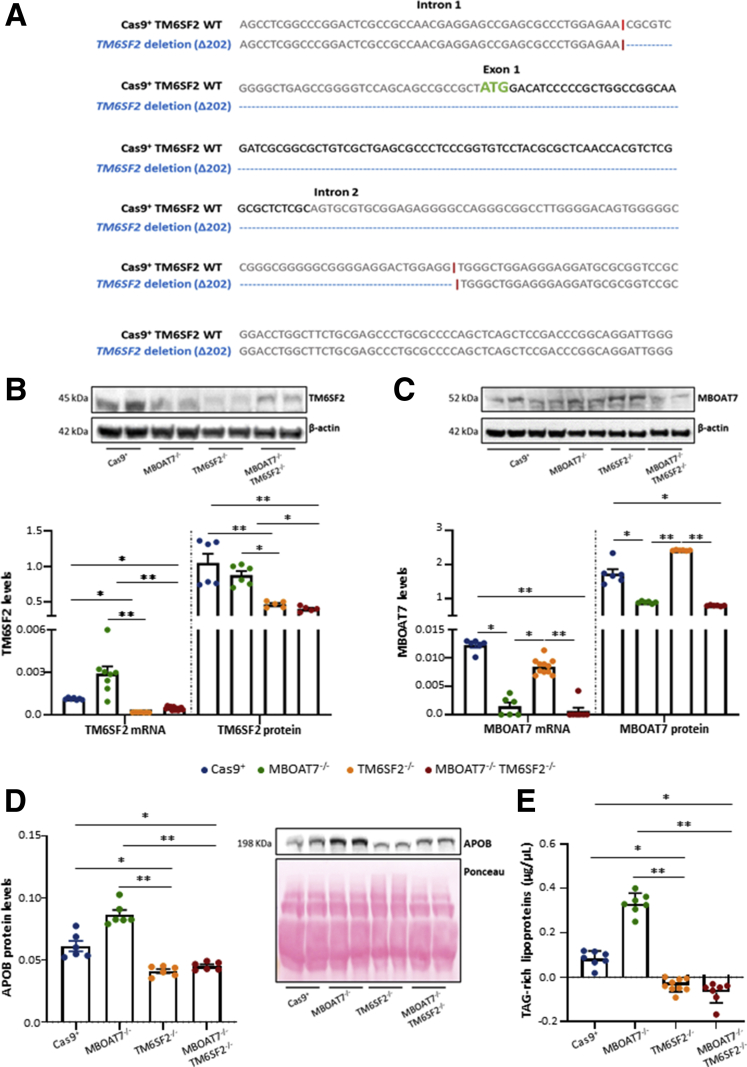
To explore whether TM6SF2 and MBOAT7 loss-of function in the context of the I148M PNPLA3 genetic background may exert an additive effect in hepatocytes in terms of fat accumulation, lipid metabolism, hepatocellular stress, and carcinogenesis, we exploited CRISPR/Cas9 technology to induce genetic deficiency of the MBOAT7 (MBOAT7-/-),11 TM6SF2 (TM6SF2-/-), or both (MBOAT7-/-TM6SF2-/-) in HepG2 cells. Sanger sequencing confirmed TM6SF2 silencing resulting from a deletion of 202 nucleotides (Δ202) cutting the ATG site in both TM6SF2-/- and MBOAT7-/-TM6SF2-/- clones compared with the wild-type reference sequence (Figure 2A). As expected, TM6SF2 messenger RNA (mRNA) and protein levels were reduced in TM6SF2-/- and MBOAT7-/-TM6SF2-/- cells compared with Cas9+ and MBOAT7-/- cells (P < .0001 at ANOVA; adjusted P < .05 vs Cas9+ and MBOAT7-/-) (Figure 2B). Likewise, the MBOAT7 expression was lower only in MBOAT7-/- and MBOAT7-/-TM6SF2-/- cells (P = .0002 at ANOVA; adjusted P < .05 vs Cas9+ and TM6SF2-/-) (Figure 2C), thus recapitulating the human condition of genetic NAFLD in which the rs641738 and the E167K variants cause a reduction of MBOAT7 and TM6SF2 levels, respectively.
Figure 2.
CRISPR/Cas9-mediated TM6SF2 ablation in HepG2 cells. (A) Schematic representation of the TM6SF2 sequence (gene ID: 53345; referred to as transcript variant 1: NM_001001524.3) highlighted the same clonal Cas9-induced indel mutations in both TM6SF2-/- and MBOAT7-/-TM6SF2-/- clones (blue) of 202 nucleotides (Δ202). The Cas9 cutting site is indicated by the symbol “|” (red), and the transcription start site (ATG) of the protein coding sequence (NP_001001524.2) is shown in green. (B) mRNA and protein expression of TM6SF2 was evaluated through reverse-transcription quantitative PCR and Western blot, respectively. TM6SF2 reduction was detected in TM6SF2-/- and MBOAT7-/-TM6SF2-/- cells. (C) MBOAT7 mRNA and protein levels were lower in MBOAT7-/- and MBOAT7-/-TM6SF2-/- cells compared with Cas9+ and TM6SF2-/- cells. (D) ApoB protein was assessed in cell supernatants by Western blot and normalized to the entire lane of the Ponceau stain. Either TM6SF2-/- or MBOAT7-/-TM6SF2-/- showed low ApoB levels. (E) TAG-rich lipoprotein secretion was measured in cell supernatants and normalized to levels of total cholesterol by using the Cholesterol Colorimetric Assay Kit–HDL and LDL/VLDL (Abcam). Both TM6SF2-/- and MBOAT7-/-TM6SF2-/- dampened TAG-rich lipoprotein release. Data were normalized to the β-actin housekeeping gene for reverse-transcription quantitative PCR and Western blot and they are expressed as means and SE. At least 3 independent experiments were conducted. Adjusted ∗P < .05 and ∗∗P < .01 vs Cas9+ and/or vs MBOAT7-/-. WT, wild-type.
To assess whether TM6SF2 deletion impacted its functional role, we evaluated both apolipoprotein B-100 (ApoB) protein levels and TAG-rich lipoprotein export in cell supernatants. MBOAT7-/- cells highly enhanced the ApoB and TAG-rich lipoproteins secretion, probably as a compensatory mechanism to remove intracellular lipids (Figure 2D and E). Conversely, both TM6SF2-/- and MBOAT7-/-TM6SF2-/- reduced ApoB levels (adjusted P < .05 vs Cas9+ and P < .01 vs MBOAT7-/-) (Figure 2D) and completely abrogated TAG-rich lipoprotein release compared with both Cas9+ and MBOAT7-/- cells (P = .0005 at ANOVA, adjusted P < .05 vs control and P < .01 vs MBOAT7-/-, respectively) (Figure 2E), thereby supporting that lipoprotein synthesis and export was affected by TM6SF2 silencing.
TM6SF2 Deletion Alters LD Size in Hepatocytes
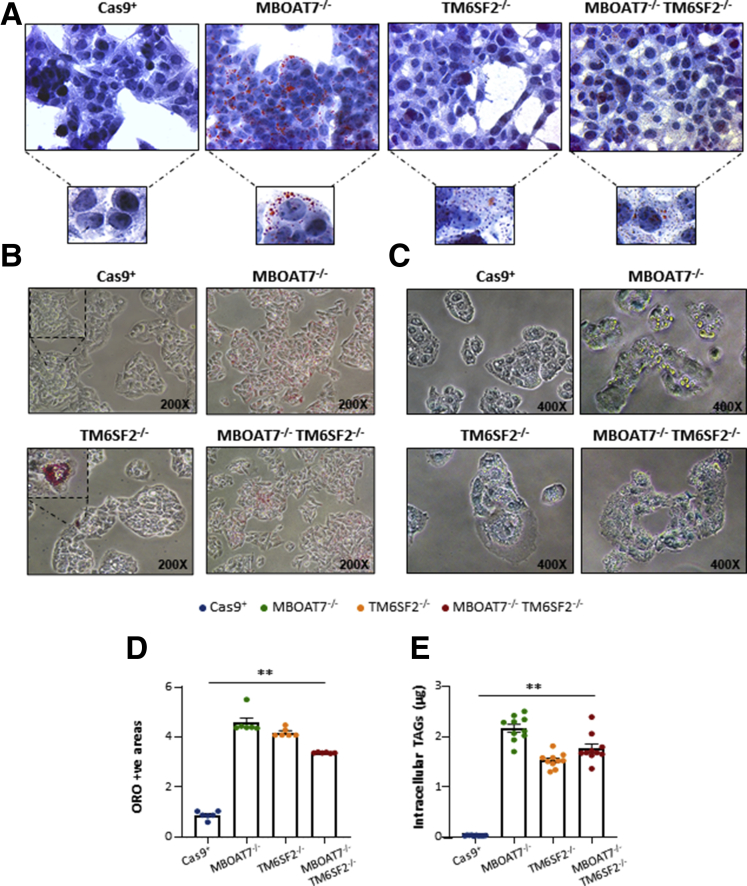
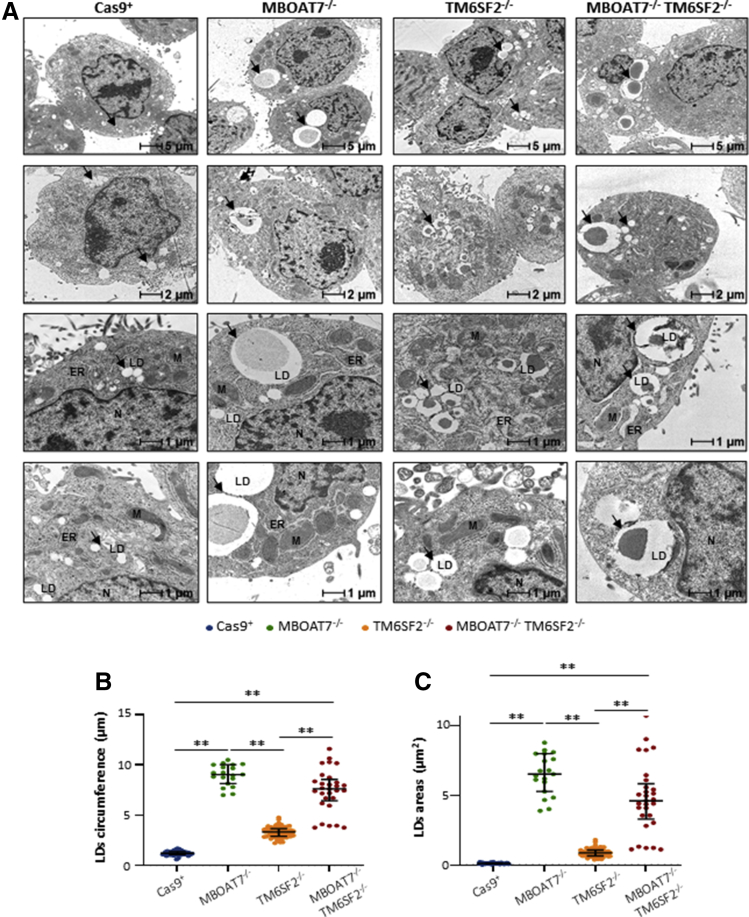
We investigated whether genetically edited clones could reliably reproduce in vitro human steatosis. We assessed intracellular fat content through Oil Red O (ORO) staining. Consistent with our recent findings,11 MBOAT7-/- cells spontaneously accumulated giant LDs (Figure 3A–C). Here, we found that TM6SF2-/- cells developed small LDs at baseline, whereas the MBOAT7-/-TM6SF2-/- clones presented a mixed pattern with either large or small LDs (Figure 3A–C), thus suggesting that TM6SF2 loss-of-function may diversely affect LD formation compared with that exerted by MBOAT7 deletion. According to the qualitative results, measurement of ORO-positive areas and intracellular TAGs showed approximately 30- to 40-fold higher enrichment of lipids in TM6SF2-/- and MBOAT7-/-TM6SF2-/- cells compared with controls (P = .0002 at ANOVA, adjusted P < .01 vs Cas9+; Figure 3D and E), while in the MBOAT7-/- cells the increase was approximately 50-fold, supporting that MBOAT7 deletion exerted the largest influence on lipid handling.
Figure 3.
TM6SF2 deletion induced spontaneous LD accumulation. (A and B) Spontaneous development of LDs in TM6SF2-/- and MBOAT7-/-TM6SF2-/- cells assessed by ORO staining (magnification, 630× and 200×, respectively). (C) Alkaline phosphatase (ALP) stained the LD surface in MBOAT7-/-, TM6SF2-/-, and MBOAT7-/-TM6SF2-/- clones, highlighting in yellow the differences in LD size for each condition. (D) ORO positive (+ve) areas were quantified by ImageJ in 10 random nonoverlapping micrographs per condition by calculating the percentage of pixels above the threshold value in respect to total pixels per area. (E) Measurement of TAG content in cell lysates. Data are expressed as means and SE. At least 3 independent experiments were conducted. Adjusted ∗∗P < .01 vs Cas9+.
Therefore, we deeply examined intracellular fat content through high-resolution transmission electron microscopy (TEM) imaging, which highlighted remarkable differences in LD size (Figure 4A). Although Cas9+ cells showed scarce and quite small lipid bodies, which were undetectable with ORO staining, the MBOAT7-/-, TM6SF2-/-, and MBOAT7-/-TM6SF2-/- clones differed in fat deposit volumes. Indeed, MBOAT7-/- showed the largest LD circumference and area (median size: 6.51 µm2 vs 0.11 µm2; adjusted P < .01 vs Cas9+ and TM6SF2-/-) (Table 3 and Figure 4B and C, respectively). Conversely, TM6SF2-/- cells had clustered areas enriched in much smaller LDs (median size: 0.87 μm2 vs 0.11 μm2; P < .0001 at ANOVA, adjusted P < .01 vs Cas9+ and MBOAT7-/-) (Table 3 and Figure 4B and C, respectively). MBOAT7-/-TM6SF2-/- cells show features between MBOAT7-/- and TM6SF2-/- cells showing areas with a mixed pattern of greater or smaller LDs (median size: 4.60 μm2 vs 0.11 μm2; P < .0001 at ANOVA, adjusted P < .01 vs Cas9+) (Table 3 and Figure 4B and C, respectively). These results support that MBOAT7 and TM6SF2 differently impact lipid handling and could lead to a diverse distribution of microvesicles and macrovesicles.
Figure 4.
TM6SF2 deficiency promotes small LD budding in hepatocytes. (A) Representative TEM images of LDs obtained by ultrathin 70-nm sections of hepatocytes. Black arrows indicate LDs. (B and C) The average of LDs’ circumference and area was calculated from 10 random non-overlapping micrographs (scalebar: 1 μm) by ImageJ. At least 3 independent experiments were conducted. Data are expressed as median and interquartile range. Adjusted ∗P < .05 and ∗∗P < .01 vs Cas9+, vs MBOAT7-/-, and vs TM6SF2-/-. ER, endoplasmic reticulum; M, mitochondria; N, nucleus.
Table 3.
Diameter, Circumference, and Areas of LDs Evaluated by TEM Analysis and Stratified According to the Genetic Background of the HepG2 Cells
| Diameter | ANOVA | Student t test | P valuea | |
|---|---|---|---|---|
| Cas9+ | 0.38 {0.33–0.43} | <0.0001 | ||
| MBOAT7-/- | 2.88 {2.59–3.18} | <0.0001 | <0.0001 | <.0001 |
| TM6SF2-/- | 1.05 {0.91–1.16} | <0.0001 | <0.0001 | <.0001 |
| MBOAT7-/-TM6SF2-/- | 2.42 {2.04–2.72} | <0.0001 | <0.0001 | <.0001 |
| Circumference, μm | ||||
|---|---|---|---|---|
| Cas9+ | 1.19 {1.05–1.35} | <0.0001 | ||
| MBOAT7-/- | 9.09 {8.13–10.0} | <0.0001 | <0.0001 | <.0001 |
| TM6SF2-/- | 3.31 {2.87–3.65} | <0.0001 | <0.0001 | <.0001 |
| MBOAT7-/-TM6SF2-/- | 7.60 {6.42–8.55} | <0.0001 | <0.0001 | <.0001 |
| Area, μm2 | ||||
|---|---|---|---|---|
| Cas9+ | 0.11 {0.09–0.14} | <0.0001 | ||
| MBOAT7-/- | 6.51 {5.26–7.98} | <0.0001 | <0.0001 | <.0001 |
| TM6SF2-/- | 0.87 {0.65–1.06} | <0.0001 | 0.003 | <.0001 |
| MBOAT7-/-TM6SF2-/- | 4.60 {3.28–5.82} | <0.0001 | <0.0001 | <.0001 |
NOTE. Values are reported as median {interquartile range}.
Bold P values are adjusted at post hoc Dunn's multiple comparison test and compared with the Cas9+ control group.
TM6SF2 Ablation Alone or Combined With MBOAT7 Impacts Lipid Composition
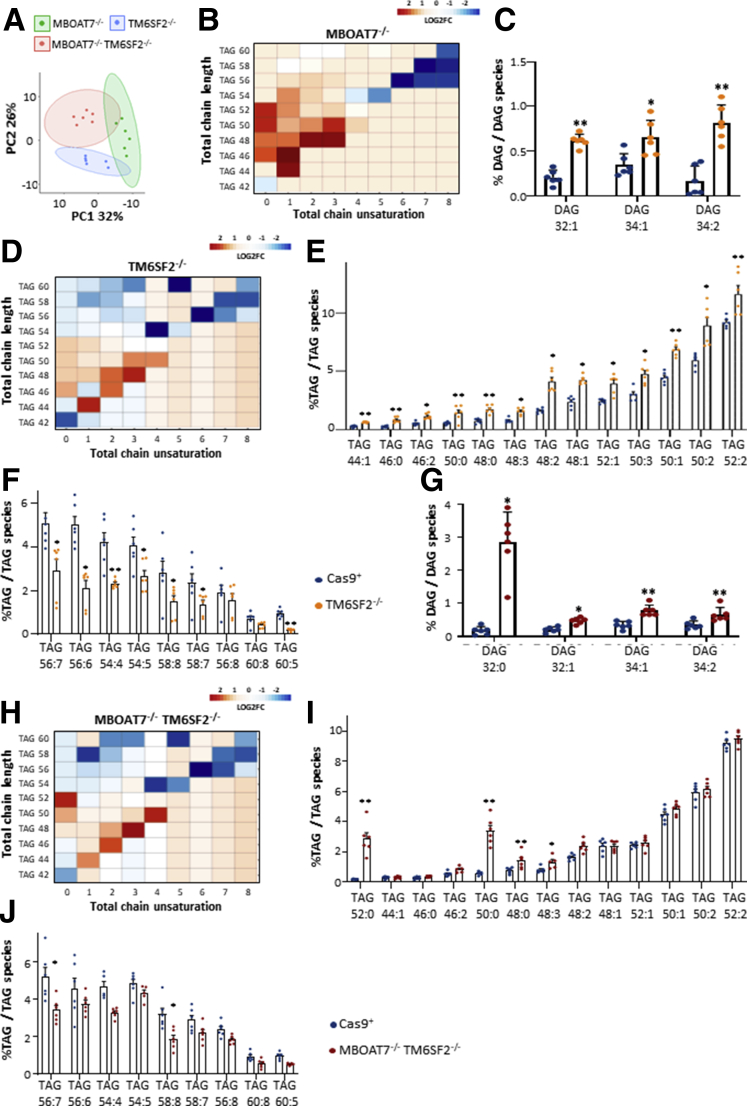
To investigate whether differences in LD dimensions are correlated with changes in lipid species, we performed a lipidomic analysis of each experimental group. Principal component analysis (PCA) identified a different lipid profile among MBOAT7-/-, TM6SF2-/-, and MBOAT7-/-TM6SF2-/- cells (Figure 5A). Specifically, levels of saturated/monounsaturated TAGs (Figure 5B and Supplementary Table 1) were laden in MBOAT7-/- cells, possibly owing to enhanced de novo lipogenesis.11 TM6SF2-/- cells were strongly enriched in highly saturated diacylglycerols (DAGs) (Figure 5C) and TAGs (Figure 5D and E and Supplementary Table 1), and less in unsaturated TAGs compared with the control group (Figure 5F and Supplementary Table 1).
Figure 5.
The impact of TM6SF2 deletion on LD composition. (A) PCA of the lipidomic profile of MBOAT7-/-, TM6SF2-/-, and MBOAT7-/-TM6SF2-/- models. (B) Heatmap of TAG species was generated by calculating the log2 fold change (log2FC) ratio between MBOAT7-/-/Cas9+ quantification. (C) Relatively enriched DAGs in TM6SF2-/- vs Cas9+. (D) Heatmaps of TAGs were generated by calculating the log2FC ratio between TM6SF2-/-/Cas9+. (E and F) Relative abundance of saturated/monounsaturated and polyunsaturated TAGs in TM6SF2-/- vs Cas9+. (G) Enrichment of DAG species in MBOAT7-/-TM6SF2-/- cells compared with Cas9+ cells. (H) Heatmaps of TAGs were generated by calculating the log2FC ratio between MBOAT7-/-TM6SF2-/-/Cas9+ quantification. (I and J) Relative abundance of saturated/monounsaturated and polyunsaturated TAGs in MBOAT7-/-TM6SF2-/- cells vs Cas9+. For heatmaps, red and blue boxes indicate overexpression or repression, respectively. For bar graphs, data are expressed as the percentage means and SE of DAG or TAG species compared with the total DAGs or TAGs. Adjusted ∗P < .05 or ∗∗P < .01 vs Cas9+.
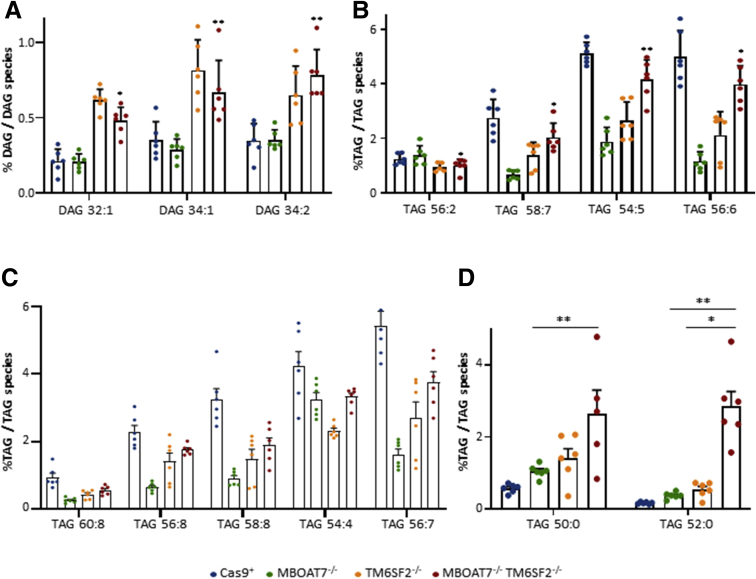
The MBOAT7-/-TM6SF2-/- model showed an in-between lipid profile among MBOAT7-/- and TM6SF2-/- cells (Figure 5A). Similar to the single-knockout cells, the compound knockout increased saturated/monounsaturated DAGs (Figure 5G) and TAGs (Figure 5H and I and Supplementary Table 1) rather than long-chain polyunsaturated TAGs compared with controls (Figure 5J and Supplementary Table 1). Notably, we found that TM6SF2 deletion rather than MBOAT7 affected the amount of DAGs in the MBOAT7-/-TM6SF2-/- cells by increasing mono-unsaturated/di-unsaturated DAG 32:1, DAG 34:1, and DAG 34:2 (Figure 6A and Supplementary Table 2). Among the unsaturated TAGs, TAG 54:5, TAG 56:2, TAG 56:6, and TAG 58:7 were influenced by a TM6SF2 genetic background (Figure 6B and Supplementary Table 2). However, the down-regulation of most of the polyunsaturated TAGs did not show a prevailing impact between MBOAT7 and TM6SF2 deficiency (Figure 6C and Supplementary Table 1).
Figure 6.
The contribution of TM6SF2 or MBOAT7 deletion on lipid composition in the compound knockout model. (A) Relative enriched DAGs in MBOAT7-/-TM6SF2-/- cells compared with MBOAT7-/-. (B–D) Relative abundance of saturated/monounsaturated and polyunsaturated TAGs in MBOAT7-/-TM6SF2-/- cells vs MBOAT7-/- and TM6SF2-/-. Data are expressed as the percentage means and SE of DAG or TAG species compared with the total DAGs or TAGs. Adjusted ∗P < .05 or ∗∗P < .01 vs MBOAT7-/- or TM6SF2-/-.
In attempt to identify whether the MBOAT7-/-TM6SF2-/- model may be characterized by peculiar TAG species, we found that the compound knockout markedly expressed the saturated TAG 50:0 and TAG 52:0 compared with Cas9+, MBOAT7-/-, and TM6SF2-/- models (Figure 6D, Supplementary Tables 1 and 2), potentially mirroring the hepatic lipid profile of NAFLD patients and the most up-regulated TAGs observed in HCC specimens.17,18
TM6SF2 Silencing Markedly Induces ER Stress in TM6SF2-Silenced Models
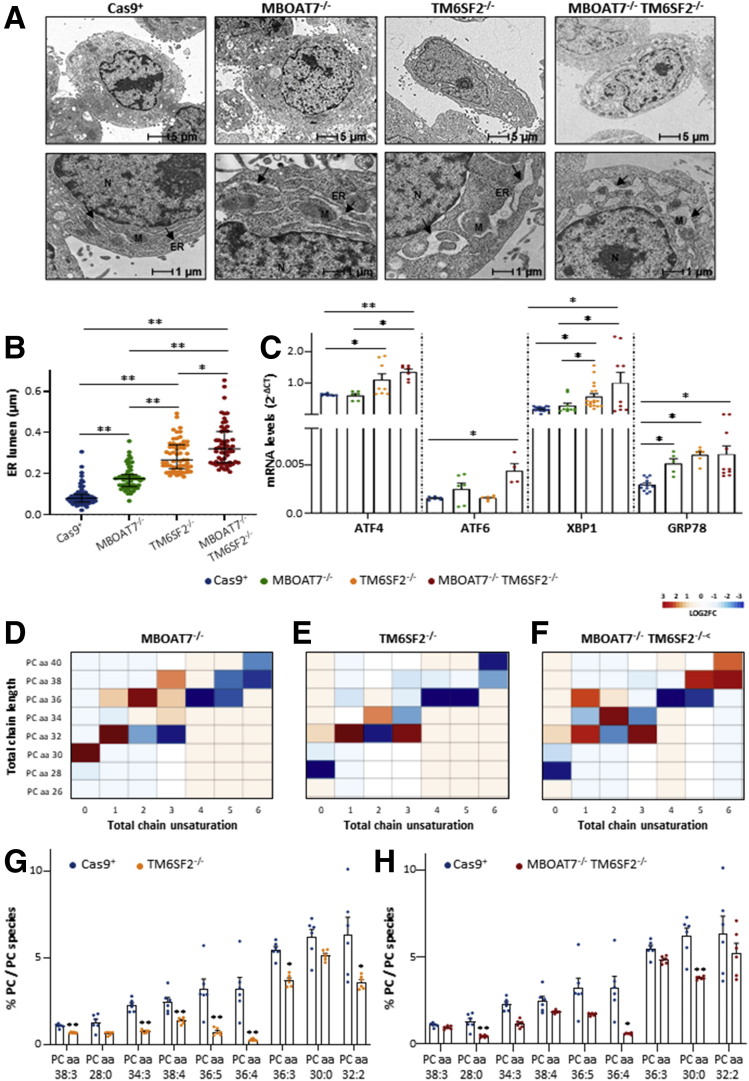
ER stress perturbs lipid metabolism, and it may play a crucial role in the NAFLD to NASH transition. Therefore, we assessed whether TM6SF2 and MBOAT7 deletion influenced ER morphology and function. We observed ultrastructural differences among each experimental group in the organization and width of the ER lumen, which are signs of cellular stress. In normal conditions, Cas9+ showed regular arrangement of the ER cisternae, whose parallel tubules appeared continuous and randomly distributed throughout the cytoplasm (Figure 7A). In the MBOAT7-/- model, we found significant enlargement of the ER lumen, but its architecture still was preserved (median width, 0.18 μm vs 0.09 μm; P < .0001 at ANOVA; adjusted P < .01 vs Cas9+) (Table 4 and Figure 7A and B, respectively) and increased GRP78 mRNA expression (P = .0002 at ANOVA; adjusted P < .05 vs Cas9+) (Figure 7C), suggesting that a mild unfolded protein response is associated with MBOAT7 deletion as a possible consequence of intracellular fat content.
Figure 7.
TM6SF2 silencing affects ER stress, morphology, and ER membrane fluidity. (A) Representative TEM images of ER cisternae obtained by ultrathin 70-nm sections of hepatocytes. Black arrows indicate ER tubules. (B) ER width was obtained by taking at least 3 measurements per ER lumen (n = 15 nonoverlapping micrographs for each condition, scalebar: 1 μm). (C) The expression of ATF4/6, XBP1 GRP78 was evaluated by reverse-transcription quantitative PCR and normalized to the β-actin housekeeping gene. (D–F) Heatmaps of PCs were generated by calculating the log2 fold change ratio between MBOAT7-/-/Cas9+, TM6SF2-/-/Cas9+, and MBOAT7-/-TM6SF2-/-/Cas9+ quantification. Red and blue boxes indicate overexpression or repression, respectively. (G and H) Relative enriched PCs in TM6SF2-/- and MBOAT7-/-TM6SF2-/- cells vs control. For reverse-transcription quantitative PCR, data are expressed as means and SE. For lipidomic analysis, data are expressed as the percentage means of PC species and SE compared with the total PC species. At least 3 independent experiments were conducted. Adjusted ∗P < .05 or ∗∗P < .01 vs Cas9+ and/or vs MBOAT7-/-. M, mitochondria; N, nucleus.
Table 4.
ER Cisternae Width and Area of Golgi Apparatus Calculated From TEM Micrographs and Stratified According to the Genetic Background of the HepG2 Cells
| ER width, μm | ANOVA | Student t test | P valuea | |
|---|---|---|---|---|
| Cas9+ | 0.09 {0.07–0.13} | <0.0001 | ||
| MBOAT7-/- | 0.18 {0.14–0.25} | <0.0001 | <0.0001 | <.0001 |
| TM6SF2-/- | 0.30 {0.23–0.37} | <0.0001 | <0.0001 | <.0001 |
| MBOAT7-/-TM6SF2-/- | 0.35 {0.27–0.44} | <0.0001 | <0.0001 | <.0001 |
| Golgi body area, μm2 | ||||
|---|---|---|---|---|
| Cas9+ | 0.007 {0.005–0.010} | <0.0001 | ||
| MBOAT7-/- | 0.026 {0.023–0.028} | <0.0001 | <0.0001 | <.0001 |
| TM6SF2-/- | 0.030 {0.027–0.033} | <0.0001 | <0.0001 | <.0001 |
| MBOAT7-/-TM6SF2-/- | 0.036 {0.033–0.039} | <0.0001 | <0.0001 | <.0001 |
NOTE. Values are reported as median {interquartile range}.
Bold P values are adjusted at post hoc Dunn’s multiple comparison test and compared with the Cas9+ control group.
Notably, the ER lumen appeared remarkably dilatated in cells lacking the TM6SF2 gene (median width, 0.30 and 0.35 μm vs 0.09 and 0.18 μm) (Table 4), showing extremely disorganized tubules and fragmented cisternae compared with both Cas9+ and MBOAT7-/- (P < .0001 at ANOVA; adjusted P < .01 vs Cas9+ and MBOAT7-/-) (Figure 7A and B). According to these morphologic changes, TM6SF2-/- clones strongly up-regulated markers of ER stress and unfolded protein response as ATF4 (P = .0008 at ANOVA; P < .05 vs Cas9+) (Figure 7C), XBP1 (P = .0001 at ANOVA; adjusted P < .05 vs Cas9+ and MBOAT7-/-) (Figure 7C), and GRP78 (P = .0002 at ANOVA; adjusted P < .05 vs Cas9+) (Figure 7C). MBOAT7-/-TM6SF2-/- cells showed an even more exacerbated breakage and enlargement of ER tubules, high local curvature of ER membranes (Figure 7A and B), and increased mRNA levels of ATF4 (P = .0008 at ANOVA; adjusted P < .05 vs MBOAT7-/- and P < .01 vs Cas9+) (Figure 7C), ATF6 (P = .03 at ANOVA; adjusted P < .05 vs Cas9+) (Figure 7C), XBP1 (P = .0001 at ANOVA; adjusted P < .05 vs Cas9+ and MBOAT7-/-) (Figure 7C), and GRP78 (P = .0002 at ANOVA; adjusted P < .05 vs Cas9+) (Figure 7C).
Lacking TM6SF2 Gene Affects Phosphatidylcholine Metabolism, Leading to Shapeless ER
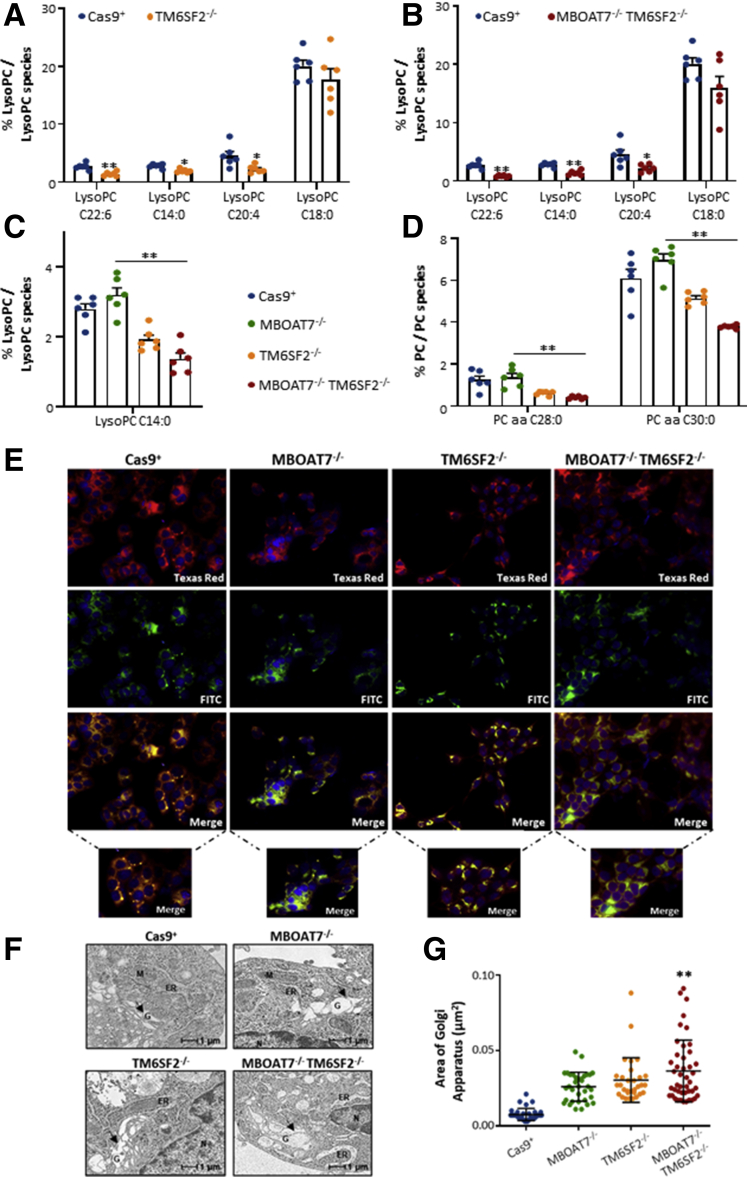
Phospholipids exert a central role to maintain ER functions, membrane fluidity, and ER-mitochondria contact sites. Deletion of the MBOAT7 gene reduced the amount of phosphatidylcholine (PC) conjugated with arachidonoyl-Coenzyme A (ie, PC 36:4, PC 38:6, PC 40:6) (Figure 7D and Supplementary Table 1), reinforcing previous data from our group.11 Here, we found that TM6SF2 silencing caused both lysophosphatidylinositol (lyso)–PC and PC depletion with either high or low side-chain saturation grade (Figures 7E–H and 8A and B, Supplementary Table 1) in both single and double knockouts compared with Cas9+. In particular, the compound knockout model dramatically reduced levels of saturated lyso-PC 14:0, PC 28:0, and PC 30:0 compared with both controls (Figure 8A and B and Supplementary Table 1) and MBOAT7-/- (Figure 8C and D and Supplementary Table 2), and this effect was amenable to a TM6SF2 shortage. Such evidence supports the pivotal role of TM6SF2 deletion to drive ER morphologic alterations and, in the double knockout, ER stress may be additively worsened by the presence of MBOAT7 ablation.
Figure 8.
Changes in ER–Golgi morphology upon MBOAT7 and/or TM6SF2 silencing. (A and B) Relative enrichment of lyso-PCs in TM6SF2-/- and MBOAT7-/-TM6SF2-/- cells vs Cas9+. (C and D) Relative enrichment of lyso-PC and PC species in MBOAT7-/-TM6SF2-/- cells vs MBOAT7-/-. (E) Representative cyto-staining of ER (Texas Red, red) and Golgi (fluorescein isothiocyanate [FITC], green) structures showing differences in ER–Golgi bodies (orange-yellow) in MBOAT7-/-, TM6SF2-/-, and MBOAT7-/-TM6SF2-/- models. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). (F) Representative TEM images of Golgi architecture obtained by ultrathin 70-nm sections of hepatocytes. Black arrows indicate Golgi (G) compartments. (G) The area of Golgi bodies was obtained by taking at least 3 measurements per Golgi lumen and compared with Cas9+ Golgi width (n = 10 nonoverlapping micrographs for each condition, scalebar: 1 μm). For lipidomic analysis, data are expressed as the percentage means of lyso-PC or PC species and SE compared with the total lyso-PC or PC species. Data are expressed as median and interquartile range for Golgi area. Adjusted ∗P < .05 or ∗∗P < .01 vs Cas9+ and/or vs MBOAT7-/-. M, mitochondria; N, nucleus.
Remarkable Changes in Golgi Compartments: A Possible Consequence of ER Stress
In an attempt to provide a detailed overview of alterations involving ER dysfunctions, we stained Cas9+, MBOAT7-/-, TM6SF2-/-, and MBOAT7-/-TM6SF2-/- cells with 3 types of dyes selective for Golgi bodies (fluorescein isothiocyanate, green), ER (Texas Red), and nucleus (4′,6-diamidino-2-phenylindole, blue) to evaluate ER–Golgi communications. In addition, we acquired 10 random micrographs at TEM to both obtain high-resolution images of Golgi architecture and to measure the area of Golgi compartments. At fluorescence microscopy, Golgi apparatus was well arranged in disc-shaped cisternae and co-localized with the ER (orange-yellow corpuscles) close to the nuclei of Cas9+ cells (Figure 8E). Conversely, recognition of the ER–Golgi complex resulted challenging in MBOAT7-/-, TM6SF2-/-, and MBOAT7-/-TM6SF2-/- models because they showed an uneven pattern of ER–Golgi staining, suggestive of architectural disorganization of these structures (Figure 8E). Consistently, evaluation of Golgi morphology by TEM showed that Golgi vesicles were approximately 4-fold enlarged in all mutated models (0.026, 0.03, and 0.036 vs 0.007 μm2) (Table 4), with the largest area observed in the compound knockout (P < .0001 at ANOVA; adjusted P < .01 vs Cas9+, MBOAT7-/-, and TM6SF2-/-) (Figure 8F and G and Table 4). Taken together, these aberrancies may be suggestive of interruption of ER–Golgi interplay in genetically edited models, possibly contributing to cell damage.
TM6SF2 Silencing Influences Mitochondrial Morphology, Number, and Oxidative Stress
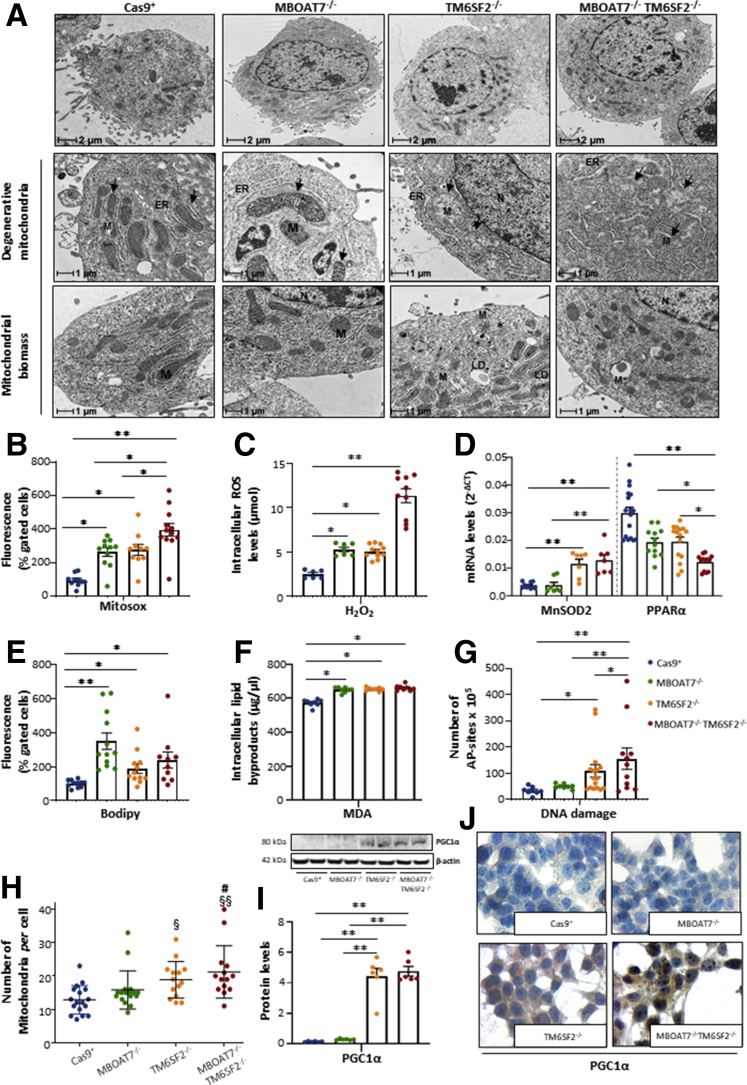
Because mitochondrial dysfunction is a hallmark of human hepatic steatosis and its progression, we analyzed mitochondrial morphology and investigated oxidative injury in our in vitro models. By using TEM, we observed features suggestive of mitochondrial damage and derangement in all knockout cell lines. In the control group, we found normal-shaped mitochondria with densely packed cristae, many of which were oriented longitudinally, and a few others were smaller in size, probably resulting from mitochondrial fusion–fission balancing (mitobiogenesis) (Figure 9A). In MBOAT7-/- cells, we found swollen and irregular mitochondrial cristae that still maintained a quite normal morphology (Figure 9A). Conversely, both TM6SF2-/- and MBOAT7-/-TM6SF2-/- cells showed several areas enriched in mitochondria with small and globular shape, loss of cistern architecture, and ultrastructural electron density, which may indicate mitochondrial failure and degeneration (Figure 9A).
Figure 9.
Alterations of mitochondrial degeneration and renewal in TM6SF2 knockout models. (A) Representative TEM images of degenerative mitochondria (M) and mitochondrial biomass obtained by ultrathin 70-nm sections of hepatocytes are indicated by black arrows. (B) ROS content was measured in live cells through MitoSOX Red reagent. (C) The H2O2 levels were measured in cell lysates through DCF ROS/RNS Colorimetric Assay Kit (Abcam). (D) The MnSOD2 and PPARα mRNA expression was assessed by reverse-transcription quantitative PCR and normalized to the β-actin housekeeping gene. (E) Lipid peroxides were quantified in live cells by BODIPY 581/591 C11. (F) The malondialdehyde (MDA) secretion was colorimetrically measured in cell supernatants following the manufacturer’s instructions. (G) Number of apurinic/apyrimidinic (AP) sites was obtained by isolating total DNA from each model. (H) The number of mitochondria per cell was counted from 15 random nonoverlapping micrographs (§P < .05 and §§P < .01 vs Cas9+; #P < .05 vs MBOAT7-/-; scalebar: 2 μm). (I) PGC1α protein levels were assessed by Western blot and normalized to β-actin. (J) Cytoplasmatic and nuclear localization of PGC1α protein was assessed in in vitro models. For the box plot, data are expressed as means and SE. Adjusted ∗P < .05 and ∗∗P < .01 vs Cas9+ and/or vs MBOAT7-/-.
In keeping with these results, MBOAT7-/-, TM6SF2-/-, and MBOAT7-/-TM6SF2-/- models hugely boosted reactive oxygen species (ROS) production compared with Cas9+ (P < .0001 at ANOVA; adjusted P < .05 vs control) (Figure 9B and C), and it was worsened in the compound knockout (adjusted P < .05 vs MBOAT7-/- and TM6SF2-/-) (Figure 9B and C). The latter augmented the expression of Manganese superoxide dismutase (MnSOD2) (P = .002 at ANOVA; P < .01 vs Cas9+ and MBOAT7-/-) (Figure 9D), possibly as a compensatory mechanism to dampen ROS overflowing. We dug deeper into ROS-induced cellular damage and found that all mutated models showed a conspicuous increase in lipid peroxidation (P < .0001 at ANOVA, adjusted P < .05 vs Cas9+; P = .002 at ANOVA; P < .05 vs Cas9+) (Figure 9E and F, respectively). Furthermore, TM6SF2-/- remarkably increased apurinic/apyrimidinic sites, the main ROS-induced DNA damage (P < .05 vs Cas9+) (Figure 9G), whose levels were higher in the double knockout (P < .01 vs Cas9+ and MBOAT7-/-) (Figure 9G). Lower Peroxisome proliferator-activated receptor alpha (PPARα) expression was observed only in the compound knockout (P < .0001 at ANOVA; adjusted P < .01 vs Cas9+; P < .05 vs MBOAT7-/- and TM6SF2-/- cells) (Figure 9D), supporting that the co-existence of MBOAT7 and TM6SF2 loss-of-function may jointly affect lipid synthesis and catabolism, contributing to progressive damage.
In parallel with the presence of degenerated mitochondria, the abundance of morphologically normal organelles was increased significantly in TM6SF2-/- (P = .003 at ANOVA; adjusted P < .05 vs Cas9+) (Figure 9A) and MBOAT7-/-TM6SF2-/- cells (P = .003 at ANOVA; adjusted P < .05 vs MBOAT7-/-; and P < .01 vs Cas9+) (Figure 9H), as well as Peroxisome proliferator-activated receptor-γ (PPARγ) coactivator-1α (PGC1α) protein levels, the master regulator of mitobiogenesis (P < .01 vs Cas9+ and MBOAT7-/-) (Figure 9I). At immunocytochemistry, PGC1α markedly localized in the cytoplasm and in several nuclei of TM6SF2-/- and MBOAT7-/-TM6SF2-/- clones compared with Cas9+ and MBOAT7-/- cells (Figure 9J), supporting its activation in response to unbalancing in mitochondrial fusion–fission events.
MBOAT7-/-TM6SF2-/- Model Switches Oxidative Respiration Toward Anaerobic Glycolysis
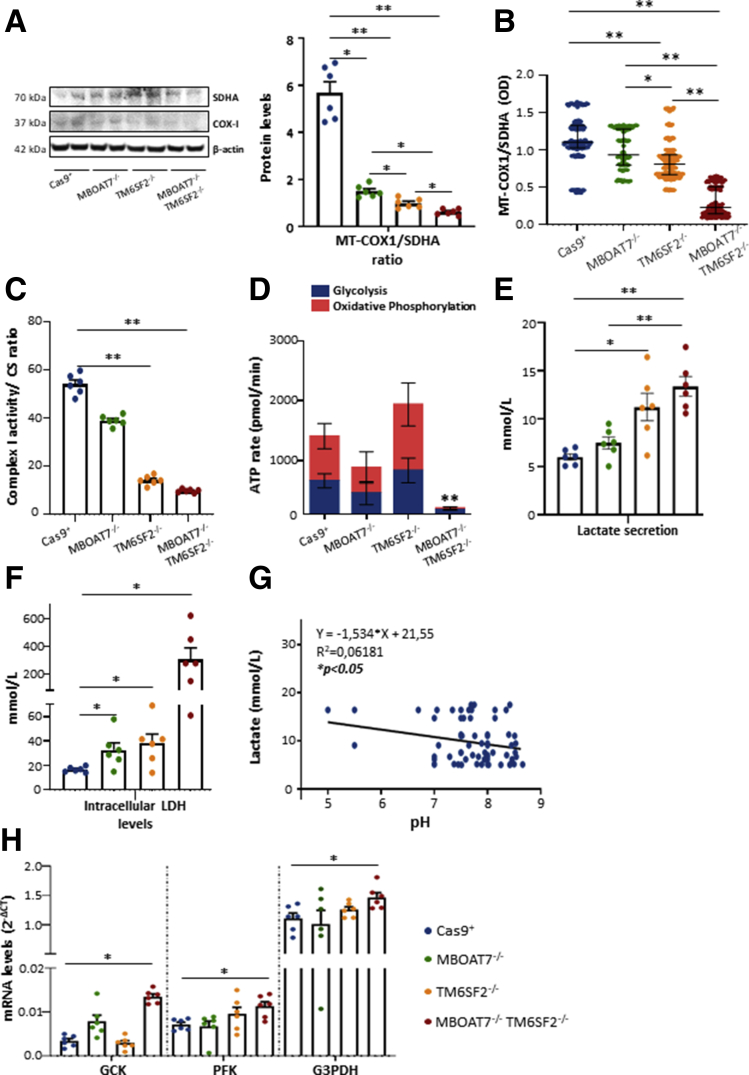
To explore mitochondrial functionality in NAFLD models, we measured the mitochondrially encoded cytochrome c oxidase subunit 1 (MT-COX1) levels, the main mitochondrially DNA–encoded subunit of the complex IV, which were normalized on succinate dehydrogenase complex flavoprotein subunit A (SDHA) levels, a nuclear-encoded subunit of complex II. In MBOAT7-/- clones, the MT-COX-1/SDHA ratio was reduced compared with control (Figure 10A). In both TM6SF2-/- and MBOAT7-/-TM6SF2-/- cells, the MT-COX-1/SDHA ratio was even lower (P < .05 and P < .01 vs Cas9+ and MBOAT7-/-) (Figure 10A). The MT-COX-1 and SDHA expression were evaluated independently by enzyme-linked immunosorbent assay, which confirmed the significant reduction of the MT-COX-1/SDHA ratio in TM6SF2-/- clones (P < .0001 at ANOVA; P < .05 and P < .01 vs Cas9+ and MBOAT7-/-) (Figure 10B) and the double knockout cells (P < .0001 at ANOVA; P < .01 vs Cas9+ and MBOAT7-/-) (Figure 10B).
Figure 10.
TM6SF2 silencing affectsoxydative phopshorylation (OXPHOS)and when combined with MBOAT7 deletion switches up to metabolic reprogramming. (A) MT-COX1 levels were evaluated by Western blot and normalized to SDHA. (B) MT-COX1 protein expression was measured by enzyme-linked immunosorbent assay (λ = 600 nm) and normalized to SDHA levels (λ = 405 nm). (C) Complex I enzymatic activity was evaluated biochemically and normalized to citrate synthase (CS). (D) The total ATP rate was obtained by Seahorse XF Analyzers in live cells. (E and F) Biochemical measurements of lactate (mmol/L) and LDH (mmol/L) were assessed in cell supernatants and lysates, respectively. (G) Inverse correlation between secreted lactate levels and pH values. (H) The mRNA levels of Glucokinase (GCK), Phosphofructokinase (PFK), and Glyceraldehyde-3-phosphate dehydrogenase (G3PDH) were evaluated by reverse-transcription quantitative PCR and normalized to the β-actin housekeeping gene. At least 3 independent experiments were conducted. For the box plot, data are expressed as means and SE. Adjusted ∗P < .05 and ∗∗P < .01 vs Cas9+ and/or vs MBOAT7-/-.
The enzymatic activity of complex I was compromised significantly in TM6SF2-/- and MBOAT7-/-TM6SF2-/- cells, suggesting that TM6SF2 silencing may directly interfere with the mitochondrial respiratory chain (P < .05 and P < .01 vs Cas9+ and MBOAT7-/-) (Figure 10C). In keeping with these results, we provided a quantitative measurement of the total adenosine triphosphate (ATP) rate derived from mitochondrial and glycolytic pathways. We observed an equal contribution of mitochondria and glycolysis to total ATP production in all cell lines, excluding the MBOAT7-/-TM6SF2-/- ones. The latter significantly reduced the total amount of ATP (P < .01 vs Cas9+, MBOAT7-/-, and TM6SF2-/-) (Figure 10D), but further showed that 87.1% of the ATP rate derived from glycolysis and only 12.9% derived from oxidative phosphorylation (Figure 10D). Consistently, by measuring lactate production, the end-product of anaerobic glycolysis, in cell supernatants, we found that TM6SF2-/- clones increased lactate release and up-regulated intracellular lactate dehydrogenase (LDH), which catalyzes the conversion of pyruvate to lactate and back (P < .05 vs control) (Figure 10E and F). No differences in lactate production were detected in the MBOAT7-/- cells, albeit it augmented LDH (P < .05 vs control) (Figure 10E and F).
In MBOAT7-/-TM6SF2-/-, lactate secretion was still more increased than the other clones (P < .01 vs Cas9+ and MBOAT7-/-) (Figure 10E) and correlated inversely with pH values (Figure 10G). The compound knockout model further showed the highest intracellular LDH levels (∼20-fold more than control; P < .05 vs Cas9+) (Figure 10F) and mRNA expression of glycolytic enzymes as Glucokinase (GCK) (adjusted P < .05 vs Cas9+) (Figure 10H), whose increasing levels were possibly owing to MBOAT7 deficiency, Phosphofructokinase (PFK), and Glyceraldehyde-3-phosphate dehydrogenase (G3PDH) (adjusted P < .05 vs Cas9+) (Figure 10H), thus supporting enhancement of the glycolytic pathway.
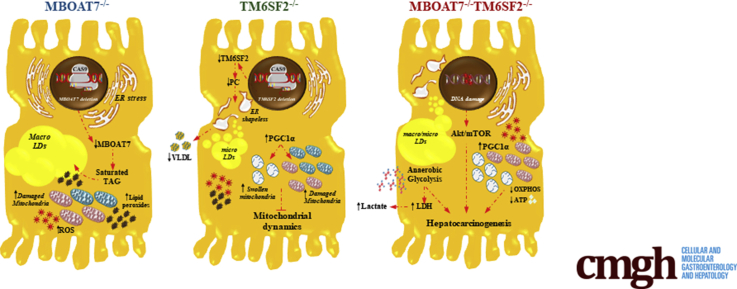
In sum, we found that the combined silencing of MBOAT7 and TM6SF2 markedly impairs mitochondrial dynamics and shift towards metabolic reprogramming, possibly contributing to progression to malignant transformation.
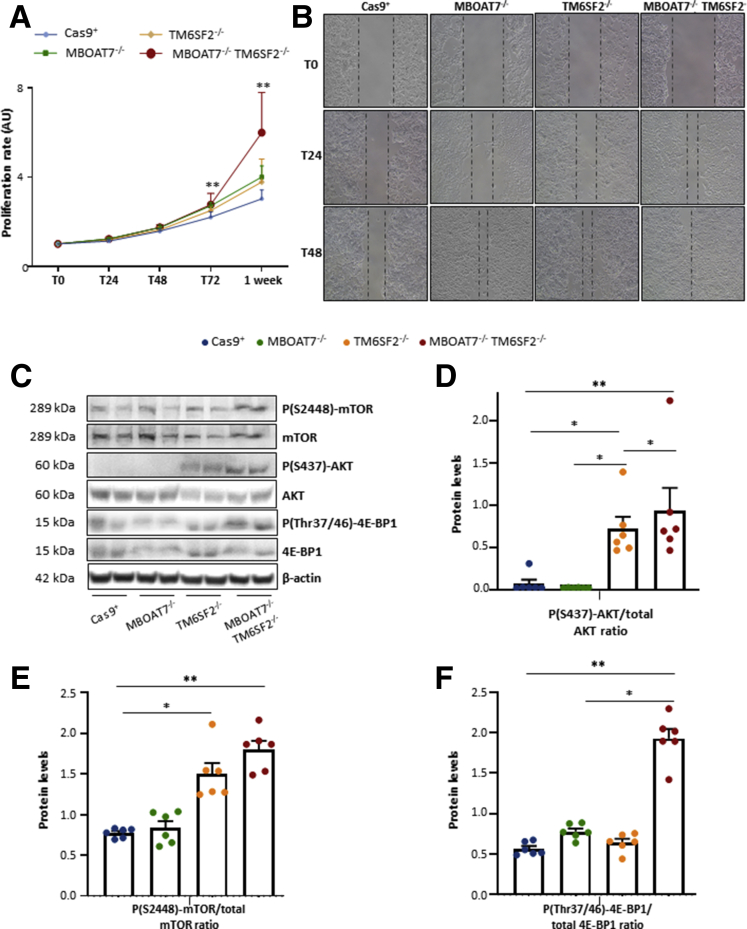
MBOAT7-/-TM6SF2-/- Cells Increase Cell Survival, Proliferation, and Invasiveness
To fulfill our hypothesis that TM6SF2 and MBOAT7 deletion in the background of PNPLA3 mutation may sustain the carcinogenic phenotype, we evaluated growth potential and migration in our models. MBOAT7-/-, TM6SF2-/-, and the compound knockout hepatocytes had a higher proliferation rate compared with Cas9+ at 72 hours (adjusted P < .01 vs control) (Figure 11A). MBOAT7-/-TM6SF2-/- clones further showed the greatest growing ability at 1 week (adjusted P < .01 vs Cas9+, MBOAT7-/-, and TM6SF2-/-) (Figure 11A). In the scratch assay, both MBOAT7-/- and TM6SF2-/- cells showed greater wound healing capacity at 24 and 48 hours compared with Cas9+, whereas MBOAT7-/-TM6SF2-/- was able to almost completely repair the scratch after just 24 hours (Figure 11B), thereby showing the largest proliferative and invasiveness power. Consistently, TM6SF2-silenced cells showed aberrant activation of the Phosphatidylinositol-3-kinase/Protein kinase B/Mammalian target of rapamycin (PI3K/Akt/mTOR) cascade in the absence of any stimuli (Figure 11C–F), with the strongest effect in the compound knockout cells, which may support its more proliferative and aggressive phenotype.
Figure 11.
TM6SF2lackingpromotes cell survival and carcinogenesis. (A) Cell growth was assessed through MTS assay for 24, 48, 72 hours and 1 week (λ = 490 nm). Data are expressed as fold increase vs control. (B) Representative images of wound healing assay were acquired at 0, 24, 48 hours (magnification, 100×). The dotted lines indicate the scratch width. (C) Phosphorylation of Akt at serine 437 residue (P[S437]-Akt), mTOR at serine 2448 residue (p[S2448]-mTOR), phosphorylation of 4E-BP1 at threonine 37/46 residues P[Thr37/46]-4E-BP1, and total Akt, mTOR, and 4E-BP1 were evaluated by Western blot. (D–F) Quantification of P(S437)-Akt/total Akt, p(S2448)-mTOR/total mTOR, and P(Thr37/46)-4E-BP1/total 4E-BP1 ratios were measured through ImageJ software and normalized to the β-actin housekeeping gene. At least 3 independent experiments were conducted. For bar graphs, data are expressed as means and SE. Adjusted ∗P < .05 and ∗∗P < .01 vs Cas9+ and/or vs MBOAT7-/-. AU, arbitrary unit.
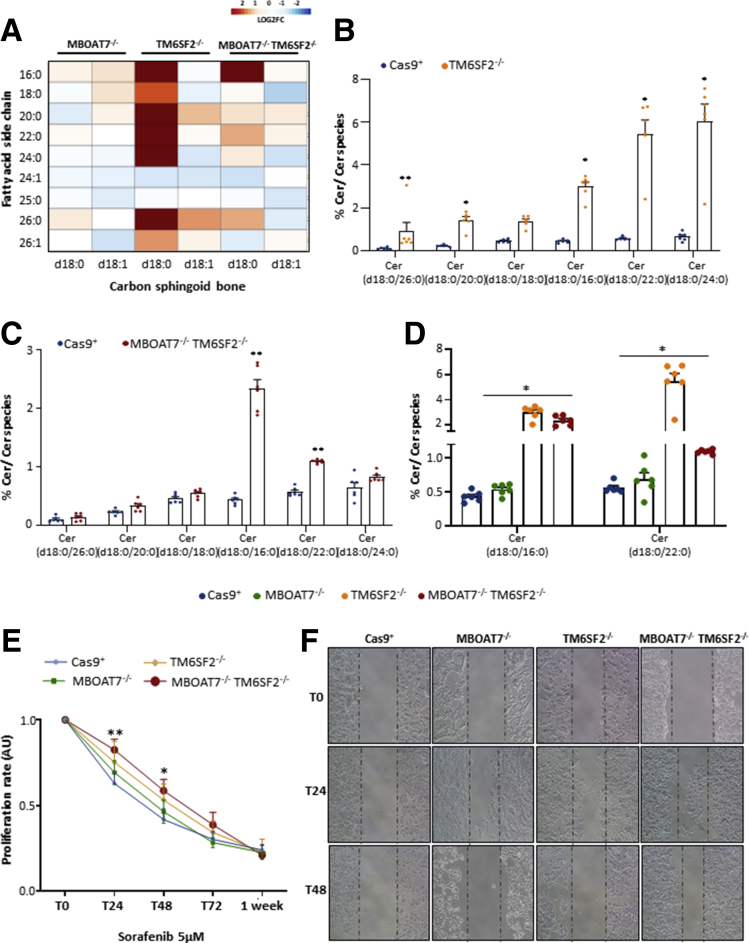
Loss of the TM6SF2 Gene Impacts the Abundance in Dihydroceramide Species
Dihydroceramides are precursors of ceramides and both exert different biological roles as modulators of cell fate. The percentage of dihydroceramides was strongly increased in TM6SF2-/- cells (Figure 12A), especially those species consisting of saturated sphingosine analog (d18:0) and incorporating long-chain fatty acids with no double bonds (adjusted P < .05 and P < .01 vs Cas9+) (Figure 12B). An in-between sphingolipid phenotype was observed in the compound knockout model (Figure 12A), in which ceramides (Cer) (d18:0/16:0) and Cer (d18:0/22:0) showed the highest levels compared with Cas9+ (P < .0001 at ANOVA, P = .0002 and P = .002, respectively) (Figure 12C) and were probably driven by TM6SF2 rather than MBOAT7 silencing (P < .0001 at ANOVA, adjusted P = .02 and P = .01 vs MBOAT7-/-) (Figure 12D). Therefore, the dramatic increase in dihydroceramides owing to TM6SF2 deletion is partially retained in the double-knockout model and may contribute to the prosurvival phenotype observed in MBOAT7-/-TM6SF2-/- cells.
Figure 12.
TM6SF2 silencing dramatically increases dihydroceramides and confers sorafenib resistance. (A) Heatmap of dihydro-Cers was generated by calculating the log2 fold change (log2FC) ratio between MBOAT7-/-/Cas9+, TM6SF2-/-/Cas9+, and MBOAT7-/-TM6SF2-/-/Cas9+ quantification. Red and blue boxes indicate overexpression or repression, respectively. (B and C) Relative abundance of dihydro-Cers in TM6SF2-/- and MBOAT7-/-TM6SF2-/- cells vs Cas9+. (D) Relative abundance of dihydro-Cers in MBOAT7-/-TM6SF2-/- cells vs MBOAT7-/-. (E) Cells were exposed to sorafenib (5 μmol/L) and cell growth was monitored through MTS assay for 0, 24, 48, 72 hours and 1 week. MBOAT7-/-TM6SF2-/- cells showed a significant resistance to sorafenib cytotoxicity at 24 and 48 hours. MTS absorbance (λ = 490 nm) was recorded at 0, 24, 48, 72 hours and 1 week. (F) Representative images of wound healing assay were acquired at 0, 24 and 48 hours (magnification, 100×) upon sorafenib (5 μmol/L) administration. The dotted lines indicate the scratch width. At least 3 independent experiments were conducted. For lipidomic analysis, data are expressed as the percentage means of dihydro-Cer species and SE compared with the total Cer species. For bar graphs, data are expressed as means and SE. Adjusted ∗P < .05 and ∗∗P < .01 vs Cas9+ or vs MBOAT7-/-. AU, arbitrary unit.
Synergic Contribution of MBOAT7 and TM6SF2 Silencing to Sorafenib Response
We investigated whether the co-presence of Cas9-induced mutations in HepG2 cells also may affect the pharmacologic response to sorafenib, a multikinase inhibitor approved for the treatment of advanced HCC. Therefore, we exposed Cas9+, MBOAT7-/-, TM6SF2-/-, and MBOAT7-/-TM6SF2-/- models either to sorafenib (5 μmol/L) or vehicle for 24, 48 and 72 hours and 1 week. Although just after 24 hours the proliferation rate of Cas9+ was reduced remarkably at MTS assay, MBOAT7-/- or TM6SF2-/- cells delayed cell death in response to sorafenib (adjusted P < .05 vs Cas9+) (Figure 12E), and survival was even higher in clones bearing both mutations (adjusted P < .01 vs Cas9+) (Figure 12E). In addition, the MBOAT7-/-TM6SF2-/- cells showed significant resistance to sorafenib cytotoxicity until 48 hours (adjusted P < .05 vs Cas9+) (Figure 12E). Similarly, after 24 hours of sorafenib (5 μmol/L) exposure, the MBOAT7-/-, TM6SF2-/-, and MBOAT7-/-TM6SF2-/- cells were able to migrate from one side to the other side of the wound healing to repair the scratch area, and the major effect was observed in the MBOAT7-/-TM6SF2-/- model (Figure 12F).
MBOAT7 and/or TM6SF2 Overexpression Rescued Metabolic and Tumorigenic Aberrancies
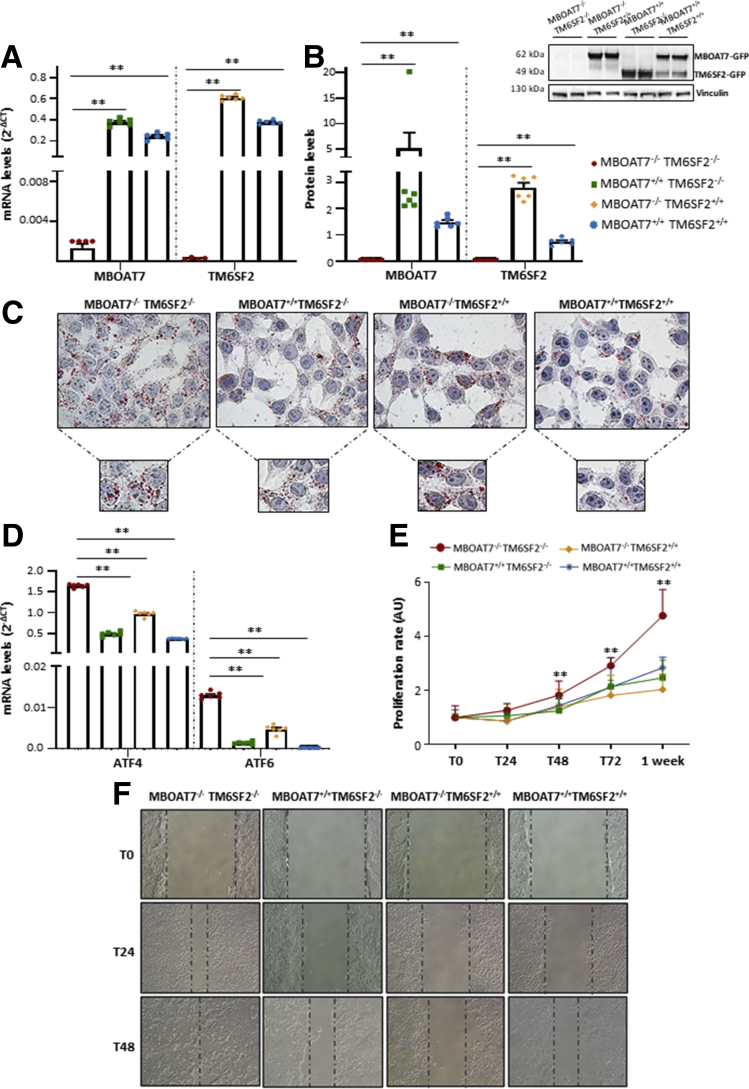
Finally, we overexpressed MBOAT7 and/or TM6SF2 in the MBOAT7-/-TM6SF2-/- model, attempting to validate the nature of metabolic aberrancies observed in hepatocytes after MBOAT7 and/or TM6SF2 silencing. Both mRNA and protein levels of MBOAT7 and TM6SF2 were strongly increased after lentiviral transfection, supporting that we obtained stable cell lines overexpressing the wild-type forms of the following: (1) MBOAT7–green fluorescent protein (GFP) but silenced for TM6SF2 gene (MBOAT7+/+TM6SF2-/-); (2) TM6SF2-GFP but without MBOAT7 (MBOAT7-/-TM6SF2+/+); or (3) both MBOAT7+/+TM6SF2+/+ (P = .001 at ANOVA, adjusted P < .01 vs MBOAT7-/-TM6SF2-/-) (Figure 13A and B).
Figure 13.
Recovery of metabolic and carcinogenic anomalies upon MBOAT7 and/or TM6SF2 overexpression. (A and B) The mRNA expression of MBOAT7 and TM6SF2 was evaluated by reverse-transcription quantitative PCR and normalized to the β-actin housekeeping gene. The protein levels of MBOAT7 and TM6SF2 tagged with GFP (MBOAT7-GFP and TM6SF2-GFP) were assessed by Western blot and normalized to the vinculin housekeeping gene. (C) LD accumulation was assessed in MBOAT7-/-TM6SF2-/-, MBOAT7+/+TM6SF2-/-, MBOAT7-/-TM6SF2+/+, and MBOAT7+/+TM6SF2+/+ by ORO staining (magnification, 630×). (D) The expression of ATF4 and ATF6 was evaluated by reverse-transcription quantitative PCR and normalized to the β-actin housekeeping gene. (E) The proliferation rate was examined through MTS assay for 0, 24, 48, 72 hours and 1 week (λ = 490 nm). (F) Representative images of wound healing assay were acquired at 0, 24 and 48 hours (magnification, 100×). The dotted lines indicate the scratch width. At least 3 independent experiments were conducted. For bar graphs, data are expressed as means and SE. Adjusted ∗P < .05 and ∗∗P < .01 vs MBOAT7-/-TM6SF2-/-.
At ORO staining, the up-regulation of MBOAT7 and/or TM6SF2 reduced lipid content compared with the compound knockout model and the largest effect on LD disposal likely was owing to the introduction of the MBOAT7 wild-type protein (Figure 13C). Notably, the MBOAT7+/+TM6SF2-/- and the MBOAT7-/-TM6SF2+/+ cells showed a mirror-effect because those overexpressing MBOAT7 but not TM6SF2 accumulated small LDs, while those overexpressing TM6SF2 but knockout for MBOAT7 gene showed large lipid bodies (Figure 13C), thereby showing a similar phenotype of TM6SF2-/- and MBOAT7-/- single-knockout models, respectively (Figure 4A). Such findings may strengthen the notion that MBOAT7 and TM6SF2 loss-of-function may induce a different pattern of fat accumulation in hepatocytes. Moreover, the reduction of lipid content in these models was accompanied by an improvement of ER stress. Indeed, the ATF4/6 mRNA levels were dramatically lower in the MBOAT7+/+TM6SF2-/- cells (P = .0004 at ANOVA, adjusted P = .003; P = .006 at ANOVA, adjusted P = .004) (Figure 13D), MBOAT7-/-TM6SF2+/+ (P = .0004 at ANOVA, adjusted P < .0001; P = .006 at ANOVA, adjusted P = .04) (Figure 13D), and in MBOAT7+/+TM6SF2+/+ models (P = .0004 at ANOVA, adjusted P = .001; P = .006 at ANOVA, adjusted P = .004) (Figure 13D) compared with the compound knockout.
Interestingly, we found that the growth rate was awfully diminished after the insertion of the MBOAT7 and/or TM6SF2 wild-type at 48 hours (P = .001 at ANOVA; adjusted P = .001; P = .005 and P = .02 vs MBOAT7-/-TM6SF2-/-) (Figure 13E), and even more at 72 hours (P = .001 at ANOVA; adjusted P = .007; P = .002 and P = .02 vs MBOAT7-/-TM6SF2-/-) (Figure 13E), and after 1 week later (P < .0001 at ANOVA; adjusted P = .0009; P = .0004 and P = .005 vs MBOAT7-/-TM6SF2-/-) (Figure 13E). Consistently, the MBOAT7+/+TM6SF2-/-, MBOAT7-/-TM6SF2+/+, and the MBOAT7+/+TM6SF2+/+ models showed a slowdown of wound healing repair, while the compound knockout cells confirmed their ability to almost close the scratch entirely after just 24 hours (Figure 13F). Therefore, preliminary results in vitro supported that the overexpression of the wild-type form of MBOAT7 and/or TM6SF2 could improve lipid accumulation and hepatocellular stress. Surprisingly, the up-regulation of these 2 genes remarkably ameliorated the proliferation rate and migration, reinforcing that both may be implicated in carcinogenic processes.
Discussion
In the present study, we performed a comprehensive analysis in a large cohort of biopsy-proven NAFLD patients (n = 1380), including 121 NAFLD-HCC individuals, to investigate the potential interactions among PNPLA3, MBOAT7, and TM6SF2 variants on liver disease severity and HCC development. Polygenic risk scores combining genetic predictors associated mainly with NAFLD have been proposed to improve diagnostic accuracy and to personalize therapeutic options.15 Here, we showed that the co-presence of the I148M PNPLA3, rs641738 close to MBOAT7, and E167K TM6SF2 SNPs were associated significantly with liver function tests and the histologic degree of steatosis, lobular inflammation, hepatocellular ballooning, and fibrosis. In addition, the risk of developing HCC in the overall cohort was approximately 2 in the presence of all 3 at-risk variants to the extent that the co-presence of the 3 SNPs was enriched significantly in NAFLD-HCC patients compared with those affected by NAFLD. Previously, the additive effect of the E167K TM6SF2 and I148M PNPLA3 variants on hepatic steatosis was explored by Xu et al19 in 158 NAFLD subjects and matched controls, in whom fatty liver was diagnosed noninvasively by Fibroscan. They found that the prevalence of NAFLD was more than 5-fold higher in patients carrying both mutations compared with patients with none or a single variant. In a multicenter biopsy-based study including 515 patients with NAFLD who were genotyped for the 3 variants, Krawczyk et al20 showed a significant association between serum aspartate aminotransferase levels and the increasing number of risk alleles, and the latter was correlated further with high HCC risk.16
We generated stable full knockouts of MBOAT7, TM6SF2, or both genes in HepG2 cells carrying the I148M PNPLA3 variant through CRISPR-Cas9 to investigate the mechanisms related to NAFLD pathogenesis in genetically edited in vitro models. We previously showed that MBOAT7 deletion reduced MBOAT7 expression and affected its enzymatic activity leading to fat accumulation in HepG2 cells,11 thus mirroring the condition observed in NAFLD carriers of the rs641738 T risk allele. Likewise, TM6SF2 silencing alone or combined with MBOAT7 deficiency led to a 50% reduction in TM6SF2 levels similarly to what was reported by Ruhanen et al,21 Smagris et al,13 and O’Hare et al,22 who induced stable TM6SF2 knockdown in HuH7 hepatoma cells, hepatic Tm6sf2 inactivation in mice, and CRISPR/Cas9-mediated tm6sf2 silencing in zebrafish, respectively. The role of TM6SF2 in lipoprotein metabolism, occurring in the ER–Golgi compartments, has been described previously in both experimental and clinical studies. In human hepatic 3-dimensional spheroids obtained from E167K donors, APOB-100 secretion was decreased.23 Smagris et al13 showed that hepatic Tm6sf2 deficiency impaired the bulk phase lipidation of VLDL. According to these findings, our TM6SF2-/- and MBOAT7-/-TM6SF2-/- models suppressed APOB and TAG-rich lipoprotein release, recapitulating features of patients carrying the E167K variant.14
To evaluate whether TM6SF2 deletion alone or combined with MBOAT7 may reliably model in vitro human hepatic steatosis, we performed an in-depth characterization of fat-storing power and the intracellular lipid profile. Both MBOAT7 and TM6SF2 silencing spontaneously developed LDs in hepatocytes, further corroborating their involvement in steatosis onset. However, the different impact on LD size according to the genetic background is currently lacking. MBOAT7 deficiency induces giant LD development, which resembles human macrovesicular steatosis, by shunting saturated PI toward the synthesis of saturated/monounsaturated TAG, thus favoring de novo lipogenesis (DNL) and perturbing membrane dynamics.11 Conversely, TM6SF2 deletion may induce the formation of smaller, clustered LDs, with a median size of 0.87 μm2 as assessed by TEM, and that may resemble microvesicular steatosis in human beings. According to our data, Smagris et al13 found that LD distribution, evaluated through ORO staining, was detectable in the smallest size range consisting of 1–2 μm2 in liver sections of Tm6sf2-/- mice. Although further studies are required to investigate the role of microvesicular steatosis in the progression of liver disease, it has been shown that the presence of microsteatosis rather than macrosteatosis correlated with hepatocellular ballooning, the presence of Mallory–Denk bodies, and mitochondrial dysfunction.24 Notably, because of the contribution of both MBOAT7 and TM6SF2 loss-of-function, the compound knockout showed a mixed pattern of LD content closely reflecting microvesicular–macrovesicular steatosis, which mainly characterizes the liver histology of NAFLD patients. The lipidomic analysis showed that TM6SF2-silenced cells mostly overexpressed TAG, incorporating saturated/monounsaturated fatty acid chains at the expense of polyunsaturated TAG, reflecting the lipidomic data obtained in experimental models21,25 and liver biopsy specimens of E167K carriers.25 In addition, the up-regulation of TAG 50:0 and TAG 52:0, lipid biomarkers featuring the serum and hepatic signature of NAFLD patients,17 and, even more, in those who developed HCC, were observed in the double knockout, thus supporting the deleterious effects caused synergically by the co-presence of MBOAT7 and TM6SF2 genetic deficiencies. Therefore, our findings, supported by conspicuous evidence in vitro, in vivo, and in human beings, suggest that TM6SF2-/- cells may trustworthily be used as a genetic NAFLD in vitro model because it fairly summarizes the phenotypic traits of patients carrying the E167K variant. TM6SF2 deletion induced the accumulation of micro-LDs compared with those developed by MBOAT7 knockout cells, and the mechanism underlying intracellular fat storage probably takes account of the increase in DAG/TAG with a high degree of saturation and, overall, from the reduction of polyunsaturated TAG besides the retention of TAG-rich lipoproteins. Regarding the MBOAT7-/-TM6SF2-/- model, it showed a pattern of LD volumes and lipid signature between MBOAT7-/- and TM6SF2-/-, thus representing the first model generated in vitro which may fully reproduce features of NAFLD individuals bearing all 3 at-risk variants.
ER and mitochondrial dysfunction are hallmarks of NAFLD because they may promote its progression to NASH and HCC. Genetic risk variants may participate actively in disease progression within hepatic cells, although it has not been investigated whether and to what extent they can lead to organelle impairment. In the present study, we showed that ER tubules and mitochondrial cristae were modestly enlarged in MBOAT7-/- cells but still maintained their morphologic architecture, and this condition was coupled with both ER/oxidative damage, probably resulting from lipotoxicity induced by the presence of large LDs.
The impact of TM6SF2 silencing on ER morphology has been described by O’Hare et al22 in human enterocytes, in small intestine, and liver of zebrafish larvae, in which TM6SF2 was acutely down-regulated. Here, we found that the distance among ER tubules was more than 3-fold larger in cells lacking the TM6SF2 gene, which also presented enhanced ER stress and a dramatic reduction in PC abundance. PCs represent one of the main components of cellular membranes and their depletion has been associated with changes in ER morphology,26 and, most recently, as hepatic lipid signature of NAFLD patients carrying the TM6SF2 T risk allele.25 Consistent with the biological role of TM6SF2, inhibition of PC production affects the amount of VLDL particles trafficking in the ER–Golgi compartments27 and it could be speculated that micro-LDs developed by TM6SF2-/- and MBOAT7-/-TM6SF2-/- models may arise from low PC abundance, whose levels impact LD expansion.28
Errors in mitochondrial dynamics have been pointed out in NAFLD human and experimental models, because they may drive both the switching from simple steatosis towards NASH and the metabolic reprogramming and malignant transformation leading to HCC. Notwithstanding, studies still are controversial, and none reported the effect of MBOAT7 or TM6SF2 silencing on mitobiogenesis. As mentioned previously, MBOAT7-/- cells affected mitochondrial morphology and oxidative stress as a possible result of the huge amount of intracellular fat accumulation, but without altering the number of mitochondria. Intriguing findings emerged in the TM6SF2 knockout cells where we found both mitochondrial degeneration and high mitochondrial biomass, suggestive of an unbalance in fusion, fission, and mitophagic processes. Alterations in the mitochondrial life cycle also were supported by increasing protein levels of PGC1α and also may take into account the loss of ER ultrastructure, which actively participates in mitochondrial dynamics and oxydative phopshorylation (OXPHOS) activity, thereby suggesting that TM6SF2 down-regulation may directly intervene to prompt mitochondrial dysfunction.
Changes in lipid composition (DAGs, TAGs, cholesterol esters) and increases in lactate production may be a sign of the Warburg effect, the metabolic shifting toward anaerobic glycolysis and mitochondrial derangement.29, 30, 31 Min et al32 showed that overexpression of the mutant I148M PNPLA3 in HuH-7 cells was associated with enhanced levels of lactate. Similarly, in our study the double knockout showed additional aspects related to mitochondrial failure because it suppressed both β-oxidation and ATP production, whose levels mostly derived from anaerobic glycolysis (87.7%), and showed the highest expression of glycolytic enzymes, lactate release, and growth potential. In addition, increased levels of dihydroceramides in the livers of obese NASH individuals have been correlated with mitochondrial failure,33 and the ratio of dihydroceramides to ceramides appears crucial to determine cell fate.34,35 Recently, Banini et al36 found that high levels of DAG, TAG, and Cer species were associated with metabolic reprogramming in NAFLD rodents expressing the I148M mutation. Here, we reported that TM6SF2 silencing heavily enhanced dihydroceramides (d18:0) binding saturated long-chain fatty acids (ie, palmitic acid [16:0]), which is consistent with the increase in shorter-chain saturated DAGs and TAGs, and suggestive of a prosurvival phenotype. However, the co-presence of MBOAT7 and TM6SF2 deletion did not induce a higher increment of dihydroceramide content, suggesting that MBOAT7 may diversely contribute to the carcinogenesis in the double knockout. As occurs in carriers of all 3 at-risk alleles which show the most increased HCC prevalence, loss of both MBOAT7 and TM6SF2 synergistically promote an aggressive phenotype involving different mechanisms, but whether the I148M PNPLA3 genetic background of the HepG2 cells participate in metabolic switching cannot be ruled out. Nonetheless, the overexpression of previously deleted genes, either MBOAT7 or TM6SF2, ameliorated not only lipid accumulation and cellular stress in the double knockout model, but also reversed the tumorigenic phenotype, suggesting that, despite the known effects of PNPLA3, both MBOAT7 and TM6SF2 genes exert a relevant role in progressive liver damage and cancer development.
Several limitations should be acknowledged in this study. First, we provided an overview of the main alterations in lipid composition and morphologic and functional derangements occurring in our in vitro models according to their genetic background. We highlighted which aspects of hepatocellular metabolism were mostly bothered by MBOAT7 or TMS6F2 deletion and which became dysregulated after silencing of both genes. However, identification of cell mediators (ie, proteins, small molecules) that orchestrate cell damage and are associated with genetic deficiency of MBOAT7 and TMS6F2 genes currently is lacking and it will be required to increase knowledge of disease pathogenesis and to introduce novel therapeutic perspectives for NAFLD management. Furthermore, the study was performed entirely in HepG2 cells, which are used commonly to study lipid metabolism in vitro and represent a suitable model for CRISPR/Cas9 technology, whose long-term use would not be applicable in primary hepatocytes. Nevertheless, the effects of MBOAT7/TM6SF2 down-regulation might be confirmed in other cell types with a different genetic background (ie, Hep3B, which are wild-type for the PNPLA3 gene), as well as the evaluation of the interplay among hepatocytes and other hepatic cell types (Kupffer cells, hepatic stellate cells) through complex culture systems (co-cultures, 3-dimensional cultures, organoids) could provide information about how these genetic-based alterations could promote advanced stages of liver injury, such as fibrosis and cirrhosis. Future validation of our results in preclinical genetic models of NAFLD and, possibly, in patients, will be crucial to shed light on drivers of disease progression and to stress the importance of genetic screening in NAFLD subjects.
To conclude, this study highlighted how MBOAT7 and TM6SF2 silencing diversely impact lipid metabolism, ER/mitochondrial dynamics, and tumorigenesis. These results may explain on one hand the impact of the I148M PNPLA3, MBOAT7 rs641738, and E167K TM6SF2 variants on steatosis development, and, on the other, how they can promote the switch to NASH up to HCC. Moreover, we proposed that MBOAT7 and TM6SF2 deletion may be decisive in developing a different macro-LD and/or micro-LD pattern in hepatocytes. If MBOAT7 deficiency predominantly induces large LD formation by affecting PI metabolism with a mechanism boosting DNL, the small LDs observed in TM6SF2-silenced cells probably involve ER dysfunction and PC depletion, which potentially affect LD swelling. We also showed that TM6SF2 loss-of-function unbalances hepatic mitochondrial biogenesis, interferes with the activity of multi-enzymatic complexes of the respiratory chain, thus extolling the oxidative and inflammatory status, and accumulates dihydro-Cer species, possibly enhancing its growth potential. Finally, preliminary in vitro results supported that the re-overexpression of MBOAT7 and/or TM6SF2 could improve lipid accumulation and hepatocellular stress and remarkably ameliorated the proliferation rate and migration, reinforcing that both may be implicated in carcinogenic processes.
In sum, we showed that the accrual of at-risk variants in PNPLA3, MBOAT7, and TM6SF2 genes predisposes to NAFLD and its progression toward cancer. Moreover, by exploiting a novel in vitro model we proposed some mechanisms through which these mutations impact disease onset and severity. Notably, the compound knockout showed matched characteristics of both single knockouts with regard to lipid composition and organelle derangement, which together may contribute to the Warburg effect, attempting to assume a prosurvival phenotype as occurs during hepatocarcinogenesis.
Patients and Methods
Overall Cohort
The overall cohort consisted of 1380 patients with NAFLD and was subdivided into the Hepatology Service cohort (n = 1259) and the NAFLD-HCC cohort (n = 121). The Hepatology Service cohort included 1259 unrelated patients of European descent who were enrolled consecutively at the Metabolic Liver Diseases outpatient service (Liver Clinic; n = 692) and bariatric surgery center (Bariatric Surgery; n = 567) at Fondazione IRCCS Cà Granda, Ospedale Maggiore Policlinico Milano (Milan, Italy). Inclusion criteria were the availability of a liver biopsy specimen for suspected NASH or severe obesity, DNA samples, and clinical data. Individuals with excessive alcohol intake (men, >30 g/d; women, >20 g/d), viral and autoimmune hepatitis, or other causes of liver disease were excluded. The study conformed to the Declaration of Helsinki and was approved by the Institutional Review Board of the Fondazione Ca’ Granda IRCCS of Milan and relevant institutions. All participants provided written informed consent.
The NAFLD-HCC patients of Italian descent (n = 121) were enrolled between January 2008 and January 2015 at the Milan, Udine, and Rome hospitals. The diagnosis of HCC was based on the European Association for the Study of the Liver–European Organization for Research and Treatment of Cancer Clinical Practice Guidelines.37 In the absence of a liver biopsy specimen, diagnosis of NAFLD-HCC required detection of ultrasonographic steatosis plus at least 1 criterion of the metabolic syndrome.
The Hepatology Service cohort and the NAFLD-HCC cohort both were stratified according to the number of risk variants, as follows: 0 for patients who had no risk alleles; 1 for the presence of 1 risk allele heterozygous or homozygous in either PNPLA3, MBOAT7, or TM6SF2; 2 for carriers who had 2 risk variants among PNPLA3, MBOAT7, or TM6SF2 in variable combinations; 3 for subjects carrying all 3 at-risk variants either heterozygous or homozygous. Demographic, anthropometric, and clinical features of the overall cohort stratified according to class enrollment criteria or the number of PNPLA3 I148M, MBOAT7 rs641738, and TM6SF2 E167K risk variants are shown in Tables 1 and 2.
Histologic Evaluation
Steatosis was divided into the following 4 categories based on the percentage of affected hepatocytes: 0, 0%–4%; 1, 5%–32%; 2, 33%–65%; and 3, 66%–100%. Disease activity was assessed according to the NAFLD activity score, with systematic evaluation of hepatocellular ballooning and necroinflammation; fibrosis also was staged according to the recommendations of the NAFLD Clinical Research Network.38 The scoring of liver biopsy specimens was performed by independent pathologists unaware of patient status and genotype.14,39 NASH was diagnosed in the presence of steatosis, lobular necroinflammation, and hepatocellular ballooning.
Genotyping
The overall cohort was genotyped for the rs738409 C>G (PNPLA3 I148M), rs58542926 C>T (TM6SF2 E167K), and rs641738 C>T MBOAT7 risk variants as previously described.10,12 Genotyping was performed in duplicate using TaqMan 5’-nuclease assays (QuantStudio 3; Thermo Fisher, Waltham, MA). Results of rs738409, rs58542926, and the rs641738 genetic frequencies were compared with those obtained in non-Finnish European healthy individuals included in the 1000 Genome project.40
CRISPR/Cas9 Gene Silencing
TM6SF2 silencing was induced by exploiting CRISPR/Cas9 genome editing after nonhomologous end joining in both HepG2 hepatoma cells (ATCC-HB-8065, which are homozygous for the I148M PNPLA3 variant, and in HepG2 knockout for the MBOAT7 gene (MBOAT7-/-)11 already available in our laboratory, allowing us to obtain a double stable model to study NAFLD. The reduced activity and expression of both MBOAT7 and TM6SF2 lead to an additive risk to develop NAFLD when combined with the I148M PNPLA3 variant,10,16,20,41,42 making the HepG2 cells an ideal model to study in vitro the impact of MBOAT7 and TM6SF2 loss-of functions coupled to the influence of I148M PNPLA3 on lipid metabolism, hepatocellular damage, and carcinogenesis.
HepG2 cells were transfected with a lentiviral vector (Edit-R Inducible Lentiviral hEF1α-Blast-Cas9 Nuclease Particles; GE Healthcare Dharmacon, Inc, Lafayette, CO) containing Cas9 under the control of doxycycline-inducible promoter and blasticidin resistance as selection marker. Concurrently, 2 single-strand DNAs were designed using the free online CRISPR Design Tool (available: http://crispr.mit.edu) (forward: 5’-AAACACGCGCTCAACCACGTCTCG-3’; reverse: 5’-CCGGCGAGACGTGGTTGAGCGCGT-3') to construct a small guide RNA (sgRNA). Double-stranded oligos were subcloned using T4 DNA ligase (ThermoFisher, Carlsbad, CA) into the pGL3-U6-sgRNA-phosphoglycerate kinase (PGK)-puromycin plasmid (Addgene plasmid 51133; available: http://n2t.net/addgene:51133; RRID: Addgene_51133; acquired from Addgene)43 and digested with Fastdigest Eco31I (ThermoFisher). The identity of the sgRNA constructs was verified by Sanger sequencing (Table 5). The sgRNA was designed to induce TM6SF2 cut by Cas9 transcription star site (ATG) in the first codifying exon of the TM6SF2 gene (isoform 1, transcript variant 1, NM_001001524.3) to induce TM6SF2 knockout altering its mRNA transcription. After doxycycline-induced Cas9 expression, HepG2/Cas9-positive clones (Cas9+) were transfected (Lipofectamine 3000 transfection reagent; ThermoFisher, Carlsbad, CA) with pGL3-U6-sgRNA-PGK-puromycin plasmid containing the sgRNA under the control of cytomegalovirus promoter (U6), which previously was subcloned in Dh5α competent cells. Positive clones carrying the TM6SF2 mutations were selected, exploiting puromycin resistance gene as selection marker (GE Healthcare Dharmacon, Inc). After single-cell clonal expansion and single-clone–derived colony formation generated by the limiting dilution, the efficiency of gene editing was tested by Sanger sequencing (approximately 100 clones per condition were screened). Potential off-target modifications were checked in the genome region of interest. We identified the same TM6SF2 deletion of 202 nucleotides (Δ202) in homozygosis in both CAS9+ and MBOAT7-/- cells-/-,11 allowing us to obtain either a single (TM6SF2-/-) or a compound knockout stable model (MBOAT7-/-TM6SF2-/-) to study genetic NAFLD in vitro. Cas9+ cells wild type in the MBOAT7 and TM6SF2 genes were used as the control group.
Table 5.
Sequence of Primers Used in Quantitative Real-Time PCR Experiments
| Forward 5’→3’ | Reverse 5’→3’ | |
|---|---|---|
| ATF4 | AAACCTCATGGGTTCTCCAG | GGCATGGTTTCCAGGTCACT |
| ATF6 | AATTCTCAGCTGATGGCTGT | TGGAGGATCCTGGTGTCCAT |
| GRP78 | CTTGCCGTTCAAGGTGGTTG | CTGCCGTAGGCTCGTTGAT |
| MnSOD2 | CAAATTGCTGCTTGTCCAAA | TCTTGCTGGGATCATTAGGG |
| PPARα | ATGGCATCCAGAACAAGGAG | TCCCGTCTTTGTTCATCACA |
| MnSOD2 | CAAATTGCTGCTTGTCCAAA | TCTTGCTGGGATCATTAGGG |
| TM6SF2 exon Ia | GGCTGCCTATGCTCTCACCTT | TGCCTCCAGCAAACACCAA |
| U6-sgRNAa | TTTCTTGGGTAGTTTGCAGTTTT | CGACTCGGTGCCACTTTT |
| XBP1 | GAAGCCAAGGGGAATGAAGT | GCCCAACAGGATATCAGACTC |
| β-actin | GCTACAGCTTCACCACCACA | AAGGAAGGCTGGAAAAGAGC |
Human primers used for Sanger sequencing.
TEM
Cas9+, MBOAT7-/-, TM6SF2-/-, and MBOAT7-/-TM6SF2-/- cells were cultured as monolayer (70%–80% confluent) and trypsinized to obtain cell suspension. Cells were fixed with an aldehyde mixture (4% paraformaldehyde + 2.5% glutaraldehyde in cacodylate buffer: pH 7.4) at 4°C overnight. After primary fixation, cells were washed repeatedly with cacodylate buffer and were postfixed in 1% osmium tetroxide (OsO4) for 2 hours in the dark. Next, samples were left in 1.5% potassium ferrocyanide dissolved in 0.1 mol/L cacodylate for 1 hour on ice. Each sample was stained with 0.5% 5-uranyl acetate in water overnight. Finally, samples were dehydrated with increasing ethanol series, embedded in an Epon resin, and polymerized in an oven at 60°C for 48 hours. Ultrathin (70–90 nm) sections were collected on nickel grids and observed with a Zeiss Leo 912 AB Omega TEM (Freiburg, Germany).44 LD size was obtained by measuring at least 4 random diameters per LD through ImageJ software (National Institutes of Health, Bethesda, MD). The mean of diameters was used to calculate the average of both circumference (C = πd) and area (A = πR2) of LDs for each condition (Table 3). Quantification of ER width was obtained through ImageJ software by acquiring at least 3 measurements per ER lumen (Table 4). Mitochondria were counted by ImageJ software in 15 random nonoverlapping micrographs containing at least a whole single cell.
ORO Staining
Cells were plated on a 6-well plate (5 × 105 cells/well) in duplicate and left overnight in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 1% L-glutamine, and 1% penicillin/streptomycin. After 24 hours, growth media was removed and cells were kept for 24 hours in quiescent medium, containing 0.5% bovine serum albumin (BSA), 1% L-glutamine, and 1% penicillin/streptomycin. The day after, we performed ORO staining, which is a soluble red powder with high affinity for neutral TAGs and lipids stored in the LDs. Quiescent medium was removed, and the 6-well plates were gently rinsed with 2 mL sterile phosphate-buffered saline 1×. Next, cells were fixed with 4% formalin for 1 hour at room temperature. After fixation, each sample was washed with sterile water and 60% isopropanol was added for 5 minutes. Concurrently, we prepared ORO working solution by mixing 3 parts of ORO stock solution (300 mg of Red Oil powder in 100 mL di-isopropanol 100%) and 2 parts of sterile water, after filtration. ORO working solution (1 mL/well) was added to each sample and left for 40 minutes. Finally, plates were rinsed with tap water, paying attention not to disrupt the monolayer. LD content was visualized in pink-red color. The ORO-positive area was quantified by ImageJ software in 10 random micrographs (magnification, 200×) by calculating the ORO-positive area as a percentage of pixels above the threshold value with respect to the total pixels per area.
Lipidomic Analysis
For lipidomic analysis, lipid classes were separated with ultra-high-pressure liquid chromatography equipped with ZORBAX Eclipse Plus C18 2.1 × 100 mm 1.8 μm columns (Agilent, Santa Clara CA), and lipid concentrations were measured by MS-QTOF (Agilent Ultra-High-Pressure Liquid Chromatography 1290 Infinity-6540 QTOF, Santa Clara, CA) in both positive (for Cer, sphingomyelins, phosphatidylethanolamines, lyso-phosphatidylethanolamines, PCs, lyso-PCs, DAGs, and TAGs) and negative electrospray ionization for PIs. Mobile phase A consists of water with 0.1% formic acid in positive mode and 5 mmol/L ammonium acetate in negative mode; mobile phase B consists of acetonitrile:isopropanol (1:1) with 0.1% formic acid in positive mode and 5 mmol/L ammonium acetate in negative mode. Analysis of the lipid profiles was performed by Mass Hunter Profinder software coupled with PCDL Manager (Agilent, Santa Clara, CA) that allows extraction, identification, and quantification of selected lipid compounds on the bases of the accurate mass m/z, retention time, and ion abundance. The software calculates the area under the curve of identified ions. For the lipid species, ions are extracted as adducts [M + H]+ and [M + Na]+ when samples were run in positive mode and as [M-H]- and [M + HAc-H]- when samples were run in negative mode (HAc = CH3COO-). All sets of samples were run together to avoid batch differences. Standard Reference Materia SRM 1950 (National Institute of Science and Technology NIST, US) was run as quality control during each run (positive and negative).
Lipids measured in positive mode concentrations were quantified using internal standards, that is, sphingomyelin (d18:1/17:0), Cer (d18:1/17:0), phosphatidylethanolamines (17:0/17:0), PC (17:0/17:0), lyso-PC 17:0, DAG (17:0/17:0), and TAG (15:/15:0/15:0) (Avanti Polar Lipids, Alabaster, AL, and Larodan, Solna, Sweden). PIs and lyso-PIs were quantified using either phosphatidylglycerol (34:0) or Cer (d18:1/17:0) as internal standards, which are visible in negative mode and have a retention time close to PI and lyso-PI. Both internal standards gave similar concentration values for PIs and lyso-PIs.
Data were analyzed as the log2 fold change ratio considering the chain length and number of double bonds (ie, degree of unsaturation). PCA (Figure 5A) was performed on the log2 fold change ratios between MBOAT7-/-, TM6SF2-/-, MBOAT7-/-TM6SF2-/-, and Cas9+ lipidomic data, using R package FactoMineR.1,45 Because PCA is an unsupervised method based on a linear combination of features, it allows evaluation of the different changes induced by the mutations on the whole lipidomic profile, even with a small number of samples.
DNA Extraction and Sanger Sequencing
DNA was extracted using the phenol/chloroform method. Next, DNA concentration and quality were assessed by a Nanodrop 1000 microvolume 42 spectrophotometer (ThermoFisher Scientific). A total of 1 μL DNA was used to verify sgRNA insertion in the pGL3-U6-sgRNA-PGK-puromycin plasmid,43 and to detect TM6SF2 mutations in exon I of the TM6SF2 sequence. Polymerase chain reaction (PCR) products were electrophoretically run on 3% agarose gel, purified with a vacuum pump, and used to perform the Sanger sequencing protocol (Big Dye mix; Applied Biosystems, Waltham, MA). Primers used for amplification and sequencing are listed in Table 5.
Gene Expression Analysis
RNA was extracted from cell cultures using TRIzol reagent (Life Technologies–ThermoFisher, Carlsbad, CA). Total RNA (1 μg) was retrotranscribed with the VILO random hexamers synthesis system (Life Technologies–ThermoFisher, Carlsbad, CA). Quantitative real-time PCR was performed by an ABI 7500 fast thermocycler (Life Technologies), using the TaqMan Universal PCR Master Mix (Life Technologies–ThermoFisher, Carlsbad, CA) and TaqMan probes for human MBOAT7, TM6SF2, GCK, PFK, and G3PDH. SYBR Green chemistry (Fast SYBR Green Master Mix; Life Technologies–ThermoFisher, Carlsbad, CA) was used for all other genes. All reactions were delivered in triplicate. Data were normalized to β-actin housekeeping gene and results were expressed as mean and standard error (SE). Primers are listed in Tables 5 and 6.
Table 6.
List of TaqMan Probes Used in Quantitative Real-Time PCR Experiments
| Probes | Catalog number |
|---|---|
| TM6SF2 | ThermoFisher Hs00403495_m1 |
| MBOAT7 | ThermoFisher Hs00383302_m1 |
| GCK | ThermoFisher Hs01564555_m1 |
| PFK | ThermoFisher Hs01036347_m1 |
| G3PDH | ThermoFisher Hs02786621_g1 |
Western Blot Analysis
Total protein lysates were extracted from 2 cell cultures using RIPA buffer containing 1 mmol/L Na-orthovanadate, 200 mmol/L phenylmethyl sulfonyl fluoride, and 0.02 μg/μL aprotinin. Samples were pooled before electrophoretic separation and all reactions were performed in duplicate. Then, equal amounts of proteins (50 μg) were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred electrophoretically to a nitrocellulose membrane (Bio-Rad, Hercules, CA), and incubated with specific antibodies overnight. At least 3 independent lots of freshly extracted proteins were used for experiments. Antibodies and concentration used are listed in Table 7.
Table 7.
List of Antibodies and Relative Dilutions Used in Western Blot and Immunocytochemistry Experiments
| Antibody | Catalog number |
|---|---|
| TM6SF2 (1:1000 WB) | ThermoFisher PA5-69304 |
| MBOAT7 (1:500 WB) | Sigma-Aldrich AV49811 |
| ApoB-100 | ThermoFisher HYB0690202 |
| P(Ser473)-Akt (1:1000 WB) | Cell Signaling 9271S |
| Akt (1:1000 WB) | Cell Signaling 2938S |
| P(Thr37/46)-4E-BP1 (1:1000 WB) | Cell Signaling 2855S |
| 4E-BP1 (1:1000 WB) | Cell Signaling 9644S |
| P(Ser2448)-mTOR (1:1000 WB) | Cell Signaling 5536S |
| mTOR (1:1000 WB) | Cell Signaling 2983S |
| PGC1-α | Novus Biologicals NBP1-04676 |
| COX-I SDHA |
Abcam ab110216 Abcam ab110216 |
| β-actin (1:5000 WB) | Abcam ab6276 |
Immunocytochemistry
A total of 1 × 106 cells were seeded on coverslips lodged in a 6-well plate in duplicate and kept overnight in DMEM containing 10% FBS, 1% L-glutamine, and 1% penicillin/streptomycin. Next, hepatocytes were fixed in 4% formalin for 1 hour and permeabilized in 0.3% Triton X-100 (Sigma-Aldrich, St Louis, MO). Cells were incubated in 5% BSA for 30 minutes and with anti-PGC1α primary antibody overnight at 4°C. Then, each sample was incubated with anti-rabbit horseradish-peroxidase–conjugated antibody, and 3,3′-diaminobenzidine was provided as chromogen. Nucleus were counterstained with hematoxylin. Finally, samples were mounted with a drop of aqueous VectaMount AQ Mounting Medium (Maravai LifeSciences, Inc, San Diego, CA).
Evaluation of MT-COX1 Expression
The MitoBiogenesis In-Cell Enzyme-Linked Immunosorbent Assay Kit (Abcam, Cambridge, UK) uses quantitative immunocytochemistry to measure MT-COX1 and nuclear-encoded SDHA protein levels in cultured cells and it was performed following the manufacturer’s instructions.46 Briefly, cells were seeded in a 96-well plate (3 × 105/well) in triplicate, fixed with 4% paraformaldehyde. and permeabilized through 1× Triton X-100. Targets of interest are detected with a highly specific, well-characterized cocktail of monoclonal antibodies, which were incubated overnight at 4°C. Then, apurinic/apyrimidinic-labeled and horseradish-peroxidase–labeled secondary antibodies are used to generate a colorimetric reaction that could be measured at 405 and 600 nm, respectively.
Evaluation of ROS and Lipid Peroxidation
Cas9+, MBOAT7-/-, TM6SF2-/-, and MBOAT7-/-TM6SF2-/- cells were cultured as monolayer. At 70%–80% of confluence, they were stained with MitoSOX Red (1.25 μmol/L; Invitrogen–ThermoFisher, Carlsbad, CA) for 30 minutes at 37°C, and subsequently harvested for fluorescence-activated cell sorter analysis (excitation, 488 nm; emission, 690/50 nm; Acea Bioscience). Increased red fluorescence was correlated with ROS formation. Data were collected from at least 20,000 cells and 6 replicates per condition. Lipid peroxidation was analyzed by staining cells with BODIPY 581/591 C11 (2 μmol/L; Thermo Fisher Scientific, Darmstadt, Germany) for 1 hour at 37°C. Cells then were harvested for fluorescence-activated cell sorter analysis (excitation, 488 nm; emission, 525/30 nm and 585/50 nm), and the shift from red to green fluorescence was used to evaluate lipid peroxidation. Data were collected from at least 20,000 cells and 6 replicates per condition.
Evaluation of Complex I Enzymatic Activity
A total of 106 cells were resuspended in proper buffer (pH 7.2). Protein extraction was performed by sonicating cell pellets at 50 W (10 s) 3 times. Lysates were centrifuged at 750 × g for 10 minutes and supernatant was recovered. The Lowry method was used for protein quantification. A Lambda 2 Perkin Elmer spectrophotometer (Waltham, MA) was used to assess enzymatic activities. Analyses were performed at specific wavelengths for each enzymatic activity after preparing proper solutions as previously described.47,48 Nicotinamide adenine dinucleotide (NADH) dehydrogenase (340 nm) consisted of the following: H2O (455 μL), K-phosphate, pH 7.5, 0.1 mol/L (340 μL), K3[Fe(CN)6] 17 mmol/L (100 μL), NADH 2 mmol/L (100 μL), and homogenate (5 μL). NADH ubiquinone 1 reductase (340 nm) consisted of the following: H2O (610 μL), K-phosphate, pH 7.5, 0.1 mol/L (200 μL), albumin 1% (100 μL), NADH 2 mmol/L (70 μL), homogenate (10 μL), sodium azide 100 mmol/L (10 μL), and CoQ10 6 mmol/L (5 μL); activity measured after rotenone administration (1 mmol/L, 5 μL) was subtracted. Citrate synthase (412 nm) consisted of the following: H2O (800 μL), disulfide, 5,5′-dithio-bis-2-nitrobenzoic acid (DTNB) 1 mmol/L (100 μL), oxaloacetic acid 10 mmol/L (50 μL), acetyl-CoA 10 mmol/L (30 μL), and homogenate (20 μL). Experiments were performed at 30°C. Analyses were performed with Perkin Elmer software. Measurements were normalized over the activity level of citrate synthase, a stable matrix mitochondrial enzyme; this latter step was performed to normalize respiratory chain activity over mitochondrial mass.
Measurement of ATP Rate
ATP production in live cells was measured by an Agilent Seahorse XF Real-Time ATP Rate Assay (Agilent Technologies), which quantifies the rate of ATP from glycolysis and mitochondria in real time. The assay uses a sequential injection of specific mitochondrial activators and inhibitors and reports multiple parameters, including glycolytic ATP production rate, mitochondrial ATP production rate, total ATP production rate, and the XF ATP rate index. Briefly, cells were plated on a Seahorse XF24-well microplate (40,000 cells per well) and on the day of the assay the cell culture growth medium was replaced by the assay medium, supplemented with glucose, pyruvate, and glutamine, and cells were incubated at 37°C for 60 minutes. The oxygen consumption rate and extracellular acidification rate were measured using the XFe24 analyzer, with 3 baseline measurements recorded before and after adding oligomycin (ATP synthase inhibitor; 1.5 μmol/L) and a mix of rotenone and antimycin A (complex I and III inhibitors, respectively; 0.5 μmol/L).
In Vitro Treatments
When specified, Cas9+, MBOAT7-/-, TM6SF2-/-, and MBOAT7-/-TM6SF2-/- were treated with either sorafenib (Santa Cruz Biotechnology, Dallas, TX) at a final concentration of 5 μmol/L or dimethyl sulfoxide (DMSO) vehicle for 24, 48, 72 hours and 1 week. Treatments were freshly prepared and administered daily.
Cell Proliferation Assay
Cas9+, MBOAT7-/-, TM6SF2-/-, and MBOAT7-/-TM6SF2-/- (2.5∗102 cells/well) were seeded in a 96-well plate in quadruplicate and incubated under normal culture conditions overnight. Cell proliferation was measured at both baseline and upon sorafenib exposure using the CellTiter96-Aqueous One Solution Cell Proliferation Assay (MTS:3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium)) kit (Promega Corporation, Fitchburg, WI) according to the manufacturer’s instructions. A dose-response curve was performed by testing 1, 2.5, 5, 10 and 50 μmol/L of sorafenib treatments to determine the minimum concentration required to evaluate cell viability. Administration of sorafenib at very low concentrations (1 and 2.5 μmol/L) was e effective to modulate cell survival, whereas the higher concentrations (10 and 50 μmol/L) showed a high mortality rate. Therefore, after a day, cells were treated with either sorafenib at a final concentration of 5 μmol/L or vehicle (DMSO), which is further consistent with the current literature.49,50
Fresh growth media with or without sorafenib was provided for 24-48-72 h and 1 week. MTS reagent (20 μL/well) was added to the cells followed by incubation for 4 hours in a 5% CO2 humidified incubator at 37°C, and the absorbance was measured at 490 nm, at 0, 24, 48, and 74 hours, and after 1 week. At least 3 independent experiments were performed.
Wound Healing Assay
Cas9+, MBOAT7-/-, TM6SF2-/-, and MBOAT7-/-TM6SF2-/- models were plated on 6-well plate (8 × 105/well) and incubated with DMEM containing 10% FBS, 1% L-glutamine, and 1% penicillin/streptomycin. A fine scratch was introduced using a sterile pipette tip in a monolayer of cells at approximately 90% confluency. Cells then were treated with sorafenib at a final concentration of 5 μmol/L or vehicle (DMSO) for 24 and 48 hours. The wounds were photographed (objective, 100×) at 24 and 48 hours. Each experiment was performed in triplicate.
Lentiviral Overexpression
MBOAT7 and/or TM6SF2 were overexpressed in MBOAT7-/-TM6SF2-/- cells through pLenti-C-mGFP-P2A-Puro lentiviral vectors, which were engineered to express a complete open reading frame (ORF) fused with a GFP tag (henceforth MBOAT7-GFP and TM6SF2-GFP). We seeded 3∗104 cells in 24-well plates and they were incubated at 37ºC and 5% CO2 overnight. Multiplicity of infection was set at 2.5 and the amount of lentiviral particles for the transduction were calculated according to the manufacturer’s instructions (OriGene, Rockville, MD). Lentiviral particles were added to prewarmed cultured media for 24 hours. To introduce MBOAT7-GFP–tagged protein, MBOAT7-/-TM6SF2-/- cells were transduced with the LENG4 (MBOAT7) Human Tagged ORF Clone Lentiviral Particle (OriGene) containing a molecular sequence that aligns with the MBOAT7 mRNA (gene accession numbers: NM_024298.2 and NP_077274.2). To insert the TM6SF2-GFP, MBOAT7-/-TM6SF2-/- cells were transfected with TM6SF2 Human Tagged ORF Clone Lentiviral Particle (OriGene) carrying the molecular sequence that aligns with the TM6SF2 mRNA (gene accession numbers: NM_001001524.2 and NP_001001524.2). Stable cell lines carrying the wild-type form of MBOAT7 (MBOAT7+/+TM6SF2-/-), TM6SF2 (MBOAT7-/-TM6SF2+/+), or both (MBOAT7+/+TM6SF2+/+) were selected using the puromycin resistance gene as a selection marker (GE Healthcare Dharmacon, Inc).
Statistical Analysis
For descriptive statistics, continuous variables were reported as means and SD or as the median and interquartile range for highly skewed biological variables. Variables with skewed distribution were logarithmically transformed before analyses. The frequencies for each risk variant subgroup were compared using the chi-squared test. Differences between groups were calculated by 1-way nonparametric ANOVA (Kruskal–Wallis), followed by post hoc t test (2-tailed) when 2 groups were compared, or the Dunn multiple comparison test when multiple groups were compared, adjusted for the number of comparisons. Lipidomic analyses were performed using the R package. P values less than .05 were considered statistically significant. Statistical analyses were performed using JMP 14.0 (SAS, Cary, NC) and Prism software (version 6, GraphPad Software, San Diego, CA).
Chemicals
Sorafenib was acquired from Santa Cruz Biotechnology (Dallas, TX). Doxycycline, anti-MBOAT7, and BSA were purchased from Sigma-Aldrich (St Louis, MO). Anti–P-Ser473 Akt, Akt, P(Thr37/46)-4E-BP1, 4E-BP1, P(Ser2448)-mTOR, and mTOR antibodies were acquired from Cell Signaling Technology (Danvers, MA). Anti–β-actin was purchased from Abcam. Anti-Cas9 and anti-PGC1α were obtained from Novus Biologicals (Littleton, CO). TM6SF2 and APOB-100 antibodies were purchased from Life Technologies–ThermoFisher Scientific (Waltham, MA). DMEM, FBS, phosphate-buffered saline, L-glutamine, penicillin/streptomycin, Trypsin/EDTA, Hank’s balanced salt solution, fast SYBR green master mix, Lipofectamine 3000 transfection reagent, blasticidin, T4 DNA ligase, Fastdigest Eco31I, ampicillin, and DH5α competent cells were obtained from Life Technologies–ThermoFisher Scientific (Waltham, MA). PCR Master Mix 2× was purchased from BiotechRabbit (Hennigsdorf, Germany). Clarity Western ECL substrate was obtained from Bio-Rad Laboratories. CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS) was purchased from Promega Corporation. The pGL3-U6-sgRNA-PGK-puromycin was a gift from Xingxu Huang (Addgene plasmid 51133; http://n2t.net/addgene:51133; RRID:Addgene_51133; acquired from Addgene).43 Puromycin and Edit-R Inducible Lentiviral hEF1α-Blast-Cas9 nuclease particles were obtained from GE Healthcare Dharmacon, Inc.51 The TAG quantification kit was purchased from BioVision, Inc (Milpitas, CA). The Cholesterol Colorimetric Assay Kit– high density lipoprotein (HDL) and Low-density lipoprotein/ very low-density lipoprotein (LDL/VLDL), Dichlorodihydrofluorescin (DCF) Reactive oxygen species/Reactive nitrogen species (ROS/RNS) Colorimetric Assay Kit, Hydrogen Peroxide Assay Kit, Lipid Peroxidation (malondialdehyde) Assay Kit (colorimetric/fluorimetric), DNA Damage Colorimetric Assay Kit (apurinic/apyrimidinic (AP) sites), and MitoBiogenesis In-Cell Enzyme-Linked Immunosorbent Assay Kit were purchased from Abcam. VectaMount AQ Mounting Medium was obtained from Maravai Life Sciences, Inc.
Acknowledgments
The authors sincerely thank Dr Podini Paola (IRCCS Ospedale San Raffaele, Milan, Italy) for the technical support provided for the transmission electron microscopy and the intellectual contribution for data analysis interpretation.
CRediT Authorship Contributions
Miriam Longo, Dr (Conceptualization: Lead; Methodology: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Marica Meroni, PhD (Writing – review & editing: Equal)
Erika Paolini, Dr (Methodology: Equal)
Veronica Erconi, Dr (Methodology: Equal)
Fabrizia Carli, Dr (Methodology: Equal)
Francesco Fortunato, Dr (Methodology: Equal)
Dario Ronchi, PhD (Supervision: Equal)
Roberto Piciotti (Methodology: Equal)
Silvia Sabatini, Dr (Methodology: Equal)
Chiara Macchi, PhD (Methodology: Equal)
Anna Alisi, Dr (Resources: Equal)
Luca Miele, Dr (Resources: Equal)
Giorgio Soardo, Dr (Resources: Equal)
Giacomo Pietro Comi, Dr (Methodology: Equal; Supervision: Equal)
Luca Valenti, Dr (Resources: Equal)
Massimiliano Ruscica, PhD (Methodology: Equal; Resources: Equal)
Anna Ludovica Fracanzani, Dr (Supervision: Equal)
Amalia Gastaldelli, Dr (Supervision: Equal)
Paola Dongiovanni, Dr (Conceptualization: Lead; Supervision: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was supported by Ricerca Corrente Fondazione IRCCS Cà Granda, Ricerca Finalizzata Ministero della Salute RF-2013-02358319 and Ricerca Finalizzata Ministero della Salute GR-2019-12370172.
Supplementary Material
References
- 1.Younossi Z.M. Non-alcoholic fatty liver disease - a global public health perspective. J Hepatol. 2019;70:531–544. doi: 10.1016/j.jhep.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 2.Day C.P. From fat to inflammation. Gastroenterology. 2006;130:207–210. doi: 10.1053/j.gastro.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 3.Byrne C.D., Targher G. NAFLD: a multisystem disease. J Hepatol. 2015;62(Suppl):S47–S64. doi: 10.1016/j.jhep.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Dongiovanni P., Romeo S., Valenti L. Genetic factors in the pathogenesis of nonalcoholic fatty liver and steatohepatitis. Biomed Res Int. 2015;2015:460190. doi: 10.1155/2015/460190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozlitina J., Smagris E., Stender S., Nordestgaard B.G., Zhou H.H., Tybjaerg-Hansen A., Vogt T.F., Hobbs H.H., Cohen J.C. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romeo S., Kozlitina J., Xing C., Pertsemlidis A., Cox D., Pennacchio L.A., Boerwinkle E., Cohen J.C., Hobbs H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buch S., Stickel F., Trepo E. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet. 2015;47:1443–1448. doi: 10.1038/ng.3417. [DOI] [PubMed] [Google Scholar]
- 8.Dongiovanni P., Stender S., Pietrelli A., Mancina R.M., Cespiati A., Petta S., Pelusi S., Pingitore P., Badiali S., Maggioni M., Mannisto V., Grimaudo S., Pipitone R.M., Pihlajamaki J., Craxi A., Taube M., Carlsson L.M.S., Fargion S., Romeo S., Kozlitina J., Valenti L. Causal relationship of hepatic fat with liver damage and insulin resistance in nonalcoholic fatty liver. J Intern Med. 2018;283:356–370. doi: 10.1111/joim.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meroni M., Longo M., Tria G., Dongiovanni P. Genetics is of the essence to face NAFLD. Biomedicines. 2021;9:1359. doi: 10.3390/biomedicines9101359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mancina R.M., Dongiovanni P., Petta S., Pingitore P., Meroni M., Rametta R., Boren J., Montalcini T., Pujia A., Wiklund O., Hindy G., Spagnuolo R., Motta B.M., Pipitone R.M., Craxi A., Fargion S., Nobili V., Kakela P., Karja V., Mannisto V., Pihlajamaki J., Reilly D.F., Castro-Perez J., Kozlitina J., Valenti L., Romeo S. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology. 2016;150:1219–1230.e6. doi: 10.1053/j.gastro.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meroni M., Dongiovanni P., Longo M., Carli F., Baselli G., Rametta R., Pelusi S., Badiali S., Maggioni M., Gaggini M., Fracanzani A.L., Romeo S., Gatti S., Davidson N.O., Gastaldelli A., Valenti L. Mboat7 down-regulation by hyper-insulinemia induces fat accumulation in hepatocytes. EBioMedicine. 2020;52:102658. doi: 10.1016/j.ebiom.2020.102658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahdessian H., Taxiarchis A., Popov S., Silveira A., Franco-Cereceda A., Hamsten A., Eriksson P., van't Hooft F. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci U S A. 2014;111:8913–8918. doi: 10.1073/pnas.1323785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smagris E., Gilyard S., BasuRay S., Cohen J.C., Hobbs H.H. Inactivation of Tm6sf2, a gene defective in fatty liver disease, impairs lipidation but not secretion of very low density lipoproteins. J Biol Chem. 2016;291:10659–10676. doi: 10.1074/jbc.M116.719955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dongiovanni P., Petta S., Maglio C., Fracanzani A.L., Pipitone R., Mozzi E., Motta B.M., Kaminska D., Rametta R., Grimaudo S., Pelusi S., Montalcini T., Alisi A., Maggioni M., Karja V., Boren J., Kakela P., Di Marco V., Xing C., Nobili V., Dallapiccola B., Craxi A., Pihlajamaki J., Fargion S., Sjostrom L., Carlsson L.M., Romeo S., Valenti L. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. 2015;61:506–514. doi: 10.1002/hep.27490. [DOI] [PubMed] [Google Scholar]
- 15.Bianco C., Jamialahmadi O., Pelusi S., Baselli G., Dongiovanni P., Zanoni I., Santoro L., Maier S., Liguori A., Meroni M., Borroni V., D'Ambrosio R., Spagnuolo R., Alisi A., Federico A., Bugianesi E., Petta S., Miele L., Vespasiani-Gentilucci U., Anstee Q.M., Stickel F., Hampe J., Fischer J., Berg T., Fracanzani A.L., Soardo G., Reeves H., Prati D., Romeo S., Valenti L. Non-invasive stratification of hepatocellular carcinoma risk in non-alcoholic fatty liver using polygenic risk scores. J Hepatol. 2021;74:775–782. doi: 10.1016/j.jhep.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donati B., Dongiovanni P., Romeo S., Meroni M., McCain M., Miele L., Petta S., Maier S., Rosso C., De Luca L., Vanni E., Grimaudo S., Romagnoli R. MBOAT7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci Rep. 2017;7:4492. doi: 10.1038/s41598-017-04991-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanders F.W.B., Acharjee A., Walker C., Marney L., Roberts L.D., Imamura F., Jenkins B., Case J., Ray S., Virtue S., Vidal-Puig A., Kuh D., Hardy R., Allison M., Forouhi N., Murray A.J., Wareham N., Vacca M., Koulman A., Griffin J.L. Hepatic steatosis risk is partly driven by increased de novo lipogenesis following carbohydrate consumption. Genome Biol. 2018;19:79. doi: 10.1186/s13059-018-1439-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z., Guan M., Lin Y., Cui X., Zhang Y., Zhao Z., Zhu J. Aberrant lipid metabolism in hepatocellular carcinoma revealed by liver lipidomics. Int J Mol Sci. 2017;18:2550. doi: 10.3390/ijms18122550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu M., Li Y., Zhang S., Wang X., Shen J., Zhang S. Interaction of TM6SF2 E167K and PNPLA3 I148M variants in NAFLD in northeast China. Ann Hepatol. 2019;18:456–460. doi: 10.1016/j.aohep.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Krawczyk M., Rau M., Schattenberg J.M., Bantel H., Pathil A., Demir M., Kluwe J., Boettler T., Lammert F., Geier A. Combined effects of the PNPLA3 rs738409, TM6SF2 rs58542926, and MBOAT7 rs641738 variants on NAFLD severity: a multicenter biopsy-based study. J Lipid Res. 2017;58:247–255. doi: 10.1194/jlr.P067454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruhanen H., Nidhina Haridas P.A., Eskelinen E.-L., Eriksson O., Olkkonen V.M., Käkelä R. Depletion of TM6SF2 disturbs membrane lipid composition and dynamics in HuH7 hepatoma cells. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:676–685. doi: 10.1016/j.bbalip.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 22.O'Hare E.A., Yang R., Yerges-Armstrong L.M., Sreenivasan U., McFarland R., Leitch C.C., Wilson M.H., Narina S., Gorden A., Ryan K.A., Shuldiner A.R., Farber S.A., Wood G.C., Still C.D., Gerhard G.S., Robishaw J.D., Sztalryd C., Zaghloul N.A. TM6SF2 rs58542926 impacts lipid processing in liver and small intestine. Hepatology. 2017;65:1526–1542. doi: 10.1002/hep.29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prill S., Caddeo A., Baselli G., Jamialahmadi O., Dongiovanni P., Rametta R., Kanebratt K.P., Pujia A., Pingitore P., Mancina R.M., Lindén D., Whatling C., Janefeldt A., Kozyra M., Ingelman-Sundberg M., Valenti L., Andersson T.B., Romeo S. The TM6SF2 E167K genetic variant induces lipid biosynthesis and reduces apolipoprotein B secretion in human hepatic 3D spheroids. Sci Rep. 2019;9:11585. doi: 10.1038/s41598-019-47737-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tandra S., Yeh M.M., Brunt E.M., Vuppalanchi R., Cummings O.W., Ünalp-Arida A., Wilson L.A., Chalasani N. Presence and significance of microvesicular steatosis in nonalcoholic fatty liver disease. J Hepatol. 2011;55:654–659. doi: 10.1016/j.jhep.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luukkonen P.K., Zhou Y., Nidhina Haridas P.A., Dwivedi O.P., Hyotylainen T., Ali A., Juuti A., Leivonen M., Tukiainen T., Ahonen L., Scott E., Palmer J.M., Arola J., Orho-Melander M., Vikman P., Anstee Q.M., Olkkonen V.M., Oresic M., Groop L., Yki-Jarvinen H. Impaired hepatic lipid synthesis from polyunsaturated fatty acids in TM6SF2 E167K variant carriers with NAFLD. J Hepatol. 2017;67:128–136. doi: 10.1016/j.jhep.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 26.Testerink N., van der Sanden M.H.M., Houweling M., Helms J.B., Vaandrager A.B. Depletion of phosphatidylcholine affects endoplasmic reticulum morphology and protein traffic at the Golgi complex. J Lipid Res. 2009;50:2182–2192. doi: 10.1194/jlr.M800660-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verkade H.J., Fast D.G., Rusiñol A.E., Scraba D.G., Vance D.E. Impaired biosynthesis of phosphatidylcholine causes a decrease in the number of very low density lipoprotein particles in the Golgi but not in the endoplasmic reticulum of rat liver. J Biol Chem. 1993;268:24990–24996. [PubMed] [Google Scholar]
- 28.Krahmer N., Guo Y., Wilfling F., Hilger M., Lingrell S., Heger K., Newman H.W., Schmidt-Supprian M., Vance D.E., Mann M., Farese R.V., Jr., Walther T.C. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14:504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalhan S.C., Guo L., Edmison J., Dasarathy S., McCullough A.J., Hanson R.W., Milburn M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism. 2011;60:404–413. doi: 10.1016/j.metabol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Wang L., Yan X., Liu Q., Yu C., Wei H., Li Y., Zhang X., He F., Jiang Y. A proton nuclear magnetic resonance metabonomics approach for biomarker discovery in nonalcoholic fatty liver disease. J Proteome Res. 2011;10:2797–2806. doi: 10.1021/pr200047c. [DOI] [PubMed] [Google Scholar]
- 31.Beyoğlu D., Idle J.R. The metabolomic window into hepatobiliary disease. J Hepatol. 2013;59:842–858. doi: 10.1016/j.jhep.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Min H.-K., Sookoian S., Pirola C.J., Cheng J., Mirshahi F., Sanyal A.J. Metabolic profiling reveals that PNPLA3 induces widespread effects on metabolism beyond triacylglycerol remodeling in Huh-7 hepatoma cells. Am J Physiol Gastrointest Liver Physiol. 2014;307:G66–G76. doi: 10.1152/ajpgi.00335.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng K.-Y., Watt M.J., Rensen S., Greve J.W., Huynh K., Jayawardana K.S., Meikle P.J., Meex R.C.R. Mitochondrial dysfunction-related lipid changes occur in nonalcoholic fatty liver disease progression. J Lipid Res. 2018;59:1977–1986. doi: 10.1194/jlr.M085613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaurasia B., Tippetts T.S., Mayoral Monibas R., Liu J., Li Y., Wang L., Wilkerson J.L., Sweeney C.R., Pereira R.F., Sumida D.H., Maschek J.A., Cox J.E., Kaddai V., Lancaster G.I., Siddique M.M., Poss A., Pearson M., Satapati S., Zhou H., McLaren D.G., Previs S.F., Chen Y., Qian Y., Petrov A., Wu M., Shen X., Yao J., Nunes C.N., Howard A.D., Wang L., Erion M.D., Rutter J., Holland W.L., Kelley D.E., Summers S.A. Targeting a ceramide double bond improves insulin resistance and hepatic steatosis. Science. 2019;365:386. doi: 10.1126/science.aav3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez-Cuenca S., Barbarroja N., Vidal-Puig A. Dihydroceramide desaturase 1, the gatekeeper of ceramide induced lipotoxicity. Biochim Biophys Acta Mol Cell Biol Lipids. 2015;1851:40–50. doi: 10.1016/j.bbalip.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 36.Banini B.A., PK D., Cazanave S., Seneshaw M., Mirshahi F., Santhekadur P.K., Wang L., Guan H.P., Oseini A., Alonso C., Bedossa P., Koduru S.V., Min H.K., Sanyal A.J. Identification of a metabolic, transcriptomic and molecular signature of PNPLA3-mediated acceleration of steatohepatitis. Hepatology. 2021;73:1290–1306. doi: 10.1002/hep.31609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.European Association for the Study of the Liver; European Organisation for Research and Treatment of Cancer EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., Ferrell L.D., Liu Y.C., Torbenson M.S., Unalp-Arida A., Yeh M., McCullough A.J., Sanyal A.J. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 39.Dongiovanni P., Petta S., Mannisto V., Mancina R.M., Pipitone R., Karja V., Maggioni M., Kakela P., Wiklund O., Mozzi E., Grimaudo S., Kaminska D., Rametta R., Craxi A., Fargion S., Nobili V., Romeo S., Pihlajamaki J., Valenti L. Statin use and non-alcoholic steatohepatitis in at risk individuals. J Hepatol. 2015;63:705–712. doi: 10.1016/j.jhep.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L., Du S., Lu L., Lin Z., Jin W., Hu D., Jiang X., Xin Y., Xuan S. The additive effects of the TM6SF2 E167K and PNPLA3 I148M polymorphisms on lipid metabolism. Oncotarget. 2017;8:74209–74216. doi: 10.18632/oncotarget.18474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luukkonen P.K., Zhou Y., Hyotylainen T., Leivonen M., Arola J., Orho-Melander M., Oresic M., Yki-Jarvinen H. The MBOAT7 variant rs641738 alters hepatic phosphatidylinositols and increases severity of non-alcoholic fatty liver disease in humans. J Hepatol. 2016;65:1263–1265. doi: 10.1016/j.jhep.2016.07.045. [DOI] [PubMed] [Google Scholar]
- 43.Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L., Wang L., Hodgkins A., Iyer V., Huang X., Skarnes W.C. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 44.Quattrini A., Previtali S., Feltri M.L., Canal N., Nemni R., Wrabetz L. Beta 4 integrin and other Schwann cell markers in axonal neuropathy. Glia. 1996;17:294–306. doi: 10.1002/(SICI)1098-1136(199608)17:4<294::AID-GLIA4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 45.Lê S., Josse J., Husson F. FactoMineR: an R package for multivariate analysis. 2008;25:18. [Google Scholar]
- 46.Chirambo G.M., van Niekerk C., Crowther N.J. The role of alkaline phosphatase in intracellular lipid accumulation in the human hepatocarcinoma cell line, HepG2. Exp Mol Pathol. 2017;102:224–229. doi: 10.1016/j.yexmp.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 47.Monzio Compagnoni G., Kleiner G., Bordoni A., Fortunato F., Ronchi D., Salani S., Guida M., Corti C., Pichler I., Bergamini C., Fato R., Pellecchia M.T., Vallelunga A., Del Sorbo F., Elia A., Reale C., Garavaglia B., Mora G., Albanese A., Cogiamanian F., Ardolino G., Bresolin N., Corti S., Comi G.P., Quinzii C.M., Di Fonzo A. Mitochondrial dysfunction in fibroblasts of multiple system atrophy. Biochim Biophys Acta Mol Basis Dis. 2018;1864:3588–3597. doi: 10.1016/j.bbadis.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 48.Bresolin N., Zeviani M., Bonilla E., Miller R.H., Leech R.W., Shanske S., Nakagawa M., DiMauro S. Fatal infantile cytochrome c oxidase deficiency: decrease of immunologically detectable enzyme in muscle. Neurology. 1985;35:802–812. doi: 10.1212/wnl.35.6.802. [DOI] [PubMed] [Google Scholar]
- 49.Jakobi K., Beyer S., Koch A., Thomas D., Schwalm S., Zeuzem S., Pfeilschifter J., Grammatikos G. Sorafenib treatment and modulation of the sphingolipid pathway affect proliferation and viability of hepatocellular carcinoma in vitro. Int J Mol Sci. 2020;21:2409. doi: 10.3390/ijms21072409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ezzoukhry Z., Louandre C., Trécherel E., Godin C., Chauffert B., Dupont S., Diouf M., Barbare J.C., Mazière J.C., Galmiche A. EGFR activation is a potential determinant of primary resistance of hepatocellular carcinoma cells to sorafenib. Int J Cancer. 2012;131:2961–2969. doi: 10.1002/ijc.27604. [DOI] [PubMed] [Google Scholar]
- 51.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.