Abstract
Background & Aims
Tumor necrosis factor alpha (TNFα) is considered a major tissue damage-promoting effector in Crohn’s disease (CD) pathogenesis. Patient-derived intestinal organoid (enteroid) recapitulates the disease-specific characteristics of the intestinal epithelium. This study aimed to evaluate the intestinal epithelial responses to TNFα in enteroids derived from healthy controls and compare them with those of CD patient-derived enteroids.
Methods
Human enteroids derived from patients with CD and controls were treated with TNFα (30 ng/mL), and cell viability and gene expression patterns were evaluated.
Results
TNFα induced MLKL-mediated necroptotic cell death, which was more pronounced in CD patient-derived enteroids than in control enteroids. Immunohistochemistry and RNA sequencing revealed that treatment with TNFα caused expansion of the intestinal stem cell (ISC) populations. However, expanded ISC subpopulations differed in control and CD patient-derived enteroids, with LGR5+ active ISCs in control enteroids and reserve ISCs, such as BMI1+ cells, in CD patient-derived enteroids. In single-cell RNA sequencing, LGR5+ ISC-enriched cell cluster showed strong expression of TNFRSF1B (TNFR2) and cyclooxygenase-prostaglandin E2 (PGE2) activation. In TNFα-treated CD patient-derived enteroids, exogenous PGE2 (10 nmol/L) induced the expansion of the LGR5+ ISC population and improved organoid-forming efficiency, viability, and wound healing.
Conclusions
TNFα increases necroptosis of differentiated cells and induces the expansion of LGR5+ ISCs. In CD patient-derived enteroids, TNFα causes LGR5+ stem cell dysfunction (expansion failure), and exogenous PGE2 treatment restored the functions of LGR5+ stem cells. Therefore, PGE2 can be used to promote mucosal healing in patients with CD.
Keywords: Crohn’s Disease, Intestinal Organoid, Tumor Necrosis Factor-Alpha, Intestinal Stem Cell, Prostaglandin E2
Abbreviations used in this paper: ANOVA, analysis of variance; CC3, cleaved caspase-3; CD, Crohn’s disease; COX, cyclooxygenase; DEG, differentially expressed gene; ER, endoplasmic reticulum; ISC, intestinal stem cell; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; NF-κB, nuclear factor kappa B; PBS, phosphate-buffered saline; PGE2, prostaglandin E2; scRNA-seq, single-cell RNA sequencing; TNF, tumor necrosis factor; TUNEL, deoxyuride-5′-triphosphate biotin nick end labeling.
Graphical abstract
Summary.
In human intestinal organoids (enteroids), TNFα induces differentiated cell necroptosis and intestinal stem cell expansion. However, TNFα-induced expansion of the LGR5+ stem cells is impaired in CD patient-derived enteroids, resulting in decreased organoid-forming efficiency and delayed wound healing.
Crohn’s disease (CD) is a chronic relapsing-remitting inflammatory disease of the intestine, which is characterized by cycles of mucosal inflammation and ulceration, followed by regeneration and restoration of the intestinal epithelium.1 The pathogenesis of CD remains unclear, and approximately 30%–50% of the patients do not respond to current medical treatments.2,3 Recent studies have identified alterations in intestinal stem cell (ISC) properties in organoids derived from patients with CD.4, 5, 6 Alteration in ISC properties can lead to impaired epithelial regeneration, barrier dysfunction, and chronic intestinal inflammation. The intestinal organoid (enteroid) model recapitulates the functional crypt-villus, consists of distinct epithelial cell types, replicates self-organizing developmental processes, and maintains the characteristics of its origin7, 8, 9 and is therefore suitable for investigating the role of ISCs and other epithelial cells.4, 5, 6,10, 11, 12
Tumor necrosis factor alpha (TNFα) is a major pro-inflammatory and tissue damage-promoting effector in CD pathogenesis, which has been supported by experimental mouse models and therapeutic effects of TNFα-neutralizing reagents.13, 14, 15 Despite extensive investigation on TNFα-induced responses in immune cells, the effect of TNFα on intestinal epithelial cells remained largely unexplored.16 TNFα mediates diverse cellular processes, such as inflammatory mediator production, cell death, cell survival, and proliferation, which may be regulated in a cell type–specific manner.17 Tumor-derived cell lines and animal models have inherent limitations in investigating the human epithelial cell-specific responses to TNFα.15 Immortalized cell lines are homogeneous, and the cells do not undergo senescence due to the presence of certain mutations.18 Moreover, human-specific biological processes could not be fully reproduced in animal models.19
Herein, we investigated the intestinal epithelial cell-specific responses to TNFα in patients with CD by using the patient-derived intestinal organoid model and attempted to identify molecules that could improve epithelial regeneration and wound healing in CD patients under TNFα-enriched conditions.
Results
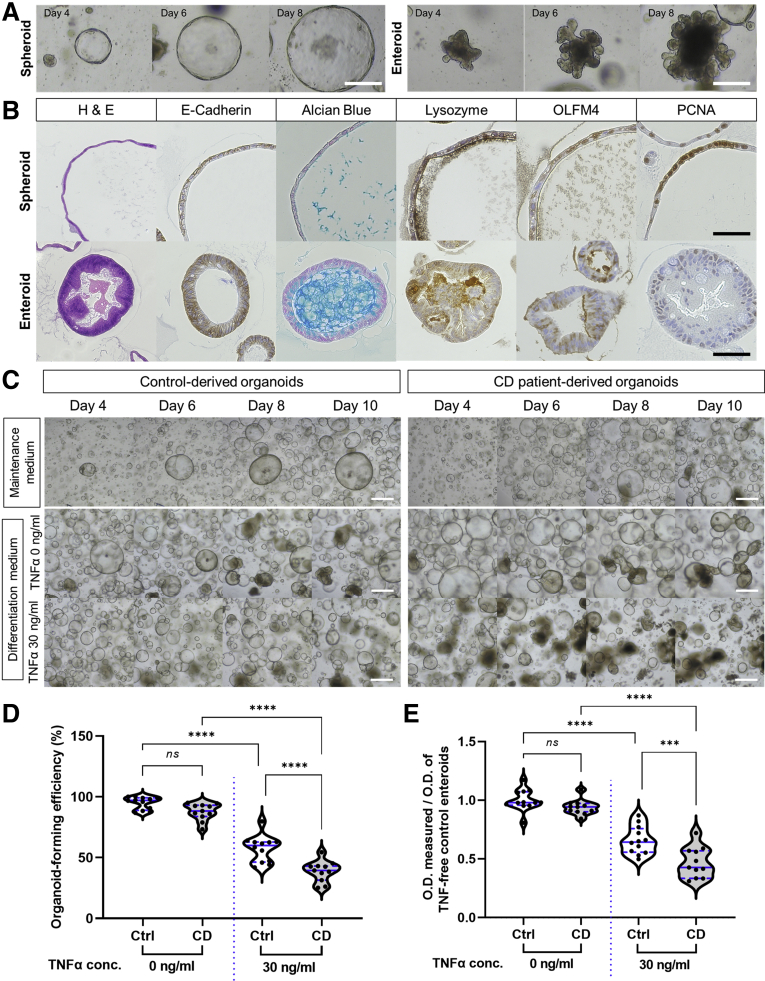
Intestinal organoids grown from the intestinal crypts of controls (n = 12) and patients with CD (n = 11) were subcultured in the maintenance medium for more than 5 passages. The intestinal organoids cultured in the maintenance medium formed spheroids and consisted of OLFM4+ ISCs and PCNA+ proliferating cells, whereas those cultured in the differentiation medium formed enteroids that contained all types of intestinal epithelial cells (Figure 1A and B). The microscopic appearance and culturing behavior of subcultured organoids derived from controls and patients with CD were undistinguishable.6,20 These phenotypic similarities could be due to sterile and non-inflammatory culture conditions.
Figure 1.
Organoid-forming efficiency and cell viability of CD patient-derived enteroids and control enteroids under TNFα-enriched conditions. (A) Representative microscopic images of spheroids and enteroids. Scale bar = 100 μm. (B) Immunohistochemical staining for epithelial cell markers in spheroids and enteroids. Scale bar = 75 μm. (C) Morphology, (D) organoid-forming efficiency, and (E) cell viability (MTT assay) of organoids derived from controls (n = 12) and patients with CD cultured under TNFα-free and -treated conditions (n = 11 each). Morphologic observations were performed using bright-field microscopy. The measured optical density (O.D.) values are expressed as a ratio based on the O.D. values of TNFα-untreated control enteroids. Scale bar = 100 μm. Assays were conducted in triplicate. Differences were assessed using one-way analysis of variance (ANOVA) with Bonferroni multiple comparison test; ∗∗∗P < .001, ∗∗∗∗ P ≤ .0001.
Previous studies have suggested that 30 ng/mL TNFα may induce cytotoxicity and distinct cellular responses in enteroids.21, 22, 23 Herein, enteroids derived from controls and patients with CD were treated with 30 ng/mL TNFα every 24 hours to evaluate epithelial cell-specific responses to TNFα. To address whether the effect of TNFα on CD patient-derived enteroids differs from that of control enteroids, morphologic changes and cell viability were assessed. TNFα-treated organoids were smaller in size and lesser in number, and no budding was observed; some of these organoids were disrupted and had a dark appearance (Figure 1C). These results are consistent with those of a previous report on murine organoid models.24 In the steady state, the organoid-forming efficiency was not different between control enteroids and CD patient-derived enteroids (100.0% ± 5.4% vs 92.3% ± 7.3%; P = .152); however, the organoid-forming efficiency of CD patient-derived enteroids treated with TNFα was significantly lower than that of control enteroids treated with TNFα (60.3% ± 11.4% vs 40.8% ± 9.0%; P < .001; Figure 1D). In the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, the cell viability of TNFα-treated CD patient-derived organoids was significantly lower than that of untreated CD patient-derived organoids (P < .001) and TNFα-treated control organoids (P < .001; Figure 1E). These results suggest that the organoid-forming efficiency and cell viability of CD patient-derived organoids were lower than those of control organoids under TNFα-enriched conditions.
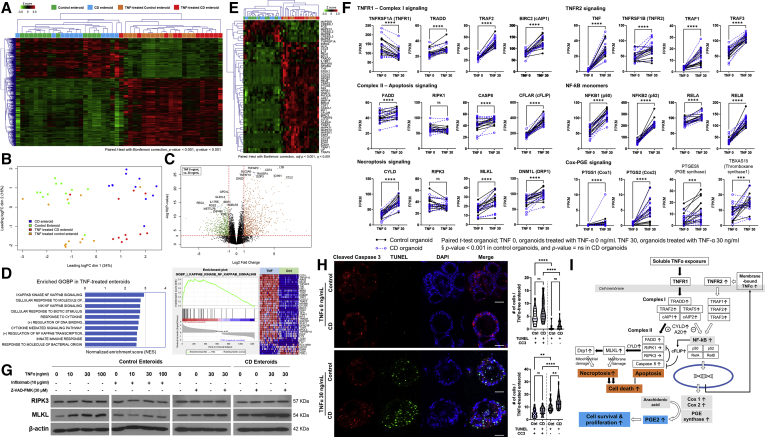
To explore the gene expression profile of TNFα-treated control and CD patient-derived enteroids, bulk RNA sequencing was performed by using paired samples of TNFα-untreated and TNFα-treated enteroids derived from healthy controls (n = 12 pairs) and patients with CD (n = 11 pairs). The hierarchical clustering heatmap of the 1247 differentially expressed genes (DEGs) showed a clear difference between TNFα-untreated and TNFα-treated enteroids (Figure 2A). Principal component analysis revealed a clear distinction between TNFα-untreated and TNFα-treated enteroids of controls and patients with CD (Figure 2B). The volcano plot shows the 10 most up-regulated and down-regulated genes, which were most responsive to TNFα (Figure 2C). As predicted, gene ontology biological process annotation for DEGs revealed that nuclear factor kappa B (NF-κB) signaling pathway was enriched in TNFα-treated enteroids (Figure 2D). Then we focused on the TNFα signaling pathway. The hierarchical clustering heatmap of TNFα signaling pathway-related genes showed a clear distinction between TNFα-untreated and TNFα-treated enteroids (Figure 2E). Exogenous soluble TNFα enhanced the expression of genes related to complex-I signaling, such as TRADD, TRAF2, and BIRC2 (cAIP1), and complex-II signaling, such as FADD and CASP8. Moreover, TNFα treatment increased the expression of CFLAR (cFLIP) and CYLD, which in turn suppressed the expression of genes associated with apoptosis (RIPK1 and RIPK3) and enhanced the expression of genes associated with necroptosis (MLKL and DNM1L) (Figure 2F). Western blotting confirmed that the MLKL protein expression increased steadily with the increasing TNFα concentration, and this effect of TNFα on MLKL protein expression was blocked by anti-TNFα monoclonal antibody (infliximab) co-treatment; however, the expression of RIPK3 protein remained unchanged. The pan-caspase inhibitor, Z-VAD-FMK, did not affect the expression of RIPK3 and MLKL proteins in TNFα-treated enteroids derived from controls and patients with CD (Figure 2G). These findings suggest that TNFα induces MLKL-mediated necroptosis rather than apoptosis in enteroids.
Figure 2.
Gene expression profile of enteroids derived from controls and patients with CD under TNFα-free and -treated conditions. (A) Hierarchical clustering heatmap and (B) principal component analysis using paired samples of TNFα-untreated and -treated enteroids derived from controls (n = 12 pairs) and patients with CD (n = 11 pairs). (C) Volcano plot of DEGs in TNFα-treated and -untreated enteroids. The 10 up- and down-regulated genes annotated with gene symbols are displayed. Volcano plot shows fold change (log2 ratio) plotted against absolute confidence (–log10P value). (D) Gene set enrichment analysis of DEGs. The top 10 significant terms of gene ontology biological process in TNFα-treated enteroids are listed, based on the normalized enrichment score. Enrichment plot of the most enriched signaling pathway (I-κB kinase/NF-κB signaling; GO:0007249) genes is illustrated. (E) Hierarchical clustering heatmap of TNFα signaling pathway and its related genes. DEGs were identified using the paired t test with Bonferroni correction (adjusted P value < .001 and Q value < 0.001). (F) Differences in expression levels (FPKM) of TNFα pathway genes in paired sample of TNFα-untreated and -treated enteroids derived from controls and patients with CD. Black dots with solid lines indicate changes in control enteroids, and blue dots with blue broken lines indicate changes in CD patient-derived enteroids. (G) Western blotting for RIPK3 and MLKL proteins in enteroids derived from controls and patients with CD. Enteroids derived from controls (n = 3) and patients with CD (n = 3) were treated with different concentrations of TNFα, infliximab, and Z-VAD-FMK. (H) Double immunofluorescence staining for CC3 and TUNEL. Distribution of CC3 (red fluorescence) and TUNEL (green fluorescence) was determined using co-immunofluorescence analysis. Cell nuclei were co-stained with DAPI (blue fluorescence). The number of cells was counted in 10 enteroids (size >100 μm) of each group. Differences were evaluated using one-way ANOVA with Bonferroni multiple comparison test. Scale bar = 50 μm; ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗ P < .0001. (I) Proposed cellular responses to TNFα in human enteroids.
We then investigated whether TNFα-induced necroptosis and apoptosis were more pronounced in enteroids derived from patients with CD. Deoxyuride-5′-triphosphate biotin nick end labeling (TUNEL) staining allows the identification of both necroptotic and apoptotic cells, whereas cleaved caspase-3 (CC3) only stains apoptotic cells.25 In TNFα-free condition, the number of TUNEL+ CC3+ apoptotic cells per enteroid was significantly higher than the number of TUNEL+ CC3– necroptotic cells per enteroid (P < .001). However, the numbers of TUNEL+ CC3+ apoptotic cells and TUNEL+ CC3– necroptotic cells per enteroid were not different between control and CD patient-derived enteroids. These apoptotic cells may be associated with physiological anoikis.26 After TNFα (30 ng/mL) treatment, the number of necroptotic cells was remarkably increased in control (P = .001) and CD patient-derived enteroids (P < .001); however, the number of necroptotic cells remained significantly higher in CD patient-derived enteroids (P = .009; Figure 2H). These data indicate that necroptosis is the primary mechanism for cell death in TNFα-treated enteroids and is significantly increased in CD patient-derived enteroids.
In TNFα-treated enteroids, paired t test showed that the expression of TNFα itself, NF-κB monomer, PTGES1/2 (cyclooxygenase [COX]1/2), and PTGES was increased compared with TNFα-untreated enteroids. TNFα-induced up-regulation of PTGES and TBSAS1 expression was more pronounced in control enteroids (Figure 2F). TNFα is produced as a transmembrane molecule (membrane-bound TNFα), which is then cleaved to produce the soluble form (soluble TNFα).27 The membrane-bound TNFα can fully activate TNFRSF1B (TNFR2) and downstream signaling.28 NF-κB and COX-prostaglandin E (PGE) signaling induces proliferation and promotes survival in most cell types (Figure 2I).29 Previously, the pleiotropic effects of TNFα have been reported in immune cells, although the underlying mechanism remains unclear.30 Our results suggest that TNFα may either promote survival of some intestinal epithelial cells or induce death in other cell types.
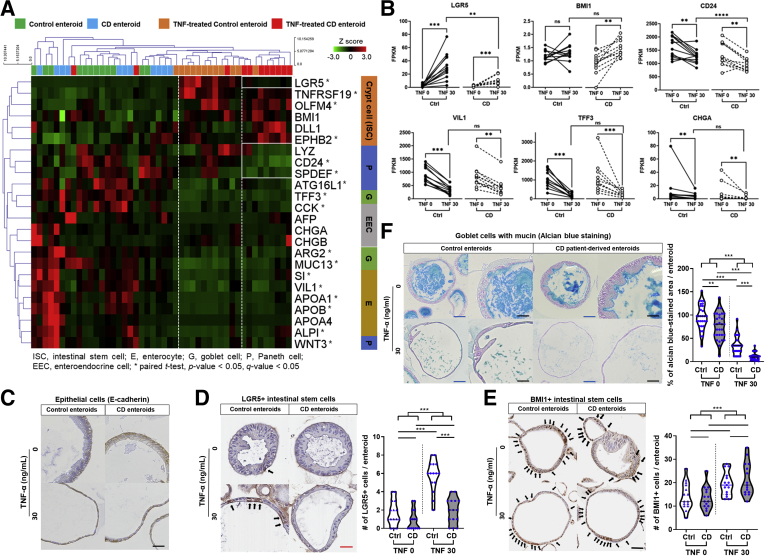
To address whether cellular responses to TNFα differ in different epithelial cell types,31 we evaluated the epithelial lineage-specific gene expression in TNFα-untreated and -treated enteroids derived from controls and patients with CD. Heatmap with hierarchical clustering identified different epithelial lineage-specific gene expression profiles (Figure 3A). In both control and CD patient-derived enteroids cultured in TNFα-free conditions, a significant increase in the expression of differentiated cell markers, such as CD24, SPDEF, ATG16L1, TFF3, CCK, ARG2, MUC13, SI, VIL1, APOA1, APOB, ALPI, and WNT3, and a significant decrease in the expression of crypt cell markers, such as LGR5, TNFRSF19, OLFM4, and EPHB2, (paired t test; P < .05 and q < 0.05) was observed. Among TNFα-treated enteroids, CD patient-derived enteroids were clustered and showed lower levels of the active ISC marker LGR5 and Paneth cell markers, such as LYZ and CD24. In paired analysis of TNFα-untreated and TNFα-treated enteroids, the expression of LGR5 was significantly higher in TNFα-treated enteroids than in TNFα-untreated enteroids (P < .001). TNFα-induced increase in expression of LGR5 was significantly lower in CD patient-derived enteroids than in control enteroids (P = .002; Figure 3B). The expression level of BMI1 in TNFα-treated CD patient-derived enteroids was significantly higher compared with TNFα-untreated CD patient-derived enteroids (P = .001); however, there was no significant difference between TNFα-untreated and TNFα-treated control enteroids. The expression level of CD24 in TNFα-treated CD patient-derived enteroids was significantly lower compared with that in TNFα-untreated CD patient-derived enteroids (P = .003) and TNFα-treated control enteroids (P < .001). Expression of the differentiated cell markers, VIL1, TFF3, and CHGA, was significantly lower in enteroids treated with TNFα; however, no difference was observed between control and CD patient-derived enteroids.
Figure 3.
TNFα treatment alters the epithelial lineage-specific gene expression in CD patient-derived enteroids. (A) Hierarchical clustering heatmap of epithelial cell lineage-specific markers and (B) differences in expression levels (FPKM) of signature genes in paired samples of TNFα-untreated and -treated enteroids derived from controls (n = 12 pairs) and patients with CD (n = 11 pairs). Wilcoxon matched-pairs signed-rank test was used to analyze differences in gene expression in paired samples (TNFα-untreated and -treated). Mann-Whitney test was used to analyze differences in gene expression between TNFα-treated control enteroids and TNFα-treated CD patient-derived enteroid; ∗P < .05, ∗∗P < .01. Immunohistochemistry for (C) E-cadherin, (D) LGR5, and (E) BMI1. The number of LGR5+ and BMI1+ cells was counted in 15 enteroids (size >100 μm) from each group (n = 3 for each group). Black arrows indicate LGR5+ or BMI1+ cells. (F) Alcian blue staining. Stained area was quantified using ImageJ and expressed as percentage of the average area stained in the TNFα-untreated control enteroids. Area was measured in 30 enteroids (size >100 μm) from each group (n = 3 for each group). Differences were evaluated using one-way ANOVA with Bonferroni multiple comparison test; ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. Blue scale bar = 100 μm, black scale bar = 50 μm, and red scale bar = 25 μm.
Immunohistochemistry confirmed that all enteroid cells were E-cadherin positive, suggesting that these cells were epithelial cells (Figure 3C). After treatment with TNFα, the number of LGR5+ cells per enteroid increased significantly only in control enteroids (P < .001), but not in enteroids derived from patients with CD (Figure 3D). The number of BMI1+ cells per enteroid was significantly higher in TNFα-treated enteroids compared with TNFα-untreated enteroids (P < .001; Figure 3E). Alcian blue stains goblet cells and mucins. The total area of alcian blue staining in TNFα-untreated enteroids was more than that in the TNFα-treated enteroids (P < .001; Figure 3F). Moreover, the alcian blue–stained area in the TNFα-treated CD-patient derived enteroids was significantly smaller than that in TNFα-treated control enteroids (P < .001). These results showed that TNFα treatment induces expansion of the ISC population and reduction of differentiated cells in the surviving enteroids. Interestingly, LGR5+ ISC expansion was observed in TNFα-treated control enteroids; however, it was impaired in the CD patient-derived enteroids.
We evaluated the expression of genes associated with ISC niche signaling pathways, including Wnt, Notch, and HIPPO signaling pathways, in TNFα-untreated and -treated enteroids. The gene expression of ISC niche signaling pathways was up-regulated in the TNFα-treated enteroids; however, the hierarchical clustering could not identify any significant differences between control and CD patient-derived enteroids (data are not shown). These results indicate that ISC properties in enteroids derived from controls and CD patients might be independent of the niche signaling and dependent on their origin.
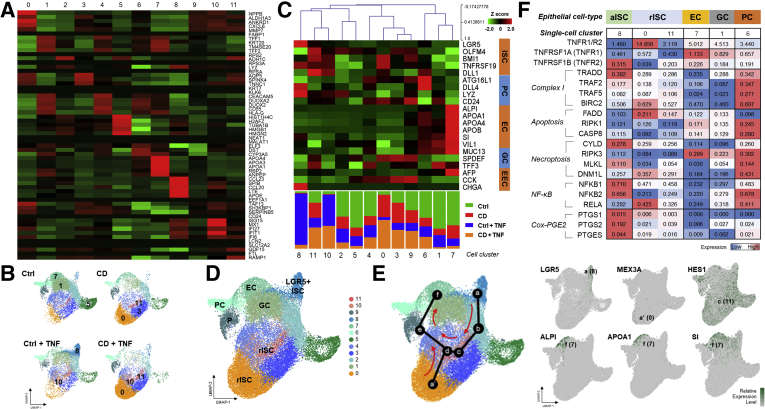
To confirm that the expansion of LGR5+ ISC is impaired in TNFα-treated CD patient-derived enteroids, single-cell RNA sequencing (scRNA-seq) was performed. A total of 43,152 individual cell transcriptomes were recovered from TNFα-untreated and TNFα-treated enteroids (control enteroids, n = 13,519; TNFα-treated control enteroids, n = 8854; CD patient-derived enteroids, n = 9560; TNFα-treated CD patient-derived enteroids, n = 11,219). After excluding samples with low-quality RNA, 29,421 cells were finally analyzed. To characterize the cellular subpopulations, the analyzed cells were divided into 12 clusters (0–11) on the basis of the presence of distinct sets of co-expressed genes (Figure 4A). The distribution of cells according to the samples and clusters is shown in the UMAP plot. The majority of cells in clusters 1, 5, and 7 originated from TNFα-untreated control enteroids, those in clusters 0, 3, and 11 from TNFα-untreated CD patient-derived enteroids, those in clusters 8 and 10 from TNFα-treated control enteroids, and those in clusters 0, 10, and 11 from TNFα-treated CD patient-derived enteroids (Figure 4B). Each single-cell cluster could be matched to the epithelial cell lineage, depending on the epithelial lineage-specific marker gene expression (Figure 4C). Cluster 8 included LGR5+ active ISCs. Clusters 0, 10, and 11 included reserve ISCs, which express BMI1, TNFRSF19 (TROY), DLL1, and MEX3A. Clusters 1, 6, and 7 were annotated with goblet cells, Paneth cells, and enterocytes (Figure 4D). The majority of cells (90.5%) in cluster 8 originated from TNFα-treated control enteroids, which confirms the expansion of LGR5+ ISC in these enteroids. In development trajectory modeling, both LGR5+ active ISCs and reserve ISCs progressed to HES1+ absorptive progenitors and differentiated into ALPI, APOA1, and SI-expressing differentiated enterocytes (Figure 4E). Interestingly, LGR5+ active ISC-enriched cluster 8 showed high expression of TNFR2, low TNFR1/TNFR2 ratio, and activation of the downstream NF-κB and COX-PGE2 pathways (Figure 4F). The scRNA-seq results indicate that TNFα induces the expansion of ISCs in the enteroids, leading to reparative proliferation and differentiation of epithelial cells and replacement of the cells lost because of TNFα-induced cytotoxicity. However, expanded ISC subpopulations differed in control and CD patient-derived enteroids; there were LGR5+ active ISCs in control enteroids and reserve ISCs, such as BMI1+ cells, in CD patient-derived enteroids.
Figure 4.
Single-cell RNA sequencing for TNFα-untreated and -treated enteroids derived from controls and patients with CD. (A) Heatmap and clustering of DEGs. Rows represent the top 5 DEGs, and columns represent clusters. (B) UMAP plot of the aggregated samples and individual samples. The number indicates a cell cluster consisting of cells mainly originating from individual sample. Ctrl, control enteroids, CD, CD patient-derived enteroids. (C) Heatmap and hierarchical clustering of intestinal epithelial lineage-specific genes based on the single-cell clusters. EC, enterocyte; EEC, enteroendocrine cell; GC, goblet cell; PC, Paneth cell. (D) UMAP plot visualizing intestinal epithelial cell types of human enteroids based on marker gene expression. EC, enterocyte; EEC, enteroendocrine cell; GC, goblet cell; PC, Paneth cell; rISC, reserve intestinal stem cell. (E) Trajectory analysis of enterocyte differentiation from LGR5+ ISCs and reserve ISCs. (F) Heatmap for TNFRSF1A (TNFR1)/TNFRSF1B (TNFR2) expression ratio and the expression level of TNFα signaling pathway-related genes according to epithelial cell types. The number represents average expression level of single-cell RNA. aISC, active intestinal stem cell.
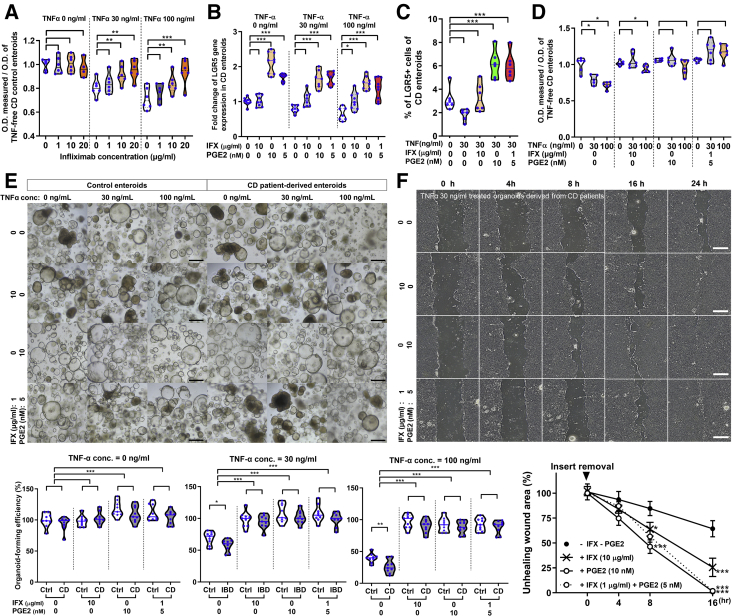
PGE2 is the most abundant COX-derived metabolite that helps in maintaining the self-renewal capacity of human ISCs via autocrine signaling.32,33 Therefore, we investigated whether exogenous PGE2 can promote LGR5+ ISC expansion and improve organoid-forming efficiency and wound healing ability in CD patient-derived enteroids under TNFα-enriched conditions. TNFα (30 ng/mL)-induced cell death was fully reversed in control enteroids at therapeutic concentrations (10 μg/mL) and partially reversed by subtherapeutic concentrations (1 μg/mL) of infliximab (Figure 5A). LGR5 mRNA levels increased significantly in CD patient-derived enteroids after incubation with 10 nmol/L PGE2 or incubation with combined low-dose infliximab (1 μg/mL) and PGE2 (5 nmol/L), regardless of TNFα treatment (P < .001 each; Figure 5B). Fluorescence-activated cell sorting analysis showed that LGR5+ cell population was increased in TNFα-treated CD patient-derived enteroids after incubation with 10 nmol/L PGE2 or incubation with combined low-dose infliximab and PGE2 (P < .001 each; Figure 5C). Infliximab treatment alone failed to increase LGR5 mRNA expression and LGR5+ cell expansion. MTT assay revealed that cell viability was not decreased in TNFα-treated CD patient-derived enteroids after treatment with infliximab, PGE2, and combined low-dose PGE2 and infliximab (Figure 5D). Infliximab and PGE2 improved the organoid-forming efficiency of TNFα-treated control and CD patient-derived enteroids. The organoid-forming efficiency of TNFα-treated enteroids treated with a combination of low-dose infliximab and PGE2 was as high as that of 10 μg/mL infliximab or 10 nmol/L PGE2 treated enteroids (Figure 5E). In the wound healing assay, delayed wound healing (cell-free gap) was improved by treatment with 10 μg/mL infliximab and 10 nmol/L PGE2. Wound healing ability was also improved by combined treatment with low-dose infliximab and PGE2, and the results were comparable with those of 10 μg/mL infliximab or 10 nmol/L PGE2 treatment (Figure 5F). These results indicate that PGE2 enhances organoid-forming efficiency and wound healing ability in CD patient-derived enteroids through LGR5+ ISC expansion. In addition, the limited effect of subtherapeutic concentrations of infliximab on organoid formation and wound healing can be compensated with low-dose PGE2 exposure.
Figure 5.
Effect of infliximab (IFX) and PGE2 on TNFα-treated CD patient-derived enteroids. (A) Effect of infliximab on viability in TNFα-treated control enteroids. Control enteroids (n = 3) were cultured in different concentrations of infliximab (0, 1, 10, and 20 μg/mL) and TNFα (0, 30, and 100 ng/mL). Assays were conducted in triplicate. Differences were evaluated using one-way ANOVA with Bonferroni multiple comparison test; ∗∗P < .01,∗∗∗P < .001. Scale bar = 200 μm. (B) Quantitative reverse transcription polymerase chain reaction for assessing LGR5 expression in CD patient-derived enteroids treated with TNFα, infliximab, and PGE2 (n = 3). Differences were evaluated using Wilcoxon signed-rank test; ∗P < .05, ∗∗∗P < .001. (C) Fluorescence-activated cell sorting analysis for LGR5+ cells in CD patient-derived enteroids treated with TNFα, infliximab, and PGE2 (n = 3). Differences were evaluated using Wilcoxon signed-rank test; ∗∗∗P < .001. (D) MTT assay for CD patient-derived enteroids treated with TNFα, infliximab, and PGE2 (n = 3). Differences were evaluated using Wilcoxon signed-rank test; ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. (E) Organoid-forming efficiency of control and CD patient-derived enteroids treated with TNFα, infliximab, and PGE2 (n = 3 each). The number of reconstituted organoids is expressed as percentage based on the value for TNFα-untreated control enteroids with no infliximab and PGE2. Differences were evaluated using Wilcoxon signed-rank test; ∗∗∗P < .001. (F) Wound healing assay for CD patient-derived enteroids treated with TNFα, infliximab, and PGE2. Non-healed wound areas were evaluated in 3 different fields. Differences were evaluated using ordinary two-way ANOVA with Bonferroni multiple comparison test; ∗P < .05, ∗∗∗P < .001. O.D., optical density.
Discussion
Our previous study showed impaired epithelial regeneration in enteroids derived from CD patients under TNFα-enriched conditions.20 Epithelial regeneration and repair are dependent on the self-renewal, proliferation, and differentiation of ISCs.34 This study demonstrates that TNFα induces the expansion of different ISC subpopulations in enteroids derived from controls and patients with CD. TNFα induces the expansion of active ISCs in control enteroids, whereas it increases the number of reserve ISCs in CD patient-derived enteroids. Impaired LGR5+ ISC expansion could be one of the reasons for impaired epithelial regeneration ability of TNFα-treated CD patient-derived enteroids.
TNFα is known for its tumor cytotoxicity.14 Herein, we observed that TNFα promotes necroptosis of intestinal epithelial cells, especially differentiated cell populations.35,36 Experiments on immortalized cell lines and animal models showed that the intestinal epithelium is susceptible to TNFR1-mediated TNFα-induced cell death.37 Our study validated the epithelial cellular response to TNFα using human enteroids. The initial response of TNFR1 upon TNFα stimulation is membrane-bound complex-I recruitment.38 Following this, TNFα-induced complex-I activates FADD/caspase-8-dependent apoptotic pathway, and the negative autocrine loop is also activated with increased CYLD expression,36,39,40 which facilitates MLKL-mediated necroptosis.36 We observed increased expression of MLKL and DNM1L in human enteroids after treatment with TNFα; however, the expression of apoptosis-associated genes, such as caspase 3, caspase 7, caspase 8, and caspase 10, was not affected. In the intestinal mucosa of patients with CD, the expression of MLKL is increased, whereas the expression of caspase-8 is reduced.35,41,42 Inflamed tissues in patients with CD showed down-regulated caspase-8 protein expression, despite elevated levels of TNFα.42 Cumulatively, these results suggest that TNFα-induced intestinal epithelial cell death occurs mainly by necroptosis rather than apoptosis. Furthermore, CD patient-derived enteroids showed increased TNFα-induced necroptotic cell death compared with control enteroids.
TNFα-induced necroptosis can cause defects in epithelial integrity, which could be reversed by expansion and differentiation of ISCs. Two populations of ISCs responsible for epithelial repair have been described: active ISCs, which are rapid cycling cells but sensitive to injury, and reserve ISCs, which are slow cycling cells but resistant to injury. LGR5 is a well-established marker of active ISCs, whereas several markers have been described for reserve ISCs, such as BMI1,43 Hopx,44 and MEX3A.45 Active and reserve ISC populations are believed to promote homeostatic epithelial renewal and regeneration after injury.46 Previously, intestinal epithelial repair was studied after acute injury after ablation of LGR5+ ISCs by irradiation, chemotherapeutic agents, or diphtheria toxin.34,47, 48, 49, 50 When active ISCs are selectively lost in acute injury models, reserve ISCs become activated and converted to active ISCs. The epithelial repair is completed as the active ISC pool is replenished by reserve ISCs and all epithelial cell types are regenerated.46 DLL1+ secretory progenitors,51 ALPI+ absorptive progenitors,52 and differentiated Paneth cells53 may also provide a source of LGR5+ ISCs in the specific environments. TNFα-induced cytotoxicity did not completely eliminate active ISCs. When the injury is limited and the active ISCs are not completely eliminated or the injury is limited to the differentiated compartment, active ISCs still have the task of repairing the damage.46 Therefore, alteration in active ISC populations under TNFα-enriched conditions might play a central role in the impairment of epithelial regeneration and wound healing in patients with CD.
In recent years, the development of intestinal organoid models has allowed deeper and more thorough investigations on ISCs and intestinal epithelial cells at single-cell level.4,5,54, 55, 56, 57 Recent studies have indicated that the properties of ISCs in CD patient-derived enteroids are altered.4 Our study is the first to identify the impairment of LGR5+ active ISC expansion in CD patient-derived enteroids under TNFα-enriched conditions, although the underlying mechanism remains unclear. This might be due to inherent dysfunction of LGR5+ ISCs of CD patients. Otherwise, the pathway regulating the conversion of reserve ISCs to active ISCs might be impaired in CD. In addition, dysfunction of Paneth cells, the major components of the ISC niche, has been implicated in CD.58 Previous studies showed that Paneth cells depletion in murine models leads to a loss of LGR5+ ISCs.59 Our results revealed a significant decrease in the expression of Paneth cell markers, LYZ and CD24, in CD patient-derived enteroids. Therefore, Paneth cell dysfunction in CD patients might affect ISC fate, including impaired LGR5+ ISC expansion.
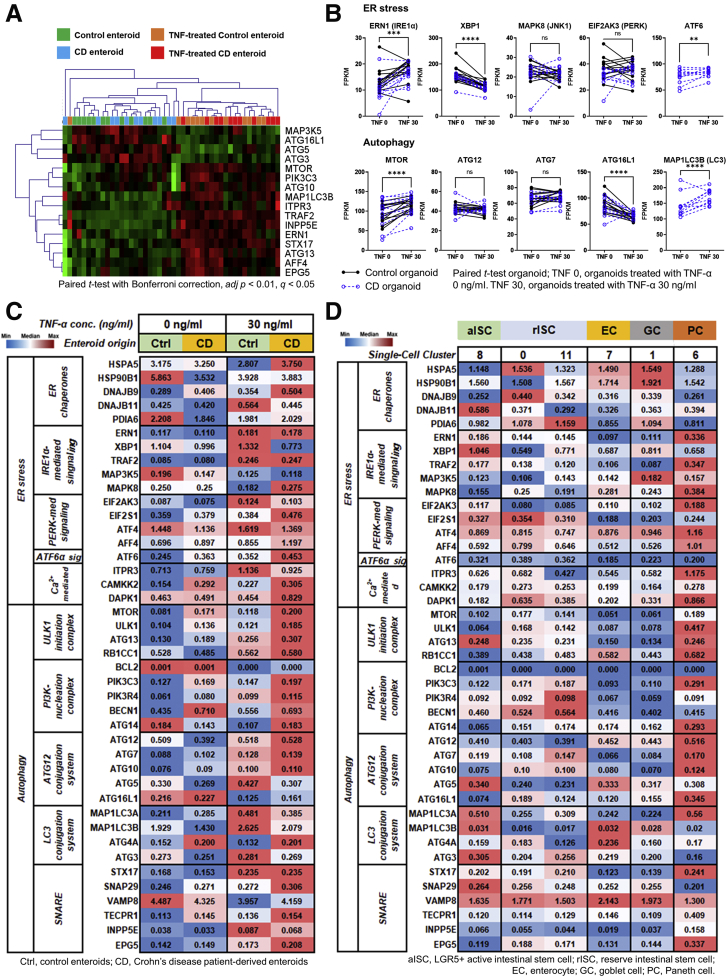
CD is associated with stress signals in the intestinal epithelium, such as endoplasmic reticulum (ER) stress and defective autophagy. These stress signals were also associated with TNF-induced necroptosis, reduced number of secretory cells, and Paneth cell dysfunction.58,60 The expression of ER stress- and autophagy-related genes was analyzed using bulk and scRNA-seq. The expression of ER stress- and autophagy-related genes was increased in TNFα-treated enteroids (Figure 6A and B). In the steady state, the expression of ER stress and autophagy genes was not significantly different between CD patient-derived enteroids and control enteroids. On the other hand, in TNFα enriched conditions, the expression of these genes was higher in CD patient-derived enteroids compared with control enteroids (Figure 6C). Furthermore, ER stress and autophagy genes were highly expressed in Paneth cells. LC3 conjugation system and SNARE genes were up-regulated in the ISCs (Figure 6D). These data indicate ER stress and autophagy genes were up-regulated in the TNFα-treated CD patient-derived enteroids, Paneth cells, and probably ISCs, which might result in the dysfunction of Paneth cells and ISCs.
Figure 6.
RNA-seq analysis for ER stress- and autophagy-related genes in control enteroids and CD patient-derived enteroids. Bulk RNA-seq. (A) Heatmap with hierarchical clustering for DEGs (paired t test with Bonferroni multiple comparison, adjusted P < .01, q < 0.05). (B) Differences in expression levels (FPKM) of ER stress- and autophagy-related genes in paired sample of TNFα-untreated and -treated enteroids derived from controls and CD patients. Single-cell RNA-seq. Expression of ER stress- and autophagy-related genes using heatmap (C) in the enteroids derived from controls and CD patients and (D) in the intestinal epithelial lineages.
The LGR5+ ISC-enriched cell cluster in scRNA-seq showed the activation of the COX2/PGE2 pathway. In human and murine studies, inhibition of COX was found to reduce the levels of PGE2 in the intestinal mucosa and inhibit the proliferation of intestinal epithelial cells.61 PGE2 interacts with the Wnt signaling pathway through PKA-dependent inhibition of GSK3β.62 A previous study reported that PGE2 promotes intestinal repair via wound-associated epithelial cells instead of LGR5+ cell recruitment.63 However, several studies have reported that PGE2 promotes the up-regulation of LGR5 expression and improves self-renewal and survival of LGR5+ ISCs via EP2-mediated signaling.32,33,64 In this study, exogenous PGE2 in the culture media improved organoid-forming efficiency and wound healing ability through the expansion of the LGR5+ ISC population in CD patient-derived enteroids treated with TNFα.
Anti-TNFα monoclonal antibody, infliximab, blocked the effects of TNFα on the control and CD patient-derived enteroids, thereby restoring the impaired cell viability and culturing behavior. In clinical practice, infliximab is regarded as the most effective therapeutic option for the treatment of CD; however, a previous study showed that 23%–46% of patients lose responsiveness to anti-TNFα agents within 12 months of treatment.65 Because low serum level of infliximab is associated with lack of clinical response, recent studies have used infliximab within the therapeutic range of 3–10 μg/mL.66 In cases of loss of response to anti-TNFα therapy, the therapeutic option is to increase the dose to raise the serum infliximab levels.67 In this study, subtherapeutic infliximab dose resulted in partial restoration of cellular response to TNFα, and it was completely restored by co-treatment with low levels of PGE2. The LGR5+ cell population in TNFα-treated CD patient-derived enteroids was increased after treatment with PGE2, but not infliximab. Taken together, our data indicate that the combination of infliximab and PGE2 may have synergistic effects on the wound healing against TNFα-induced cytotoxicity.
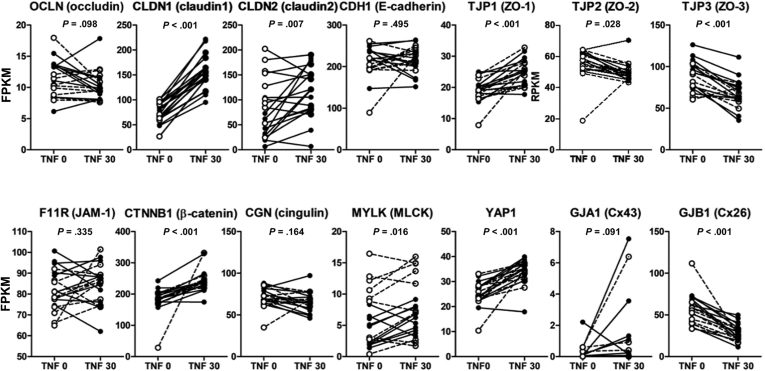
Impaired epithelial differentiation due to high TNFα levels could result in epithelial barrier dysfunction.68,69 Mucus acts as a primary physical barrier that prevents the adhesion or invasion of the intestinal microbiota. Previous studies have reported that loss of goblet cells and changes in mucin expression are associated with TNFα.70 Our study confirmed that TNFα prevents goblet cell differentiation and decreases mucus production in CD patient-derived enteroids. In the intestinal epithelial monolayer, TNFα-induced barrier defects are associated with increased expression of MLCK and CLDN-2,71 which is verified in enteroids treated with TNFα. Furthermore, TNFα-treated enteroids showed decreased levels of TJP2 and TJP3 and increased levels of CLDN1, TJP1, CTNNB1, YAP1, and GJB1 (Figure 7).
Figure 7.
Changes in the expression of epithelial intracellular junctional molecules in TNFα-untreated and -treated enteroids. FPKM of paired TNFα-untreated and -treated enteroids derived from controls and patients with CD were analyzed using the paired t test. Black dotted lines indicate changes in control organoids, and white dotted and dashed lines indicate changes in CD patient-derived enteroids.
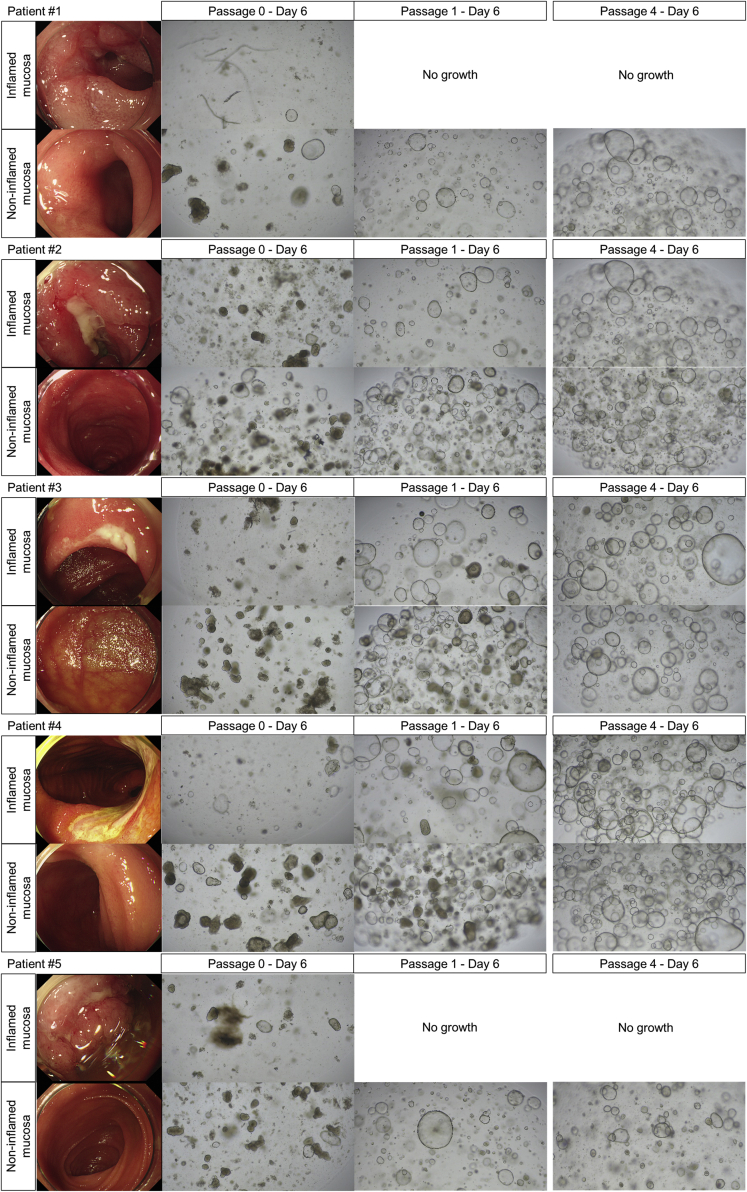
The limitation of this study is that the organoid culture system does not take into account the effects of the intestinal microbiota, dietary components, and mucosal immune system. The intestinal microbiota and dietary components contribute to the fine-tuning of the survival and differentiation of ISCs.37 In addition, considering the heterogeneity of the human samples, the number of enteroids used in the current study tends to be small. Our bulk RNA-seq data were obtained with the average depth of coverage of 100×. When we assumed equal within-group coefficient of variation of 0.5, the target effect size as 2, target false-positive rate (α) as 0.05, and desired power as 0.9, the number of samples was calculated as 11. We expected that the difference in gene expression with an effect size greater than 2 may be identified with 90% statistical power using 11–12 samples for each group. In addition, our scRNA-seq data were obtained from 43,152 individual cell transcriptomes. The number of cells is expected to be sufficient for a comparative analysis in scRNA-seq data; however, only 1 representative patient-derived and 1 control enteroid were used. Nevertheless, scRNA-seq results supported our bulk RNA-seq analysis and the main findings. Finally, CD enteroids were generated by using the crypts isolated from the non-inflamed mucosa. In our previous study, the generation of organoids from patient biopsy samples was attempted.20 Before the publication of this study, we attempted to establish an intestinal organoid of the inflamed mucosa near the ulcer. However, the isolated crypts were sparse, damaged, short, and broken. The subsequent organoid formation was ineffective. When the organoids were subcultured for more than 3 passages, there were no differences in the morphology and culturing behavior of the intestinal organoids derived from inflamed and non-inflamed mucosa of patients with CD. Therefore, we cultured organoids by using intestinal crypts isolated from macroscopically non-inflamed mucosa. In addition, considering that the turnover of the intestinal epithelium is every 3–5 days, subcultured organoids (>3 passages) derived from inflamed or non-inflamed mucosa of patients with CD may not affect the experiments to evaluate our hypothesis (Figure 8). Although the intestinal crypts can be isolated from inflamed mucosa or active ulcer margin in patients with CD, the successful long-term culture rate was low compared with those isolated from non-inflamed mucosa.
Figure 8.
Intestinal organoid generation and subculture derived from inflamed or non-inflamed mucosa of patients with CD.
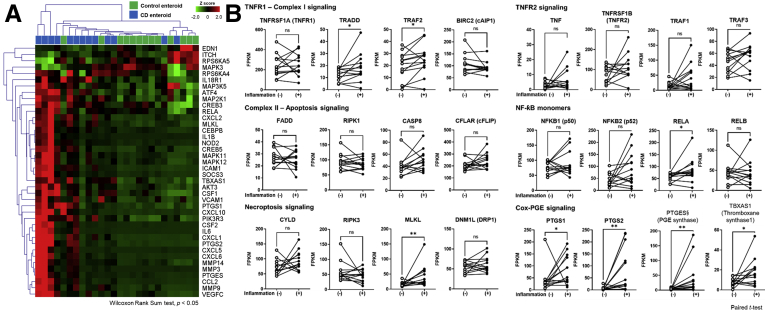
Previously, we have performed RNA-seq to evaluate genome-wide gene expression pattern in the mucosa of CD patients by using 13 pairs of inflamed and non-inflamed mucosa obtained from the same patients with active CD.72 Using these RNA-seq data, the expression pattern of the TNFα signaling pathway and COX2/PGE2 axis was analyzed. As predicted, the expression of TNFα signaling genes was increased in inflamed mucosa compared with non-inflamed mucosa of patients with CD (Figure 9). Like our organoid model, MLKL expression was significantly increased in the inflamed mucosa compared with the non-inflamed mucosa in patients with CD; however, the expression of apoptosis-related gene was not increased in the inflamed mucosa of CD. The expression of Cox-PGE2 pathway genes, including PTGS1, PTGS2, PTGES, and TBXAS1, was significantly increased in the inflamed mucosa compared with the non-inflamed mucosa in patients with CD. These results indicate MLKL-mediated necroptosis and COX-PGE2 pathway may be activated in the inflamed mucosa in patients with active CD. Furthermore, these observations support that the patient-derived enteroid model can recapitulate the response of human intestinal epithelial responses in patients with CD.
Figure 9.
TNFα signaling pathway and its related genes expression in the inflamed and non-inflamed mucosa obtained from the same patients with active CD. (A) Hierarchical clustering heatmap of TNFα signaling pathway and its related genes in 13 pairs of inflamed and non-inflamed mucosa obtained from the same patients with active CD. DEGs were identified by using the Wilcoxon matched-pairs signed-rank test (P < .05). (B) Differences in expression levels (FPKM) of TNFα pathway genes in paired sample of inflamed and non-inflamed mucosa of same patients with active CD. Differences were evaluated using paired t test; ∗P < .05, ∗∗P < .01.
Patient-derived enteroids recapitulate disease-specific characteristics of intestinal epithelial cells. In human enteroids, TNFα induces necroptotic cell death and reparative epithelial cell proliferation through LGR5+ ISC expansion. However, in CD patient-derived enteroids, TNFα-induced LGR5+ ISC expansion is impaired, resulting in decreased organoid-forming efficiency and delayed wound healing. Because self-renewal is the main feature of ISCs, we can describe the failure of ISC expansion as the dysfunction of LGR5+ ISCs. Exogenous PGE2 promoted the expansion of LGR5+ ISCs and improved organoid-forming efficiency and wound healing in TNFα-treated CD patient-derived enteroids. The combination of low-dose infliximab and PGE2 showed synergistic effects on wound healing in CD patient-derived enteroids. Therefore, PGE2 could be a promising therapeutic candidate for improving the repair and regeneration of intestinal epithelium in patients with CD. Further studies and clinical trials are required to evaluate the therapeutic efficacy of PGE2 in CD.
Methods
Sample Collection
To establish human intestinal organoids (enteroids), human small intestinal samples were obtained from controls (n = 12) and patients with CD (n = 11) by using biopsy forceps during single-balloon enteroscopy at the Samsung Medical Center, Seoul, Korea, between November 2016 and December 2018. Patients with CD were diagnosed according to the practice guidelines.73 The characteristics of the enrolled patients are listed in Table 1. In patients with CD, at least 4 biopsy specimens were obtained at least 5 cm away from the ulcers. Control patients underwent single-balloon enteroscopy for the evaluation of submucosal tumor (n = 8; ectopic pancreas [n = 3], lipoma [n = 2], gastrointestinal stromal tumor [n = 2], neuroendocrine tumor [n = 1]) and unexplained abdominal symptoms (irritable bowel syndrome, n = 4). The median age was 43 years (range, 18–72 years), and 9 patients were male. At least 4 biopsy specimens were collected from the normal mucosa of the small intestine.
Table 1.
Characteristics of Enrolled Patients With Crohn’s Disease
| CD #1 | CD #2 | CD #3 | CD #4 | CD #5 | CD #6 | CD #7 | CD #8 | CD #9 | CD #10 | CD #11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age, y | 22 | 43 | 28 | 32 | 33 | 20 | 22 | 48 | 21 | 28 | 32 |
| Gender | Male | Female | Male | Male | Male | Male | Male | Male | Female | Male | Male |
| Montreal classification (n) | |||||||||||
| Age at diagnosis (A1/A2/A3) | A1 | A2 | A2 | A2 | A2 | A2 | A2 | A3 | A2 | A2 | A2 |
| Location (L1/L2/L3) | L3 | L3 | L3 | L2 | L2 | L2 | L3 | L3 | L2 | L2 | L3 |
| Upper gastrointestinal modifier (L4) | - | - | - | - | L4 | - | L4 | - | - | - | - |
| Behavior (B1/B2/B3) | B1 | B2 | B2 | B1 | B2 | B2 | B2 | B1 | B1 | B2 | B2 |
| Perianal disease modifier (p) | p | - | - | p | p | - | p | - | - | p | - |
| Medication at sampling | |||||||||||
| 5-aminosalicylic acid | - | + | - | - | - | - | - | + | - | - | - |
| Glucocorticoids | + | - | - | - | - | + | + | - | - | - | - |
| Immunomodulator | + | + | + | - | + | + | + | - | + | + | + |
| Infliximab | - | + | + | + | - | - | - | + | - | - | |
| Adalimumab | - | - | - | - | + | - | - | + | - | - | - |
| Ustekinumab | - | - | - | - | - | - | - | - | - | + | + |
| Endoscopic finding | |||||||||||
| Mucosal healing | - | - | + | + | - | - | - | - | - | - | - |
| Active ulcer | + | + | - | - | + | + | + | + | + | + | + |
This study was approved by the Institutional Ethical Committee of the Samsung Medical Center (IRB No. 2016-02-022). All samples were taken with informed consent from the enrolled individuals.
Crypt Isolation From Biopsies
The intestinal crypts were isolated from endoscopic biopsy specimens as described previously.8,9,74,75 Briefly, endoscopic biopsy specimens were incubated in phosphate-buffered saline (PBS) with 10 mmol/L EDTA (Thermo Fischer Scientific, San Jose, CA) and 1 mmol/L dithiothreitol (Thermo Fischer Scientific) at 4°C on a rocker at 50 rpm for 30 minutes. The supernatant, containing villi and debris, was decanted and discarded. Intestinal crypts were obtained by adding fresh PBS to the pellet, followed by vortexing for 30 seconds. The supernatant containing crypts were collected and filtered through a 70-μm cell strainer (Corning, Bedford, MA). This procedure was repeated 3 times. The fractions were combined and centrifuged at 200g at 4°C for 2 minutes, and the pellet was resuspended in basal medium (advanced Dulbecco modified Eagle medium/F12 [Thermo Fisher Scientific]) supplemented with antibiotic–antimycotic solution (Thermo Fischer Scientific), 10 mmol/L HEPES (Thermo Fisher Scientific), GlutaMAX (Thermo Fisher Scientific), 1× N2 (Thermo Fisher Scientific), 1× B27 (Thermo Fisher Scientific), and 1 mmol/L N-acetylcysteine (Sigma-Aldrich, St Louis, MO). The suspension was centrifuged at 200g at 4°C for 2 minutes, and the pellet was resuspended in the basal medium.
Three-Dimensional Intestinal Crypt Culture
Intestinal crypts were cultured three-dimensionally as described previously.8,9,74,75 Briefly, the isolated crypts were pelleted with 3 quick spins, resuspended in Matrigel (Corning), and plated in 48-well culture plates (Corning). After incubation at 37°C for 15 minutes, 250 μL of maintenance medium (50% Wnt3a-conditioned medium [ATCC#CRL-2647, Manassas, VA]), 50% 2× basal medium supplemented with 50 ng/mL recombinant human epidermal growth factor (Sigma-Aldrich), 100 ng/mL recombinant human noggin (R&D Systems, Minneapolis, MN), 500 ng/mL recombinant human R-spondin 1 (PeproTech, Cranbury, NJ), 10 mmol/L nicotinamide (Sigma-Aldrich), 10 μmol/L p160ROCK inhibitor (Y27632; Selleckchem, Houston, TX), 10 μmol/L p38 MAP kinase inhibitor (SB202190; Sigma-Aldrich), and 10 nmol/L PGE2 (Cayman Chemical, Ann Arbor, MI). GSK3 inhibitor (CHIR99021; Stemgent, Cambridge, MA) was added to the medium during the first 2 days.
Organoid Subculture and Maintenance
After culturing the cells for 7 days, the organoids in the Matrigel were mechanically disrupted by pipetting. The dissociated organoids were washed with 10 mL of basal medium and centrifuged at 200g at 4°C for 30 seconds. The pellet was resuspended in 2 mL of cell dissociation buffer (Thermo Fisher Scientific) and incubated in a water bath at 37°C for 5 minutes. The cell pellet was resuspended in Matrigel and plated in 48-well culture plates (Corning). After incubation at 37°C for 15 minutes, 250 μL of maintenance medium was added. The medium was changed every 2 days, and organoids were passaged at a ratio of 1:2 to 1:4 on day 7.
Enteroids Differentiation
After 4–6 passages, most of the organoids in the maintenance medium formed uniform spheroids and could be passaged stably for a long time. To recapitulate the structure and function of the intestinal epithelium, the spheroids were cultured in the differentiation medium (maintenance medium without Wnt3A conditional medium, SB202190, nicotinamide, and PGE2).8 The intestinal organoids cultured in differentiation medium (enteroids) had visually sharp borders (buddings) along their basolateral (anti-luminal) side and irregular, thick walls composed of epithelial cells.76 The differentiation medium was changed every 2 days, and enteroids were cultured for 7–12 days.
Organoid-Forming Efficiency
The intestinal organoids were cultured in the maintenance medium for 2 days to obtain a stable number of organoids before inducing organoid differentiation because TNFα is known to induce cytotoxicity in certain cells, leading to cell death. After 2 days, the maintenance medium was replaced with the differentiation medium and was replenished every 2 days. Different concentrations of human recombinant TNFα (R&D Systems) were added to the culture medium every 24 hours. The number of organoids formed depends on the density of the seeded cells.77 Therefore, the number of organoids was counted at 2 days (before treatment with TNFα) and 9–10 days (after treatment with TNFα) after embedding under an inverted microscope (Leica Microsystems, Wetzlar, Germany). Organoid-forming efficiency was calculated as follows: organoid-forming efficiency (%) = (number of organoids at 9–10 days/number of organoids at 2 days) × 100.
MTT Assay
Ten microliters of MTT (Sigma-Aldrich) was added to each well of the culture plates and incubated for 3 hours until purple precipitate was visible. After adding 100 μL of the detergent reagent, the organoids were incubated at room temperature in the dark for 2 hours, and the absorbance was recorded at 570 nm. The optical density of organoids at 570 nm was expressed as percentage based on the values for TNFα-free control enteroids.
Two-Dimensional Human Intestinal Organoid Culture and Wound Healing Assay
Each well of the 24-well plate was coated with 50 μL of 0.8 mg/mL Matrigel diluted in the basal organoid medium at 37°C for 1 hour. For two-dimensional culture, organoids in the three-dimensional culture were enzymatically digested into single cells using TrypLE Express (Thermo Fisher Scientific). The digested cells were resuspended in the basal medium and seeded into Matrigel-coated wells.
To perform the wound healing assay, 5 × 104 cells were seeded in wells of a 24-well plate containing inserts of CytoSelect 24-Well Wound Healing Assay (Cell Biolabs, San Diego, CA). We cultured two-dimensional organoid monolayers in maintenance medium until confluence was reached. The inserts were then carefully removed to produce 0.9-mm-diameter wounds, and fresh differentiation medium was added to each well.78 The wound was monitored using a phase-contrast microscope at 0, 4, 8, 16, and 24 hours (Leica Microsystems). The area of the non-healed wound was measured in 3 different regions. The unhealed wound area was expressed as percentage based on the values for TNFα-free control enteroids.
Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction
Total RNA was extracted from intestinal organoids using the RNeasy Mini Kit (Qiagen, Hilden, Germany). One-step quantitative polymerase chain reaction was performed using the One Step PrimeScript III RT-qPCR Mix (Takara, Kusatsu, Japan) with the following primers: LGR5 primer (forward: 5′-CCTGCTTGACTTTGAGGAAGACC-3′, reverse: 5′-ACACATTGGGGGTAGGAACA-3′).
RNA Sequencing
Total RNA was extracted from enteroids using RNeasy Mini Kit (Qiagen). RNA-seq was conducted using total RNA samples with >10 μg of RNA and an integrity number >8. cDNA libraries were sequenced with HiSeq2500 using the 100 base pair paired-end mode. Reads from files in the FASTQ format were mapped to the hg19 human reference genome using HISAT 2.2.0, with default parameters. Raw read counts mapped to genes were measured by using the BAM format file in HTSeq version 0.12.3 to quantify transcript abundance. The coding genes were selected, and DEGs analysis was conducted with EdgeR (version 3.28.1). Raw read counts were normalized to the trimmed mean of M-values. Unsupervised hierarchical clustering analysis with the Euclidean distance and complete linkage algorithm was used to create a heatmap with the associated dendrogram.
Single-Cell RNA-Seq
Intestinal organoids treated with and without 30 ng/ml TNF were mechanically disrupted by vigorous pipetting and then enzymatically digested into single cell suspension using TrypLE Express (Thermo Fisher Scientific). Bar-coded sequencing libraries were prepared and sequenced on a HiSeq X Ten system, targeting 10,000 per each sample. Reads were aligned to a human reference genome (GRCh38-3.0.0) and processed using the CellRanger 3.1.0 pipeline (10X Genomics, Pleasanton, CA).
The raw gene expression matrix was filtered using the Seurat R package (version 4.0.3) and selected according to the following criteria: cells with >200 genes and <20% of mitochondrial gene expression in UMI counts. Cells that passed the filtering criteria were integrated and clustered. Intestinal epithelial cell type was annotated on the cluster depending on the presence of the differential expression of the cell marker identified in the database.79 When several cell markers were expressed significantly in a single cluster, the cell type was assigned on the basis of the number of expressing cell markers or well-established marker expressions.
Developmental Trajectories Analysis
Slingshot was used to illustrate lineage differentiation within all clusters.80 The start point was set at cluster 8 (LGR5+ ISC) and cluster 0 (reserve ISC). The endpoint was inferred by pseudotime ordering along the trajectory of the differentiated cell-enriched clusters.
Immunohistochemistry
After removing the culture medium, the organoids were washed with PBS and fixed in cold 4% paraformaldehyde at room temperature for 30 minutes. The fixed organoids were washed with PBS and embedded in HistoGel (Thermo Fisher Scientific). HistoGel blocks were used for paraffin embedding and sectioned for histologic and immunohistochemical analyses. Histologic evaluation was performed using hematoxylin-eosin–stained sections. After heating-induced epitope retrieval with citrate buffer, immunohistochemistry was performed using antibodies against E-cadherin (1:100; Abcam, Cambridge, UK), OLFM4 (1:200; Cell Signaling, Danvers, MA), lysozyme (1:200; Thermo Fisher Scientific), CC3 (1:200; Cell Signaling), LGR5 (1:400; Abcam), and BMI1 (1:800; Abcam).
TUNEL Assay
Apoptosis-associated DNA fragmentation was detected by TUNEL assay using the In Situ Cell Death Detection Kit (Merck, Darmstadt, Germany). Positive control sections were incubated with 10 U/mL recombinant DNase I, and negative control sections were processed in the same manner without the terminal transferase enzyme.
Double Immunofluorescence for CC3 and TUNEL
Primary rabbit antibodies specific for CC3 and a goat anti-rabbit secondary antibody conjugated to Alexa 594 fluorochrome (red fluorescence) were used to localize CC3. To assess DNA fragmentation, the same sections were processed for TUNEL staining using fluorescein-labeled dUTP (green fluorescence). Cell nuclei were stained with DAPI (blue fluorescence). The number of CC3+ and TUNEL+ cells was measured in 10 organoids (size >100 μm) selected from TNFα-untreated and -treated enteroids derived from controls and patients with CD.
Alcian Blue Assay
After heating-induced epitope retrieval with citrate buffer, the paraffin-embedded organoids were fixed with 3% acetic acid, stained with alcian blue solution (Sigma-Aldrich) for 30 minutes, and washed with distilled water. The stained area was quantified using ImageJ and expressed as percentage relative to the average stained area in the TNFα-untreated control enteroids.
Flow Cytometry
To prepare single-cell suspensions, enteroids were incubated with TrypLE Express in the water bath at 37°C for 30 minutes. The dissociated cells were washed with basal medium and collected by filtering through a 40-μm strainer. Cells (1 × 105) were incubated with the human Lgr5/GPR49 antibody (R&D Systems) in fluorescence-activated cell sorting buffer for 30 minutes. Unbound antibodies were washed out, and the cells were incubated with Alexa Fluor 488-conjugated anti-mouse immunoglobulin G secondary antibody (Thermo Fisher Scientific) for 30 minutes. The cells were then washed, resuspended in fluorescence-activated cell sorting buffer, and analyzed on the FACS Aria III instrument (BD Biosciences, Franklin Lakes, NJ) to evaluate LGR5+ ISCs population.
Acknowledgments
The author thanks Prof Martín G. Martín, (Department of Pediatrics, Mattel Children’s Hospital and the David Geffen School of Medicine, University of California Los Angeles), Prof Matthias Stelzner (Department of Surgery, Veterans Affairs Greater Los Angeles Healthcare System), and Prof James C. Y. Dunn (Division of Pediatric Surgery, Department of Surgery, Stanford University), who taught and trained me in constructing intestinal organoid models despite their busy schedules.
CRediT Authorship Contributions
Chansu Lee (Conceptualization: Supporting; Data curation: Lead; Formal analysis: Lead; Methodology: Equal; Writing – original draft: Lead)
Minae An (Formal analysis: Lead; Methodology: Equal; Visualization: Equal)
Je-Gun Joung (Data curation: Equal; Formal analysis: Equal; Visualization: Equal)
Park Woong-Yang (Data curation: Equal; Formal analysis: Equal; Methodology: Equal)
Dong Kyung Chang (Funding acquisition: Supporting; Supervision: Equal)
Young-Ho Kim (Funding acquisition: Lead; Supervision: Lead)
Sung Noh Hong, MD, PhD (Conceptualization: Lead; Data curation: Equal; Formal analysis: Supporting; Funding acquisition: Lead; Investigation: Equal; Methodology: Equal; Supervision: Lead; Writing – original draft: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (2019R1A2C2010404 and 2019R1I1A01062205) and Future Medicine 20∗30 Project of the Samsung Medical Center, Seoul, South Korea.
References
- 1.Pizarro T.T., Stappenbeck T.S., Rieder F., Rosen M.J., Colombel J.F., Donowitz M., Towne J., Mazmanian S.K., Faith J.J., Hodin R.A., Garrett W.S., Fichera A., Poritz L.S., Cortes C.J., Shtraizent N., Honig G., Snapper S.B., Hurtado-Lorenzo A., Salzman N.H., Chang E.B. Challenges in IBD research: preclinical human IBD mechanisms. Inflamm Bowel Dis. 2019;25(Suppl 2):S5–S12. doi: 10.1093/ibd/izz075. [DOI] [PubMed] [Google Scholar]
- 2.Torres J., Mehandru S., Colombel J.F., Peyrin-Biroulet L. Crohn’s disease. Lancet. 2017;389:1741–1755. doi: 10.1016/S0140-6736(16)31711-1. [DOI] [PubMed] [Google Scholar]
- 3.Ordás I., Eckmann L., Talamini M., Baumgart D.C., Sandborn W.J. Ulcerative colitis. Lancet. 2012;380:1606–1619. doi: 10.1016/S0140-6736(12)60150-0. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki K., Murano T., Shimizu H., Ito G., Nakata T., Fujii S., Ishibashi F., Kawamoto A., Anzai S., Kuno R., Kuwabara K., Takahashi J., Hama M., Nagata S., Hiraguri Y., Takenaka K., Yui S., Tsuchiya K., Nakamura T., Ohtsuka K., Watanabe M., Okamoto R. Single cell analysis of Crohn’s disease patient-derived small intestinal organoids reveals disease activity-dependent modification of stem cell properties. J Gastroenterol. 2018;53:1035–1047. doi: 10.1007/s00535-018-1437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dotti I., Mora-Buch R., Ferrer-Picón E., Planell N., Jung P., Masamunt M.C., Leal R.F., Martín de Carpi J., Llach J., Ordás I., Batlle E., Panés J., Salas A. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut. 2017;66:2069–2079. doi: 10.1136/gutjnl-2016-312609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howell K.J., Kraiczy J., Nayak K.M., Gasparetto M., Ross A., Lee C., Mak T.N., Koo B.K., Kumar N., Lawley T., Sinha A., Rosenstiel P., Heuschkel R., Stegle O., Zilbauer M. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology. 2018;154:585–598. doi: 10.1053/j.gastro.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 8.Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 9.Lei N.Y., Jabaji Z., Wang J., Joshi V.S., Brinkley G.J., Khalil H., Wang F., Jaroszewicz A., Pellegrini M., Li L., Lewis M., Stelzner M., Dunn J.C., Martín M.G. Intestinal subepithelial myofibroblasts support the growth of intestinal epithelial stem cells. PLoS One. 2014;9:e84651. doi: 10.1371/journal.pone.0084651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnauts K., Verstockt B., Ramalho A.S., Vermeire S., Verfaillie C., Ferrante M. Ex vivo mimicking of inflammation in organoids derived from patients with ulcerative colitis. Gastroenterology. 2020;159:1564–1567. doi: 10.1053/j.gastro.2020.05.064. [DOI] [PubMed] [Google Scholar]
- 11.DeHaan R.K., Sarvestani S.K., Huang E.H. Organoid models of colorectal pathology: do they hold the key to personalized medicine? a systematic review. Dis Colon Rectum. 2020;63:1559–1569. doi: 10.1097/DCR.0000000000001806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aslam M.N., McClintock S.D., Attili D., Pandya S., Rehman H., Nadeem D.M., Jawad-Makki M.A.H., Rizvi A.H., Berner M.M., Dame M.K., Turgeon D.K., Varani J. Ulcerative colitis-derived colonoid culture: a multi-mineral-approach to improve barrier protein expression. Front Cell Dev Biol. 2020;8:577221. doi: 10.3389/fcell.2020.577221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braegger C.P., Nicholls S., Murch S.H., Stephens S., MacDonald T.T. Tumour necrosis factor alpha in stool as a marker of intestinal inflammation. Lancet. 1992;339:89–91. doi: 10.1016/0140-6736(92)90999-j. [DOI] [PubMed] [Google Scholar]
- 14.Sedger L.M., McDermott M.F. TNF and TNF-receptors: from mediators of cell death and inflammation to therapeutic giants—past, present and future. Cytokine Growth Factor Rev. 2014;25:453–472. doi: 10.1016/j.cytogfr.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 15.Delgado M.E., Brunner T. The many faces of tumor necrosis factor signaling in the intestinal epithelium. Genes Immun. 2019;20:609–626. doi: 10.1038/s41435-019-0057-0. [DOI] [PubMed] [Google Scholar]
- 16.Rieder F., Brenmoehl J., Leeb S., Scholmerich J., Rogler G. Wound healing and fibrosis in intestinal disease. Gut. 2007;56:130–139. doi: 10.1136/gut.2006.090456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leppkes M., Roulis M., Neurath M.F., Kollias G., Becker C. Pleiotropic functions of TNF-α in the regulation of the intestinal epithelial response to inflammation. Int Immunol. 2014;26:509–515. doi: 10.1093/intimm/dxu051. [DOI] [PubMed] [Google Scholar]
- 18.Rhim J.S. Development of human cell lines from multiple organs. Ann N Y Acad Sci. 2000;919:16–25. doi: 10.1111/j.1749-6632.2000.tb06863.x. [DOI] [PubMed] [Google Scholar]
- 19.Kim J., Koo B.K., Knoblich J.A. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol. 2020:1–14. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee C., Hong S.N., Kim E.R., Chang D.K., Kim Y.H. Epithelial regeneration ability of Crohn’s disease assessed using patient-derived intestinal organoids. Int J Mol Sci. 2021:22. doi: 10.3390/ijms22116013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y.S., Morgan M.J., Choksi S., Liu Z.G. TNF-induced activation of the Nox1 NADPH oxidase and its role in the induction of necrotic cell death. Mol Cell. 2007;26:675–687. doi: 10.1016/j.molcel.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 22.Csiszar A., Smith K.E., Koller A., Kaley G., Edwards J.G., Ungvari Z. Regulation of bone morphogenetic protein-2 expression in endothelial cells: role of nuclear factor-kappaB activation by tumor necrosis factor-alpha, H2O2, and high intravascular pressure. Circulation. 2005;111:2364–2372. doi: 10.1161/01.CIR.0000164201.40634.1D. [DOI] [PubMed] [Google Scholar]
- 23.Zhang S., Liu Y., Zhang X., Zhu D., Qi X., Cao X., Fang Y., Che Y., Han Z.C., He Z.X., Han Z., Li Z. Prostaglandin E(2) hydrogel improves cutaneous wound healing via M2 macrophages polarization. Theranostics. 2018;8:5348–5361. doi: 10.7150/thno.27385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grabinger T., Luks L., Kostadinova F., Zimberlin C., Medema J.P., Leist M., Brunner T. Ex vivo culture of intestinal crypt organoids as a model system for assessing cell death induction in intestinal epithelial cells and enteropathy. Cell Death Dis. 2014;5:e1228. doi: 10.1038/cddis.2014.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen S., Ling Y., Yang W., Shen J., Li C., Deng W., Liu W., Liu K. Necroptosis is a key mediator of enterocytes loss in intestinal ischaemia/reperfusion injury. J Cell Mol Med. 2017;21:432–443. doi: 10.1111/jcmm.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullen T.F., Forrest S., Campbell F., Dodson A.R., Hershman M.J., Pritchard D.M., Turner J.R., Montrose M.H., Watson A.J. Characterization of epithelial cell shedding from human small intestine. Lab Invest. 2006;86:1052–1063. doi: 10.1038/labinvest.3700464. [DOI] [PubMed] [Google Scholar]
- 27.Black R.A., Rauch C.T., Kozlosky C.J., Peschon J.J., Slack J.L., Wolfson M.F., Castner B.J., Stocking K.L., Reddy P., Srinivasan S., Nelson N., Boiani N., Schooley K.A., Gerhart M., Davis R., Fitzner J.N., Johnson R.S., Paxton R.J., March C.J., Cerretti D.P. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 28.Grell M., Douni E., Wajant H., Löhden M., Clauss M., Maxeiner B., Georgopoulos S., Lesslauer W., Kollias G., Pfizenmaier K., Scheurich P. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell. 1995;83:793–802. doi: 10.1016/0092-8674(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 29.Luo J.L., Kamata H., Karin M. IKK/NF-kappaB signaling: balancing life and death—a new approach to cancer therapy. J Clin Invest. 2005;115:2625–2632. doi: 10.1172/JCI26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wajant H., Pfizenmaier K., Scheurich P. Tumor necrosis factor signaling. Cell Death Differ. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- 31.Ruder B., Atreya R., Becker C. Tumour necrosis factor alpha in intestinal homeostasis and gut related diseases. Int J Mol Sci. 2019;20:1887. doi: 10.3390/ijms20081887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Kharusi M.R., Smartt H.J., Greenhough A., Collard T.J., Emery E.D., Williams A.C., Paraskeva C. LGR5 promotes survival in human colorectal adenoma cells and is upregulated by PGE2: implications for targeting adenoma stem cells with NSAIDs. Carcinogenesis. 2013;34:1150–1157. doi: 10.1093/carcin/bgt020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee B.C., Kim H.S., Shin T.H., Kang I., Lee J.Y., Kim J.J., Kang H.K., Seo Y., Lee S., Yu K.R., Choi S.W., Kang K.S. PGE2 maintains self-renewal of human adult stem cells via EP2-mediated autocrine signaling and its production is regulated by cell-to-cell contact. Sci Rep. 2016;6:26298. doi: 10.1038/srep26298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metcalfe C., Kljavin N.M., Ybarra R., de Sauvage F.J. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14:149–159. doi: 10.1016/j.stem.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 35.Günther C., Martini E., Wittkopf N., Amann K., Weigmann B., Neumann H., Waldner M.J., Hedrick S.M., Tenzer S., Neurath M.F., Becker C. Caspase-8 regulates TNF-α-induced epithelial necroptosis and terminal ileitis. Nature. 2011;477:335–339. doi: 10.1038/nature10400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moquin D.M., McQuade T., Chan F.K. CYLD deubiquitinates RIP1 in the TNFalpha-induced necrosome to facilitate kinase activation and programmed necrosis. PLoS One. 2013;8:e76841. doi: 10.1371/journal.pone.0076841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delgado M.E., Grabinger T., Brunner T. Cell death at the intestinal epithelial front line. FEBS J. 2016;283:2701–2719. doi: 10.1111/febs.13575. [DOI] [PubMed] [Google Scholar]
- 38.Wajant H., Siegmund D. TNFR1 and TNFR2 in the control of the life and death balance of macrophages. Front Cell Dev Biol. 2019;7:91. doi: 10.3389/fcell.2019.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J., Stirling B., Temmerman S.T., Ma C.A., Fuss I.J., Derry J.M., Jain A. Impaired regulation of NF-kappaB and increased susceptibility to colitis-associated tumorigenesis in CYLD-deficient mice. J Clin Invest. 2006;116:3042–3049. doi: 10.1172/JCI28746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brenner D., Blaser H., Mak T.W. Regulation of tumour necrosis factor signalling: live or let die. Nat Rev Immunol. 2015;15:362–374. doi: 10.1038/nri3834. [DOI] [PubMed] [Google Scholar]
- 41.Welz P.S., Wullaert A., Vlantis K., Kondylis V., Fernández-Majada V., Ermolaeva M., Kirsch P., Sterner-Kock A., van Loo G., Pasparakis M. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 2011;477:330–334. doi: 10.1038/nature10273. [DOI] [PubMed] [Google Scholar]
- 42.Pierdomenico M., Negroni A., Stronati L., Vitali R., Prete E., Bertin J., Gough P.J., Aloi M., Cucchiara S. Necroptosis is active in children with inflammatory bowel disease and contributes to heighten intestinal inflammation. Am J Gastroenterol. 2014;109:279–287. doi: 10.1038/ajg.2013.403. [DOI] [PubMed] [Google Scholar]
- 43.Sangiorgi E., Capecchi M.R. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeda N., Jain R., LeBoeuf M.R., Wang Q., Lu M.M., Epstein J.A. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barriga F.M., Montagni E., Mana M., Mendez-Lago M., Hernando-Momblona X., Sevillano M., Guillaumet-Adkins A., Rodriguez-Esteban G., Buczacki S.J.A., Gut M., Heyn H., Winton D.J., Yilmaz O.H., Attolini C.S., Gut I., Batlle E. Mex3a marks a slowly dividing subpopulation of Lgr5+ intestinal stem cells. Cell Stem Cell. 2017;20:801–816.e807. doi: 10.1016/j.stem.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bankaitis E.D., Ha A., Kuo C.J., Magness S.T. Reserve stem cells in intestinal homeostasis and injury. Gastroenterology. 2018;155:1348–1361. doi: 10.1053/j.gastro.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tao S., Tang D., Morita Y., Sperka T., Omrani O., Lechel A., Sakk V., Kraus J., Kestler H.A., Kühl M., Rudolph K.L. Wnt activity and basal niche position sensitize intestinal stem and progenitor cells to DNA damage. Embo J. 2015;34:624–640. doi: 10.15252/embj.201490700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian H., Biehs B., Warming S., Leong K.G., Rangell L., Klein O.D., de Sauvage F.J. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rees W.D., Tandun R., Yau E., Zachos N.C., Steiner T.S. Regenerative intestinal stem cells induced by acute and chronic injury: the saving grace of the epithelium? Front Cell Dev Biol. 2020;8:583919. doi: 10.3389/fcell.2020.583919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitt M., Schewe M., Sacchetti A., Feijtel D., van de Geer W.S., Teeuwssen M., Sleddens H.F., Joosten R., van Royen M.E., van de Werken H.J.G., van Es J., Clevers H., Fodde R. Paneth cells respond to inflammation and contribute to tissue regeneration by acquiring stem-like features through SCF/c-Kit signaling. Cell Rep. 2018;24:2312–2328.e2317. doi: 10.1016/j.celrep.2018.07.085. [DOI] [PubMed] [Google Scholar]
- 51.van Es J.H., Sato T., van de Wetering M., Lyubimova A., Yee Nee A.N., Gregorieff A., Sasaki N., Zeinstra L., van den Born M., Korving J., Martens A.C.M., Barker N., van Oudenaarden A., Clevers H. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tetteh P.W., Basak O., Farin H.F., Wiebrands K., Kretzschmar K., Begthel H., van den Born M., Korving J., de Sauvage F., van Es J.H., van Oudenaarden A., Clevers H. Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell. 2016;18:203–213. doi: 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 53.Yousefi M., Li N., Nakauka-Ddamba A., Wang S., Davidow K., Schoenberger J., Yu Z., Jensen S.T., Kharas M.G., Lengner C.J. Msi RNA-binding proteins control reserve intestinal stem cell quiescence. J Cell Biol. 2016;215:401–413. doi: 10.1083/jcb.201604119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugimoto S., Ohta Y., Fujii M., Matano M., Shimokawa M., Nanki K., Date S., Nishikori S., Nakazato Y., Nakamura T., Kanai T., Sato T. Reconstruction of the human colon epithelium in vivo. Cell Stem Cell. 2018;22:171–176.e175. doi: 10.1016/j.stem.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 55.Martin J.C., Chang C., Boschetti G., Ungaro R., Giri M., Grout J.A., Gettler K., Chuang L.S., Nayar S., Greenstein A.J., Dubinsky M., Walker L., Leader A., Fine J.S., Whitehurst C.E., Mbow M.L., Kugathasan S., Denson L.A., Hyams J.S., Friedman J.R., Desai P.T., Ko H.M., Laface I., Akturk G., Schadt E.E., Salmon H., Gnjatic S., Rahman A.H., Merad M., Cho J.H., Kenigsberg E. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 2019;178:1493–1508.e1420. doi: 10.1016/j.cell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rees W.D., Stahl M., Jacobson K., Bressler B., Sly L.M., Vallance B.A., Steiner T.S. Enteroids derived from inflammatory bowel disease patients display dysregulated endoplasmic reticulum stress pathways, leading to differential inflammatory responses and dendritic cell maturation. J Crohns Colitis. 2020;14:948–961. doi: 10.1093/ecco-jcc/jjz194. [DOI] [PubMed] [Google Scholar]
- 57.Shimizu H., Suzuki K., Watanabe M., Okamoto R. Stem cell-based therapy for inflammatory bowel disease. Intest Res. 2019;17:311–316. doi: 10.5217/ir.2019.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adolph T.E., Tomczak M.F., Niederreiter L., Ko H.J., Bock J., Martinez-Naves E., Glickman J.N., Tschurtschenthaler M., Hartwig J., Hosomi S., Flak M.B., Cusick J.L., Kohno K., Iwawaki T., Billmann-Born S., Raine T., Bharti R., Lucius R., Kweon M.N., Marciniak S.J., Choi A., Hagen S.J., Schreiber S., Rosenstiel P., Kaser A., Blumberg R.S. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Larabi A., Barnich N., Nguyen H.T.T. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy. 2020;16:38–51. doi: 10.1080/15548627.2019.1635384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bjarnason I., Scarpignato C., Holmgren E., Olszewski M., Rainsford K.D., Lanas A. Mechanisms of damage to the gastrointestinal tract from nonsteroidal anti-inflammatory drugs. Gastroenterology. 2018;154:500–514. doi: 10.1053/j.gastro.2017.10.049. [DOI] [PubMed] [Google Scholar]
- 62.Tanaka K., Suemasu S., Ishihara T., Tasaka Y., Arai Y., Mizushima T. Inhibition of both COX-1 and COX-2 and resulting decrease in the level of prostaglandins E2 is responsible for non-steroidal anti-inflammatory drug (NSAID)-dependent exacerbation of colitis. Eur J Pharmacol. 2009;603:120–132. doi: 10.1016/j.ejphar.2008.11.058. [DOI] [PubMed] [Google Scholar]
- 63.Miyoshi H., VanDussen K.L., Malvin N.P., Ryu S.H., Wang Y., Sonnek N.M., Lai C.W., Stappenbeck T.S. Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium. Embo J. 2017;36:5–24. doi: 10.15252/embj.201694660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olsen Hult L.T., Kleiveland C.R., Fosnes K., Jacobsen M., Lea T. EP receptor expression in human intestinal epithelium and localization relative to the stem cell zone of the crypts. PLoS One. 2011;6:e26816. doi: 10.1371/journal.pone.0026816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ding N.S., Hart A., De Cruz P. Systematic review: predicting and optimising response to anti-TNF therapy in Crohn’s disease—algorithm for practical management. Aliment Pharmacol Ther. 2016;43:30–51. doi: 10.1111/apt.13445. [DOI] [PubMed] [Google Scholar]
- 66.Vande Casteele N., Herfarth H., Katz J., Falck-Ytter Y., Singh S. American Gastroenterological Association Institute technical review on the role of therapeutic drug monitoring in the management of inflammatory bowel diseases. Gastroenterology. 2017;153:835–857.e836. doi: 10.1053/j.gastro.2017.07.031. [DOI] [PubMed] [Google Scholar]
- 67.Qiu Y., Chen B.L., Mao R., Zhang S.H., He Y., Zeng Z.R., Ben-Horin S., Chen M.H. Systematic review with meta-analysis: loss of response and requirement of anti-TNFα dose intensification in Crohn’s disease. J Gastroenterol. 2017;52:535–554. doi: 10.1007/s00535-017-1324-3. [DOI] [PubMed] [Google Scholar]
- 68.Mashukova A., Wald F.A., Salas P.J. Tumor necrosis factor alpha and inflammation disrupt the polarity complex in intestinal epithelial cells by a posttranslational mechanism. Mol Cell Biol. 2011;31:756–765. doi: 10.1128/MCB.00811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Al-Sadi R., Guo S., Ye D., Rawat M., Ma T.Y. TNF-alpha modulation of intestinal tight junction permeability is mediated by NIK/IKK-alpha axis activation of the canonical NF-kappaB pathway. Am J Pathol. 2016;186:1151–1165. doi: 10.1016/j.ajpath.2015.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McElroy S.J., Prince L.S., Weitkamp J.H., Reese J., Slaughter J.C., Polk D.B. Tumor necrosis factor receptor 1-dependent depletion of mucus in immature small intestine: a potential role in neonatal necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol. 2011;301:G656–G666. doi: 10.1152/ajpgi.00550.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma T.Y., Boivin M.A., Ye D., Pedram A., Said H.M. Mechanism of TNF-{alpha} modulation of Caco-2 intestinal epithelial tight junction barrier: role of myosin light-chain kinase protein expression. Am J Physiol Gastrointest Liver Physiol. 2005;288:G422–G430. doi: 10.1152/ajpgi.00412.2004. [DOI] [PubMed] [Google Scholar]
- 72.Hong S.N., Joung J.G., Bae J.S., Lee C.S., Koo J.S., Park S.J., Im J.P., Kim Y.S., Kim J.W., Park W.Y., Kim Y.H. RNA-seq reveals transcriptomic differences in inflamed and noninflamed intestinal mucosa of Crohn’s disease patients compared with normal mucosa of healthy controls. Inflamm Bowel Dis. 2017;23:1098–1108. doi: 10.1097/MIB.0000000000001066. [DOI] [PubMed] [Google Scholar]
- 73.Ye B.D., Jang B.I., Jeen Y.T., Lee K.M., Kim J.S., Yang S.K. Diagnostic guideline of Crohn’s disease. Korean J Gastroenterol. 2009;53:161–176. [PubMed] [Google Scholar]
- 74.Lahar N., Lei N.Y., Wang J., Jabaji Z., Tung S.C., Joshi V., Lewis M., Stelzner M., Martín M.G., Dunn J.C. Intestinal subepithelial myofibroblasts support in vitro and in vivo growth of human small intestinal epithelium. PLoS One. 2011;6:e26898. doi: 10.1371/journal.pone.0026898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kelly D., Kotliar M., Woo V., Jagannathan S., Whitt J., Moncivaiz J., Aronow B.J., Dubinsky M.C., Hyams J.S., Markowitz J.F., Baldassano R.N., Stephens M.C., Walters T.D., Kugathasan S., Haberman Y., Sundaram N., Rosen M.J., Helmrath M., Karns R., Barski A., Denson L.A., Alenghat T. Microbiota-sensitive epigenetic signature predicts inflammation in Crohn’s disease. JCI Insight. 2018;3:e122104. doi: 10.1172/jci.insight.122104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stelzner M., Helmrath M., Dunn J.C., Henning S.J., Houchen C.W., Kuo C., Lynch J., Li L., Magness S.T., Martin M.G., Wong M.H., Yu J. A nomenclature for intestinal in vitro cultures. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1359–G1363. doi: 10.1152/ajpgi.00493.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fujimichi Y., Otsuka K., Tomita M., Iwasaki T. An efficient intestinal organoid system of direct sorting to evaluate stem cell competition in vitro. Sci Rep. 2019;9:20297. doi: 10.1038/s41598-019-55824-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin J.Y., Lo K.Y., Sun Y.S. A microfluidics-based wound-healing assay for studying the effects of shear stresses, wound widths, and chemicals on the wound-healing process. Sci Rep. 2019;9:20016. doi: 10.1038/s41598-019-56753-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Franzen O., Gan L.M., Bjorkegren J.L.M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database (Oxford) 2019;2019:baz046. doi: 10.1093/database/baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]