Abstract
Oxidative stress-associated endothelial damage is the initiation factor of cardiovascular disease, and protein posttranslational modifications play critical roles in this process. Bcl-2-associated athanogene 3 (BAG3) is a molecular chaperone regulator of the BAG family, which interacts with various proteins and influences cell survival by activating multiple pathways. BAG3 undergoes posttranslational modifications; however, research evaluating BAG3 acetylation and its regulatory mechanism is lacking. In addition, the interacting protein and regulatory mechanism of BAG3 in oxidative stress-associated endothelial damage remain unclear. Here, key molecular interactions and protein modifications of BAG3 were identified in oxidative stress-associated endothelial damage. Endothelial-specific BAG3 knockout in the mouse model starkly enhances oxidative stress-associated endothelial damage and vascular remodeling, while BAG3 overexpression in mice significantly relieves this process. Mechanistically, poly(ADP-ribose) polymerase 1 (PARP1), causing oxidative stress, was identified as a novel physiological substrate of BAG3. Indeed, BAG3 binds to PARP1's BRCT domain to promote its ubiquitination (K249 residue) by enhancing the E3 ubiquitin ligase WWP2, which leads to proteasome-induced PARP1 degradation. Furthermore, we surprisingly found that BAG3 represents a new substrate of the acetyltransferase CREB-binding protein (CBP) and the deacetylase Sirtuin 2 (SIRT2) under physiological conditions. CBP/SIRT2 interacted with BAG3 and acetylated/deacetylated BAG3's K431 residue. Finally, deacetylated BAG3 promoted the ubiquitination of PARP1. This work reveals a novel regulatory system, with deacetylation-dependent regulation of BAG3 promoting PARP1 ubiquitination and degradation via enhancing WWP2, which is one possible mechanism to decrease vulnerability of oxidative stress in endothelial cells.
Keywords: BAG3, PARP1, Ubiquitination, WWP2, Acetylation, SIRT2
Abbreviations: BAG3, Bcl-2-associated athanogene 3; PARP1, poly ADP-ribose polymerase 1; CBP, CREB-binding protein; SIRT2, Sirtuin 2; ROS, reactive oxygen species; AngII, angiotensin II; BAG3-eKO, endothelial specific knockout; BAG3-TG, BAG3 transgenic; HE, hematoxylin and eosin; HUVECs, Human umbilical vein endothelial cells; DMEM, Dulbecco's modified Eagle medium; FBS, fetal bovine serum; SD, standard deviation; CHX, cycloheximide; P300, E1A-binding protein, 300 kDa; PCAF, P300/CBP-associated factor
Graphical abstract
Highlights
-
•
Endothelial-specific BAG3 knockout in mice aggravates oxidative stress endothelial injury.
-
•
BAG3 transgenic mice relieves oxidative stress endothelial injury.
-
•
BAG3 promotes ubiquitination at the K249 residue of PARP1 via mobilization of the E3 ubiquitin ligase WWP2.
-
•
CBP/SIRT2 interacted with BAG3 and acetylated/deacetylated BAG3's K431 residue.
-
•
Deacetylated BAG3 promoted the ubiquitination of PARP1.
1. Introduction
Cardiovascular diseases have become one of the main threats to human health, and vascular endothelial damage is the initial event in cardiovascular disease [[1], [2], [3], [4], [5]]. One of the most important causes of endothelial damage is oxidative stress-induced overactivation of PARP1, which promotes the consumption of NAD mainly from the cytosolic NAD+ pool and the shutdown of processes that require energy [[6], [7], [8]]. Currently, PARP1 inhibitors are used for the clinical treatment of cancer [9,10]; however, cardiovascular-related PARP1 inhibitors have poor clinical effects, as determined in a phase-2 evaluation [11], suggesting the regulatory mechanisms of PARP1 in cardiovascular disease are incompletely understood. Indeed, PARP1 undergoes post-translational modifications, and our previous research revealed that the Nedd4 family member WWP2 is the specific E3 ubiquitin ligase of PARP1 [12,13]. However, the precise mechanism of WWP2-PARP1 interaction remains unclear.
BAG3, a co-chaperone comprising a Bcl-2-associated athanogene (BAG) domain, belongs to the BAG protein family [[14], [15], [16]]. All the BAG protein family share a C-terminal conserved region, referred to as the BAG domain [17,18]. BAG3 interacts with many proteins and affects cell survival by activating multiple pathways, and it also plays important roles in apoptosis, cell adhesion, cytoskeleton remodeling and autophagy [[19], [20], [21]]. Previous evidences suggest that BAG3 is involved in regulating the contractile force and calcium homeostasis in ventricular myocytes [[22], [23], [24], [25], [26], [27]]. However, the function and regulatory mechanisms of BAG3 in oxidative stress-associated endothelial damage remain unclear. In addition, BAG3 undergoes post-translational modifications [28], but BAG3 acetylation and the associated regulatory mechanisms are undefined.
Here, a critical role for BAG3 in oxidative stress-associated endothelial damage was revealed. We firstly established BAG3 endothelial specific knockout mice and found significantly enhanced oxidative stress-associated vascular endothelial damage and post-injury remodeling. By contract, mice overexpressing BAG3 showed a relieved condition. Mechanistically, PARP1 was identified as a new BAG3 substrate under physiological conditions. BAG3 binds to PARP1's BRCT domain and promotes its ubiquitination (K249 residue) by enhancing the E3 ubiquitin ligase WWP2, which leads to proteasomal PARP1 degradation. Surprisingly, we found that BAG3 was a new substrate of the acetyltransferase CBP and the deacetylase SIRT2. CBP/SIRT2 interacted with BAG3 and acetylated/deacetylated the BAG3-K431 residue. Finally, deacetylated BAG3 could promote the ubiquitination of PARP1. Therefore, we can indicate that one possible mechanism by which BAG3 decreases vulnerability to oxidative stress in endothelial cells is represented by PARP1 ubiquitination and degradation.
2. Materials and methods
2.1. BAG3 knockout and overexpression in mice
Conditional vascular endothelium-specific genotypes, including TekCre+, BAG3Fl/Fl (BAG3-eKO) and TekCre−, BAG3Fl/Fl (BAG3-eWT), as well as BAG3-WT and BAG3-TG mice (CAG promoter) were obtained from Shanghai Model Organisms Science and Technology Development. BAG3 knockdown and overexpression efficiencies were assessed by immunoblot (Fig. 1E). Eight to ten-week old male specific pathogen-free (SPF) mice were examined in all assays. In the Ang II and NaCl infusion mouse model, BAG3-eWT, BAG3-eKO, BAG3-WT and BAG3-TG mice (6 animals/group) were randomly assigned to groups, administered anesthesia with 2% isoflurane in oxygen (1500 ml/min) by inhalation, underwent an incision in the middle scapular region and received subcutaneous implantation of an osmotic minipump (Alzet) as directed by the manufacturer. For endothelial damage and vascular remodeling induction, Ang II (1.5 mg/kg/day) was infused for 2 weeks via the ALZET minipump (model 2002) at 0.5 μl/h [12]. The mice underwent euthanasia by decapitation after isoflurane anesthesia. Assays involving animals had approval from the Animal Subjects Committee of China Medical University (permission number: 2019037), following the Guide for the Care and Use of Laboratory Animals outlined by the US National Institutes of Health (NIH Publication No. 85-23, revised 1985).
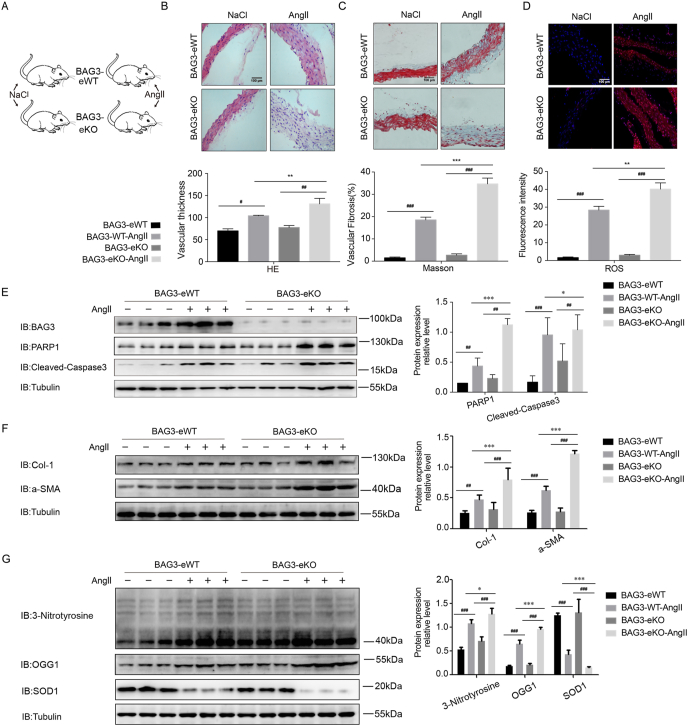
Fig. 1.
BAG3 endothelial specific knockout mice have aggravated Ang II-induced vascular endothelial damage and post-injury remodeling
(A) BAG3-eWT and BAG3-eKO mice were treated without or with Ang II (1.5 mg/kg/day) by the subcutaneous route at 0.5 μl/h for 14 days. Euthanasia was carried out by cervical dislocation after the 14-day treatment. (B) H&E, (C) Masson and (D) DHE staining was carried out for assessing vascular thickening, fibrosis and oxidative stress injury, respectively. Scale bar, 100 μm. Data are mean ± SD (n = 6; ***P < 0.001, two-way ANOVA with post hoc Bonferroni test). (E) Vascular tissue samples from BAG3-eWT and BAG3-eKO mice infused without or with Ang II for 14 days were examined by immunoblot for BAG3, PARP1 and cleaved caspase-3. Data are mean ± SD (n = 6; **P < 0.01, two-way ANOVA with post hoc Bonferroni test). (F) Col-1 and a-SMA protein amounts assessed by immunoblot. Data are mean ± SD (n = 6; **P < 0.01, two-way ANOVA with post hoc Bonferroni test). (G) 3-Nitrotyrosine, OGG1 and SOD1 protein amounts assessed by immunoblot. Data are mean ± SD (n = 6; **P < 0.01, two-way ANOVA with post hoc Bonferroni test).
2.2. Histopathology
Vascular tissue samples underwent formalin (4%) fixation for 4 h, and were embedded in paraffin and sectioned at 5 μm. This was followed by xylene dewaxing, rehydration with graded ethanol, and staining with H&E and Masson's trichrome reagent (G1340; Solarbio, China). Frozen vascular tissue samples were sectioned and stained with dihydroethidium (DHE) (C1300-2, Applygen Technologies, China).
2.3. Cell treatments
Human umbilical vein endothelial cell (HUVEC) culture was carried out in F-12K medium containing 10% fetal bovine serum (FBS) (HyClone), heparin (0.1 mg/ml) and ECGS (0.05 mg/ml). HEK293T cell culture used high-glucose Dulbecco's modified eagle medium (DMEM) containing 10% FBS (HyClone). HEK293T cells and HUVECs were provided by ATCC (a nonprofit organization that collects, stores, and distributes standard reference cell lines) and grown in a humid environment with 5% CO2 at 37 °C.
2.4. ROS quantitation HUVECs
Supplementary Table 1 includes the ROS Assay Kits applied in this study (S0033; Beyotime Biotechnology, China). An ROS Assay Kit was utilized for intracellular ROS detection. Post-treatment, PBS-washed HUVECs underwent incubation with 2,7-DCFH-DA (10 mM) in serum-free medium for 1 h at 37 °C in the dark. After washing with PBS, an Olympus fluorescence microscope was utilized for analysis (excitation at 595 nm).
2.5. TUNEL of HUVECs
The TUNEL assay was carried out using a kit obtained from Jiangsu KeyGEN Biotech (China) as directed by the product's booklet. Post-treatment, HUVECs underwent fixation (4% formalin for 20 min), permeabilization (1% Triton X-100 for 10 min) and DNase I treatment (37 °C for 30 min). Next, the specimens were successively incubated with terminal deoxynucleotidyl transferase (TDT, 60 min) and streptavidin-fluorescein-labeled droplets (30 min) at 37 °C shielded from light. DAPI counterstaining was carried out for 10 min at ambient also in the dark, followed by analysis under an Olympus fluorescence microscope (excitation at 488 nm).
2.6. Plasmids and other reagents
Supplementary Table 2 shows all constructs applied in this work. Lipofectamine 3000 (Invitrogen, USA), Highgene (Applygen, China) and jetPRIME (Polyplus, France) were utilized, respectively, for transfection as directed by each manufacturer. shRNAs underwent transfection using a lentiviral vector. Supplementary Table 1 summarizes the antibodies applied in this study. The proteasome suppressor MG132 (A2585; 20 μM in DMSO) and cycloheximide (CHX, GC17198; 100 μM in DMSO) were from GlpBio (USA). Ang II (A9525; Sigma, USA) was utilized at 10 μM in DMSO.
2.7. PARP1 ubiquitination analysis
Cell lysis was carried out with 1% SDS buffer (Tris pH7.5 containing EDTA [0.5 mM] and DTT [1 mM]) by a 10-min boiling. Before lysis, cells underwent transfection for 48 h with HA-linked (HA-ubiquitin), full-length human Myc-PARP1, mutant Myc-PARP1 and additional plasmids, respectively. The lysates underwent successive incubations with anti-Myc (B26302; Biotool) immunoprecipitation magnetic beads (12 h at 4 °C). Immunoprecipitation of endogenous proteins was performed by using anti-PARP1 antibodies (1 μg/mg of cell lysate, 4 °C) for 2–3 h and protein A/G (30 μl) immunoprecipitation magnetic beads (12 h at 4 °C). Ubiquitinated PARP1 was detected with anti-HA antibodies.
2.8. Co-immunoprecipitation and immunoblot
Mouse vascular tissue samples and cells underwent lysis with Flag lysis buffer (Thermo Fisher Scientific, USA) containing protease inhibitors (B14002, Bimake). Then, lysates mixed with 30 μl of anti-Flag/Myc Affinity Gel (B23102/B26302; Biotool) were incubated for 12 h at 4 °C. Immunoprecipitation of endogenous proteins was performed by using respective antibodies (1 μg of antibody/mg of lysate) for 2–3 h and protein A/G (30 μl) immunoprecipitation magnetic beads for 12 h at 4 °C. Separation of immunoprecipitation complexes utilized SDS-PAGE. Next, electro-transfer onto PVDF membranes was performed. Then, the samples were incubated with 5% bovine serum albumin (1 h at room temperature) and underwent successive incubations with primary (4 °C, overnight) and secondary (room temperature, 1 h) antibodies. Protein expression was quantitated with Image J v1.46 (National Institutes of Health, USA); normalization was performed with GAPDH and Tubulin.
2.9. Identification of BAG3 interacting proteins
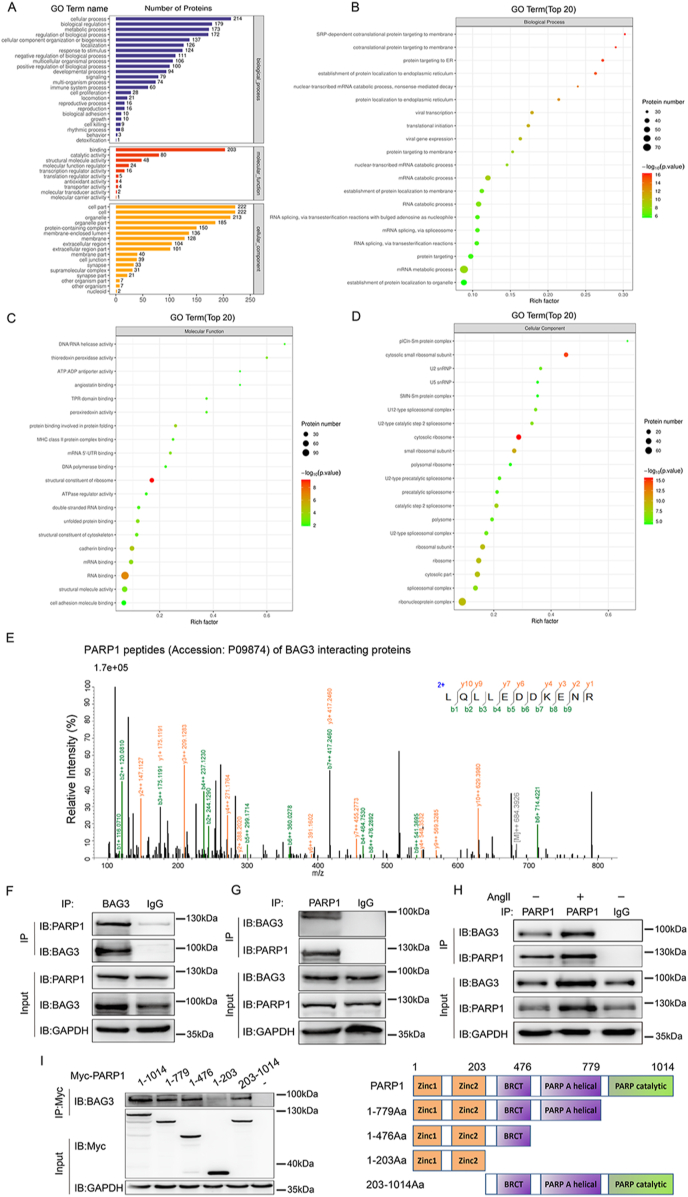
BAG3 expression system are HEK293T cells and the cells underwent transfection for 48 h with full-length human Flag-BAG3 (Vector: GV219; TAG: 3Flag; Species: Human) and cells underwent lysis with lysis buffer (Thermo Fisher Scientific, USA) containing protease inhibitors (B14002, Bimake). Then, lysates mixed with 30 μl of anti-Flag Affinity Gel (B23102; Biotool) were incubated for 12 h at 4 °C. And separation of immunoprecipitation complexes utilized SDS-PAGE. Next, the sample was electrophoresed and cut the gel for mass spectrometry (Thermo Scientific ™ Q Exactive HF-X) to identify the proteins interacting with Flag-BAG3, which was completed in Applied Protein Technology Co., Ltd., (Shanghai, China). The names and peptides of proteins interacting with Flag-BAG3 are shown in Supplementary Table 3 and Fig. 3A–D.
Fig. 3.
Mass spectrometry analysis of BAG3-interacting proteins, identifying PARP1 as a new BAG3 substrate
(A) Mass spectrometry-detected BAG3-interacting proteins in HEK293T cells were assessed by Gene Ontology enrichment, with the most significant pathways related to protein binding. **P < 0.01, Fisher's exact test. (B-D) Mass spectrometry-detected BAG3-interacting proteins in HEK293T cells were assessed for Biological Process, Molecular Function and Cellular Component enrichment to screen physiological substrates of BAG3. **P < 0.01, Fisher's exact test. (E) The spectrograms showed mass spectroscopy-identified PARP1 peptides (Accession: P09874; Description: Poly [ADP-ribose] polymerase 1 OS=Homo sapiens OX = 9606 GN = PARP1 PE = 1 SV = 4) of BAG3 interacting proteins in HEK293T cells. (F–G) The interaction of endogenous BAG3 with PARP1 in HUVECs was assessed by immunoprecipitation with the indicated antibodies. (H) HUVECs were treated without or with Ang II, and BAG3's interaction with PARP1 was examined by immunoprecipitation with anti-PARP1 antibody and by Western blot with anti-BAG3 antibody. (I) Full-length Myc-PARP1 or a truncated Myc-PARP1 plasmid was transfected into HUVECs, and total lysate was examined by immunoprecipitation using anti-Myc antibodies, with subsequent immunoblot utilizing anti-BAG3 antibodies.
2.10. Identification of SIRT2 interacting proteins
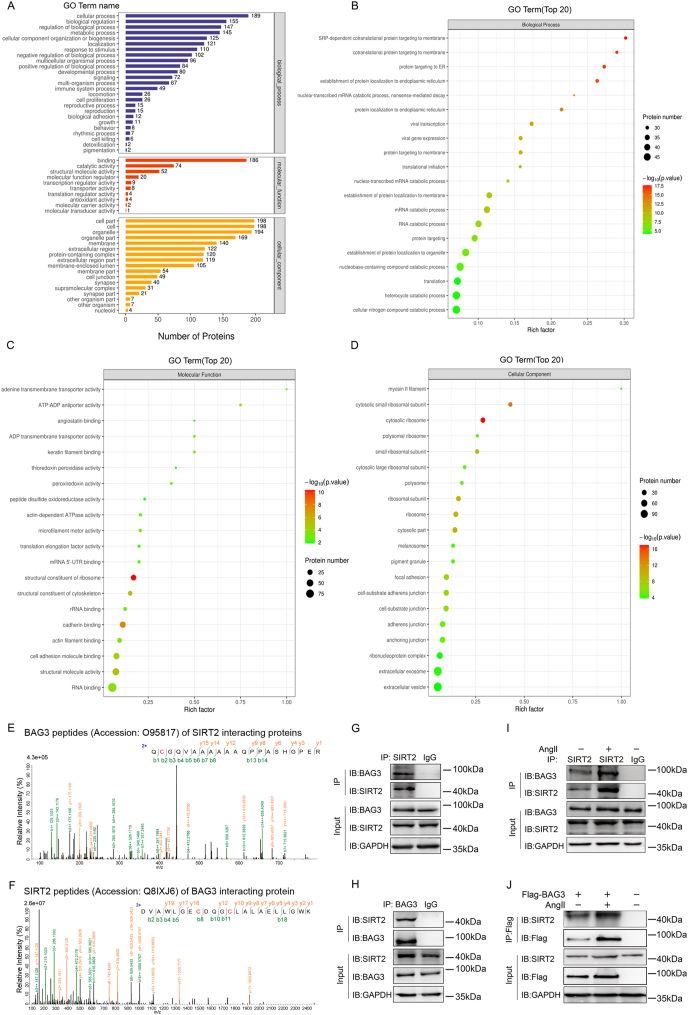
SIRT2 expression system are HEK293T cells and the cells underwent transfection for 48 h with full-length human Myc-SIRT2 (Vector: PCMV; TAG: Myc; Species: Human) and cells underwent lysis with lysis buffer (Thermo Fisher Scientific, USA) containing protease inhibitors (B14002, Bimake). Then, lysates mixed with 30 μl of anti-Myc Affinity Gel (B26302; Biotool) were incubated for 12 h at 4 °C. And separation of immunoprecipitation complexes utilized SDS-PAGE. Next, the sample was electrophoresed and cut the gel for mass spectrometry (Thermo Scientific ™ Q Exactive HF-X) to identify the proteins interacting with Myc-SIRT2, which was completed in Applied Protein Technology Co., Ltd., (Shanghai, China). The names and peptides of proteins interacting with Myc-SIRT2 are shown in Supplementary Table 4 and Fig. 5A–D.
Fig. 5.
Mass spectrometry identifies BAG3 as a new SIRT2 substrate
(A) Mass spectrometry analysis to identify SIRT2-interacting proteins in HEK293T cells. The resulting SIRT2-interacting proteins were assessed by Gene Ontology enrichment analysis. The pathways with highest significance were related to protein binding. **P < 0.01, Fisher's exact test. (B-D) Mass spectrometry-detected SIRT2 interacting proteins in HEK293T cells were assessed for Biological Process, Molecular Function and Cellular Component enrichment analyses to screen physiological substrates of SIRT2. **P < 0.01, Fisher's exact test. (E) The spectrograms showed mass spectroscopy-identified BAG3 peptides (Accession: O95817; Description: BAG family molecular chaperone regulator 3 OS=Homo sapiens OX = 9606 GN = BAG3 PE = 1 SV = 3) of SIRT2 interacting proteins in HEK293T cells. (F) The spectrograms showed mass spectroscopy-identified SIRT2 peptides (Accession: Q8IXJ6; Description: NAD-dependent protein deacetylase sirtuin-2 OS=Homo sapiens OX = 9606 GN = SIRT2 PE = 1 S V = 2) of BAG3 interacting proteins in HEK293T cells. (G-H) Interaction of endogenous SIRT2 and BAG3 in HUVECs determined by immunoprecipitation with the indicated antibodies. (I) HUVECs underwent incubation without or with Ang II. Then, BAG3's interaction with SIRT2 was determined by immunoprecipitation with anti-SIRT2 antibodies, followed by immunoblot with anti-BAG3 antibodies. (J) Interaction between BAG3 and SIRT2 in presence or absence of Ang II determined by immunoprecipitation with anti-Flag-beads in HUVECs, followed by immunoblot with anti-SIRT2 antibodies.
2.11. Statistical analysis
Data are mean ± standard deviation (SD). The F- and Brown–Forsythe tests were carried out to examine homogeneity of variance for two and ≥3 groups, respectively. The Shapiro–Wilk test was utilized to assess whether the data had normal or skewed distribution. Student's t-test and Welch's t-test were performed to compare parameters showing equal and unequal variances, respectively (2 groups). One-way and two-way ANOVA were carried out for comparing one and two factors among multiple groups, respectively, followed by the Bonferroni test. Data were analyzed with SPSS 22.0 (SPSS, USA). P < 0.05 indicated statistical significance.
3. Results
3.1. BAG3 endothelial specific knockout mice show significantly aggravated Ang II-associated vascular endothelial damage and post-injury remodeling
BAG3 is a molecular chaperone regulator of the BAG family, which interacts with various proteins and influences cell survival by activating multiple pathways [[19], [20], [21], [22], [23], [24]]. Previous studies have been shown that BAG3 is highly up-regulated in many tumor cells in the context of oxidative stress, and increased BAG3 plays an important role in the occurrence and development of tumors [[29], [30], [31]]. Oxidative stress is an initiation factor of vascular endothelial damage, however, the role of BAG3 in oxidative stress-associated endothelial damage remain unclear. Therefore, we firstly examined BAG3's function during oxidative stress-related mouse vascular endothelial damage. We generated BAG3 endothelial specific knockout (BAG3-eKO) mice (Fig. 1A), and found that compared with BAG3-eWT mice, BAG3-eKO counterparts displayed markedly enhanced Ang II-related vascular endothelial damage and post-injury remodeling, including vascular thickening (Fig. 1B), vascular fibrosis (Fig. 1C), and vascular ROS generation (Fig. 1D). Additionally, compared with BAG3-eWT mice, BAG3-eKO mice showed remarkably elevated levels of the Ang II-induced vascular damage proteins PARP1 and cleaved caspase-3. And we found that a more striking difference in cleaved caspase-3 levels was related to control tissues without Ang II treatment. As cleaved caspase-3 is a very important marker of apoptosis, which indicated that BAG3-eKO mice were also undergoing apoptosis without Ang II treatment (Fig. 1E). Compared with BAG3-eWT mice, BAG3-eKO mice showed remarkably elevated levels of the Ang II-induced vascular fibrosis proteins Col-1 and a-SMA (Fig. 1F). In addition, compared with BAG3-eWT mice, BAG3-eKO mice showed remarkably elevated amounts of the Ang II-related oxidative stress factors 3-nitrotyrosine and OGG1. Furthermore, our experimental data showed an important result that compared with BAG3-eWT mice, the amounts of the Ang II-related antioxidative stress protein SOD1 were marked reduced in BAG3-eKO mice. As SOD1 is a very important antioxidant enzyme, which further indicated that BAG3-eKO decreased antioxidative capability in mice (Fig. 1G).
These findings revealed that BAG3 protected against vascular endothelial damage and post-injury remodeling, and BAG3-eKO mice had aggravated endothelial damage and post-injury vascular remodeling.
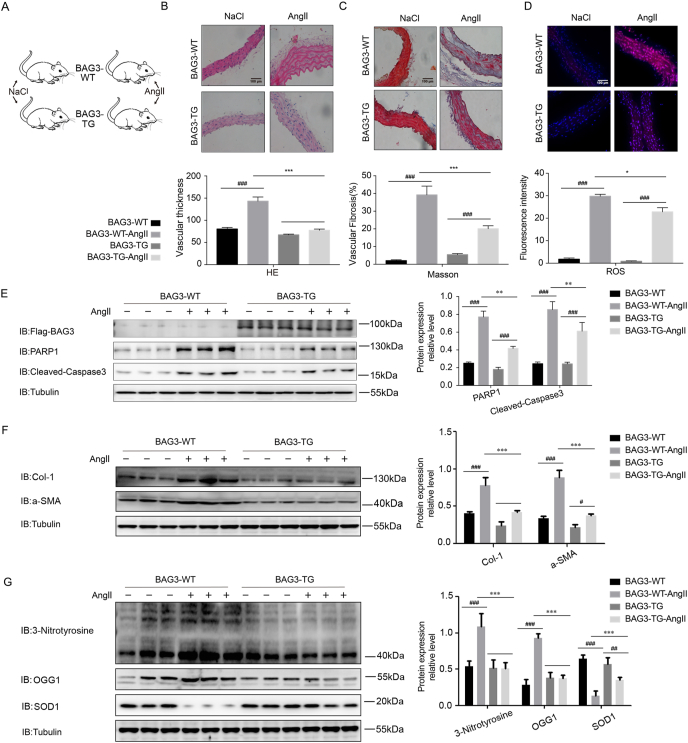
3.2. BAG3 transgenic mice show significantly relieved Ang II-induced vascular endothelial damage and post-injury remodeling
Next, we tested the effect of BAG3 overexpression in the mouse model of vascular endothelial damage and post-injury remodeling. Therefore, we constructed BAG3 transgenic (BAG3-TG) mice (Fig. 2A). The results showed that compared with BAG3-WT mice, BAG3-TG mice had significantly alleviated vascular thickening (Fig. 2B), vascular fibrosis (Fig. 2C), and vascular ROS generation (Fig. 2D) after Ang II-induced vascular endothelial damage. Additionally, compared with BAG3-WT mice, BAG3-TG mice had significantly reduced the levels of Ang II-induced vascular damage factors PARP1 and cleaved caspase-3 (Fig. 2E). Compared with BAG3-WT mice, BAG3-TG mice had significantly reduced levels of Ang II-induced vascular fibrosis proteins Col-1 and a-SMA (Fig. 2F). In addition, compared with BAG3-WT mice, BAG3-TG mice had significantly reduced levels of 3-nitrotyrosine and OGG1, and remarkably increased levels of SOD1 (Fig. 2G).
Fig. 2.
BAG3 transgenic mice show relieved Ang II-associated vascular endothelial damage and post-injury remodeling
(A) BAG3-WT and BAG3-TG mice were administered Ang II (1.5 mg/kg/day) or not by the subcutaneous route at 0.5 μl/h for 14 days. Euthanasia was carried out by cervical dislocation after the 14-day treatment. B) H&E, (C) Masson and (D) DHE staining was carried out for assessing vascular thickening, fibrosis and oxidative stress injury, respectively. Scale bar, 100 μm. Data are mean ± SD (n = 6; ***P < 0.001, two-way ANOVA with post hoc Bonferroni test). (E) Vascular tissue samples from BAG3-WT and BAG3-TG mice infused without or with Ang II for 14 days were examined by immunoblot for BAG3, PARP1 and cleaved caspase-3. Data are mean ± SD (n = 6; **P < 0.01, two-way ANOVA with post hoc Bonferroni test). (F) Col-1 and a-SMA protein amounts assessed by immunoblot. Data are mean ± SD (n = 6; **P < 0.01, two-way ANOVA with post hoc Bonferroni test). (G) 3-Nitrotyrosine, OGG1 and SOD1 protein amounts assessed by immunoblot. Data are mean ± SD (n = 6; **P < 0.01, two-way ANOVA with post hoc Bonferroni test).
Thus, we further proved that BAG3 was a protective factor against vascular endothelial damage and post-injury remodeling, and BAG3-TG mice had alleviated endothelial damage and remodeling after injury.
3.3. Mass spectrometry analysis of BAG3-interacting proteins reveals PARP1 as a new BAG3 substrate
After clarifying the function of BAG3 in vascular endothelial damage and post-injury remodeling, we next explored the key regulatory mechanism by which BAG3 affects endothelial dysfunction. Therefore, HEK293T cells underwent transfection for full-length human Flag-BAG3 (Vector: GV219; TAG: 3Flag; Species: Human). And co-immunoprecipitation was carried out to identify the proteins interacting with BAG3 by mass spectrometry. Some representative names and peptides of proteins interacting with BAG3 were shown in Supplementary Table 3. And according to the BAG3 interacting proteins identified by mass spectrometry, Gene Ontology, Biological Process, Molecular Function and Cellular Component enrichment analyses were shown in Fig. 3A–D. Interestingly, we found the oxidative stress injury protein PARP1 maybe a new key potential interacting protein of BAG3 (Supplementary Table 3). Therefore, we extracted the spectrograms, which showed PARP1 peptides identified by mass spectrometry (Accession: P09874; Description: Poly [ADP-ribose] polymerase 1 OS=Homo sapiens OX = 9606 GN = PARP1 PE = 1 SV = 4) of BAG3 interacting proteins (Fig. 3E).
Next, we verified the interaction between BAG3 and PARP1 through biological experiments. Endogenous immunoprecipitation was demonstrated by using anti-BAG3 antibody to verify the BAG3's interaction with PARP1 (Fig. 3F). In addition, endogenous immunoprecipitation was demonstrated by using anti-PARP1 antibody to verify the PARP1's interaction with BAG3 (Fig. 3G). Moreover, we determined that the interaction between BAG3 and PARP1 is maintained in presence of Ang II (Fig. 3H).
Furthermore, we identified which domain of PARP1 interacting with BAG3. Common domains of PARP1 included Zinc1-Zinc2 domain, BRCT domain, PARP A helical domain and PARP catalytic domain. Our results showed that BAG3 bound to the BRCT domain of PARP1 (PARP1-BRCT) (Fig. 3I).
These results suggested PARP1 as a novel interacting protein of BAG3, which bound to the BRCT domain of PARP1 and may be involved in vascular endothelial damage by regulating PARP1.
3.3.1. BAG3 promotes proteasome pathway degradation of PARP1 by polyubiquitination modification
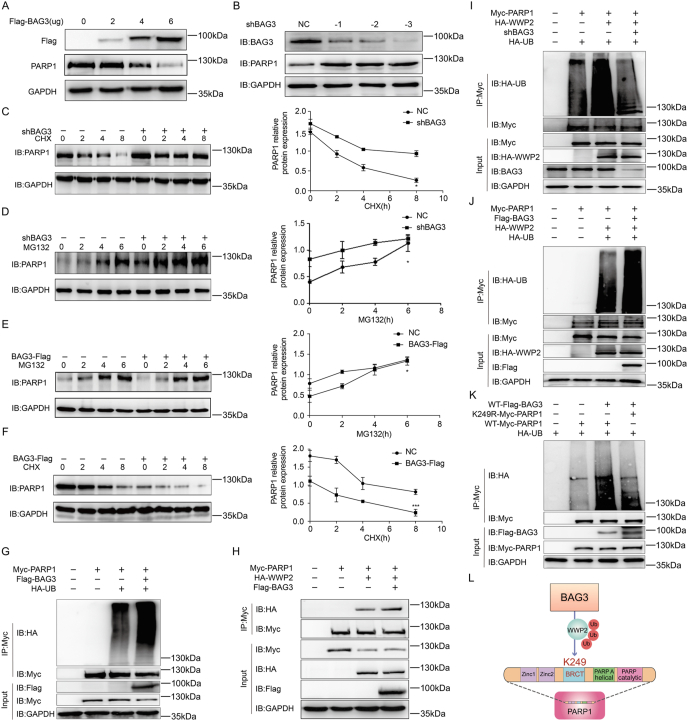
We next explored the key regulatory mechanism of BAG3's effect on PARP1. Indeed, an increasing gradient of BAG3 overexpression was associated with a progressive reduction of endogenous PARP1 levels (Fig. 4A). Moreover, endogenous PARP1 amounts were increased after BAG3 knockdown in HUVECs (Fig. 4B). Since the shBAG3 -3 fragment yielded the greatest knockdown efficiency, all subsequent experiments used stable knockdown cell lines generated based on this shBAG3 fragment. These results confirmed that BAG3 could promote the degradation of PARP1.
Fig. 4.
BAG3 induces PARP1-K249 ubiquitination by WWP2
(A) Varying amounts of Flag-BAG3 plasmid were transfected into HUVECs, and PARP1 expression in these cells was detected by Western blot. (B) BAG3 expression levels were measured in NC, shBAG3 -1, shBAG3 -2, and shBAG3 -3 lentivirus HUVECs. (C) NC or shBAG3 HUVECs underwent subsequent treatment with CHX for different lengths of time, and the degradation speed (slope) of PARP1 was determined by immunoblot. Experiments were repeated thrice; *P < 0.05, two-way ANOVA with post hoc Bonferroni test. (D) NC or shBAG3 HUVECs underwent subsequent treatment with MG132 for different lengths of time, and the rate and slope of PARP1 protein accumulation were determined by immunoblot. Experiments were repeated thrice; *P < 0.05, two-way ANOVA with post hoc Bonferroni test. (E) Overexpressing Flag-control or Flag-BAG3 HUVECs were administered MG132 for different lengths of time, and the rate and slope of PARP1 protein accumulation were determined by immunoblot. Experiments were repeated thrice; *P < 0.05, two-way ANOVA with post hoc Bonferroni test. (F) Overexpressing Flag-control or Flag-BAG3 HUVECs were administered CHX for different lengths of time, and the degradation speed (slope) of PARP1 was determined by immunoblot. Experiments were repeated thrice; ***P < 0.001, two-way ANOVA with post hoc Bonferroni test. (G) HUVECs were co-transfected with Myc-PARP1, Flag-BAG3 and HA-UB, and ubiquitinated PARP1 underwent immunoprecipitation with anti-Myc antibodies, followed by immunoblot detection with anti-HA antibodies. (H) HUVECs were co-transfected with Myc-PARP1, HA-WWP2 and Flag-BAG3, and interacting PARP1 and WWP2 were immunoprecipitated with anti-Myc antibodies, followed by immunoblot detection with anti-HA antibodies. (I) NC or shBAG3 HUVECs were co-transfected with Myc-PARP1 and HA-WWP2, then transfected with HA-UB. Ubiquitinated PARP1 underwent immunoprecipitation with anti-Myc antibodies, followed by immunoblot detection with anti-HA antibodies. (J) HUVECs were co-transfected with Myc-PARP1, Flag-BAG3 and HA-WWP2, then transfected with HA-UB. Ubiquitinated PARP1 underwent immunoprecipitation with anti-Myc antibodies, followed by immunoblot detection with anti-HA antibodies. (K) HUVECs were co-transfected with Flag-WT-BAG3, Myc-PARP1-K249R, or Myc-WT-PARP1 and HA-UB. Ubiquitinated PARP1 underwent immunoprecipitation with anti-Myc antibodies, followed by immunoblot detection with anti-HA antibodies. (L) Schematic model showing the potential function of BAG3 in promoting PARP1 ubiquitination by enhancing the E3 ubiquitin ligase WWP2.
To determine whether BAG3 affects PARP1 degradation by suppressing PARP1 synthesis or inducing PARP1 proteasome degradation, we analyzed the effects of CHX and MG132 (protein synthesis and proteasome inhibitors, respectively). NC cells displayed substantially higher degradation speed (slope) of PARP1 after CHX treatment, whereas the shBAG3 group maintained a low degradation speed of PARP1 over time (Fig. 4C). In addition, PARP1 protein accumulation was faster upon MG132 administration in NC cells in comparison with shBAG3 cells (Fig. 4D). Furthermore, both the rate and slope of PARP1 protein accumulation were lower in the BAG3-overexpressing group after MG132 treatment in comparison with the Flag control group (Fig. 4E). Additionally, the degradation speed (slope) of PARP1 were higher in the BAG3-overexpression group after CHX treatment, compared with the Flag control group (Fig. 4F). We next confirmed that exogenous expression of BAG3 promoted the ubiquitination of PARP1 (Fig. 4G), which was consistent with PARP1 degradation via the proteasome.
The above findings indicated PARP1 was a new interacting protein of BAG3, which bound to its BRCT domain and induced proteasomal PARP1 degradation.
3.3.2. BAG3 enhances PARP1-K249 ubiquitination by WWP2
BAG3, a co-chaperone protein, has no ability to directly induce the polyubiquitination of PARP1. Therefore, BAG3 would enhance a E3 ubiquitin ligase, which promotes PARP1 polyubiquitination, and subsequently induce proteasome pathway degradation of PARP1. Our previous research revealed that WWP2 is the key E3 ubiquitination ligase of PARP1 [12,13]. Therefore, there might be a potential functional relationship between BAG3 and WWP2, that is, BAG3 promotes the polyubiquitination and degradation of PARP1 by WWP2. Indeed, this study found that compared with the control cells, the level of interaction between WWP2 and PARP1 was increased in cells that were transfected with Flag-BAG3, confirming that BAG3 regulates the interaction between E3 ubiquitination ligase WWP2 and PARP1 (Fig. 4H). Consistent with the above results, the level of PARP1 ubiquitination by WWP2 was low in shBAG3 cells (Fig. 4I) and higher in cells overexpressing exogenous BAG3 (Fig. 4J).
In order to obtain direct evidence that BAG3 promotes PARP1 ubiquitination by WWP2, we sought to identify the ubiquitination sites of PARP1 modulated by BAG3. We found BAG3 enhanced the ubiquitination level of PARP1-WT but bot PARP1-K249R, suggesting BAG3 induced PARP1 ubiquitination through K249 (Fig. 4K).
The above results thus far showed that BAG3 interacts with the BRCT domain of PARP1 and induces PARP1 ubiquitination at K249 through WWP2. These findings provide us with novel insights to find new BAG3 regulatory pathways (Fig. 4L).
3.4. Mass spectrometry analysis of SIRT2-interacting proteins reveals BAG3 as a new SIRT2 substrate
After confirming that BAG3 interacts with the BRCT domain of PARP1 and induces PARP1 ubiquitination at K249 through WWP2, we next explored the mechanism by which BAG3 is regulated to exert its key regulatory effects. It is known that BAG3 undergoes posttranslational modifications [28]; however, studies evaluating BAG3 acetylation and the associated regulatory mechanism are lacking.
Therefore, we continue to search the representative names and peptides of proteins interacting with BAG3 in Supplementary Table 3. Surprisingly, we found that SIRT2, a key enzyme in protein deacetylation modification, may interact with BAG3 (Supplementary Table 3).
As a deacetylase, SIRT2 can deacetylation regulate physiological substrates and affect biological function of its downstream proteins [[32], [33], [34]]. Next, HEK293T cells underwent transfection for full-length human Myc-SIRT2 (Vector: PCMV; TAG: Myc; Species: Human). And co-immunoprecipitation was carried out to identify the proteins interacting with SIRT2 by mass spectrometry. Some representative names and peptides of proteins interacting with SIRT2 were shown in Supplementary Table 4. And according to the SIRT2 interacting proteins identified by mass spectrometry, Gene Ontology, Biological Process, Molecular Function and Cellular Component enrichment analyses were shown in Fig. 5A–D. Interestingly, we also found BAG3 maybe a new key potential interacting protein of SIRT2 (Supplementary Table 4). Therefore, we extracted the spectrograms, which showed BAG3 peptides identified by mass spectrometry (Accession: O95817; Description: BAG family molecular chaperone regulator 3 OS=Homo sapiens OX = 9606 GN = BAG3 PE = 1 SV = 3) of SIRT2 interacting proteins (Fig. 5E). Convincingly, we also extracted the spectrograms (Supplementary Table 3), which showed SIRT2 peptides identified by mass spectrometry (Accession: Q8IXJ6; Description: NAD-dependent protein deacetylase sirtuin-2 OS=Homo sapiens OX = 9606 GN = SIRT2 PE = 1 SV = 2) of BAG3 interacting proteins (Fig. 5F).
Furthermore, we verified the interaction between SIRT2 and BAG3 through biological experiments. This interaction was demonstrated by co-immunoprecipitation of endogenous SIRT2 and BAG3 (Fig. 5G). And this interaction was also demonstrated by co-immunoprecipitation of endogenous BAG3 and SIRT2 (Fig. 5H). Moreover, we determined that the interaction between SIRT2 and BAG3 is maintained in presence of Ang II (Fig. 5I–J).
3.4.1. SIRT2/CBP regulates the acetylation state of BAG3
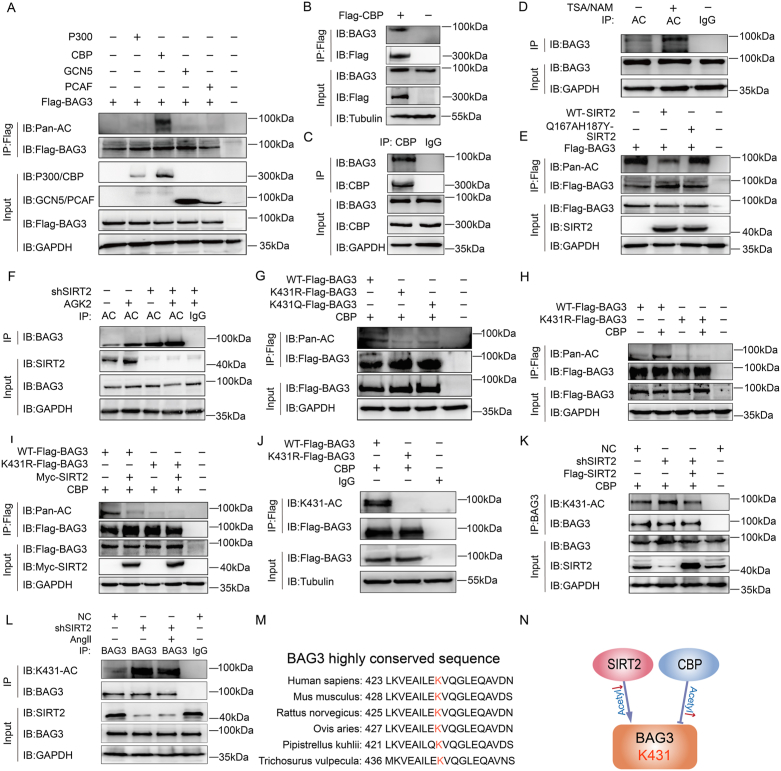
The identification of BAG3 as a SIRT2 interaction partner prompted us to identify factors involved in the regulation of BAG3 acetylation. Acetyltransferase is an enzyme that acetylates histone, weakens the tight binding ability between histone and DNA and promotes transcription [35,36]. Protein acetylation was first found to occur mainly on histones in the nucleus and participate in the regulation of gene transcription. With the increase of the sensitivity of protein mass spectrometry, more non histone proteins were found to undergo acetylation modification. According to previous studies, in mammalian cells, acetyltransferases mainly include P300 (E1A-binding protein, 300 kDa), CREB-binding protein (CBP), P300/CBP-associated factor (PCAF) and GCN5 (KAT2A) [37,38]. Therefore, to identify the responsible acetyltransferase of BAG3, we separately transfected four acetyltransferases, i.e., P300, CBP, PCAF and GCN5 into cells.
Overexpression of only CBP among the four remarkably increased the acetylation level of BAG3 (Fig. 6A). Moreover, both exogenous and endogenous CBP interacted with BAG3 (Fig. 6B–C). These results confirmed that BAG3 was acetylated by the CBP acetyltransferase. Next, we found that the joint administration of trichostatin A (TSA) and nicotinamide (NAM), which are common deacetylase suppressors repressing histone deacetylases (HDACs), including HDAC I and III, as well as Sirtuin-related deacetylases, increased BAG3's acetylation levels (Fig. 6D).
Fig. 6.
SIRT2 and CBP deacetylates and acetylates BAG3, respectively, on BAG3-K431
(A) HUVECs were co-transfected with Flag-P300, Flag-GCN5, Flag-CBP, or Flag-P300/CBP-associated factor (PCAF) and Flag-BAG3, and BAG3 acetylation levels were determined by immunoprecipitation with anti-Pan-AC antibodies. (B) The interaction between exogenous Flag-CBP and endogenous BAG3 in HUVECs was detected by immunoprecipitation, followed by immunoblot with anti-BAG3 antibodies. (C) The interaction between endogenous CBP and BAG3 in HUVECs was assessed by immunoprecipitation, followed by immunoblot. (D) Nicotinamide (NAM) (5 mM, 4 h) and trichostatin A (TSA) (0.5 μM, 16 h) were used to improve protein acetylation levels, and acetylated BAG3 in HUVECs was immunoprecipitated with anti-BAG3 antibodies. (E) HUVECs were co-transfected with Flag-WT-SIRT2 or Flag-SIRT2-Q167AH187Y (Mut) and Flag-BAG3, and BAG3 acetylation was detected by immunoprecipitation using anti-Pan-AC antibodies. (F) Normal control (NC) or shSIRT2 HUVECs underwent subsequent incubation with or without AGK2 (10 μM, 24 h), and acetylated BAG3 underwent immunoprecipitation with anti-BAG3 antibodies. (G) HUVECs were co-transfected with Flag-WT-BAG3, Flag-BAG3-K431R, or Flag-BAG3-K431Q and Flag-CBP, and acetylated BAG3 underwent immunoprecipitation with anti-Pan-AC antibodies. (H) HUVECs were co-transfected with BAG3-WT or BAG3-K431R and Flag-CBP, and acetylated BAG3 underwent immunoprecipitation with anti-Pan-AC antibodies. (I) HUVECs co-transfected with Flag-WT-BAG3, Flag-BAG3-K431R, or Myc-SIRT2 and Flag-CBP, and acetylated Flag-BAG3 was detected by immunoprecipitation with anti-Pan-AC antibodies. (J) Flag-WT-BAG3 or Flag-BAG3-K431R was individually transfected in HUVECs, and immunoprecipitated Flag-BAG3 was assessed with site-specific K431 acetylation antibodies against BAG3 (BAG3-K431AC). (K) NC or shSIRT2 HUVECs underwent subsequent transfection with Flag-SIRT2, and BAG3 acetylation was examined by immunoprecipitation with anti-BAG3-K431AC antibodies. (L) ShSIRT2 HUVECs were constructed, then treated without or with Ang II as indicated. BAG3 acetylation was assessed by immunoprecipitation with anti-BAG3 antibodies, followed by immunoblot with anti-BAG3-K431AC antibodies. (M) Alignment of sequences surrounding K431 in BAG3 homologs from various species. The acetylated lysine residue at BAG3-431 is depicted as bold and red. (N) Schematic model showing the potential function of the SIRT2 deacetylase in regulating BAG3. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We next tested whether BAG3 is a direct substrate of SIRT2. Overexpression of SIRT2-WT decreased exogenous BAG3 acetylation, while a catalytically inactive SIRT2 mutant (Q167AH187Y) did not affects cells upon transfection (Fig. 6E). We then examined normal control (NC)- and short hairpin (sh) SIRT2-treated cells, which were incubated with or without 20 μmol/L AGK2, a commonly used SIRT2-specific inhibitor. BAG3 acetylation levels were higher in shSIRT2-treated and AGK2-treated cells compared with NC-treated cells (Fig. 6F), further supporting the conclusion that BAG3 is a novel physiological substrate of SIRT2.
3.4.2. SIRT2/CBP deacetylates/acetylates the K431 residue of BAG3
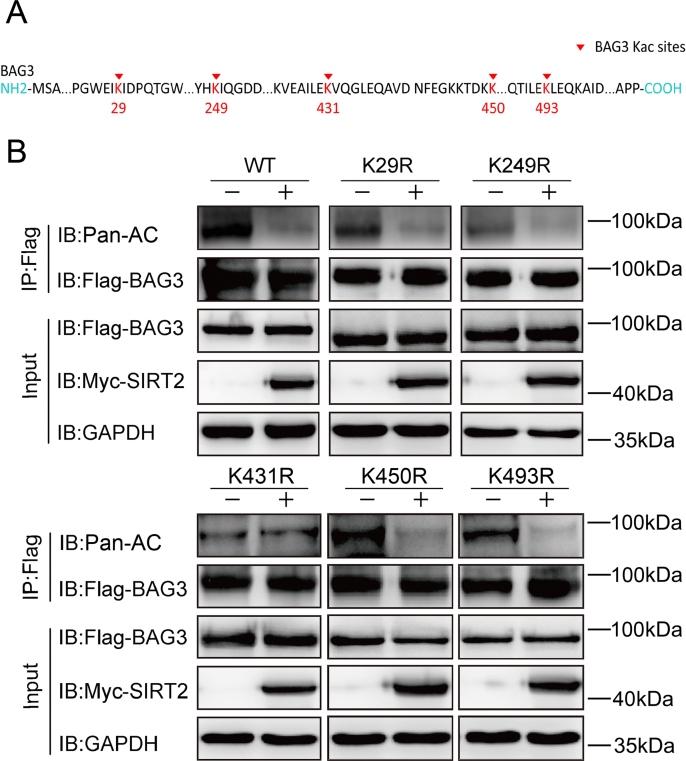
In order to provide direct evidence that SIRT2/CBP deacetylates/acetylates BAG3, we sought to identify the specific site of BAG3 that is deacetylated/acetylated by SIRT2/CBP. Therefore, we analyzed the effect of individual mutations of BAG3 acetylation sites identified by mass spectrometry. Each of the five identified putative acetylation sites was mutated to arginine (R) through site-directed mutagenesis, thus producing mutants that cannot be acetylated. Only mutating lysine (K) 431 to R reduced BAG3 acetylation, and the BAG3 mutant with an arginine substitution at residue K431 showed similar levels of BAG3 acetylation with or without SIRT2 overexpression (Fig. S1A–B). This indicated K431 as a major site for BAG3 regulation by SIRT2.
We then probed the role of the BAG3-K431 site in the regulation of BAG3 acetylation. Replacing K431 with either K431R (unacetylable) or an acetyl-mimetic glutamine (K431Q) dramatically decreased the overall BAG3 acetylation level compared with the WT protein, indicating that K431 was indeed an important acetylation site in BAG3 (Fig. 6G). We then transfected WT- or K431R-mutant BAG3 plasmids along with the Flag-control and Flag-CBP plasmids, respectively. Arginine substitution in K431R suppressed BAG3 acetylation, in presence or absence of additional CBP, while WT-BAG3 acetylation level was elevated with additional CBP (Fig. 6H). Consistent with these findings, BAG3 acetylation was suppressed when the BAG3-K431R mutant plasmid was co-transfected with either the Myc control or Myc-tagged SIRT2 plasmid, whereas BAG3-WT acetylation level was decreased by additional SIRT2 (Fig. 6I). BAG3-K431 acetylation was next examined with antibodies specific to ectopically produced BAG3-WT without recognizing the BAG3-K431R mutant (Fig. 6J). In addition, the level of exogenous K431-acetylated BAG3 was increased in shSIRT2-treated cells in comparison with the NC group (Fig. 6K). Moreover, the level of Ang II-induced K431-acetylated BAG3 remained unchanged in shSIRT2 cells, but restored in SIRT2-expressing cells, confirming that SIRT2 regulated BAG3-K431 deacetylation (Fig. 6L). Interestingly, BAG3-K431 is evolutionarily conserved among mammals (Fig. 6M).
Taken together, the results described above confirmed that the deacetylase SIRT2 and the acetyltransferase CBP interact with BAG3 and, respectively, deacetylates and acetylates BAG3 at the K431 site (Fig. 6N).
3.5. Deacetylation-dependent regulation of BAG3 promotes PARP1 ubiquitination and degradation, inhibiting Ang II-induced oxidative stress endothelial damage
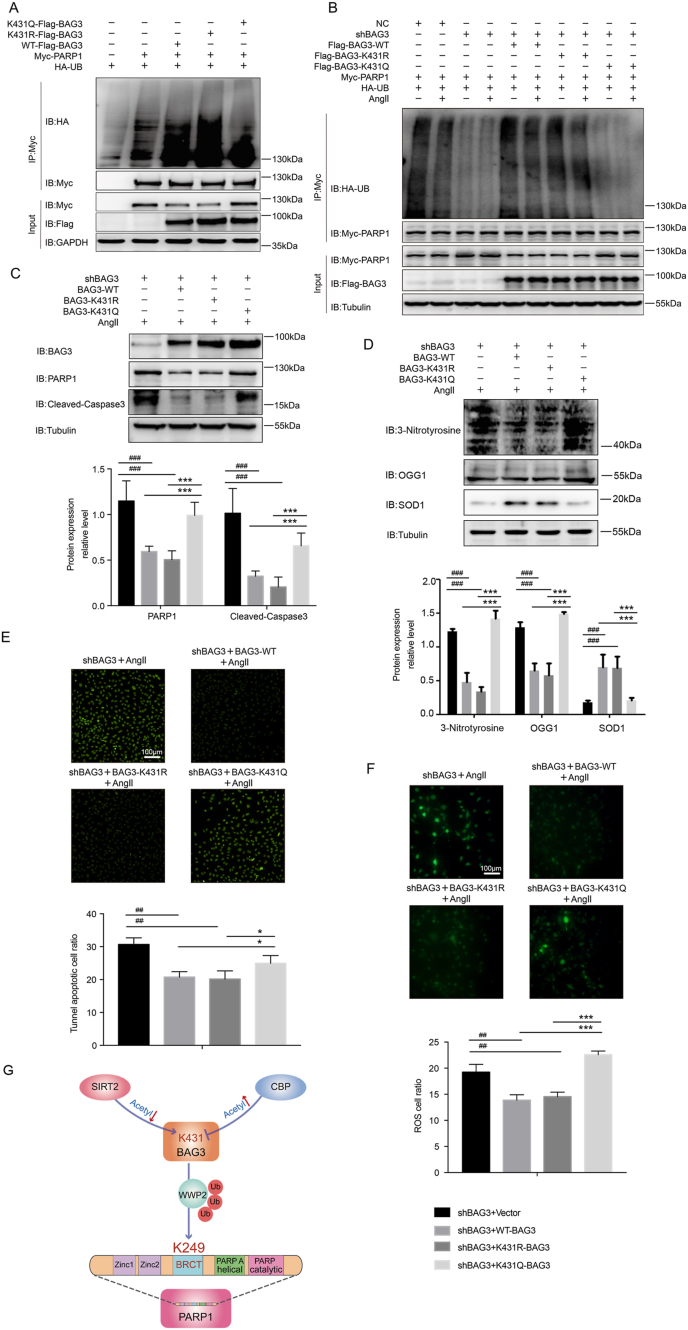
According to the above result, we demonstrated that BAG3 interacts with the BRCT domain of PARP1 and induces PARP1 ubiquitination at K249 through WWP2. In addition, we confirmed that the acetyltransferase CBP acetylates BAG3 at the K431 site. And the deacetylase SIRT2 deacetylates BAG3 at the K431 site. Moreover, we verified that the ubiquitination and degradation of PARP1 depend on the deacetylation state of BAG3.
The results showed that BAG3-WT or inactivation of the K431 residue of BAG3 (BAG3-K431R) (deacetylation regulated by SIRT2) promoted the ubiquitination of PARP1, but not BAG3-K431Q (acetylation regulated by CBP and inhibition of SIRT2 regulation) (Fig. 7A). Furthermore, after Ang II treatment, BAG3 knockdown decreased PARP1 ubiquitination, and supplementation of BAG3-WT or inactivation of the K431 residue of BAG3 (BAG3-K431R) promoted PARP1 ubiquitination; meanwhile, supplementation of BAG3-K431Q could not induce PARP1 ubiquitination (Fig. 7B). Thirdly, BAG3 knockdown in endothelial cells with BAG3-WT or BAG3-K431R, but not BAG3-K431Q, significantly reduced the levels of Ang II-associated vascular endothelial cell damage proteins PARP1 and cleaved caspase-3 (Fig. 7C), and Ang II-related oxidative stress biomarkers 3-nitrotyrosine and OGG1, and markedly elevated the amounts of the antioxidative stress biomarker SOD1 (Fig. 7D). In addition, supplementation of BAG3-knockdown endothelial cells with BAG3-WT or BAG3-K431R, but not BAG3-K431Q, significantly reduced vascular endothelial cell apoptosis (Fig. 7E) and ROS generation (Fig. 7F).
Fig. 7.
Deacetylation-dependent regulation of BAG3 promotes PARP1 ubiquitination and degradation, inhibiting Ang II-induced oxidative stress endothelial damage.
(A) HUVECs co-transfected with Flag-BAG3-K431Q, Flag-BAG3-K431R, Flag-WT-BAG3, or Myc-PARP1 and HA-UB. Ubiquitinated PARP1 underwent immunoprecipitation with anti-Myc antibodies, followed by immunoblot detection with anti-HA antibodies. (B) NC or shBAG3 HUVECs were co-transfected with Myc-PARP1, HA-UB, and Flag-WT-BAG3, Flag-BAG3-K431R, or Flag-BAG3-K431Q. The cells were then treated without or with Ang II. PARP1 ubiquitination was examined by immunoprecipitation with anti-Myc antibodies, followed by immunoblot detection with anti-HA antibodies. (C) ShBAG3 HUVECs underwent transfection with BAG3-WT, BAG3-431R, or BAG3-431Q, then treated without or with Ang II. The expression levels of BAG3, PARP1 and cleaved caspase-3 were assessed by immunoblot with various antibodies, with tubulin as a reference. Experiments were repeated thrice; **P < 0.01, two-way ANOVA with post hoc Bonferroni test. (D) The expression levels of 3-nitrotyrosine, OGG1 and SOD1 were assessed by immunoblot with various antibodies, with tubulin as a reference. Experiments were repeated thrice; **P < 0.01, two-way ANOVA with post hoc Bonferroni test.(E–F) The TUNEL assay was performed, and ROS production was evaluated in Ang II-treated shBAG3 HUVECs transfected with BAG3-WT, BAG3-431R, or BAG3-431Q. Experiments were repeated thrice; **P < 0.01, ***P < 0.001, two-way ANOVA with post hoc Bonferroni test. (G) Schematic model showing the potential function of deacetylated BAG3 in promoting PARP1 ubiquitination by enhancing the E3 ubiquitin ligase WWP2.
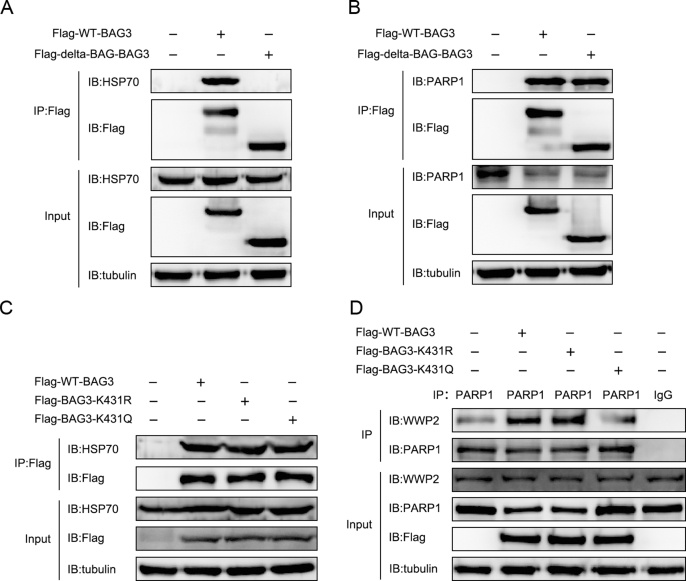
In addition, since HSP70 is the main interacting protein of BAG3, we will figure out whether HSP70 plays a role in the ubiquitination modification of PARP1 by deacetylated BAG3. Our results showed that BAG3-WT, but not Delta-BAG-BAG3 interacted with HSP70 (Fig. S2A), which is consistent with previous studies. In addition, we found that both BAG3-WT and Delta-BAG-BAG3 interacted with PARP1, and there was no significant change in the levels of interaction (Fig. S2B), suggesting that the interaction between BAG3 and PARP1 may not depend on HSP70.
Furthermore, our results showed that both BAG3-WT, inactivation of the K431 residue of BAG3 (BAG3-K431R) and activation of the K431 residue of BAG3 (BAG3-K431Q) interacted with HSP70, and there was no significant change in the levels of interaction (Fig. S2C), suggesting that the acetylation and deacetylation of BAG3 may not affect the interaction between BAG3 and HSP70. Interestingly, BAG3-WT or BAG3-K431R increased the interaction between E3 ubiquitination ligase WWP2 and PARP1, but not BAG3-K431Q (Fig. S2D). Therefore, the above results suggested that BAG3 increases the interaction between WWP2 and PARP1, and this effect may be independent of HSP70.
Taken together, these results confirmed that BAG3 interacts with PARP1's BRCT domain and induces PARP1 ubiquitination at K249 through WWP2, and SIRT2/CBP interact with BAG3 to deacetylate/acetylate BAG3-K431; in addition, deacetylation-dependent regulation of BAG3 promotes PARP1 ubiquitination and degradation, which is one possible mechanism to decrease vulnerability of oxidative stress in endothelial cells, thereby inhibiting oxidative stress-induced endothelial damage.
4. Discussion
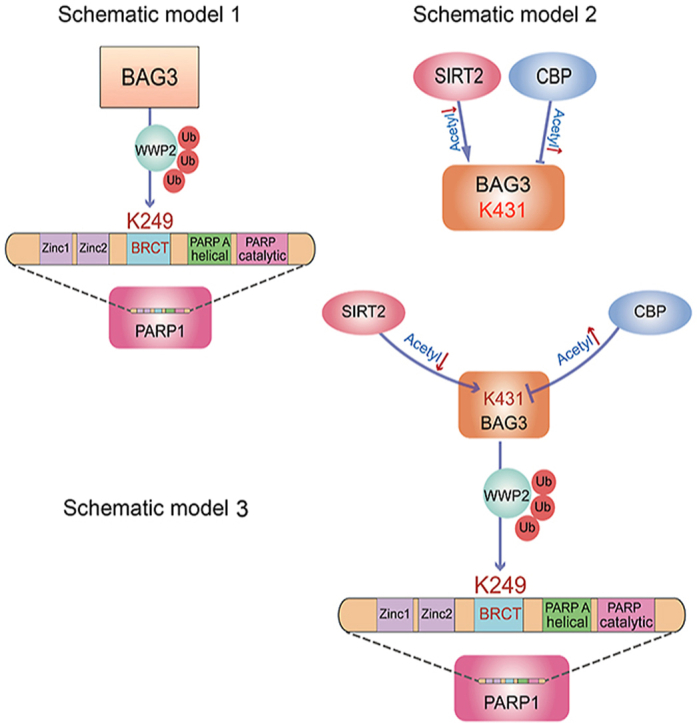
The main findings of this study are that, firstly, we constructed BAG3 endothelial specific knockout mice, which showed significantly worsened oxidative stress-related vascular endothelial damage as well as post-injury remodeling. In addition, we constructed BAG3 transgenic mice, which relieved this phenomenon. Mechanistically, the oxidative stress injury protein PARP1 was identified as a new BAG3 substrate. BAG3 interacts with PARP1's BRCT domain and promotes its ubiquitination at K249 by enhancing WWP2, an E3 ubiquitin ligase (Schematic model 1). Although BAG3 undergoes posttranslational modifications [39], reports evaluating its acetylation and regulatory mechanism are lacking. Surprisingly, we found that BAG3 is a novel physiological substrate of the acetyltransferase CBP and the deacetylase SIRT2. CBP/SIRT2 interacted with BAG3, and acetylated/deacetylated the BAG3-K431 residue, respectively (Schematic model 2). Finally, deacetylated BAG3 promoted the ubiquitination of PARP1, which is one possible mechanism to decrease vulnerability of oxidative stress in endothelial cells, thereby inhibiting oxidative stress-induced endothelial damage (Schematic model 3). Thus, identifying a new modification complex may help further design potential therapeutic approaches for cardiovascular disease.
BAG3 is a stress-induced protein and a molecular chaperone regulator of the BAG family, which interact with various proteins and influence cell survival by activating multiple pathways, making BAG3 involving in the regulation of many major biological processes including apoptosis and development, cytoskeleton arrangement and autophagy [15,16,[19], [20], [21], [22], [23], [24], [25]]. Under physiological condition, BAG3 in mammals mainly assist the 70 kDa heat-shock protein (HSP70) in protein folding [40]. But their role as oncogenes is becoming increasingly obvious, more and more cancers have been confirmed regulated by BAG3 [[41], [42], [43]]. BAG3 is expressed most significantly in the heart and skeletal muscles, and it was reported that BAG3 plays an important role in heart disease [44]. In our study, BAG3 was found to be deacetylated by SIRT2 then promoted the ubiquitination of PARP1, thereby inhibiting oxidative stress-induced endothelial damage.
The currently known types of post-translational modifications of PARP1 include ubiquitination, methylation, phosphorylation and acetylation [12,13,[45], [46], [47]]. The smyd2-related PARP1 methylation target, Lysine 528, was confirmed by LC-MS/MS and Edman degradation [45]. The receptor tyrosine kinase c-Met interacts with and phosphorylates the tyrosine 907 of PARP1 [46]. MORC2, a chromatin remodeling enzyme involved in DNA damage response, stabilizes PARP1 by increasing its acetylation at lysine 949 under control of the acetyltransferase nat10, blocking PARP1 ubiquitination on the same residue, followed by subsequent binding by E3 ubiquitin Enzyme CHFR for degradation [47]. In our study, BAG3 binds to PARP1's BRCT domain to promote its ubiquitination (K249 residue) leading to the degradation of PARP1, which is one possible mechanism to decrease vulnerability of oxidative stress in endothelial cells.
In this study, we found BAG3 promoted PARP1 ubiquitination by enhancing WWP2. WWP2 represents an important member of the Nedd4-like protein family of E3 ubiquitin ligases, which regulate various physiological processes by ubiquitinating substrates on specific lysine residues [[48], [49], [50]]. As an important regulator of the cardiovascular system, WWP2 mediates pathological cardiac fibrosis by regulating TGFβ/Smad signaling [51]. Moreover, phosphorylation at the C-terminal domain of S1P1 leads to poly-ubiquitinylation by WWP2, as the key to maintain vascular integrity and resist vascular leak–inducing agents [52]. More importantly, we previously demonstrated PARP1 is the key cardiovascular injury factor that undergoes ubiquitination by WWP2 [12,13]. Therefore, this study further refined our theory and confirmed that BAG3 is a key regulatory protein of WWP2 ubiquitination-modified PARP1.
An important finding is that BAG3 is a newly discovered substrate of SIRT2 deacetylase. BAG3 is a stress-induced protein and a member of the BAG family of proteins [[19], [20], [21], [22], [23], [24],53]. As a deacetylase, SIRT2 can induce post-translational modifications of its downstream proteins and affect the occurrence and development of a variety of diseases [[32], [33], [34],54,55]. SIRT2 directly deacetylates key factors affecting NFATc2 and inhibits its activity, thereby suppressing pathological myocardial hypertrophy [54]. In addition, SIRT2 reduces the degradation of BUBR1 by inhibiting the acetylation of BUBR1, which maintains the normal pumping function of the heart [55]. Here, we found that SIRT2 deacetylated BAG3 at K431 residue. In addition, BAG3, deacetylated by SIRT2, promoted PARP1 ubiquitination, which is one possible mechanism to decrease vulnerability of oxidative stress in endothelial cells and decreased endothelial damage as well as remodeling after injury.
We firstly found out deacetylated BAG3 can promote the ubiquitination of PARP1 at K249. Since BAG3 has a wide range of effects on downstream proteins, we suggested choosing the ubiquitination site (K249) of PARP1 which was regulated by BAG3 as the possible theoretical basis of targeted drugs in the future. Further studies are warranted to explore whether other BAG family members are involved in oxidative stress-associated endothelial damage and subsequent vascular remodeling. Moreover, whether BAG3 protects from atherosclerosis and hypertension deserves further attention.
Sources of funding
The present study was funded by the National Natural Science Foundation of China (82171571, 82171572, 81900372, 81970211 and 81900355) and National Postdoctoral Innovative Talents Support Program (BX2021376).
Author contributions
N.Z., Y.Z., Y.S. and L.C. performed study conception and design. N.Z., Y.Z., B.W., YM.Z., W.M. and Q.M. carried out animal experiments. N.Z., Y.Z., S.Y., S.L., C.S., Z.C. and J.X. performed Co-IP and immunoblot. Y.Z. performed immunohistochemistry. N.Z., Y.Z., J.L. and X.H. carried out data analysis. N.Z. and Y.Z. wrote the first manuscript draft, which was revised by Y.S., and L.C.
Data and materials availability
The data supporting the findings of this study are available in this article and its supplementary files, or from the corresponding authors on request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are grateful to Prof. Qunying Lei (Fudan University, Shanghai, China) who provided the Flag-P300, Flag-CBP and Myc-GCN5 plasmids, and to Prof. Weiguo Zhu (Shenzhen University, Shenzhen, China) who provided the Flag-PCAF plasmid, and to Prof. Huaqin Wang (China Medical University, Shenyang, China) who provided the Flag-Delta-BAG-BAG3 plasmid.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102238.
Contributor Information
Liu Cao, Email: lcao@cmu.edu.cn.
Yingxian Sun, Email: yxsun@cmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1.
Acetylation sites of BAG3 modulated by SIRT2
(A) Mass spectroscopy revealed potential acetylation sites of BAG3 utilized by SIRT2. (B) Flag-WT-BAG3, Flag-BAG3-K29R, Flag-BAG3-K249R, Flag-BAG3-K431R, Flag-BAG3-K450R, or Flag-BAG3-K493R was transfected, and BAG3 acetylation was examined by immunoprecipitation with anti-Flag beads, followed by immunoblot with anti-Pan-AC antibodies.
Fig. S2.
BAG3 increases the interaction between WWP2 and PARP1 independent of HSP70
(A) Flag-WT-BAG3 or Flag-Delta-BAG-BAG3 was transfected, and the interaction between Flag-WT-BAG3 or Flag-Delta-BAG-BAG3 and HSP70 was examined by immunoprecipitation with anti-Flag beads, followed by immunoblot detection with anti-HSP70 antibodies. (B) Flag-WT-BAG3 or Flag-Delta-BAG-BAG3 was transfected, and the interaction between Flag-WT-BAG3 or Flag-Delta-BAG-BAG3 and PARP1 was examined by immunoprecipitation with anti-Flag beads, followed by immunoblot detection with anti-PARP1 antibodies. (C) Flag-WT-BAG3, Flag-BAG3-K431R or Flag-BAG3-K431Q was transfected, and the interaction between Flag-WT-BAG3, Flag-BAG3-K431R or Flag-BAG3-K431Q and HSP70 was examined by immunoprecipitation with anti-Flag beads, followed by immunoblot detection with anti-HSP70 antibodies. (D) Flag-WT-BAG3, Flag-BAG3-K431R or Flag-BAG3-K431Q was transfected, and the interaction between PARP1 and WWP2 was examined by immunoprecipitation with anti-PARP1 antibodies, followed by immunoblot detection with anti-WWP2 antibodies.
References
- 1.Münzel Thomas, Camici Giovanni G., Maack Christoph, Bonetti Nicole R., Fuster Valentin, Kovacic Jason C. Impact of oxidative stress on the heart and vasculature: Part 2 of a 3-Part Series. J Am Coll Cardiol. 2017;70(2):212–229. doi: 10.1016/j.jacc.2017.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sabbatinelli Jacopo, Prattichizzo Francesco, Olivieri Fabiola, Procopio Antonio Domenico, Rippo Maria Rita, Giuliani Angelica. Where metabolism meets senescence: focus on endothelial cells. Front Physiol. 2019;10:1523. doi: 10.3389/fphys.2019.01523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Suowen, Ilyas Iqra, Little Peter J., Li Hong, Kamato Danielle, Zheng Xueying, et al. Endothelial dysfunction in atherosclerotic cardiovascular diseases and beyond: from mechanism to pharmacotherapies. Pharmacol Rev. 2021;73(3):924–967. doi: 10.1124/pharmrev.120.000096. [DOI] [PubMed] [Google Scholar]
- 4.Li Yuanbin, Liang Pengfei, Jiang Bimei, Tang Yuting, Liu Xuanyou, Liu Meidong, et al. CARD9 promotes autophagy in cardiomyocytes in myocardial ischemia/reperfusion injury via interacting with Rubicon directly. Basic Res Cardiol. 2020;115(3):29. doi: 10.1007/s00395-020-0790-6. [DOI] [PubMed] [Google Scholar]
- 5.Mason Fleur E., Pronto Julius Ryan D., Alhussini Khaled, Maack Christoph, Voigt Niels. Cellular and mitochondrial mechanisms of atrial fibrillation. Basic Res Cardiol. 2020;115(6):72. doi: 10.1007/s00395-020-00827-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hans Chetan P., Zerfaoui Mourad, Naura Amarjit S., Catling Andrew, Hamid Boulares A. Differential effects of PARP inhibition on vascular cell survival and ACAT-1 expression favouring atherosclerotic plaque stability. Cardiovasc Res. 2008;78(3):429–439. doi: 10.1093/cvr/cvn018. [DOI] [PubMed] [Google Scholar]
- 7.Henning Robert J., Bourgeois Marie, Harbison Raymond D. Poly(ADP-ribose) polymerase (PARP) and PARP inhibitors: mechanisms of action and role in cardiovascular disorders. Cardiovasc Toxicol. 2018;18(6):493–506. doi: 10.1007/s12012-018-9462-2. [DOI] [PubMed] [Google Scholar]
- 8.Jagtap Prakash, Szabó Csaba. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov. 2005;4(5):421–440. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 9.Mateo J., Lord C.J., Serra V., Tutt A., Balmaña J., Castroviejo-Bermejo M., et al. A decade of clinical development of PARP inhibitors in perspective. Ann Oncol. 2019;30(9):1437–1447. doi: 10.1093/annonc/mdz192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu H., Wei M., Xu J., Hua J., Liang C., Meng Q., et al. PARP inhibitors in pancreatic cancer: molecular mechanisms and clinical applications. Mol Cancer. 2020;19(1):49. doi: 10.1186/s12943-020-01167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Jing, Li Jingyan, Hu Yuehuai, Guo Zhen, Sun Duanping, Wang Panxia, et al. Chrysophanol protects against doxorubicin-induced cardiotoxicity by suppressing cellular PARylation. Acta Pharm Sin B. 2019;9(4):782–793. doi: 10.1016/j.apsb.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Naijin, Zhang Ying, Wu Boquan, You Shilong, Sun Yingxian. Role of WW domain E3 ubiquitin protein ligase 2 in modulating ubiquitination and Degradation of Septin4 in oxidative stress endothelial damage. Redox Biol. 2020;30:101419. doi: 10.1016/j.redox.2019.101419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Naijin, Zhang Ying, Qian Hao, Wu Shaojun, Cao Liu, Sun Yingxian. Selective targeting of ubiquitination and degradation of PARP1 by E3 ubiquitin ligase WWP2 regulates isoproterenol-induced cardiac remodeling. Cell Death Differ. 2020;27(9):2605–2619. doi: 10.1038/s41418-020-0523-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang Xi, Bogomolovas Julius, Trexler Christa, Chen Ju. The BAG3-dependent and -independent roles of cardiac small heat shock proteins. JCI Insight. 2019;4(4) doi: 10.1172/jci.insight.126464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doong H., Price J., Kim Y.S., Gasbarre C., Probst J., Liotta L.A., et al. CAIR-1/BAG-3 forms an EGF-regulated ternary complex with phospholipase C-gamma and Hsp70/Hsc70. Oncogene. 2000;19(38):4385–4395. doi: 10.1038/sj.onc.1203797. [DOI] [PubMed] [Google Scholar]
- 16.Carra Serena, Seguin Samuel J., Landry Jacques. HspB8 and Bag3: a new chaperone complex targeting misfolded proteins to macroautophagy. Autophagy. 2008;4(2):237–239. doi: 10.4161/auto.5407. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs A.T., Marnett L.J. Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc Chem Res. 2010;43(5):673–683. doi: 10.1021/ar900286y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doong H., Vrailas A., Kohn E.C. What's in the 'BAG'?--A functional domain analysis of the BAG-family proteins. Cancer Lett. 2002;188(1–2):25–32. doi: 10.1016/s0304-3835(02)00456-1. [DOI] [PubMed] [Google Scholar]
- 19.Chakraborty Debapriya, Felzen Vanessa, Hiebel Christof, Stürner Elisabeth, Perumal Natarajan, Manicam Caroline, et al. Enhanced autophagic-lysosomal activity and increased BAG3-mediated selective macroautophagy as adaptive response of neuronal cells to chronic oxidative stress. Redox Biol. 2019;24:101181. doi: 10.1016/j.redox.2019.101181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gamerdinger Martin, Hajieva Parvana, Kaya A Murat, Wolfrum Uwe, Ulrich Hartl F., Behl Christian. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28(7):889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gamerdinger Martin, Kaya A Murat, Wolfrum Uwe, Clement Albrecht M., Behl Christian. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 2011;12(2):149–156. doi: 10.1038/embor.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin Thomas G., Myers Valerie D., Dubey Praveen, Dubey Shubham, Perez Edith, Moravec Christine S., et al. Cardiomyocyte contractile impairment in heart failure results from reduced BAG3-mediated sarcomeric protein turnover. Nat Commun. 2021;12(1):2942. doi: 10.1038/s41467-021-23272-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myers Valerie D., Gerhard Glenn S., McNamara Dennis M., Tomar Dhanendra, Madesh Muniswamy, Kaniper Scott, et al. Association of variants in BAG3 with cardiomyopathy outcomes in african American individuals. JAMA Cardiol. 2018;3(10):929–938. doi: 10.1001/jamacardio.2018.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruparelia Avnika A., McKaige Emily A., Williams Caitlin, Schulze Keith E., Fuchs Margit, Oorschot Viola, et al. Metformin rescues muscle function in BAG3 myofibrillar myopathy models. Autophagy. 2020:1–17. doi: 10.1080/15548627.2020.1833500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hishiya Akinori, Kitazawa Toshio, Takayama Shinichi. BAG3 and Hsc70 interact with actin capping protein CapZ to maintain myofibrillar integrity under mechanical stress. Circ Res. 2010;107(10):1220–1231. doi: 10.1161/CIRCRESAHA.110.225649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura Kenichi, Ooms Astrid, Graf-Riesen Kathrin, Kuppusamy Maithreyan, Unger Andreas, Schuld Julia, et al. Overexpression of human BAG3 P209L in mice causes restrictive cardiomyopathy. Nat Commun. 2021;12(1):3575. doi: 10.1038/s41467-021-23858-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordan Elizabeth, Peterson Laiken, Ai Tomohiko, Asatryan Babken, Bronick Lucas, Brown Emily, et al. Evidence-based assessment of genes in dilated cardiomyopathy. Circulation. 2021;144(1):7–19. doi: 10.1161/CIRCULATIONAHA.120.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Ping, Cong Xiaoji, Liao Shengjie, Jia Xinglong, Wang Xiaomin, Dai Wei, et al. Global identification of phospho-dependent SCF substrates reveals a FBXO22 phosphodegron and an ERK-FBXO22-BAG3 axis in tumorigenesis. Cell Death Differ. 2021 Jul 2 doi: 10.1038/s41418-021-00827-7. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felzen V., Hiebel C., Koziollek-Drechsler I., Reissig S., Wolfrum U., Kogel D., et al. Estrogen receptor alpha regulates non-canonical autophagy that provides stress resistance to neuroblastoma and breast cancer cells and involves BAG3 function. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosati A., Graziano V., De Laurenzi V., Pascale M., Turco M.C. BAG3: a multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011;2:e141. doi: 10.1038/cddis.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falco A., Rosati A., Festa M., Basile A., De Marco M., d'Avenia M., et al. BAG3 is a novel serum biomarker for pancreatic adenocarcinomas. Am J Gastroenterol. 2013;108:1178–1180. doi: 10.1038/ajg.2013.128. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Manchao, Du Wuying, Acklin Scarlett, Jin Shengkai, Xia Fen. SIRT2 protects peripheral neurons from cisplatin-induced injury by enhancing nucleotide excision repair. J Clin Invest. 2020;130(6):2953–2965. doi: 10.1172/JCI123159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Naijin, Zhang Ying, Wu Boquan, Wu Shaojun, You Shilong, Lu Saien, et al. Deacetylation-dependent regulation of PARP1 by SIRT2 dictates ubiquitination of PARP1 in oxidative stress-induced vascular injury. Redox Biol. 2021;47:102141. doi: 10.1016/j.redox.2021.102141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Xiaoyan, Park Seong-Hoon, Chang Hsiang-Chun, Shapiro Jason S., Vassilopoulos Athanassios, Sawicki Konrad T., et al. Sirtuin 2 regulates cellular iron homeostasis via deacetylation of transcription factor NRF2. J Clin Invest. 2017;127(4):1505–1516. doi: 10.1172/JCI88574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narita T., Weinert B.T., Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat Rev Mol Cell Biol. 2019;20(3):156–174. doi: 10.1038/s41580-018-0081-3. [DOI] [PubMed] [Google Scholar]
- 36.Shen Y., Wei W., Zhou D.X. Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci. 2015;20(10):614–621. doi: 10.1016/j.tplants.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 37.You Z., Jiang W.X., Qin L.Y., Gong Z., Wan W., Li J., et al. Requirement for p62 acetylation in the aggregation of ubiquitylated proteins under nutrient stress. Nat Commun. 2019;10(1):5792. doi: 10.1038/s41467-019-13718-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin R., Tao R., Gao X., Li T., Zhou X., Guan K.L., et al. Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Mol Cell. 2013;51(4):506–518. doi: 10.1016/j.molcel.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li N., Du Z.-X., Zong Z.-H., Liu B.-Q., Li C., Zhang Q., et al. PKCδ-mediated phosphorylation of BAG3 at Ser187 site induces epithelial-mesenchymal transition and enhances invasiveness in thyroid cancer FRO cells. Oncogene. 2013;32(38):4539–4548. doi: 10.1038/onc.2012.466. [DOI] [PubMed] [Google Scholar]
- 40.Mariotto E., Viola G., Zanon C., Aveic S. A BAG's life: every connection matters in cancer. Pharmacol Ther. 2020;209:107498. doi: 10.1016/j.pharmthera.2020.107498. [DOI] [PubMed] [Google Scholar]
- 41.Rosati A., Graziano V., De Laurenzi V., Pascale M., Turco M.C. BAG3: a multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011;2(4):e141. doi: 10.1038/cddis.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobs A.T., Marnett L.J. Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc Chem Res. 2010;43(5):673–683. doi: 10.1021/ar900286y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Behl C. Breaking BAG: the Co-chaperone BAG3 in health and disease. Trends Pharmacol Sci. 2016;37(8):672–688. doi: 10.1016/j.tips.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 44.Kirk J.A., Cheung J.Y., Feldman A.M. Therapeutic targeting of BAG3: considering its complexity in cancer and heart disease. J Clin Invest. 2021;131(16) doi: 10.1172/JCI149415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piao Lianhua, Kang Daechun, Suzuki Takehiro, Masuda Akiko, Dohmae Naoshi, Nakamura Yusuke, et al. The histone methyltransferase SMYD2 methylates PARP1 and promotes poly(ADP-ribosyl)ation activity in cancer cells. Neoplasia. 2014;16(3):257–264. doi: 10.1016/j.neo.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du Yi, Yamaguchi Hirohito, Wei Yongkun, Hsu Jennifer L., Wang Hung-Ling, Hsu Yi-Hsin, et al. Blocking c-Met-mediated PARP1 phosphorylation enhances anti-tumor effects of PARP inhibitors. Nat Med. 2016;22(2):194–201. doi: 10.1038/nm.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Lin, Li Da-Qiang. MORC2 regulates DNA damage response through a PARP1-dependent pathway. Nucleic Acids Res. 2019;47(16):8502–8520. doi: 10.1093/nar/gkz545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maddika Subbareddy, Kavela Sridhar, Rani Neelam, Reddy Palicharla Vivek, Pokorny Jenny L., Sarkaria Jann N., et al. WWP2 is an E3 ubiquitin ligase for PTEN. Nat Cell Biol. 2011;13(6):728–733. doi: 10.1038/ncb2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang Lan, Zhang Ling, Wei Wei, Jin Xueling, Wang Ping, Tong Yufeng, et al. A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol Cell. 2014;55(4):537–551. doi: 10.1016/j.molcel.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 50.Mokuda Sho, Nakamichi Ryo, Matsuzaki Tokio, Ito Yoshiaki, Sato Tempei, Miyata Kohei, et al. Wwp2 maintains cartilage homeostasis through regulation of Adamts5. Nat Commun. 2019;10(1):2429. doi: 10.1038/s41467-019-10177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Huimei, Moreno-Moral Aida, Pesce Francesco, Devapragash Nithya, Mancini Massimiliano, Ling Heng Ee, et al. WWP2 regulates pathological cardiac fibrosis by modulating SMAD2 signaling. Nat Commun. 2019;10(1):3616. doi: 10.1038/s41467-019-11551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin Oo Myat, Chang Sung-Hee, Thangada Shobha, Wu Ming-Tao, Rezaul Karim, Blaho Victoria, et al. Engagement of S1P₁-degradative mechanisms leads to vascular leak in mice. J Clin Invest. 2011;121(6):2290–2300. doi: 10.1172/JCI45403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Domínguez Fernando, Cuenca Sofía, Bilińska Zofia, Toro Rocío, Villard Eric, Barriales-Villa Roberto, et al. Dilated cardiomyopathy due to BLC2-associated athanogene 3 (BAG3) mutations. J Am Coll Cardiol. 2018;72(20):2471–2481. doi: 10.1016/j.jacc.2018.08.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarikhani M., Maity S., Mishra S., Jain A., Tamta A.K., Ravi V., et al. SIRT2 deacetylase represses NFAT transcription factor to maintain cardiac homeostasis. J Biol Chem. 2018;293:5281–5294. doi: 10.1074/jbc.RA117.000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.North B.J., Rosenberg M.A., Jeganathan K.B., Hafner A.V., Michan S., Dai J., et al. SIRT2 induces the checkpoint kinase BubR1 to increase lifespan. EMBO J. 2014;33:1438–1453. doi: 10.15252/embj.201386907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.