Abstract
Since the beginning of the COVID-19 pandemic, the development of effective vaccines against this pathogen has been a priority for the scientific community. Several strategies have been developed including vaccines based on recombinant viral protein fragments. The receptor-binding domain (RBD) in the S1 subunit of S protein has been considered one of the main targets of neutralizing antibodies. In this study we assess the potential of a vaccine formulation based on the recombinant RBD domain of SARS-CoV-2 expressed in the thermophilic filamentous fungal strain Thermothelomyces heterothallica and the hepatitis B virus (HBV) core protein. Functional humoral and cellular immune responses were detected in mice. To our knowledge, this is the first report on the immune evaluation of a biomedical product obtained in the fungal strain T. heterothallica. These results together with the intrinsic advantages of this expression platform support its use for the development of biotechnology products for medical purpose.
Keywords: COVID, SARS CoV-2, Vaccine, Protein, C1, Immunogenicity
1. Introduction
Since the beginning of the COVID-19 pandemic, caused by the coronavirus SARS-CoV-2, the development of an effective vaccine against this pathogen that can be efficiently manufactured rapidly and affordably on a global scale, has become a priority for the scientific community. Several strategies have been developed including vaccines based on recombinant viral protein fragments [1].
The coronavirus S protein plays essential roles in virus binding, fusion and entry to the cells; therefore, it has constituted an important target in the development of vaccine candidates. The receptor-binding domain (RBD) in the S1 subunit of the S protein recognizes and binds to the receptor angiotensin-converting enzyme 2 (ACE2) on the cells. This protein fragment has been considered one of the main targets of neutralizing antibodies [2].
In this study, we aimed at assessing a vaccine formulation based on combining a recombinant RBD domain of SARS-CoV-2 and the hepatitis B virus (HBV) core protein.
The RBD evaluated in the present study (hereinafter RBDf), was produced in the filamentous fungal strain Thermothelomyces heterothallica (formerly Myceliophthora thermophila). This thermophilic strain, originally isolated from an alkaline soil in Russia and nicknamed C1, grows under a broad range of fermentation conditions at different production scales [3]. It has been used to produce industrial enzymes and several heterologous human and animal proteins where it has proven to be a high-yield, low-cost, and rapidly scalable expression platform. Additional advantages of this expression system are the higher amount of human-type glycan structures in expressed proteins as compared to yeast and typical filamentous fungi as well as a shorter development and production cycle in comparison with the CHO cell line [3]. The production of biomedical products in C1 to be used in humans is an innovative strategy that is still waiting for thorough validation.
Recently, Aebischer and coworkers evidenced the immunogenicity in mice and cattle of the N-terminal region of the Gc envelope protein from the Schmallenberg virus (SBV), produced from the C1 fungal system [4]. SBV is transmitted by midges and predominately infects ruminants [5]. The protein induced protective immune responses in both animal models and conferred complete clinical protection after a single shot immunization in mice when presented on lumazine synthase particles [4].
The adjuvant capacity of the hepatitis B core protein (HBc) has been previously documented [6]. For instance, the intranasal and subcutaneous co-administration of a multi-antigenic formulation comprising a recombinant protein of human immunodeficiency virus (HIV) −1 and the main structural antigens of hepatitis B virus (HBV) induced a strong Th1 specific response against the HIV component [7]. Moreover, an intranasal vaccine against chronic HBV disease using the HBc stimulated a Th1 response against hepatitis B surface antigen [8], [9]. This adjuvant capacity has been related to the presence of bacterial RNA associated with the C-terminal arginine-rich domain of HBc [10].
In this work, we look into the antigenicity of the recombinant RBDf protein. Moreover, we evaluated in BALB/c mice the immunogenicity induced by the vaccine preparation RBDf HBc after the combined intranasal and subcutaneous inoculation.
Functional humoral and cellular immune responses, the latter one measured by the IFNγ levels secreted by spleen cells after in vitro stimulation, were detected. To our knowledge, this is the first report on the immune evaluation of a vaccine candidate to be used in humans, expressed from the thermophilic filamentous fungal strain Thermothelomyces heterothallica (C1 strain). These results, together with the intrinsic advantages of the C1 expression platform, pave the way for the development of larger quantities of more affordable biotechnology products based on protein molecules for medical use in humans.
2. Materials and methods
2.1. Recombinant protein
The gene coding for the RBDf protein was cloned into an expression vector between the bgl8 promoter and chi1 terminator. The CBH1 signal sequence was added to the N-terminus. The construct was transformed into the fungus Thermothelomyces heterothallica (formerly Myceliophthora thermophila), C1 strain DNL155, and it was integrated into the bgl8 locus. The RBDf protein has been expressed at greater than 2 g/l levels in 120 h in fed-batch fermentation and it was purified using a CaptureSelect™ C-tag affinity resin coupled to a camelid single-chain antibody, called NbSyn2, reaching a high level of purity [11]. The protein sequence has 204 aa and starts after the first N-glycosylation site (RBD amino acids 333–527 of the S2 protein). It includes the four amino acids (E-P-E-A) 'C-tag' at the C-terminal after the GGGGS linker.
The hepatitis B virus core protein (subtype adw, genotype A) expressed in Escherichia coli was obtained with more than 95% purity at the Center for Genetic Engineering and Biotechnology production facilities (CIGB, Havana, Cuba) as described [12].
The RBD protein expressed in Human Embryonic Kidney cells (HEK293) cells (hereinafter CRBDH6_HEK) was obtained with a high purity level at the Center of Molecular Immunology (CIM, Havana, Cuba).
All proteins were well-characterized by ESI-MS analysis as described [11].
2.2. Human sera
Human sera were collected in Havana during the 2020–2021 COVID-19 pandemic. Convalescent sera were coded SH 34–39, 41 and 45. SH 36, 37, and 41 belonged to asymptomatic patients while the rest of the sera were collected from patients with a mild clinical presentation. Negative sera were collected from persons without COVID-19 history and were coded N1, 3, 7, and 9.
2.3. SDS-PAGE and Western blotting
Protein samples were analyzed by SDS-PAGE and Western blotting as described by Lazo et al., 2007 [13]. Six micrograms of each sample were loaded per lane in the presence or absence of 10 mM 2-mercaptoethanol as reducing agent. A homemade monoclonal antibody developed in mice against the SARS-CoV-2 RBD protein (MAb S7, produced by the Research-Development Department of the Sancti Spiritus Center for Genetic Engineering and Biotechnology (CIGBSS)) was used for the Western blotting.
2.4. Interaction ACE2-RBDf by Surface Plasmon Resonance
The Surface Plasmon Resonance (SPR) experiments were performed using a BIACORE X (GE Health-care) at 25 °C. Murine Fc molecule fused with ACE2 (mFc-ACE2) was immobilized by capture on a Protein A biosensor chip (GE Health-care) according to the manufacturer’s protocol. The immobilization of the ligand was through flow cell 1 (FC1), while the FC2 was used as the reference cell. The running buffer was PBS, pH 7.2 (10 µl/min). The real-time response of the RBDf over the immobilized mFc-ACE2 was recorded by triplicate, in a single-cycle mode experiment. It was analyzed a concentration range from 15 to 1000 nM, allowing association for 120 s, while dissociation took place for another 120 s after sample injection. The chip was re-generated using glycine buffer, pH 2. The equilibrium dissociation constant (binding affinity, KD) was estimated with the BIAevaluation® software (GE Health-care) fitting the experimental data to the Langmuir 1:1 interaction model, with drifting baseline. At least five curves were taken into account for calculations. The human recombinant Epidermal Growth Factor (hrEGF) expressed from yeast S. cerevisiae was used as a negative control.
2.5. Reactivity of human sera with the RBDf protein by Enzyme-linked immunosorbent assay (ELISA)
Briefly, polystyrene plates with 96 wells (Costar, USA) were coated with the Mab S7 (10 μg/ml); after blocking and washing RBDf at 5 μg/ml was added to the plate. Serial dilutions of sera were incubated and later, an anti-human IgG-peroxidase conjugate was added. After washing, the substrate solution was added. The reaction was stopped, and an automated ELISA reader recorded the absorbance at 492 nm.
2.6. Immunological evaluation in BALB/c mice
Female BALB/c mice, aged 9–10 weeks, were vaccinated at days 0, 14 and 35. The immunization schedule consisted on 6 groups of ten animals each exploring subcutaneous and intranasal routes (Table 1 ). The intranasal preparations did not include alum while preparations administered by subcutaneous route included alum at 1.4 mg/ml.
Table 1.
Groups of the immunization schedule.
| Group | Immunogen | Adjuvant | Volume | Administration route |
|---|---|---|---|---|
| 1 | RBDf HBc alum SC | alum | 100 μl | Subcutaneous |
| 2 | RBDf HBc IN | – | 25 μl | Intranasal |
| 3 | RBDf HBc alum SC/IN | alum | 100 μl | Subcutaneous |
| – | 25 μl | Intranasal | ||
| 4 | RBDf HBc alum SC/IN MIX | alum | 100 μl | Subcutaneous |
| – | 25 μl | Intranasal | ||
| 5 | CRBDH6_HEK HBc alum SC/IN | alum | 100 μl | Subcutaneous |
| – | 25 μl | Intranasal | ||
| 6 | HBc alum SC/IN | alum | 100 μl | Subcutaneous |
| – | 25 μl | Intranasal |
Ten animals per group received three doses at days 0, 14, and 35. HBc: Hepatitis B virus core protein; SC: subcutaneous; IN: intranasal; MIX: simple mixture. RBDf: RBD protein expressed in the fungus Thermothelomyces heterothallica ; CRBDH6_HEK: RBD expressed in HEK293 cells.
Groups 1, 2, 3 and 5 were immunized with the RBD associated with the HBc protein. For the RBD-HBc association the RBD was mixed with citric acid and sodium phosphate dibasic solutions, to favor the protonation of the protein. The HBc was mixed with sodium phosphate dibasic solution. The HBc protein solution was dripped on the RBDf solution and shaken. The association between both molecules was assessed by Dynamic Light Scattering (data not shown).
Group 1 was vaccinated by the subcutaneous route with 25 μg of the RBDf and 5 μg of HBc using aluminum hydroxide as adjuvant (RBDf HBc alum SC). A second group was vaccinated by the intranasal route with 25 μg of the RBDf and 5 μg of HBc without adjuvant (RBDf HBc IN). The third group was inoculated simultaneously by both administration routes, intranasal and subcutaneous. Mice received 12.5 μg of the RBDf and 2.5 μg of HBc by both routes (RBDf HBc alum SC/IN). The fourth group received, by both administration routes, the same vaccine preparation as the third one but the RBDf and the HBc were mixed and no association reaction was performed (RBDf HBc alum SC/IN MIX). A Fifth group assayed the RBD expressed in HEK293 cells. Animals were inoculated with 12.5 μg of the CRBDH6_HEK and 2.5 μg of HBc by both administration routes (CRBDH6_HEK HBc alum SC/IN). A negative control group or placebo was immunized with 2.5 μg of HBc by both administration routes (HBc alum SC/IN).
The volumes for subcutaneous and intranasal administrations were adjusted to 100 μl and 25 μl per dose, respectively.
2.6.1. Humoral immune response
2.6.1.1. Enzyme-linked immunosorbent assay
Animals were bled at days 13, 21, 28, 34, 42, 49, and 56. Anti-RBD IgG levels were measured by ELISA. Briefly, 96 wells-polystyrene plates (Costar, USA) were coated with the RBDf (5 μg/ml); after blocking and washing two-fold serial dilutions of sera were incubated and later, an anti-mouse IgG-peroxidase conjugate (Amersham-Pharmacia, UK) was added. After proper incubation and washing, a substrate solution was added to the plates. An automated ELISA reader recorded the absorbance at 492 nm. A value of absorbance was considered positive if it was two-fold higher than that of the control preparation.
2.6.1.2. Molecular neutralization: ACE2-RBD inhibition ELISA
Serum pools per group were used in the next humoral immune evaluation experiments. Briefly, polystyrene plates with 96 wells (Costar, USA) were coated with mFc-ACE2 molecule at 5 μg/ml. Later, plates were blocked with skim milk. In unison, serial dilutions of serum pools were incubated with RBD human Fc molecule –HRPO conjugate. Some wells were incubated only with the HRPO conjugate (maximum signal) and the negative control wells, only with the dilution buffer. After washing, the mix was incubated in the plate and a TMB substrate solution was added. An automated ELISA reader recorded the absorbance at 450 nm (A450). The ACE2-RBD binding inhibition percent was defined as (1-A450 (sample) / A450 (maximum signal) *100.
2.6.1.3. Viral neutralization
Neutralization antibody titers were detected by a traditional virus microneutralization assay (MN50) using SARS-CoV-2 (CUT20 https://doi.org/10-2025/Cuba/2020 (D614G) strain or hCoV-19/CUBA/DC05/2021 (Delta) strain). Vero E6 cells (2 × 104 per well) were seeded in 96-well plates one night before use. Animals’ sera were inactivated at 56 °C for 30 min. The samples were prepared by two-fold serial serum pools dilutions in the Eagle’s Minimal Essential Medium (MEM, Gibco, UK) containing 2 % (v/v) fetal bovine serum (Capricorn, Germany). SARS-CoV-2 Cuban strain mentioned above at 100 TCID50 was incubated in the absence or presence of diluted sera. Afterward, Vero E6 cells were overlaid with virus suspensions. The cells were inspected for signs of cytopathogenic effects at 96 h post-infection and later were stained with neutral red (Sigma, USA). After three washes neutral red was dissolved in lysis solution and OD540 was detected. The cut-off was calculated as half of the control cell OD values. The neutralization titer was defined as the highest serum dilution showing an OD value greater than the cut-off. The viral neutralizing titers (VNT50) were calculated as the highest serum dilution at which 50 % of the cells remain intact according to neutral red incorporation in the control wells (no virus added).
2.6.2. Cell-mediated immunity
Thirty-four days after the last dose, three to five mice per group were splenectomized under aseptic conditions. The splenocytes were processed as described elsewhere [14]. The cells (2.5 x105 cells per well) were cultured in 96-well round-bottom plates with the RBDf at 10 μg/ml. Concanavalin A (ConA; Sigma, St. Louis, MO, USA) was used as assay positive control. The culture supernatants of splenocytes previously stimulated with the RBDf protein were analyzed for INFγ concentrations by ELISA for mouse IFNγ kit (Mabtech, Nacía, Sweden).
3. Results
3.1. Characterization of the RBDf protein
3.1.1. SDS-PAGE and Western blotting characterization
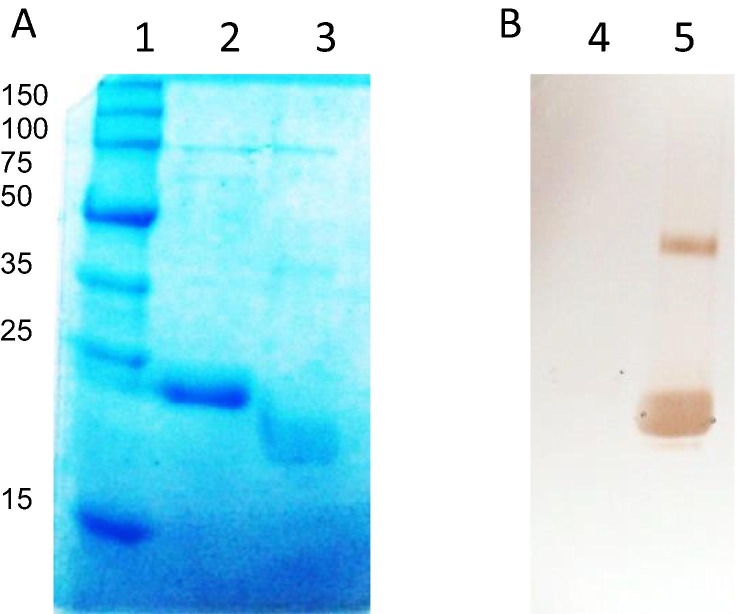
SDS-PAGE analysis of the RBDf protein under reducing conditions shows a unique band migrating at around 22 kDa (Fig. 1 , lane 2) agreeing very well with the theoretical molecular weight value of 22.5 kDa according to amino acid sequence.
Fig. 1.
Analysis of the RBDf protein. A, SDS-PAGE 12.5%; B, Western blotting with the MAb S7. Lanes: 1, molecular weight marker; 2, 4, RBDf under reducing conditions; 3, 5, RBDf under non-reducing conditions.
In addition, the sample under non-reducing conditions (Fig. 1, lane 3) shows a diffuse band with lower molecular weight corresponding to the N-glycosylated monomer.
A monoclonal antibody specific to the SARS-CoV-2 RBD protein was used for the immune-identification of the RBDf protein in reducing and non-reducing conditions by a Western blotting (Fig. 1B). The results indicate that the MAb recognition depends on the formation of the disulphide bonds and the conformation of the molecule. Furthermore, an inmuno-identified additional band, migrating at high molecular weight (around 40 kDa), suggests the formation of dimmers of the RBDf protein under non-reducing conditions (Fig. 1B, lane 5).
3.1.2. Interaction ACE2-RBDf
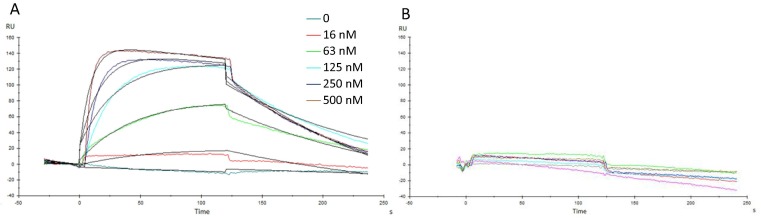
The interaction between RBD and ACE2 was evaluated by Surface Plasmon Resonance measurement. Fig. 1 shows curves of Resonance Units (RU) corresponding to different RBDf protein concentrations assayed (Fig. 2 A). The protein used as negative control did not display a detectable response, regardless of the protein concentration in the sample (Fig. 2B).
Fig. 2.
Surface Plasmon Resonance analysis from RBDf interacting with mFc-ACE2 cell receptor immobilized by capture in a Protein A sensor chip. (A) Sensorgrams corresponding to one of the replicates of the protein dissolved in PBS, pH = 7.2, (B) Non-related protein expressed in S. cerevisiae used as a negative control for the interaction with the immobilized mFc-ACE2 cell receptor. Black lines in panel A are the curves of the Langmuir 1:1 fitting, with drifting baseline (Chi2 ≤ 1.85). RU: response units.
The association rate for RBDf protein, estimated through the experimental data fitting to Langmuir 1:1 model, was 2.7 ± 0.8 × 105 M−1-s−1, while dissociation rate was 8.0 ± 0.7 × 10−6 s−1. The equilibrium was reached between 25 and 30 s, with a dissociation constant KD = 27.2 ± 9.9 × 10−9 M.
3.1.3. Recognition of RBDf by human sera
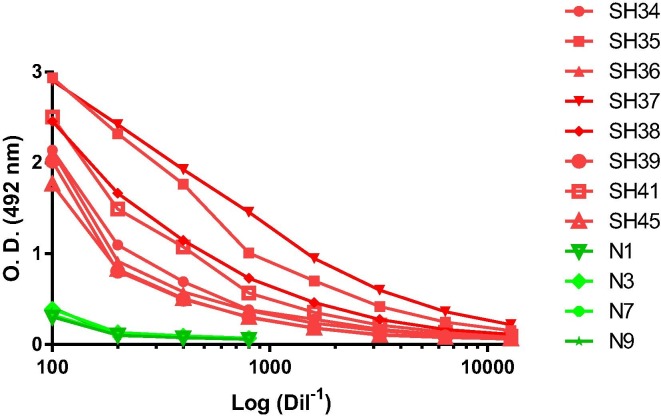
The recognition of RBDf by human IgG antibodies, from COVID-19-convalescent subjects, was analyzed by ELISA. Four negative human sera were included as controls. Fig. 3 shows that the eight positive human sera evaluated recognized the protein while the negative sera showed low levels of reaction.
Fig. 3.
Reactivity of convalescent human sera against the RBDf protein, measured by ELISA. SH 34–39, 41 and 45: convalescents human sera; N1, 3, 7 and 9: negative human sera.
3.2. Immunological evaluation in BALB/c mice
To evaluate the immune response induced by the RBDf protein, BALB/c mice were immunized with different preparations by subcutaneous and intranasal administration routes (Table 1). As a control, the RBD expressed in the HEK293 cell line was also evaluated [11]. The HBV core protein was co-administered with the RBD proteins, taking into account its immune-enhancement capacity previously described. A control group included the simple mixture of both proteins. All the formulations administered by the subcutaneous route included alum as adjuvant.
3.2.1. Humoral immune response
The humoral immune response (IgG) against the RBDf protein, induced in immunized mice, was assayed by ELISA. Fig. 4 shows the IgG anti-RBD, at day 49 of the study (fifteen days after the third dose).
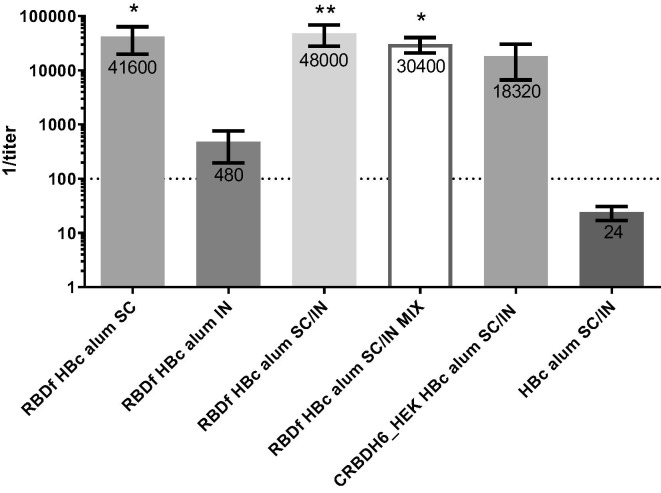
Fig. 4.
IgG anti-RBDf response in mice assayed by ELISA. The chart shows means of titers with standard error media (n = 5). The statistical analysis was performed using Kruskal–Wallis and Dunn’s multiple comparison tests (*p < 0.05; **p < 0.01). All the statistical differences are shown and were detected only compared to the placebo group (HBc alum SC/IN). SC: subcutaneous; IN: intranasal; MIX: simple mixture. Numbers on top of the bars are the antibody titer means. The dashed line indicates the cutoff value.
Mice immunized with the preparation RBDf HBc alum, administrated simultaneously by both immunization routes (RBDf HBc alum SC/IN) developed the highest antibody titers, having statistical differences compared to the placebo group (p < 0.01). In addition, animals immunized only by the subcutaneous route and those immunized with the simple mixture RBDf HBc by the combination of subcutaneous and intranasal routes, showed significantly higher titers of IgG anti-RBDf compared to the placebo group (p < 0.05). Mice immunized with the formulation CRBDH6_HEK alum SC/IN developed IgG levels without statistical differences with respect to the placebo group. No statistical differences were detected among the groups immunized with the different vaccine preparations.
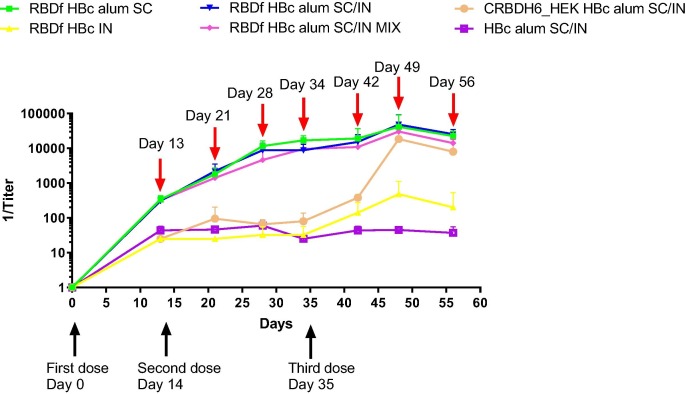
To evaluate the humoral response kinetics, the anti-RBDf IgG titers were determined 13 days after the first dose and 7 days after the remaining doses, by ELISA. All groups of mice immunized by subcutaneous route developed a humoral immune response (Fig. 5 ). The IgG titer reached the maximum values fifteen days after the third dose (day 49 of the study) and decreased 7 days later.
Fig. 5.
Kinetics of anti-RBDf IgG titers in vaccinated mice. Immunized animals were bleed every seven days and titers against RBDf were determined by ELISA. The chart shows the mean of titer plus standard deviations of each group, by bleeding day. Red arrow: bleeding days; Black arrow: dose day. SC: subcutaneous; IN: intranasal; MIX: simple mixture.
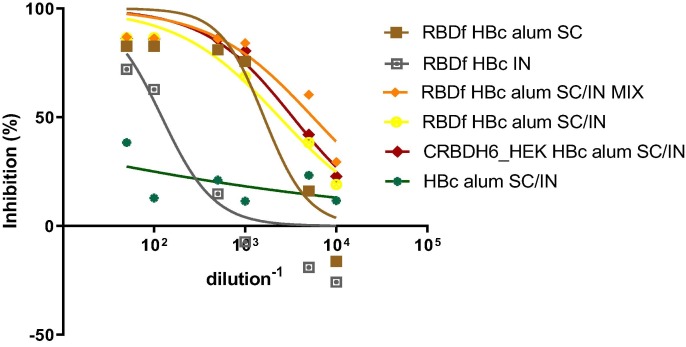
The functionality of the humoral immune response was assayed using one sera pool per group. Firstly, an RBD-ACE2 binding inhibition ELISA was performed. Secondly, the neutralizing capacity of sera was tested by a SARS-CoV-2 virus neutralization test. Table 2 and Fig. 6 show the results achieved with serum pools of the different groups assayed.
Table 2.
Functionality of the humoral immune response.
| Group | mFc-ACE2 inhibition (%) |
mFc-ACE2 inhibition titer (ID50) |
Neutralization Titer (D614G strain) |
Neutralization Titer (Delta strain) |
|---|---|---|---|---|
|
83 | 1607 | 640 | 160 |
|
63 | 122 | ND | ND |
|
87 | 2376 | 320 | 160 |
|
86 | 5535 | 1280 | 160 |
|
86 | 3330 | 320 | 112 |
|
0 | 0 | ND | ND |
Serum pools collected fifteen days after the third dose (day 49 of the study) were evaluated in an inhibition ELISA using murine Fc ACE2 (mFc-ACE2) and RBD human Fc molecules. The neutralization titers were determined against SARS-CoV-2, using the VERO E6 cell line. ID50 is the serum dilution where 50% of binding inhibition is attained. ND: non-determined. SC: subcutaneous; IN: intranasal; MIX: simple mixture.
Fig. 6.
ACE2-RBD binding inhibition curves. SC: subcutaneous; IN: intranasal; MIX: simple mixture.
The highest neutralization titer was achieved in the group of animals immunized with RBDf HBc alum SC/IN MIX vaccine preparation. The serum pools of groups immunized with formulations including RBDf and CRBDH6_HEK in equal conditions (Groups 3 and 5), showed similar neutralization titers and ID50.
3.2.2. Cell-mediated immunity
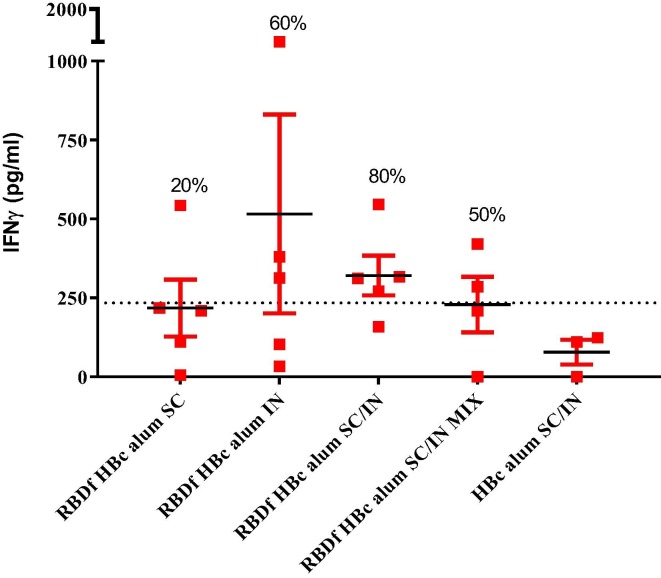
The cellular immune response was evaluated thirty-four days after the third dose (day 69 of the study), measuring the IFNγ levels secreted by spleen cells after in vitro stimulation with the RBDf protein (Fig. 7 ). Eighty percent of animals immunized with the vaccine preparation RBDf HBc alum SC/IN showed a positive IFNγ response (321 ± 141 μg/mL). Despite the absence of humoral response, three out of five mice inoculated by the intranasal route showed cytokine secretion after the stimulation of their splenocytes with the RBD protein (516 ± 705 μg/mL). No mice in the placebo group showed positive levels of IFNγ secretion.
Fig. 7.
Cell-mediated immune response induced in mice. Thirty-four days after the third dose, spleen cells from the animals were stimulated in vitro with the RBDf and the IFNγ concentration in the resulting culture supernatants was determined by ELISA. The chart shows mean ± standard error media (n = 3–5). The dashed line indicates the cutoff value (two times the average of the placebo group); Percentages of animals with positive IFNγ secreting cells response are shown. SC: subcutaneous; IN: intranasal; MIX: simple mixture.
4. Discussion
Several studies support the use of the RBD as a vaccine candidate against COVID-19 [15]. Yang and colleagues showed that a recombinant RBD-based vaccine candidate was able to elicit robust RBD-specific neutralizing antibody response in mice, rabbits and non-human primates [16]. In addition, it protected non-human primates against SARS-CoV-2 challenge. The recombinant RBD was produced using insect cells and the Bac-to-Bac baculovirus expression system [16]. Similarly, a SARS-CoV-2 RBD protein, expressed in the yeast Pichia pastoris induced in mice high levels of IgG antibodies and strong neutralizing antibody titers. Moreover, the vaccine induced IFNγ, IL-6, and IL-10 secretion by stimulated splenocytes [17]. Expi293F human cells have also been used to express a recombinant a RBD fused to the Fc fragment of human IgG. The RBD-Fc protein elicited in mice high titers of RBD-specific antibodies with strong neutralizing activity against SARS-CoV2 [18].
In the present study, we assessed the receptor-binding domain of the SARS-CoV-2 S protein expressed in the fungal C1-system (coded RBDf). Among expression systems for producing recombinant proteins, fungal-based systems are characterized by high-levels of secreted proteins. According to Dyadic®, C1 stable cell line expressed the RBD, after fermentation optimization, at a level of approximately 2–3 g/l in five days [19]. In the case of monoclonal antibody production, C1 expression system is able to produce three to four batches of mAbs in the same time as one bath in CHO cells, reaching a production level of 3.1 g/l/day [19]. Verma and coworkers reviewed antibody molecules expression in bacterial, yeast, insect and mammalian expression systems [20], evidencing lower production levels than those described in fungal C1-system. In general, yeast expression systems produce higher yields than E. coli. and the later higher than mammalian cells. For instance, a fragment of the B72.3 antibody has been expressed in E. coli culture medium up to 450 mg/l in fermentations; however, the yield of B72.3 fragment was 4 mg/l in CHO cells. Similarly, anti-CD7 antibody was expressed at 0.25 mg/l in E.coli, but when same fragments were expressed in Pichia pastoris their yields were increased to 60 mg/l [20].
Moreover, fungal systems become a good choice since they share important processes for the protein expression with other eukaryotic organisms [21]. The fungal C1-expression system has been previously used for the heterologous protein production of diverse industrial enzymes and a human IgG antibody against tumor necrosis factor-alpha (TNFα) [3], as well as several other human proteins including IgG1, IgG4 mAbs, Fabs, bi and tri-specific antibodies, virus-like particles (VLP’s) and hemagglutinin (HA) [19].
Previously, the RBDf was characterized by mass spectrometry using an in-solution buffer-free digestion protocol [22]. Authors concluded that the protein has three predominant glycoforms, Man4, Man4A1 and Man5A1, the last one being predominant. In addition, the electrospray ionization mass spectrometry permitted full-sequence coverage and confirmed the integrity of the N- and C-terminal ends and the correct conformation of the four native disulfide bonds [22].
The first piece of evidence of the RBDf protein functionality was the high-affinity interaction with the ACE2 receptor determined through a BIACORE system. The estimated affinity was in the nanomolar range, in compliance with previous studies concerning the RBD-ACE2 pair [23], [24], [25], [26]. This finding suggests that the RBDf protein expressed in the fungus T. heterothallica could elicit an effective antibody response, capable of blocking virus entry and replication by blocking RBD-ACE2 interaction in real SARS-CoV2 infection events.
The recognition of the RBDf by several convalescent sera was another piece of evidence of the resemblance of the recombinant protein to the viral RBD. It is known that the RBD region is the target of the antibody response in infected subjects; in fact, it is responsible for the neutralizing activity in convalescent human sera [27]. So, this result points out RBDf as an antigen capable of inducing a functional humoral immune response.
The evaluation of the immune response induced in mice by the RBDf included the co-administration of the core protein of the hepatitis B virus. It has been reported that the HBc promotes Th1 immunomodulation of the immune response to co-administered antigens, including the HBV surface antigen, after subcutaneous [28] or nasal [29] inoculations. This adjuvant activity has been related to the nucleic acid content within the particle, which interacts with the Toll-like receptor-3 [30].
This study evaluated two types of HBc-including vaccine preparations: one, favoring the association between the RBDf and the HBc and the other, just mixing both molecules. The electrostatic interaction between the RBD and HBc molecules should favor the uptake of both antigens by dendritic cells, their maturation, and selective polarization toward a Th1 response. However, the IgG humoral immune response induced by both vaccine preparations was similar, even the levels of the RBD-ACE2 binding inhibition and the neutralizing antibody titers were slightly higher in the group of animals immunized with the simple mix. Probably, the protonation of the RBD, carried out to guarantee the RBD-HBc association, affected to some extent, the conformation of the RBD protein and/or the association between both molecules affected the accessibility of some epitopes in the RBD protein.
The mucosal administration route for COVID-19 vaccine candidates has been previously evaluated in different animal models. In the present study we evaluated a vaccine preparation composed only of the HBc and the RBDf protein. Du and coworkers evaluated in mice an RBD-based subunit vaccine using the intranasal administration [31]. These authors included aluminium oxyhydroxide gel in the formulation. Unlike our results, they found functional humoral, systemic and mucosal immunity. It is known that alum cannot be used in mucosal vaccines [32] possibly due to its toxicity and that so far, there is not a mucosal adjuvant licensed for human use. This unfeasible procedure could be the cause of the different results.
The IgG antibody titers induced by both proteins, the RBDf and the RBD expressed in mammalians HEK293 cells, were high and similar. Likewise, the functionality of the humoral immune response, measured by the ACE2-RBD binding inhibition assay and the virus neutralization test, was similar. Neutralizing antibody titers were around two to eight times lower against Delta strain. Similar titer-fall has been previously described [33]. Nevertheless, all these results indicate antigenic similarities between both molecules, the RBDf and CRBDH6_HEKs, and support the fungal C1 expression system for the production of biotherapeutics.
Only a few T cell epitopes have been identified in the RBD region. Recently, Low and coworkers described a conserved and immunodominant CD4 T cell epitope within the RBD [34]. Here, we detected IFNγ-secretion positive responses, after the stimulation of spleen cells from immunized animals, with the RDBf protein. This cytokine has known antiviral and antimicrobial functions and influences multiple cells and cellular activities [35]. Although only five animals per group were evaluated and no statistical test was performed, the group immunized with the vaccine preparation RBDf HBc alum SC/IN, showed the highest percent of animals with positive IFNγ secreting cells response (80%). This result suggests a positive influence of the molecules association on the induction of cell-mediated immunity. Surprisingly, 60% of those immunized by the intranasal route were considered positive. The induction of cellular instead of humoral immune response has been previously described, after the evaluation in mice of vaccine candidates against Dengue virus using a mucosal administration route [36]. However, future studies assessing the immune response induced by the RBDf administered by the intranasal route, should evaluate not only serum IgG but also mucosal IgA and IgG response.
In conclusion, the present study offers additional evidence of the feasibility of the RBD region as a vaccine antigen against COVID-19 and supports the use of the C1-expression system for the development of vaccines, drugs, and biopharmaceuticals reagents for human health. Future studies should focus on the demonstration of protection after the challenge with the live coronavirus, the absence of immune-enhancement, and the induction of a functional immune response against different viral variants.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors especially thank researchers Dionne Casillas-Casanova, Ailyn de la Caridad Ramón Sánchez, as well as the Civilian Defense Scientific Research Center, for their contribution in the microneutralization assays.
References
- 1.Kwok H.F. Review of Covid-19 vaccine clinical trials - A puzzle with missing pieces. Int J Biol Sci. 2021;17(6):1461–1468. doi: 10.7150/ijbs.59170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salvatori G., Luberto L., Maffei M., Aurisicchio L., Roscilli G., Palombo F., et al. SARS-CoV-2 SPIKE PROTEIN: an optimal immunological target for vaccines. J Transl Med. 2020;18(1):222. doi: 10.1186/s12967-020-02392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visser H., Joosten V., Punt P.J., et al. Development of a mature fungal technology and production platform for industrial enzymes based on a Myceliophthora thermophila isolate, previously known as Chrysosporium lucknowense C1. Ind Biotechnol. 2011 Jun;7(3):214–223. [Google Scholar]
- 4.Aebischer A, Wernike K, König P, et al. Development of a Modular Vaccine Platform for Multimeric Antigen Display Using an Orthobunyavirus Model. Vaccines (Basel) 2021 Jun;159(6):651. doi: 10.3390/vaccines9060651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beer M., Conraths F.J., Van der poel W.H.M. 'Schmallenberg virus'–a novel orthobunyavirus emerging in Europe. Epidemiol Infect. 2013;141(1):1–8. doi: 10.1017/S0950268812002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guillén G., Aguilar J.C., Dueñas S., Hermida L., Guzmán M.G., Penton E., et al. Virus-Like Particles as vaccine antigens and adjuvants: application to chronic disease, cancer immunotherapy and infectious disease preventive strategies. Procedia Vaccinol. 2010;2(2):128–133. [Google Scholar]
- 7.Iglesias E., Thompson R., Carrazana Y., Lobaina Y., García D., Sánchez J., et al. Coinoculation with hepatitis B surface and core antigen promotes a Th1 immune response to a multiepitopic protein of HIV-1. Immunol Cell Biol. 2006;84(2):174–183. doi: 10.1111/j.1440-1711.2005.01408.x. [DOI] [PubMed] [Google Scholar]
- 8.Betancourt A.A., Delgado C.A.G., Estévez Z.C., Martínez J.C., Ríos G.V., Aureoles-Roselló S.R.M., et al. Phase I clinical trial in healthy adults of a nasal vaccine candidate containing recombinant hepatitis B surface and core antigens. Int J Infect Dis. 2007;11(5):394–401. doi: 10.1016/j.ijid.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Al Mahtab M., Akbar S.M.F., Aguilar J.C., Guillen G., Penton E., Tuero A., et al. Treatment of chronic hepatitis B naïve patients with a therapeutic vaccine containing HBs and HBc antigens (a randomized, open and treatment controlled phase III clinical trial) PLoS ONE. 2018;13(8):e0201236. doi: 10.1371/journal.pone.0201236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lobaina Y., Hardtke S., Wedemeyer H., Aguilar J.C., Schlaphoff V. In vitro stimulation with HBV therapeutic vaccine candidate Nasvac activates B and T cells from chronic hepatitis B patients and healthy donors. Mol Immunol. 2015;63(2):320–327. doi: 10.1016/j.molimm.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Espinosa L.A., Ramos Y., Andújar I., Torres E.O., Cabrera G., Martín A., et al. In-solution buffer-free digestion allows full-sequence coverage and complete characterization of post-translational modifications of the receptor-binding domain of SARS-CoV-2 in a single ESI-MS spectrum. Anal Bioanal Chem. 2021;413(30):7559–7585. doi: 10.1007/s00216-021-03721-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palenzuela D., Nuñez L., Roca J., et al. Purification of the Recombinant Hepatitis B Core Antigen, and Its Potential Use for the Diagnosis of Hepatitis B Virus Infection. Biotecnología Aplicada. 2002;19:138–142. [Google Scholar]
- 13.Lazo L., Hermida L., Zulueta A., Sánchez J., López C., Silva R., et al. A recombinant capsid protein from Dengue-2 induces protection in mice against homologous virus. Vaccine. 2007;25(6):1064–1070. doi: 10.1016/j.vaccine.2006.09.068. [DOI] [PubMed] [Google Scholar]
- 14.Lazo L., Gil L., López C., Valdés I., Blanco A., Pavón A., et al. A vaccine formulation consisting of nucleocapsid-like particles from Dengue-2 and the fusion protein P64k-domain III from Dengue-1 induces a protective immune response against the homologous serotypes in mice. Acta Trop. 2012;124(2):107–112. doi: 10.1016/j.actatropica.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Valdes-Balbin Y., Santana-Mederos D., Paquet F., Fernandez S., Climent Y., Chiodo F., et al. Molecular Aspects Concerning the Use of the SARS-CoV-2 Receptor Binding Domain as a Target for Preventive Vaccines. ACS Cent Sci. 2021;7(5):757–767. doi: 10.1021/acscentsci.1c00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J., Wang W., Chen Z., et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586(7830):572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 17.Pollet J., Chen W.H., Versteeg L., et al. SARS-CoV-2 RBD219-N1C1: A yeast-expressed SARS-CoV-2 recombinant receptor-binding domain candidate vaccine stimulates virus neutralizing antibodies and T-cell immunity in mice. Hum Vaccin Immunother. 2021 Apr;13:1–11. doi: 10.1080/21645515.2021.1901545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z., Xu W., Xia S., Gu C., Wang X., Wang Q., et al. RBD-Fc-based COVID-19 vaccine candidate induces highly potent SARS-CoV-2 neutralizing antibody response. Signal Transduct Target Ther. 2020;5(1):282. doi: 10.1038/s41392-020-00402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyadic Thermophilic Filamentous Fungus (C1 platform) Recombinant Production of Glycoprotein Antigen Vaccines A&OTPP. C1 Technology a transformational platform that transcends the limits of legacy protein production technologies. 2021. PEGS Conference May. 2021 [Google Scholar]
- 20.Verma R., Boleti E., George A.J.T. Antibody engineering: comparison of bacterial, yeast, insect and mammalian expression systems. J Immunol Methods. 1998;216(1-2):165–181. doi: 10.1016/s0022-1759(98)00077-5. [DOI] [PubMed] [Google Scholar]
- 21.Gomes A.R., Byregowda S.M., Veeregowda B.M., Balamurugan V. Belamara nahallyMuniveera ppa Veeregowda, Vinayagamurthy Balamurugan. An Overview of Heterologous Expression Host Systems for the Production of Recombinant Proteins. Adv Animal Veterinary Sci. 2016;4(7):346–356. [Google Scholar]
- 22.Espinosa LA, Ramos Y, Andújar I, et al. In-solution buffer-free digestion for the analysis of SARS-CoV-2 RBD proteins allows a full sequence coverage and detection of post-translational modifications in a single ESI-MS spectrum. Anal Bioanal Chem. 2021 Dec;413(30):7559–7585. doi: 10.1007/s00216-021-03721-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang JA, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Zhang Y, Wu L, et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan J, Ge J, Yu JA, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 26.Sui J, Deming M, Rockx B, et al. Effects of human anti-spike protein receptor binding domain antibodies on severe acute respiratory syndrome coronavirus neutralization escape and fitness. J Virol. 2020;88(23):13769–13780. doi: 10.1128/JVI.02232-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J., Liang B., Chen C., Wang H., Fang Y., Shen S., et al. SARS-CoV-2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID-19. Nat Commun. 2021;12(1):1813. doi: 10.1038/s41467-021-22034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riedl P., Buschle M., Reimann J., Schirmbeck R. Binding immune-stimulating oligonucleotides to cationic peptides from viral core antigen enhances their potency as adjuvants. Eur J Immunol. 2002;32:1709–1716. doi: 10.1002/1521-4141(200206)32:6<1709::AID-IMMU1709>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 29.Lobaina Y., Trujillo H., García D., Gambe A., Chacon Y., Blanco A., et al. The effect of the parenteral route of administration on the immune response to simultaneous nasal and parenteral immunizations using a new HBV therapeutic vaccine candidate. Viral Immunol. 2010;23(5):521–529. doi: 10.1089/vim.2010.0024. [DOI] [PubMed] [Google Scholar]
- 30.Riedl P., Stober D., Oehninger C., Melber K., Reimann J., Schirmbeck R. Priming Th1 immunity to viral core particles is facilitated by trace amounts of RNA bound to its arginine-rich domain. J Immunol. 2002;168(10):4951–4959. doi: 10.4049/jimmunol.168.10.4951. [DOI] [PubMed] [Google Scholar]
- 31.Du Y., Xu Y., Feng J., Hu L., Zhang Y., Zhang B.o., et al. Intranasal administration of a recombinant RBD vaccine induced protective immunity against SARS-CoV-2 in mouse. Vaccine. 2021;39(16):2280–2287. doi: 10.1016/j.vaccine.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moschos S.A., Bramwell V.W., Somavarapu S., Alpar H.O. Adjuvant synergy: the effects of nasal coadministration of adjuvants. Immunol Cell Biol. 2004;82(6):628–637. doi: 10.1111/j.0818-9641.2004.01280.x. [DOI] [PubMed] [Google Scholar]
- 33.Hassan A.O., Shrihari S., Gorman M.J., Ying B., Yaun D., Raju S., et al. An intranasal vaccine durably protects against SARS-CoV-2 variants in mice. Cell Rep. 2021;36(4):109452. doi: 10.1016/j.celrep.2021.109452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Low J.S., Vaqueirinho D., Mele F., et al. Clonal analysis of immunodominance and cross-reactivity of the CD4 T cell response to SARS-CoV-2. Science. 2021;372(6548):1336–1341. doi: 10.1126/science.abg8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haji Abdolvahab M., Moradi-kalbolandi S., Zarei M., Bose D., Majidzadeh-A K., Farahmand L. Potential role of interferons in treating COVID-19 patients. Int Immunopharmacol. 2021;90:107171. doi: 10.1016/j.intimp.2020.107171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lazo Vázquez L., Gil González L., Marcos López E., Pérez Fuentes Y., Cervetto de Armas L., Brown Richards E., et al. Evaluation in Mice of the Immunogenicity of a Tetravalent Subunit Vaccine Candidate Against Dengue Virus Using Mucosal and Parenteral Immunization Routes. Viral Immunol. 2017;30(5):350–358. doi: 10.1089/vim.2016.0150. [DOI] [PubMed] [Google Scholar]