Abstract
Objectives
To: 1. Describe the frequency of viral RNA detection in stools in a cohort of patients infected with SARS-CoV-2, and 2. Perform a systematic review to assess the clearance time in stools of SARS-CoV-2.
Methods
We conducted a prospective cohort study in two centers between March and May 2020. We included SARS-CoV-2 infected patients of any age and severity. We collected seriated nasopharyngeal swabs and stool samples to detect SARS-CoV-2. After, we performed a systematic review of the prevalence and clearance of SARS-CoV-2 in stools (PROSPERO-ID: CRD42020192490). We estimated prevalence using a random-effects model. We assessed clearance time by using Kaplan–Meier curves.
Results
We included 32 patients; mean age was 43.7 ± 17.7 years, 43.8% were female, and 40.6% reported gastrointestinal symptoms. Twenty-five percent (8/32) of patients had detectable viral RNA in stools. The median clearance time in stools of the cohort was 11[10–15] days. Systematic review included 30 studies (1392 patients) with stool samples. Six studies were performed in children and 55% were male. The pooled prevalence of viral detection in stools was 34.6% (twenty-four studies, 1393 patients; 95%CI:25.4–45.1); heterogeneity was high (I2:91.2%, Q:208.6; p ≤ 0.001). A meta-regression demonstrates an association between female-gender and lower presence in stools (p = 0.004). The median clearance time in stools was 22 days (nineteen studies, 140 patients; 95%CI:19–25). After 34 days, 19.9% (95%CI:11.3–29.7) of patients have a persistent detection in stools.
Conclusions
Detection of SARS-CoV-2 in stools is a frequent finding. The clearance of SARS-CoV-2 in stools is prolonged and it takes longer than nasopharyngeal secretions.
Keywords: Diarrhea, 2019-nCoV-2, Coronavirus, COVID-19, SARS-CoV-2, Novel coronavirus
Abstract
Objetivos
1. Evaluar la detección de ARN viral en deposiciones y su tiempo de excreción en una cohorte de pacientes con SARS-CoV-2; 2. Realizar una revisión sistemática para evaluar el tiempo de excreción en deposiciones del SARS-CoV-2.
Métodos
Estudio de cohorte prospectiva en dos centros entre marzo-mayo del 2020. Incluimos pacientes infectados con SARS-CoV-2 de cualquier edad y gravedad. Recolectamos secreciones nasofaríngeas y deposiciones en forma seriada para detectar SARS-CoV-2. También realizamos una revisión sistemática de la prevalencia y excreción del SARS-CoV-2 en deposiciones (PROSPERO-ID: CRD42020192490). Estimamos la prevalencia usando un modelo de efectos aleatorios. Evaluamos el tiempo de excreción usando curvas Kaplan-Meier.
Resultados
Incluimos 32 pacientes; edad media 43 ± 17,7 años, 43,8% eran mujeres y 40,6% reportaron síntomas gastrointestinales. Veinticinco por ciento (8/32) tenían ARN viral detectable en deposiciones. La mediana de excreción en deposiciones fue 11 [10-15] días. La revisión sistemática incluyó 30 estudios (1.392 pacientes) con muestras en deposiciones. Seis estudios fueron realizados en niños y 55% eran hombres. La prevalencia estimada de detección viral en deposiciones fue de 34,6% (24 estudios, 1.393 pacientes; IC 95%: 25,4-45,1); la heterogeneidad fue elevada (I2: 91,2%; Q: 208,6; p ≤ 0,001). Una metarregresión demostró una asociación entre el género femenino y menor prevalencia en deposiciones (p = 0,004). La mediana de tiempo de excreción fueron 22 días (19 estudios, 140 pacientes; IC 95%: 19-25). Tras 34 días, 19,9% (IC 95%: 11,3-29,7) de los pacientes tenían detección persistente en deposiciones.
Conclusiones
La detección de SARS-CoV-2 en deposiciones es un hallazgo frecuente. La excreción del SARS-CoV-2 en deposiciones es prolongada y es más tardía que en secreciones nasofaríngeas.
Palabras clave: Diarrea, 2019-nCoV-2, Coronavirus, COVID-19, SARS-CoV-2, Nuevo coronavirus
Introduction
The current pandemic of a novel Severe Acute Respiratory Syndrome coronavirus, named SARS-CoV-2, has constituted a new threat worldwide.1 More than 13,165,000 cumulative cases have been confirmed in 188 countries across five continents, reaching a mortality rate of 4.9%.2
The novel coronavirus was first described in patients with pneumonia, and the most common symptoms included fever, dry cough, and dyspnea.3 Gastrointestinal symptoms are less common and include diarrhea, nausea, vomiting, and abdominal discomfort. The prevalence of these symptoms varies significantly among different studied populations, which can be mild at the early onset, but is frequently followed by typical respiratory manifestations.4 A recent systematic review showed that 17.6% of individuals infected with SARS-CoV-2 presented with gastrointestinal symptoms; the most frequent being anorexia 26.8%, diarrhea 12.5%, nausea and vomiting 10.2%, and abdominal pain 9.2%.5 The presence of gastrointestinal symptoms is frequent in hospitalized patients, and these became more pronounced as the severity of the disease increased.6, 7
Several reports have shown the detection of SARS-CoV-2 RNA in stools for prolonged periods, suggesting that the virus could be transmitted through the digestive tract.8 The viral detection in stools is variable among several studies, with frequencies ranging between 15.3% and 70.3%.5, 8, 9, 10 Interestingly, recent publications have described enterocyte viral invasion and detection of viable virus in stools, suggesting that the fecal-oral route could provide a source of transmission.4, 10
To date, the relevance of viral presence in stools and length of shedding has not been established adequately. In this study, we aimed to assess the viral presence and clearance of SARS-CoV-2 in stools of infected Chilean patients. We also performed a systematic review to establish the frequency of SARS-CoV-2 detection in stools, and median clearance time in nasopharyngeal secretions and stools.
Material and methods
Cohort study: baseline characteristics and sample collection
Patient clinical–epidemiological data and clinical samples were collected after informed written consent was obtained. This study was reviewed and approved by the Scientific Ethics Committee of Pontificia Universidad Católica de Chile (PUC) (ID 16-066).
All patients diagnosed with COVID19 were eligible to participate upon consent, including hospitalized individuals and outpatients, of any age and severity. The study protocol did not have any exclusion criteria. Between March 5 and May 28, 2020, patients diagnosed as positive for SARS-CoV-2 were recruited at two centers, which are part of the Healthcare Network of the Faculty of Medicine, Pontificia Universidad Católica de Chile: The Red de Salud UC-Christus Hospital (Santiago, Chile) and the Clínica UC San Carlos de Apoquindo (Santiago, Chile). The diagnosis of SARS-CoV-2 form a nasopharyngeal swab (NPS) specimen was confirmed by a qRT-PCR Light Mix Modular Wuhan CoV-2 RdRP-gene (TibMolBiol, ROCHE), performed at the Institutional Infectious Diseases and Molecular Virology Laboratory (Santiago, Chile).
For clinical information, we performed an interview and a survey to characterize the presence and timing of symptoms. We recorded the physical examination and primary laboratory data obtained during medical evaluation. All the patients were characterized by sociodemographic data and comorbidities. We collected NPS and stool or rectal swabs between days 2 to 35 after the onset of symptoms and tested for the presence of SARS-CoV-2 by qRT-PCR. All the information was registered in an anonymous database where sensitive information was excluded. The stages of this process, collecting, transporting and processing samples are illustrated in Fig. 1 . Additional data of RNA extraction and viral genome detection are described in Supplementary Methods.
Figure 1.
Stages of collection, transporting, and processing of samples from SARS-CoV-2 infected patients. NSP: nasopharyngeal swabs and aspirates.
Data analysis of the cohort study
We assessed the normality assumption by means of the Shapiro–Wilk test. Descriptive statistics were obtained using the mean and standard deviation (SD) for continuous variables with normal distribution and non-normal distributions were described with the median and interquartile range. In the case of categorical variables, the proportions and percentage were estimated.
Systematic review of the Viral RNA presence in stools
We performed a systematic review to establish the prevalence of viral detection in stools and median clearance time among previous published data and our cohort. This systematic review with meta-analysis was registered on PROSPERO (CRD42020192490) and followed a prespecified analysis plan. This study is reported in accordance with the Preferred Reporting Items for a Review and Meta-analysis (PRISMA) guidelines.11
Eligibility criteria
Eligible studies had to include patients diagnosed with COVID-19, regardless of age and gender. The diagnosis of COVID-19 had to be based on a compatible clinical history and molecular evidence with a qRT-PCR for SARS-CoV-2. We included studies reporting the frequency of positive viral RNA testing in stool. We considered stool samples at any time of the course of the disease.
We included studies that reported the main outcome, regardless of the design (case-reports, case-series, descriptive cases, case-control studies, cross-sectional studies, cohort studies and randomized controlled trials). We excluded studies performed in vitro, animal models, or lacking evidence of SARS-CoV-2 infection from this systematic review.
Search strategy and selection process
We conducted an electronic search from December 1, 2019, to June 3, 2020 in MEDLINE (via PubMed) database. We used included terms related to SARS-CoV-2 and stools in the search strategy. References of studies searched were also included. We hand searched (up to June 3, 2020) preprint servers (bioRxiv, medRxiv, and SSRN) and coronavirus resource centers of The Lancet, JAMA, and New England Journal of Medicine. We did not limit our search by language. Two investigators (LAD and DR) independently screened the titles and abstracts to ascertain whether each study met the eligibility criteria. The full texts of the identified eligible articles were then evaluated to determine whether they should be included in the analysis. Disagreements between the two reviewers were resolved by consensus. In case of persistent disagreement, arbitration by a third reviewer (JO) settled the discrepancy.
Data collection and risk of bias assessment
Two authors (LAD and DR) independently extracted data from the studies included using forms specially designed for this purpose. The following data describing the design of the study were extracted: number of participants, age, gender, ethnicity, hospitalization, presence of viral RNA in stools, clearance time and presence of gastrointestinal symptoms. Discrepancies were resolved by a third reviewer (JO). Two investigators (LAD and DR) independently assessed the risk of bias of each included study with the Appraisal tool for Cross-Sectional Studies (AXIS) checklist for cross-sectional studies, the Institute of Health Economics (IHE) checklist tool for Case Series, and the Newcastle-Ottawa Scale (NOS) for case-control studies and cohort studies, and the Cochrane Risk of Bias Tool for randomized trials and quasi-experimental studies.
Outcomes
The main outcome was the prevalence of detection of viral RNA in stools of COVID-19 patients. This data was registered as an absolute number and percentage of the total cases. The secondary outcome was the clearance time (defined as the days between onset of symptoms and a negative test result for viral RNA in stools).
Systematic review analysis
We estimated prevalence or event rates in the form of a proportion (with confidence interval 95%). Proportions were pooled using random-effects models. Only those studies with a sample size of at least 10 patients were included in the meta-analysis to decrease bias caused by sampling error.12 We used Q statistic and I 2 to quantify heterogeneity. We planned a subgroup analysis according to age, gender, ethnicity, presence of gastrointestinal symptoms and hospitalization. We stratified age in three subgroups: pediatrics (patients with less than 18 years old), adults (patients over 18 years old), and general (patients of all ages). Ethnicity was defined per region of origin (Asian versus not-Asian studies). We stratified gender by predominant gender (the gender with a prevalence higher than 50%). Presence of gastrointestinal symptoms was arbitrarily grouped in high (50% or more) and low (lesser than 50%) prevalence of gastrointestinal symptoms. In the case of studies with very low or very high proportion, we used a double arcsine transformation.13 Small study effect was evaluated with a funnel plot. Random effects meta-regression was used to examine whether baseline characteristics covariates explained heterogeneity of proportions between studies.14 As such, subgroup analysis should be considered as exploratory. Clearance of viral RNA in stools was defined as the time between onset of symptoms and the last detection of SARS-CoV-2 in stools. We registered the disaggregated data from studies that reported clearance in NPS and stools for each participant. Based on this data, we performed Kaplan–Meier survival curves to estimate viral clearance in NPS and stools. We compared clearance in NPS and stools curves with log-rank test. The analyses were done with the use of STATA (version 16, StataCorp, College Station, Texas) and R software for statistical computing version 3.6.1.15 We included the STATA metaprop command to perform meta-analyses.16
Grading of evidence
The quality of evidence for the outcomes was graded with the GRADE framework.
Role of the funding source
The funding source only provided support for the analysis of the nasopharyngeal and stool samples. The researchers did not receive a fee or other incentives for the systematic review process.
Results
Gastrointestinal symptoms and stool detection in our cohort study
We included 32 patients recruited between the 3rd March 2020 and 28th May 2020. Median age was 43.7 ± 17.7 years and 14 (43.8%) were females. Nineteen (59.4%) patients were hospitalized and 6 (31.6%) required invasive mechanical ventilation. One of our patients died at the end of this study (mortality rate of 3.1%).
In our cohort, the most common extraintestinal manifestations were fever (90.6%), cough (81.3%), myalgia (50%), headache (50%), dyspnea (40.6%), anosmia (37.5%), chills (34.4%), ageusia (28.1%), asthenia (25%) and odynophagia (18.8%). In addition, thirteen patients (40.6%) referred to gastrointestinal symptoms, including: diarrhea (37.5%), nausea (12.5%), abdominal pain (6.3%), vomiting (3.1%), and constipation (3.1%). The median NPS and stool samples obtained were 3 [2–4] and 2 [1–3], respectively. After diagnosis, Viral RNA was detected in respiratory secretions of 28 patients (87.5%), and in stools of 8 (25%) patients. Three patients (37.5%) with viral detection in stools presented with gastrointestinal symptoms: three of them declared to have diarrhea. Six out of 19 individuals that were hospitalized had at least one gastrointestinal symptom during infection. Participant characteristics and gastrointestinal symptoms are summarized in Table 1 . Positive stool samples were detected at different time points (days 4–18)(Supplementary material). The median clearance time in stools was 11 [10–15] days.
Table 1.
Baseline characteristics of our cohort according to outpatient or hospitalized care.
| Global (%) (N = 32) | Outpatient care (%) (N = 13) | Hospitalized patients (%) (N = 19) | P value | |
|---|---|---|---|---|
| Age (mean) | 43.7 ± 17.7 | 38.0 ± 13.6 | 47.7 ± 19.4 | 0.131 |
| Gender | ||||
| Male | 18 (56.2) | 5 (38.5) | 13 (68.4) | 0.093 |
| Female | 14 (43.8) | 8 (61.5) | 6 (31.6) | |
| Gastrointestinal symptoms | ||||
| Diarrhea | 12 (37.5) | 6 (46.2) | 6 (31.6) | 0.403 |
| Constipation | 1 (3.1) | 0 (0) | 1 (5.3) | 0.401 |
| Nausea | 4 (12.5) | 2 (15.4) | 2 (10.5) | 0.683 |
| Vomiting | 1 (3.1) | 1 (7.7) | 0 (0) | 0.219 |
| Abdominal pain | 2 (6.3) | 0 (0) | 2 (10.5) | 0.227 |
| SARS-CoV-2 detection in stools | 8 (25) | 4 (30.8) | 4 (21.1) | 0.533 |
| Mechanical ventilation | – | – | 6 (31.6) | – |
| Deaths | 1 (3.1) | 0 (0) | 1 (5.3) | 0.401 |
Systematic review of SARS-CoV-2 detection in stools
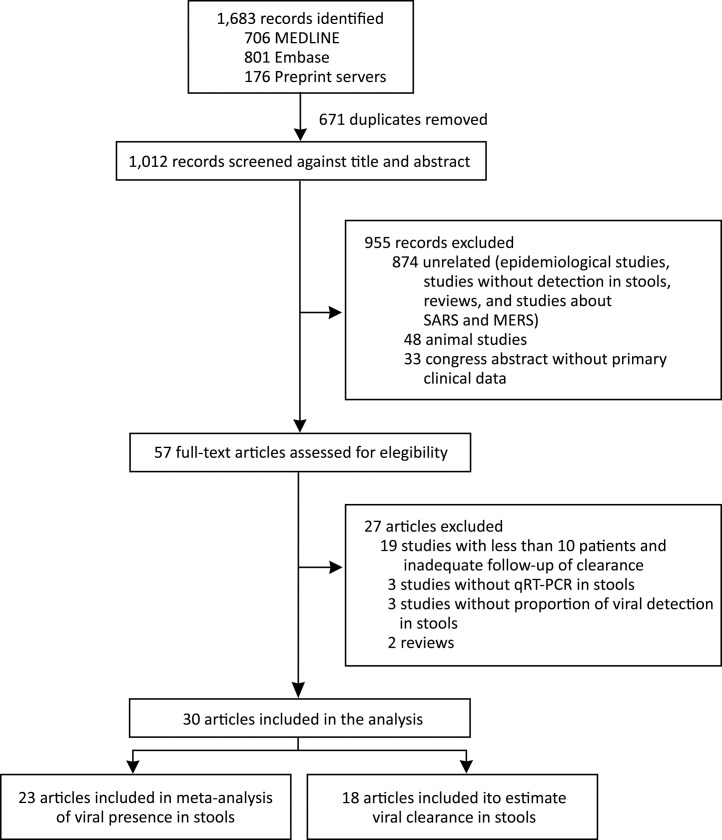
We identified 1683 records and 671 were duplicated. After a screening process against title and abstract, we obtained 59 full-text articles that were assessed for eligibility. We finally selected 30 studies for our systematic review from 6 countries (China, France, Germany, Hong Kong, Singapore, and the United States). We included 23 articles in the meta-analysis of prevalence of SARS-CoV-2 detection in stools and 18 articles were used to estimate clearance in stools. We also included data of the current study in both analyses. The selection process is described in Fig. 2 . All studies were observational in design; no randomized trials were identified. Among the 30 studies, 14 were case series and 16 cohort studies. The study with the largest number of stool samples included a total of 273 cases.17 The risk of bias assessment results is presented in supplementary material.
Figure 2.
Study selection for the systematic review.
The pooled studies included a total of 1392 patients that were tested by SARS-CoV-2 qRT-PCR in stools. Fifty-five percent were male and 6 studies were performed exclusively in children. Twenty-two out of 30 (73.3%) studies included only hospitalized patients. The grade of evidence was considered very low and low, since all the information emerged from case series and cohort studies, respectively. We performed subgroup analyses per age, gender, ethnicity, presence of gastrointestinal symptoms, and hospitalization. Table 2 summarizes the main baseline characteristics summary of each study.
Table 2.
Main characteristics of studies included in the analysis.
| First author | Country | Date | Population | Gender (female) (%) | Exclusively hospitalized patients | Number of patients with stool samples | Follow-up of clearance in stools |
|---|---|---|---|---|---|---|---|
| Chen C29 | China | March 2020 | General | 8 (36.36) | Yes | 19 | Yes |
| Chen Y27 | China | April 2020 | Adult | 27 (64.29) | Yes | 42 | Yes |
| Chen W49 | China | February 2020 | Adult | Not reported | Yes | 28 | Yes |
| Cheung KS5 | Hong Kong | April 2020 | Adult | 32 (54.24) | No | 59 | No |
| Dreher M50 | Germany | April 2020 | Adult | 17 (34) | Yes | 15 | No |
| Han C51 | China | April 2020 | Adult | 115 (55.8) | Yes | 22 | No |
| Huang JT17 | China | June 2020 | Adult | 157 (51) | Yes | 273 | No |
| Jiang X52 | China | April 2020 | General | 2 (66.7) | No | 3 | No |
| Jiehao C53 | China | February 2020 | Pediatrics | 6 (60) | Yes | 6 | Yes |
| Kujawski S54 | USA | March 2020 | Adult | 4 (33.3) | No | 10 | Yes |
| Li H55 | China | February 2020 | Adult | 18 (62.1) | No | 29 | Yes |
| Lescure FX56 | France | March 2020 | Adult | 2 (40) | Yes | 5 | No |
| Pan Y57 | China | February 2020 | Not reported | Not reported | No | 17 | No |
| Wang W10 | China | February 2020 | General | 66 (32) | No | 153 | No |
| Wei XS58 | China | March 2020 | Adult | 56 (66.6) | Yes | 84 | No |
| Wolf GK59 | Germany | February 2020 | Pediatrics | 2 (66) | Yes | 3 | Yes |
| Wu Y33 | China | March 2020 | Adult | 35 (47.3) | Yes | 74 | Yes |
| Xiao F 139 | China | January 2020 | Not reported | Not reported | No | 28 | No |
| Xiao F 260 | China | February 2020 | General | 32 (43.8) | Yes | 73 | No |
| Xing YH61 | China | March 2020 | Pediatrics | 1 (33.3) | Yes | 3 | Yes |
| Xiong XL62 | China | May 2020 | Pediatrics | 94 (38.52) | No | 105 | No |
| Xu Y63 | China | April 2020 | Pediatrics | 4 (40) | Yes | 10 | Yes |
| Yang Z64 | China | February 2020 | Adult | 1 (33.3) | Yes | 3 | Yes |
| Young BE65 | Singapore | March 2020 | Adult | 9 (50) | Yes | 8 | Yes |
| Zautner AE66 | Germany | April 2020 | Adult | 6 (35.3) | Yes | 12 | No |
| Zhang J67 | China | February 2020 | Adult | 7 (50) | Yes | 14 | No |
| Zeng L68 | China | March 2020 | Pediatrics (Newborns) | 14 (42.4) | Yes | 33 | Yes |
| Zhang W 132 | China | February 2020 | Not reported | Not reported | Yes | 39 | No |
| Zhang W 232 | China | February 2020 | Not reported | Not reported | Yes | 139 | No |
| Zheng S69 | China | February 2020 | Adult | 38 (39.6) | Yes | 83 | No |
Detection of viral RNA in stools
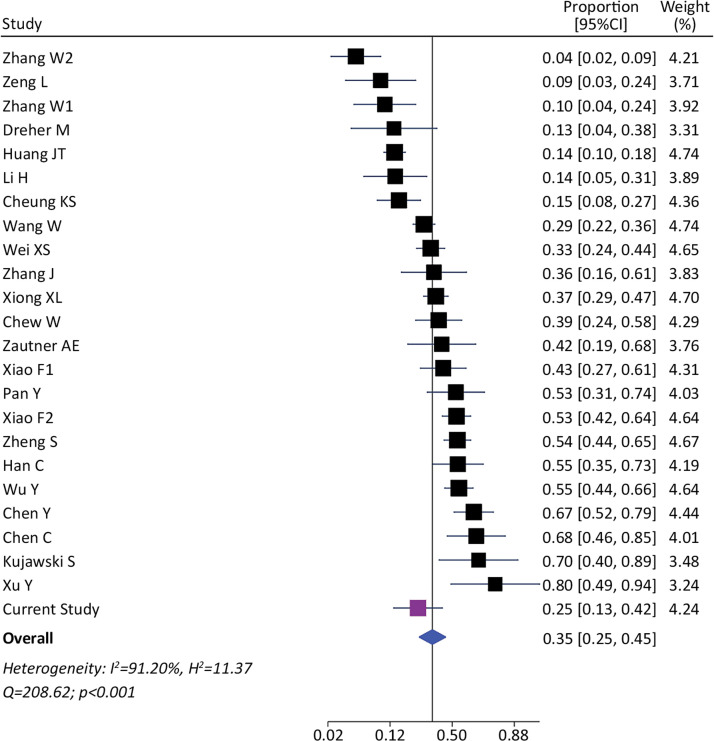
We identified 1393 patients from 23 studies and our current study, which tested the presence of viral RNA in stools (including our cohort). The pooled prevalence of viral detection in stools was 34.6% (twenty-four studies, 1393 patients; 95%CI: 25.4–45.1); the heterogeneity among studies was high (I 2: 91.2%, Q: 208.6; p ≤ 0.001) (Fig. 3 ). The funnel plot of this analysis is provided in supplementary material (Fig. 1S). Subgroup analysis by gender showed a prevalence of 30% in female-predominant studies (six studies, 509 patients; 95%CI: 15.0–51.0) and 42% in male-predominant studies (seven studies, 208 patients; 95%CI: 29.0–57.0). However, there was no significant differences between subgroups (p = 0.30), and heterogeneity was high [(I 2: 92.8%, Q: 66.9; p ≤ 0.001) and (I 2: 73.7%, Q: 19.2; p ≤ 0.001), respectively]. Subgroup analysis by age, ethnicity, presence of gastrointestinal symptoms and hospitalization did not identify relevant differences to global pooled prevalence (Supplementary material). The heterogeneity was high in those four subgroups analysis. We performed a meta-regression of 8 studies including age, gender, and presence of gastrointestinal symptoms. The I 2 was 60.6% and the R 2 was 63.1%, with a p-value = 0.038 of the model. For each increase of 1% of female in the sample, proportion of SARS-CoV-2 detection in stools decreased a 0.5% (p = 0.004). The age and presence of gastrointestinal symptoms were not associated to increase in the proportion of SARS-CoV-2 detection in stools (p = 0.816 and p = 0.103, respectively).
Figure 3.
Forest plots of prevalence of viral detection in stools from SARS-CoV-2 infected patients.
Clearance time of viral RNA in stools
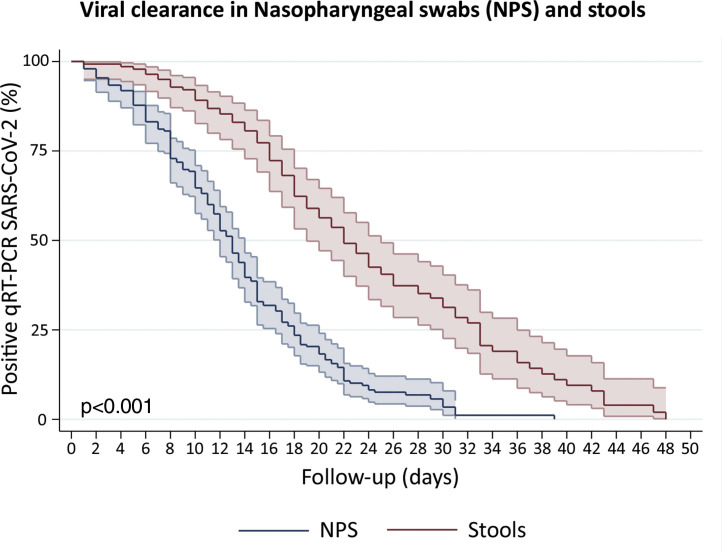
We included patients with viral detection in stool samples from 18 studies and our current study to assess viral clearance in stools. From a total of 442 patients, 423 were tested in stools and only 140 had an adequate prospectively follow-up. We also included 197 patients with nasopharyngeal samples follow-up. The percentage of viral absence in stools at the end of follow up was 72.6% (95%CI: 64.39–79.51). The median clearance time of viral RNA detection in NPS and stools was 13 days (95%CI: 12–14) and 22 days (95%CI: 19–25), respectively (Fig. 4 ). At day 10, only 10.8% (95%CI: 6.7–17.3) of patients presented virus clearance in stools. Interestingly, after 34 days, the 19.9% (95%CI: 11.3–29.7) of patients still had a persistent detection of viral RNA in stools.
Figure 4.
Survival curves to estimate clearance of SARS-CoV-2 in Nasopharyngeal swabs (NPS) and stools. We disaggregated information for each patient to obtain individualized data. Clearance was estimated considering the time between onset of symptoms and the last viral detection in NPS or stools. Survival estimates were performed using Kaplan–Meier curves and we compared both curves with log-rank test.
Discussion
In this study, we analyzed general clinical data from 32 SARS-CoV-2 infected patients. In the first stage, we found that 40.6% of the studied population presented with gastrointestinal symptoms: 37.5% reported diarrhea, 12.5% nausea, 6.3% abdominal pain, 3.1% vomiting, and 3.1% constipation. A significant percentage of our cohort required hospitalization 59.4%, reaching a mortality of 3.1%. We determined that 25% of patients had detectable stool viral RNA during illnesses. Posteriorly, we conducted a systematic review from 30 studies and the data obtained from our cohort phase. The systematic review of the evidence found that the proportion of viral RNA detection in stools is 34.6%, with a median clearance time in stools of 22 days (95%CI 19–25).
In our cohort, diarrhea and nausea were the most prevalent symptoms, highlighting and confirming the diverse clinical manifestation induced by this novel coronavirus.5, 18, 19, 20, 21 The pooled prevalence of SARS-CoV-2 detection in stools was 34.6%, which reflects gastrointestinal shedding in a considerable number of patients. On the other hand, there is an important number of patients without SARS-CoV-2 detection in stools. The differences among detection in stools could potentially be explained by time since onset of symptoms and collection of stool samples, inadequate transport/storage conditions, differences in sample processing, or characteristics inherent to the virus or patients.
The SARS and SARS-CoV-2 strains share approximately 78% identity at the amino acid level.22 It has been proposed that SARS could enter into cells using the Angiotensin-converting Enzyme 2 (ACE2) as a receptor, which is widely distributed on the respiratory and gastrointestinal tract, could potentially allow the entry of the virus in the intestinal tract.23 Hence, given their similarity in tropism, SARS-CoV-2 could also enter cells by similar mechanism.24, 25 Moreover, since ACE2 modulates intestinal inflammation, it is plausible that SARS-CoV-2 infection could disrupt the ACE2 function, promoting fecal-oral transmission.26
Remarkably, the Kaplan–Meier clearance curves showed a median clearance time in NPS of 13 days (95%CI: 12–14), which was significantly lower than clearance in stools (p < 0.001). In fact, viral RNA detection in stools could occur for at least another nine (for 22 days or more) in the half proportion of individuals tested (Fig. 4). These findings suggest a longer shedding period in the gastrointestinal tract, which is in agreement with reports from cohort studies in Hong Kong, Macau, Beijing and Germany,27, 28, 29, 30 and a case study in Italy.31
The identification of prolonged SARS-CoV-2 RNA shedding in rectal swabs was reported as early as mid-February 2020 by Zhang et al.32 Therefore, the potential of viral transmission through the oral-fecal route has been cautioned since then, and recent additional evidence supports this idea. First, it was reported that viral RNA can be detected in fecal samples of infected individuals for up to 11.2 days after respiratory samples became negative and up to 47 days after symptom onset33; secondly, virus RNA was found at high concentrations in stool, and occasionally viral sub genomic messenger RNA (only present in replicating cells) can also be detected in these specimens; third, endoscopic studies of SARS-CoV-2 infected individuals revealed esophageal bleeding, erosions and ulcers in one patient and viral RNA was detected in different gastrointestinal tissues of 3 individuals.34 Fourth, several studies demonstrated the presence of viral RNA in untreated wastewater from the Netherlands, Italy, Australia, and Chile.35, 36, 37, 38 Interestingly, two studies demonstrated that viral isolation from feces was possible.10, 39 Additionally, two recent studies reported the infection and replication of SARS-CoV-2 on intestinal organoids.40, 41 Altogether, our current study and previous reports provide evidence strongly suggesting that SARS-CoV-2 can infect and possibly replicate in the gastrointestinal tract, supporting the oral-fecal route of SARS-CoV-2 as a potential source for transmission. This highlights the need to exercise increased caution when handling and disposing of stool samples, and when conducting endoscopic procedures in SARS-CoV-2 infected individuals.
SARS-CoV-2 detection could also be helpful in monitoring and tracking SARS-CoV-2 circulation, especially during massive outbreaks with limited testing capacity. Interestingly, a recent study from Spain demonstrated that SARS-CoV-2 detection in wastewater could anticipate COVID-19 occurrence in a low prevalence area.42 On the other hand, in a study carried out in Santiago, Chile (a high populated city), the SARS-CoV-2 detection on wastewater also demonstrated a prediction of outbreak in some neighborhoods.38 Therefore, the epidemiological information from SARS-CoV-2 testing in untreated wastewater could significantly impact the surveillance of the viral circulation of SARS-CoV-2 and its variants in the future.43
The limitations of the cohort study were the limited number of patients followed-up and the difficult in obtaining daily stool samples. In addition, there is not a gold standard of SARS-CoV-2 infection and SARS-CoV-2 qRT-PCR has sensitivity near to 70% in NPS.44 This sensitivity could be even lower according to a recent study, reaching 47.3% in real life and underestimating detection of SARS-CoV-2 in organs, fluids, and stools.45 Also, we identified a high heterogeneity among the studies to estimate proportion of SARS-CoV-2 RNA detection in stools. We performed several analyses to elucidate the cause of this heterogeneity including a meta-regression. We identified that gender can importantly contribute to this high heterogeneity. This interesting observation could be explained by the higher expression of ACE2 and a less robust T cell-mediated immunity in male patients.46, 47 However, this heterogeneity could also be explained by important differences and quality among prior published studies. Future research questions include the establishment of additional data to support the fecal-oral transmission and the assessment of the long-term impact of SARS-CoV-2 infection in the gastrointestinal tract. Finally, although anal swabs to detect SARS-CoV-2 have been performed as a screening tool in countries such as Indonesia, the sensitivity of this technique is low (36.7%).48 Therefore, we require more sensitive anal-swabs-based tests to recommend SARS-CoV-2 detection in anal swabs as a screening tool.
In conclusion, this cohort study and systematic review demonstrated that detection of SARS-CoV-2 in stools is a frequent finding. The clearance of SARS-CoV-2 in stools is significantly prolonged as compared to clearance in nasopharyngeal secretions. The heterogeneity in the systematic reviews were high and could be partially explained by gender and methodological deficiencies of several included studies. Further data is required to support the plausibility of fecal-oral infective route.
Authors’ contributors
AR and RAM are responsible for the overall content of the project and the manuscript submitted as guarantors of the project. AR, RAM, LAD, TGS, DR, and JO had full access to all the data in the study. LAD and TGS take responsibility for the integrity of the data and the accuracy of the data analysis. AR, RAM, LAD, and TGS take the final decision to submit for publication. Study concept and design: AR, RAM, MF, MA, CC, LAD, RC, GR, LO. Acquisition of data: LAD, TGS, EFL, GHV, EiS, ErS, AT, SV, MO, JC, DR, JO, SB, VG, AG, MFS. Experiments and qRT-PCR analysis: RAM, MF, TGS, JL, LIA, EiS. Analysis or interpretation of data: RAM, AR, LAD, TGS, EFL, JC, LIA, EiS, RC, CP, HM, AE, DR, JO, GR. Drafting of the manuscript: LAD, TGS, EFL, RAM, AR, JC, MP. Critical revision of the manuscript for relevant intellectual content: LAD, TGS, DR, JO, EFL, JC, JL, LIA, EiS, SB, VG, AG, MFS, MF, CC, MA, CP, AE, RC, HM, GR, LO, SV, ErS, AT, MO, PG, MP, RAM, AR. Statistical analysis: EFL, RC, LAD, TGS.
Financial support
Protocols and the study set-up used for this study were based on influenza virus studies established in part with the support of the FONDECYT 1161971 grant from the Agencia Nacional de Investigación y Desarrollo (ANID) from Chile, the FLUOMICS Consortium (NIAID grant U19AI135972) funded by NIH and with funding from CRIP (Center for Research on Influenza Pathogenesis), an NIH funded Center of Excellence for Influenza Research and Surveillance (CEIRS, contract number HHSN272201400008C) to RAM.
Conflict of interest
The authors reported no potential conflict of interest.
Acknowledgement
Valentina Riquelme, Centro UC - Síndrome de Down, Escuela de Medicina, Pontificia Universidad Católica de Chile, for her contribution with illustrations. Alonso Rioseco MD, Director of Clinica UC San Carlos, Olga Martinez, Chief-Coordinator of Nursery, Lorena Solís, Pediatric Nurse coordinator (Clinica UC San Carlos, Red de Salud UC Christus), Lorena Troncoso, Nurse coordinator, Alejandra Muñoz, nurse supervisor (Intensive Care Unit, Clinica UC San Carlos), Camila Noceda, Nurse (Clínica UC San Carlos, Red de Salud UC Christus), Jaime Labarca, Department of Infectious Diseases, and Patricia García, Department of Clinical Laboratory, for their support related to the recruitment process for this study.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.gastrohep.2021.12.009.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal–oral transmission. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong Cohort and systematic review and meta-analysis. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan L., Mu M., Yang P., Sun Y., Wang R., Yan J., et al. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional multicenter study. Am J Gastroenterol. 2020 doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Díaz L.A., García-Salum T., Fuentes-López E., Ferrés M., Medina R.A., Riquelme A., et al. Symptom profiles and risk factors for hospitalization in patients with SARS-CoV-2 and COVID-19: a large cohort from South America. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling Y., Xu S.B., Lin Y.X., Tian D., Zhu Z.Q., Dai F.H., et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J (Engl) 2020 doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sultan S., Altayar O., Siddique S.M., Davitkov P., Feuerstein J.D., Lim J.K., et al. AGA Institute rapid review of the gi and liver manifestations of COVID-19 meta-analysis of international data, and recommendations for the consultative management of patients with COVID-19. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liberati A., Altman D.G., Tetzlaff J., Mulrow C., Gøtzsche P.C., Ioannidis J.P.A., et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700. doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin L. Bias caused by sampling error in meta-analysis with small sample sizes. PLOS ONE. 2018;13:e0204056. doi: 10.1371/journal.pone.0204056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barendregt J.J., Doi S.A., Lee Y.Y., Norman R.E., Vos T. Meta-analysis of prevalence. J Epidemiol Community Health. 2013;67:974–978. doi: 10.1136/jech-2013-203104. [DOI] [PubMed] [Google Scholar]
- 14.Thompson S.G., Higgins J.P.T. How should meta-regression analyses be undertaken and interpreted? Stat Med. 2002;21:1559–1573. doi: 10.1002/sim.1187. [DOI] [PubMed] [Google Scholar]
- 15.Core Team R.R. R Foundation for Statistical Computing; Vienna, Austria: 2013. A language and environment for statistical computing. [Google Scholar]
- 16.Nyaga V.N., Arbyn M., Aerts M. Metaprop: a Stata command to perform meta-analysis of binomial data. Arch Public Health. 2014;72:39. doi: 10.1186/2049-3258-72-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J.-T., Ran R.-X., Lv Z.-H., Feng L.-N., Ran C.-Y., Tong Y.-Q., et al. Chronological changes of viral shedding in adult inpatients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miri S.M., Roozbeh F., Omranirad A., Alavian S.M. Panic of buying toilet papers: a historical memory or a horrible truth? Systematic review of gastrointestinal manifestations of COVID-19. Hepat Mon. 2020;20:e102729. [Google Scholar]
- 19.Rodriguez-Morales A.J., Cardona-Ospina J.A., Gutiérrez-Ocampo E., Villamizar-Peña R., Holguin-Rivera Y., Escalera-Antezana J.P., et al. Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Travel Med Infect Dis. 2020:101623. doi: 10.1016/j.tmaid.2020.101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Y, Liu X, Xiong L, Cai K. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2: a systematic review and meta-analysis. J Med Virol; n/a. [DOI] [PMC free article] [PubMed]
- 21.Li L-q, Huang T, Wang Y-q, Wang Z-p, Liang Y, Huang T-b, et al. COVID-19 patients’ clinical characteristics, discharge rate, and fatality rate of meta-analysis. J Med Virol; n/a. [DOI] [PMC free article] [PubMed]
- 22.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020:94. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang W., Feng Z., Rao S., Xiao C., Xue X., Lin Z., et al. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020 doi: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- 24.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., et al. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. 2020 2020.01.22.914952. [Google Scholar]
- 25.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashimoto T., Perlot T., Rehman A., Trichereau J., Ishiguro H., Paolino M., et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., et al. The presence of SARS-CoV-2 RNA in feces of COVID-19 patients. J Med Virol. 2020 doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 28.Lo I.L., Lio C.F., Cheong H.H., Lei C.I., Cheong T.H., Zhong X., et al. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int J Biol Sci. 2020;16:1698–1707. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C., Gao G., Xu Y., Pu L., Wang Q., Wang L., et al. SARS-CoV-2-positive sputum and feces after conversion of pharyngeal samples in patients with COVID-19. Ann Intern Med. 2020 doi: 10.7326/M20-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 31.Nicastri E., D’Abramo A., Faggioni G., De Santis R., Mariano A., Lepore L., et al. Coronavirus disease (COVID-19) in a paucisymptomatic patient: epidemiological and clinical challenge in settings with limited community transmission, Italy February 2020. Euro Surveill. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.11.2000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microb Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020 doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin L., Jiang X., Zhang Z., Huang S., Zhang Z., Fang Z., et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020 doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- 35.Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol Hepatol. 2020 doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci Total Environ. 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ampuero M., Valenzuela S., Valiente-Echeverría F., Soto-Rifo R., Barriga G.P., Chnaiderman J., et al. SARS-CoV-2 detection in sewage in Santiago Chile – preliminary results. medRxiv. 2020 2020.07.02.20145177. [Google Scholar]
- 39.Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., et al. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis J. 2020:26. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020 doi: 10.1126/science.abc1669. eabc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 42.Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daughton C.G. Wastewater surveillance for population-wide Covid-19: The present and future. Sci Total Environ. 2020;736:139631. doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection – challenges and implications. N Engl J Med. 2020;383:e38. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- 45.Kortela E., Kirjavainen V., Ahava M.J., Jokiranta S.T., But A., Lindahl A., et al. Real-life clinical sensitivity of SARS-CoV-2 RT-PCR test in symptomatic patients. PLOS ONE. 2021;16:e0251661. doi: 10.1371/journal.pone.0251661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W., Single-cell R.N.A. expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. bioRxiv. 2020 2020.01.26.919985. [Google Scholar]
- 47.Takahashi T., Wong P., Ellingson M., Lucas C., Klein J., Israelow B., et al. Sex differences in immune responses to SARS-CoV-2 that underlie disease outcomes. medRxiv: the preprint server for health sciences. 2020 2020.06.06.20123414. [Google Scholar]
- 48.Abdullah M., Sudrajat D.G., Muzellina V.N., Kurniawan J., Rizka A., Utari A.P., et al. The value of anal swab RT-PCR for COVID-19 diagnosis in adult Indonesian patients. BMJ Open Gastroenterol. 2021:8. doi: 10.1136/bmjgast-2020-000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen W., Lan Y., Yuan X., Deng X., Li Y., Cai X., et al. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect. 2020;9:469–473. doi: 10.1080/22221751.2020.1732837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dreher M., Kersten A., Bickenbach J., Balfanz P., Hartmann B., Cornelissen C., et al. The characteristics of 50 hospitalized COVID-19 patients with and without ARDS. Dtsch Arztebl Int. 2020;117:271–278. doi: 10.3238/arztebl.2020.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han C., Duan C., Zhang S., Spiegel B., Shi H., Wang W., et al. Digestive symptoms in COVID-19 patients with mild disease severity: clinical presentation stool viral RNA testing, and outcomes. Am J Gastroenterol. 2020;115:916–923. doi: 10.14309/ajg.0000000000000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang X., Luo M., Zou Z., Wang X., Chen C., Qiu J. Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J Med Virol. 2020 doi: 10.1002/jmv.25941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiehao C., Jin X., Daojiong L., Zhi Y., Lei X., Zhenghai Q., et al. A case series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kujawski S.A., Wong K.K., Collins J.P., Epstein L., Killerby M.E., Midgley C.M., et al. First 12 patients with coronavirus disease 2019 (COVID-19) in the United States. medRxiv. 2020 doi: 10.1038/s41591-020-0877-5. 2020.03.09.20032896. [DOI] [PubMed] [Google Scholar]
- 55.Li H., Wang Y., Ji M., Pei F., Zhao Q., Zhou Y., et al. Transmission routes analysis of SARS-CoV-2: a systematic review and case report. Front Cell Dev Biol. 2020;8:618. doi: 10.3389/fcell.2020.00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lescure F.-X., Bouadma L., Nguyen D., Parisey M., Wicky P.-H., Behillil S., et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infectious Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei X.-S., Wang X., Niu Y-R., Ye L.-L., Peng W.-B., Wang Z.-H., et al. Diarrhea is associated with prolonged symptoms and viral carriage in corona virus disease 2019. Clin Gastroenterol Hepatol. 2020;18:1753–1759. doi: 10.1016/j.cgh.2020.04.030. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolf G.K., Glueck T., Huebner J., Muenchhoff M., Hoffmann D., French L.E., et al. Clinical and epidemiological features of a family cluster of symptomatic and asymptomatic severe acute respiratory syndrome coronavirus 2 infection. J Pediatr Infect Dis Soc. 2020 doi: 10.1093/jpids/piaa060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xing Y.H., Ni W., Wu Q., Li W.J., Li G.J., Wang W.D., et al. Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J Microbiol Immunol Infect. 2020;53:473–480. doi: 10.1016/j.jmii.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiong X.L., Wong K.K., Chi S.Q., Zhou A.F., Tang J.Q., Zhou L.S., et al. Comparative study of the clinical characteristics and epidemiological trend of 244 COVID-19 infected children with or without GI symptoms. Gut. 2020 doi: 10.1136/gutjnl-2020-321486. [DOI] [PubMed] [Google Scholar]
- 63.Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Z., Li G., Dai X., Liu G., Li G., Jie Y. [Three cases of novel coronavirus pneumonia with viral nucleic acids still positive in stool after throat swab detection turned negative] Chin J Dig. 2020;40 E002. [Google Scholar]
- 65.Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323:1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zautner AE, Bauswein M, Dörschug A, Blaschk S, Lugert R, Ellenrieder V, et al. High proportion of SARS-CoV-2 RNA persistence in IgG-positive patients with COVID-19. Available at SSRN: https://ssrn.com/abstract=3605269 2020.
- 67.Zhang J., Wang S., Xue Y. Fecal specimen diagnosis 2019 novel coronavirus-infected pneumonia. J Med Virol. 2020;92:680–682. doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeng L., Xia S., Yuan W., Yan K., Xiao F., Shao J., et al. Neonatal early-onset infection with SARS-CoV-2 in 33 neonates born to mothers with COVID-19 in Wuhan, China. JAMA Pediatrics. 2020;174:722–725. doi: 10.1001/jamapediatrics.2020.0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China January–March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.