Abstract
The coronavirus has posed a serious threat to the world since its discovery in Wuhan in 2019. Beta, gamma, delta, and the final omicron variants have emerged as a result of several mutations in the virion structure. The Australian Omicron S protein variant contains 37 mutations out of a total of 67 mutations. According to preliminary data from South Africa, Omicron variant infection is not associated with any particular symptoms. The purpose of this research was to determine how changes in the structure of the S protein alter the protein's interaction with the ACE2 receptor. The Omicron variant stimulates the immune response more than the wild strain.
Keywords: Omicron, SARS-CoV-2, COVID-19, Mutation, Variant
1. Introduction
The globe is still on high COVID-19 alert over two years after the onset of the SARS-CoV-2 epidemic, which has claimed the lives of over 5 million people. Weekly, if not daily, reports of new SARS-CoV-2 mutations have been made [1,2]. Poor public health infrastructure, poor vaccination rates, and the existence of a significant number of immunocompromised persons with weakened immune defenses and a higher susceptibility to infection might provide an ideal ground for the development of novel variations [3,4]. In addition, RNA viruses are highly susceptible to mutations due to the inexistence repairing mechanisms or very rudimentary repairing during RNA synthesis. Travel restrictions have been imposed by a number of nations for visitors coming in or departing from southern African countries. South Africa informed the WHO on November 24th, 2021, that a new SARS-CoV-2 strain, had been discovered. B.1.1.529 was initially discovered in Botswana on November 11th, 2021, and in South Africa on November 14th, 2021. This variant contains many mutations that may open up a great deal of speculation about upcoming problems [5,6]. The Omicron variant's spike protein has at least 30 amino acid mutations, three tiny deletions, and one short insertion. The receptor binding domain contains 15 of the 30 amino acid mutations [7,8]. This variant has been found at higher rates than earlier outbreaks, suggesting that it may have a growth advantage. It is yet unknown if infection with the Omicron form causes more severe illness. Assessment of illness severity is challenging due to the minimal number of cases linked to the Omicron variants [9,10]. According to preliminary data from South Africa, Omicron variant infection is not associated with any particular symptoms, and some individuals are asymptomatic, as is the case with other variations [11,12].
2. Materials and methods
From the GISAID website, 16 strains of the SARS-CoV-2 carrying the code (B.1.1.529) of the Omicron variant were selected. Information of cases have been registered in Botswana, South Africa, Hong Kong and Australia. Spike protein was chosen because it binds to the angiotensin-converting enzyme 2 (ACE2) which is essential for viral entry into the host cell [13,14].
For the process of evaluating, identifying mutations, and examining the structural changes in the protein, the registered strain in Australia was chosen. The identified strain in Wuhan, ID (6VXX), was used as a template reference from PDB. The Australian Omicron S protein variant contains 37 mutations. Table 1 shows description of wild and mutant amino acid residues.
Table 1.
Alteration, deletion and insertion of amino acids in omicron variant according to the Wuhan strain. del: deletion amino acid. ins: insertion amino acid. AA: amino acid.
| Type | Residue | Wild AA | Omicron AA | Type | Residue | Wild AA | Omicron AA |
|---|---|---|---|---|---|---|---|
| H69del | 69 | Histidine | – | T478K | 478 | Threonine | Lysine |
| A67V | 67 | Alanine | Valine | E484A | 484 | Glutamate | Alanine |
| V70del | 70 | Valine | – | Q493R | 493 | Glutamine | Arginine |
| T95I | 95 | Threonine | Isoleucine | G496S | 496 | Glycine | Serine |
| G142D | 142 | Glycine | Aspartate | Q498R | 498 | Glutamine | Arginine |
| V143del | 143 | Valine | – | N501Y | 501 | Asparagine | Tyrosine |
| Y144del | 144 | Tyrosine | – | Y505H | 505 | Tyrosine | Histidine |
| Y145del | 145 | Tyrosine | – | T547K | 547 | Threonine | Lysine |
| N211del | 211 | Asparagine | – | D614G | 614 | Aspartate | Glycine |
| L212I | 212 | Leucine | Isoleucine | H655Y | 655 | Histidine | Tyrosine |
| ins214EPE | 214 | – | Glutamate, Proline Glutamate | N679K | 679 | Asparagine | Lysine |
| G339D | 339 | Glycine | Aspartate | P681H | 681 | Proline | Histidine |
| S371L | 371 | Serine | Leucine | N764K | 764 | Asparagine | Lysine |
| S373P | 373 | Serine | Proline | D796Y | 796 | Aspartate | Tyrosine |
| S375F | 375 | Serine | Phenylalanine | N856K | 856 | Asparagine | Lysine |
| K417 N | 417 | Lysine | Asparagine | Q954H | 954 | Glutamine | Histidine |
| N440K | 440 | Asparagine | Lysine | N969K | 969 | Asparagine | Lysine |
| G446S | 446 | Glycine | Serine | L981F | 981 | Leucine | Phenylalanine |
| S477 N | 477 | Serine | Asparagine |
3. Constructed 3D structure of the Australian Omicron S protein
In the current study, we used two approaches to construct missing loops in the S protein (6VXX): the PyMOL Builder tool and the ModLoop Queue online site. According to the reference S protein isolated from Wuhan state, the Australian Omicron S protein 3D structure was constructed using PyMol.
4. Molecular docking between chain A of S protein and ACE2
HDOCK and PatchDock servers were used to investigate the interaction of S protein with the ACE2 (1R42) receptor. Based on the RMSDs and docking scores, the top five models were chosen.
5. Immunity system simulation
Using in silico immunological simulations, the immunogenicity and immune response profile were further defined. In this investigation, immunological simulations were performed using the C-ImmSim server, which is an agent-based model with a position-specific scoring matrix (PSSM) [15,16].
6. Applied RBD mutations
RBD-ACE2 (PDB: 6M0J) was subjected to more scrutiny. Since the RBD is the primary section of S protein due to its relationship with the (ACE2) receptor, we assigned mutations to it and noted the effect of those changes in the form of interaction. 15 of the 37 protein S mutations were carried out inside the RBD residues (333–526). To compare the distances between the wild and the mutated strains, we assessed the distance between atoms in the docking residues (RBD-ACE2). The immunogenicity of RBD-ACE2 wild and mutant strains was determined using the C-ImmSim server.
7. Results
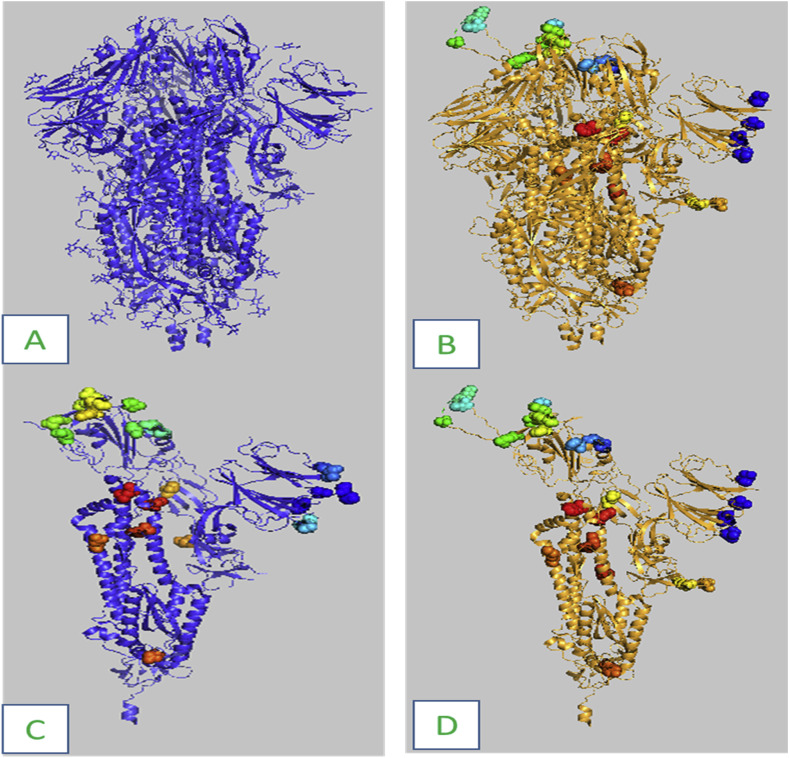
The residues created using Builder PyMOL and ModLoop in the current investigation had no discernible differences. There are obvious conformational changes between wild type and Omicron S protein variants, fig. 1 .
Fig. 1.
Cartoon representation of S protein. A: Wild strain. B: Omicron variant strain shows the 37 mutation residues in different color dots. C: Chain A of wild strain shows wild residues. D: Mutated chain A of Omicron variant.
8. Predict molecular docking of chain A and ACE2 receptor
The output models were evaluated after the ACE2 receptor was uploaded to chain A of S protein. The best 5 models were chosen after examining the docking outcomes and calculating the RMSD. Fig. 2 depicts the ACE2 receptor's ability to bind with the A chain of wild-type and Omicron S proteins. The protein model quality was measured between 3.0 ≤ LGscore <5.0. The LGscore for wild S protein is (4.615) while in the omicron variant was higher (4.450). On the other hand, the MaxSub score were (0.355) and (0.333) respectively. The energy docking score of the omicron variant is higher than the wild type of all predicted models. On the contrary, for the RMSD, a discrepancy was observed in the proposed models for the two types, as shown in Table 2 .
Fig. 2.
The best 5 docking models of S protein and ACE2 receptor. 3D green structure is the wild type of chain A. 3D magenta structure is the omicron chain A.
Table 2.
The docking energy score and RMSD in all predict models of wild and omicron variant's S protein.
| Rank | Model 1 | Model 2 | Model 3 | Model 4 | Model 5 |
|---|---|---|---|---|---|
| Wild S protein (Docking score) | −245.91 | −245.69 | −242.93 | −236.36 | −235.79 |
| Omicron S variant (Docking score) | −275.32 | −249.02 | −247.10 | −246.39 | −241.10 |
| Wild RMSD | 268.66 | 263.14 | 243.94 | 285.37 | 249.59 |
| Omicron RMSD | 266.54 | 243.85 | 323.89 | 213.96 | 300.68 |
9. Immunogenicity analysis
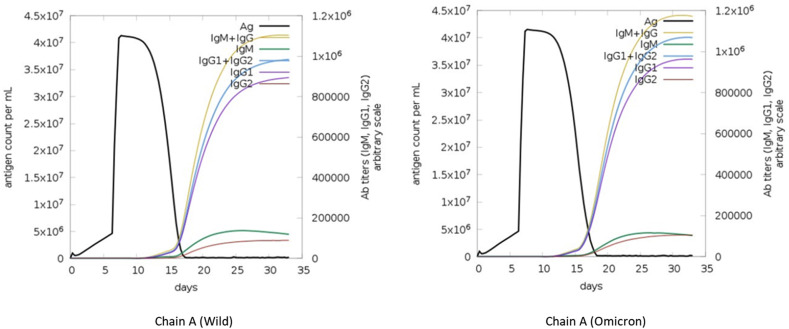
The immunogenicity and the immune response of the predicted molecular docking of (chain A-ACE2) were determined using immunoinformatics for both wild (6VXX) and omicron variant. Despite having equal immunological characteristics, there was a distinct difference between the wild type and the omicron when the outcomes of the immune response were analyzed. The duration of the immune response has been shorter for the omicron variant. As a result of the immunogenicity, the antibody titers are greater for the omicron variant including (IgM + IgG), (IgG1+IgG2), and IgG1 alone, Fig. 3 . There is an elevation in the count of active B-lymphocytes between the wild and the mutant type, yet they are comparable in terms of presenting on class-II, internalized Ag, replicating, and being anergic, Fig. 4 . Count of T-regulatory memory lymphocyte is higher in the S protein wild type and on the contrary for the count of T-regulatory not memory lymphocytes. The activity and resting of omicron reaches 97 (cells per mm3) while it reaches 87(cells per mm3) for wild type. The regulatory T-cell (not memory) is higher for omicron, Fig. 5 . The number of cytokines and interleukins stimulated by the S protein is similar in the both types. IFN-g and TGF-b are the highest types stimulated by the immune system for both types of S protein. IFN-γ of the wild type is 620,000 ng/ml and 630,000 ng/ml for omicron, Fig. 6 .
Fig. 3.
Immunogenicity and immune response against wild and omicron S protein. Ag count per ml and all Ab titers through 30 days of infection. Peaks of (IgM + IgG), (IgG1+IgG2), and IgG1 of omicron is higher than of wild type.
Fig. 4.
B-lymphocytes population per entity-state, showing counts for active, presenting on class-II, internalized the Ag, duplicating and anergic. The number of presenting class II in omicron is higher but the count activity is less than wild type.
Fig. 5.
Immunogenicity and immune response against wild and omicron S protein. CD4 T-regulatory lymphocytes count. Both total, memory and per entity-state counts are plotted. The activity and resting of omicron reaches 97 (cells per mm3) while it reaches 87(cells per mm3) for the wild type. Regulatory T-cell not memory is higher for omicron.
Fig. 6.
Concentration of cytokines and interleukins. D in the inset plot is danger signal. IFN-γ of the wild type is 620,000 ng/ml and 630,000 ng/ml for omicron.
10. Performed RBD-ACE2 mutations and immunity response
PDB (6M0J) is the molecular interaction between RBD and ACE2. 15 mutations were constructed on the wild type to predict new omicron variant (B.1.1.529). These mutations are: E484A, G339D, G446S, G496S, K417 N, N440K, N501Y, Q493R, Q498R, S371L, S373P, S375F, S477 N, T478K, and Y505H. 10 out of 15 of these mutations are closer to the docking region. As a result of these changes, the shape of the micron variant differs significantly from the original type, which might lead to a change in the strength of the fusion between RBD and ACE2, Fig. 7 . In terms of the immune response, our research found that various characteristics, such as Ab titers amount, IgM, IgG1 and IgG2 arbitrary scale, CD8 T-cytotoxic lymphocytes count per entity-state. There is no significant difference in account of antibodies produced and stimulated by the antigens per ml between wild type and omicron variants, Fig. 8 . Anergic CD8 T-cell peak reaches 1180 cells/mm3 while it is 1390 cells/mm3 for omicron variant, Fig. 9 . T-regulatory lymphocyte count (CD4), memory and per entity-state (non-memory) counts reach 88 and 86 cells/mm3, respectively for wild type. TR memory and non-memory cells reach 91 and 89 cells/mm3, respectively for omicron variant, Fig. 10 . The levels of cytokines and interleukins, including as IFN-, IL-12, TGF-b, and IL-10, are similar in wild and omicron variants, Fig. 11 .
Fig. 7.
Applied 15 mutations on the RBD-ACE2 on the wild (6M0J). RBD: light brown color. ACE2: cyan color. A: RBD-ACE2 wild (6MOJ) shows 15 residues in res dots. B: RBD-ACE2 omicron variant shows 15 mutated resides in different colors.
Fig. 8.
Immunogenicity and immune response against wild and omicron of RBD protein. Ag count per ml and all Ab titers through 30 days of infection. No significant difference in account of Ab produced and stimulated by the Ag/ml between wild type and omicron variants.
Fig. 9.
CD8 T-cytotoxic lymphocytes count per entity-state, showing counts for active, presenting on class-II, internalized the Ag, duplicating and anergic. Anergic CD8 T-cell peak reaches 1180 cells/mm3 while it is 1390 cells/mm3 for omicron variant.
Fig. 10.
Immunogenicity and immune response against wild and omicron of RBD protein. CD4 T-regulatory lymphocytes count. Both total, memory and per entity-state counts are plotted. TR regulatory memory and non-memory cells reache 88 and 86 cells/mm3, respectively for wild type. TR memory and non-memory cells reach 91 and 89 cells/mm3, respectively for omicron.
Fig. 11.
Concentration of cytokines and interleukins. D in the inset plot is danger signal. IFN-γ, IL-12, TGF-b and IL-10 are similar between wild and omicron variants.
11. Discussion
The rapid appearance of a new coronavirus strain alarmed the globe, particularly because it included several mutations in all of its proteins [17]. The virus's efficacy and the severity of illness symptoms are both severely impacted by the abrupt shift brought on by mutations. So far, no unusual trends in the number of deaths or viral efficacy have been seen with the new Omicron type [18,19]. The fact that this strain affects youngsters more than adults is noteworthy, but happily, the number of deaths is modest, suggesting that this strain is the weakest, as future studies will reveal. According to the WHO, omicron has a greater propensity to spread than prior strains [20].
There is one significant issue: the number of mutations in Omicron varies, starting at 62 in South Africa and increasing to 67 in the strain isolated from Australia that we are studying. The S protein in the omicron variant has 37 mutations out of a total of 67 mutations. In addition to the replacement mutations that arise in each strain, we've seen a loss of certain residues and an insertion of new base pairs. Because the S protein is involved in the insertion of viral RNA into the host cell via the ACE2 receptor, it was chosen as the subject of our research.
In the current study, we presented 37 out of 67 total mutations in the viral proteins. Based on the reference strain isolated from Wuhan, the protein S model of the Omicron variation was created. Mutations result in a distinct alteration in the structure of the protein. To determine the effect of the degree of fusion on the mutant and wild type, the docking stage between S protein and the ACE2 receptor was required. In all of the predicted docking models, the Omicron variant had a higher fusion energy than the wild strain. Table 1 shows that the degree of RMSD differed between the two types.
We demonstrated in this study that the novel strain activates the immune system and produces a distinct immunological response, as opposed to the wild strain, which has higher stimulation for both IgG and IgM.
To examine the influence of the Omicron variation on vaccination efficacy and outbreak infections, especially in those who have received booster doses, more laboratory and epidemiological research is needed. Vaccination, on the other hand, is expected to continue to provide protection against hospitalization and mortality, and vaccines will continue to play an important part in the COVID-19 pandemic's containment.
15 of the 37 mutations are found in RBD region of the S protein. This region is well recognized for its involvement in docking with the ACE2 receptor. We demonstrated a noticeable difference in the form of the mutant strain's RBD compared to the original strain in this study. The degree of immunological response in the Omicron strain differs significantly from that in the original strain.
Individuals over 60 years of age who are not completely vaccinated or do not have confirmation of past SARS-COV-2 infection, as well as those with underlying health issues, should postpone travel, according to the WHO [21,22].
Because Omicron has been designated as a Variant of Concern [23], WHO recommends that countries improve surveillance and sequencing of cases, share genome sequences on publicly available databases like GISAID [24], report initial cases or clusters to WHO [25,26], and conduct field investigations and laboratory assessments to better understand if Omicron has different transmission or disease characteristics, or has an impact on vaccine effectiveness.
Despite the Omicron strain's increased dissemination, we believe it is the weakest of the ancient strains, and that it will exhaust the virus, causing its symptoms to revert to those of a common cold, as did the ancestors of the Human coronavirus 229E (HCoV-229E).
12. Conclusions
Omicron variant has an increase in the number of mutations daily. One of the strain of the omicron has 67 mutations in all proteins. 37 out of 67 presented in the S protein 15 out of 37 of S mutation protein found in the RBD region. This study revealed that mutations have conformational changes in the S protein shape. The Omicron variant stimulates the immune response more than the wild strain. To effectively contain this pandemic, a global concerted effort including government agencies, pharmaceutical, and biotech firms, as well as academic and healthcare institutions, is necessary.
Author statement
Ali A. Dawood: Data acquisition; Data analysis / interpretation; Drafting manuscript; Critical revision of manuscript; Statistical analysis; Securing funding; Admin; technical; or material; support; Supervision; Final approval.
Acknowledgments
The author send thankful letter University of Mosul for documenting this work.
Data availability
No data was used for the research described in the article.
References
- 1.Ansumana R., Sankoh O., Zumla A. Effects of disruption from COVID-19 on antimalarial strategies. Nat. Med. 2020;26:1334–1336. doi: 10.1038/s41591-020-1047-5. [PubMed] [CrossRef] [Google Scholar] [DOI] [PubMed] [Google Scholar]
- 2.Gisaid 2021. https://www.gisaid.org/
- 3.Inzaule S.C., Tessema S.K., Kebede Y., Ogwell Ouma A.E., Nkengasong J.N. Genomic-informed pathogen surveillance in Africa: opportunities and challenges. Lancet Infect. Dis. 2021;21:e281–e289. doi: 10.1016/S1473-3099(20)30939-7. [PMC free article] [PubMed] [CrossRef] [Google Scholar] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karim F., Gazy I., Cele S., Zungu Y., Krause R., Bernstein M., Khan K., et al. HIV status alters disease severity and immune cell responses in beta variant SARS-CoV-2 infection wave. Elife. 2021;10 doi: 10.7554/eLife.67397. e67397, [PubMed] [CrossRef] [Google Scholar] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawood A. Identification of CTL and B-cell epitopes in the nucleocapsid phosphoprotein of COVID-19 using immunoinformatics. Microbiol. J. 2021;83(1):78–86. doi: 10.15407/microbiolj83.01.078. [DOI] [Google Scholar]
- 6.Lapidus S., Liu F., Casanovas-Massana A., Dai Y., Huck J.D., Lucas C., Klein J., et al. Plasmodium infection induces cross-reactive antibodies to carbohydrate epitopes on the SARS-CoV-2 Spike protein. medRxiv. 2021 doi: 10.1101/2021.05.10.21256855. [Preprint] May 12:2021.05.10.21256855. [CrossRef] [Google Scholar] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawood A. Perspectives into what will happen after the end of COVID-19 pandemic. OAJMB. 2021;6(4):1–2. doi: 10.23880/oajmb-16000206. [DOI] [Google Scholar]
- 8.Martin D.P., Weaver S., Tegally H., San J.E., Shank S.D., Wilkinson E., et al. The emergence and ongoing convergent evolution of the SARS-CoV-2 N501Y lineages. Cell. 2021;184:5189–5200. doi: 10.1016/j.cell.2021.09.003. 30. e7. [CrossRef] [Google Scholar] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawood A., Altobje M., Alnori H. Compatibility of the ligand binding sites in the spike glycoprotein of COVID-19 with those in the aminopeptidase and the caveolins 1, 2 proteins. Res. J. Pharm. Technol. 2021;14(9):4760–4766. doi: 10.52711/0974-360X.2021.00828. [DOI] [Google Scholar]
- 10.Nkengasong J.N., Ndembi N., Tshangela A., Raji T. COVID-19 vaccines: how to ensure Africa has access. Nature. 2020;586:197–199. doi: 10.1038/d41586-020-02774-8. [PubMed] [CrossRef] [Google Scholar] [DOI] [PubMed] [Google Scholar]
- 11.Nachega J.B., Sam-Agudu N.A., Mellors J.W., Zumla A., Mofenson L.M. Scaling up covid-19 vaccination in Africa - lessons from the HIV pandemic. N. Engl. J. Med. 2021;385:196–198. doi: 10.1056/NEJMp2103313. [PMC free article] [PubMed] [CrossRef] [Google Scholar] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawood A. New variant of SARS-CoV-2 in South Africa. Prog Med Sc. 2021;5(1):1–2. doi: 10.5455/pms.20211013. [DOI] [Google Scholar]
- 13.Nachega J.B., Sam-Agudu N.A., Masekela R., van der Zalm M.M., Nsanzimana S., Condo J., et al. Addressing challenges to rolling out COVID-19 vaccines in African countries [PMC free article] [PubMed] [CrossRef] [Google Scholar] Lancet Global Health. 2021;9:e746–e748. doi: 10.1016/S2214-109X(21)00097-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nachega J.B., Kapata N., Sam-Agudu N.A., Decloedt E.H., Katoto P.D.M.C., et al. Minimizing the impact of the triple burden of COVID-19, tuberculosis and HIV on health services in sub-Saharan Africa. Int. J. Infect. Dis. Mar. 2021;20(21):S1201–S9712. doi: 10.1016/j.ijid.2021.03.038. 00256-3. [CrossRef] [Google Scholar] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawood A. Glycosylation, ligand binding sites and antigenic variations between membrane glycoprotein of COVID-19 and related coronaviruses. Vacunas. 2021;22(1):1–9. doi: 10.1016/j.vacun.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersen E., McCloskey B., Hui D.S., Kock R., Ntoumi F., Memish Z.A., et al. COVID-19 travel restrictions and the International Health Regulations - call for an open debate on easing of travel restrictions. Int. J. Infect. Dis. 2020;94:88–90. doi: 10.1016/j.ijid.2020.04.029. [PMC free article] [PubMed] [CrossRef] [Google Scholar] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawood A., Altobje M., Alrassam Z. Vol. 83. 2021. pp. 82–92. (Molecular Docking of SARS-CoV-2 Nucleocapsid Protein with Angiotensin-Converting Enzyme II. Mikrobio Zhu). 2. [DOI] [Google Scholar]
- 18.Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., et al. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22:757–773. doi: 10.1038/s41576-021-00408-x. [PMC free article] [PubMed] [CrossRef] [Google Scholar] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson E., Giovanetti M., Tegally H., San J.E., Lessells R., Cuadros D., et al. A year of genomic surveillance reveals how the SARS-CoV-2 pandemic unfolded in Africa. Science. 2021;374:423–431. doi: 10.1126/science.abj4336. [PubMed] [CrossRef] [Google Scholar] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WHO Classification of omicron (B.1.1.529): SARS-CoV-2 variant of concern. 2021. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
- 21.WHO Tracking of SARS-COV-2 variants. 2021. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- 22.WHO WHO 2nd Global consultation on assessing the impact of SARS-CoV-2 variants of concern on Public health interventions. 2021. https://www.who.int/publications/m/item/2nd-global-consultation-on-assessing-the-impact-of-sars-cov-2-variants-of-concern-on-public-health-interventions
- 23.WHO Policy and technical considerations for implementing a risk-based approach to international travel in the context of COVID-19. 2021. https://www.who.int/news-room/articles-detail/policy-and-technical-considerations-for-implementing-a-risk-based-approach-to-international-travel-in-the-context-of-covid-19
- 24.WHO COVAX announces additional deals to access promising COVID-19 vaccine candidates; plans global rollout starting Q1 2021. 2021. https://www.who.int/news/item/18-12-2020-covax-announces-additional-deals-to-access-promising-covid-19-vaccine-candidates-plans-global-rollout-starting-q1-2021
- 25.WHO Working for global equitable access to COVID-19 vaccines. 2021. https://www.who.int/initiatives/act-accelerator/covax
- 26.WHO Less than 10% of African countries to hit key COVID-19 vaccination goal. 2021. https://www.afro.who.int/news/less-10-african-countries-hit-key-covid-19-vaccination-goal
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.