Abstract
We present a new class of DNA-based nanoswitches that, upon enzymatic repair, could undergo a conformational change mechanism leading to a change in fluorescent signal. Such folding-upon-repair DNA nanoswitches are synthetic DNA sequences containing O6-methyl-guanine (O6-MeG) nucleobases and labelled with a fluorophore/quencher optical pair. The nanoswitches are rationally designed so that only upon enzymatic demethylation of the O6-MeG nucleobases they can form stable intramolecular Hoogsteen interactions and fold into an optically active triplex DNA structure. We have first characterized the folding mechanism induced by the enzymatic repair activity through fluorescent experiments and Molecular Dynamics simulations. We then demonstrated that the folding-upon-repair DNA nanoswitches are suitable and specific substrates for different methyltransferase enzymes including the human homologue (hMGMT) and they allow the screening of novel potential methyltransferase inhibitors.
Keywords: conformational change mechanism, DNA nanoswitches, DNA nanotechnology, DNA repair enzymes, triplex DNA
Introduction
The genetic information inside cells is protected against DNA damage by multi-enzymatic DNA repair mechanisms, such as base excision, nucleotide excision repair,[1] or a direct damage reversal by O6-methylguanine-DNA-methyltransferases (abbreviated here as AGTs).[2–4] The latter is a class of evolutionarily conserved biocatalysts, able to directly and irreversibly remove alkyl groups at the O6-position of guanines on DNA in a single SN2-like reaction mechanism. These enzymes, indeed, represent the major factor in contrasting the effects of alkylating agents that form such adducts.[2] On the other hand, human methyltransferase (hMGMT) activity impacts the alkylating agent-based chemotherapy in cancer cells.[2,5] For this reason, different hMGMT inactivators/inhibitors are usually employed in combination with this kind of chemotherapy.[2,6]
Because of the clinical relevance of AGTs, assays able to measure their activity in a reliable and rapid way are needed.[7] Most of the assays developed so far are based on the use of oligonucleotides carrying radioactive O6-methylguanine (O6-MeG) groups and chromatographic separations.[8–11] These are, however, time-consuming, tedious, and unsafe. Alternative fluorescence-based assays that allow simple and fast detection of hMGMT activity have been recently developed. For example, Kool and co-workers designed a chemosensor that couples fluorescence change to the bond-breaking step occurring during repair activity.[12,13] Other approaches employing optically-labelled DNA aptamers,[11] DNA-based electrochemical sensors[14] or O6-MeG-containing double strands DNA (dsDNA) oligonucleotides in competition with fluorescent substrates[4,15–18] were also proposed. While these latter systems provide several advantages including ease of use and high sensitivity, there is still an urgent need for finding new strategies to achieve efficient activity-based monitoring of DNA repair enzymes that can be versatile enough to be suitable for a wide range of repair activities.
Recently, DNA nanoswitches have emerged as a new class of programmable probes that allow the sensitive and rapid detection of a wide range of molecular targets.[19] DNA nanoswitches usually undergo a binding-induced conformational change that can provide a measurable output in the presence of a specific input. A number of strategies employing DNA nanoswitches have been developed for the detection of different targets including pH,[20,21] metal ions,[22] small molecules,[23,24] proteins,[25,26] or specific antibodies.[27,28] Critically, because their signaling is linked to a change in the physics of the DNA probe induced by the recognition with a target, such conformational switching sensors are generally highly specific and selective.[29]
Motivated by the above considerations, we demonstrate here the rational design of a new class of DNA nanoswitches, here named folding-upon-repair DNA nanoswitches, that can be conveniently applied for monitoring DNA repair activity. As an applicative example, we initially focused on methyltransferase enzymes and designed a fluorescent-labelled DNA nanoswitch containing O6-MeG that, upon enzymatic repair, could undergo a conformational change associated with a change in the output signal. This approach allows the direct measurement of the activity of, in principle, any methyltransferase enzyme thus opening the possibility to develop an easy methodology for the high-throughput determination of methyltransferase enzyme inhibitors.
Results and Discussion
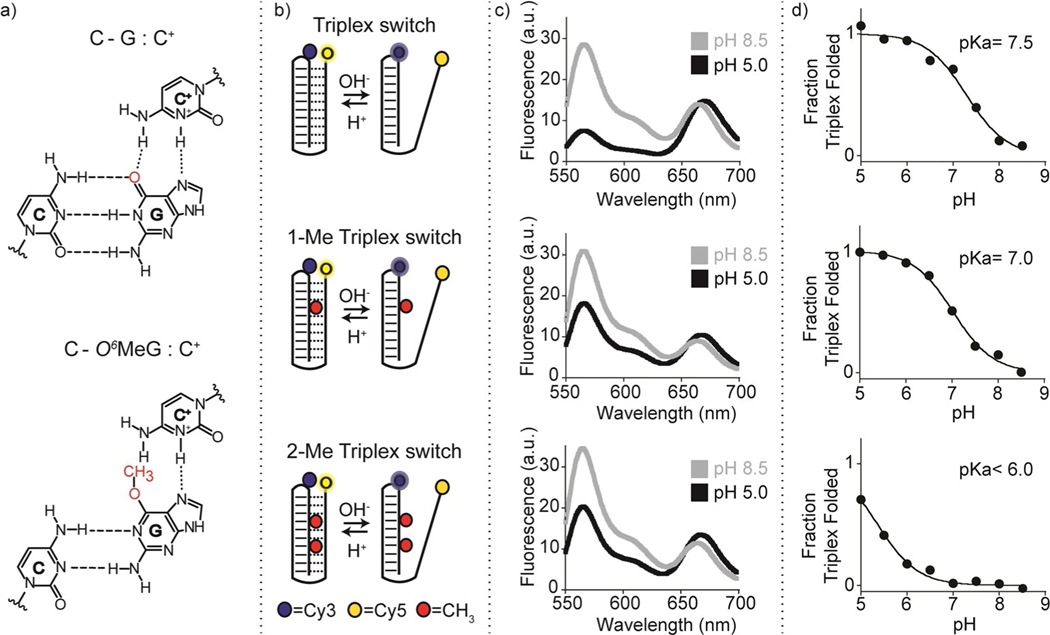
Our strategy to design programmable nucleic acid nanoswitches for the detection of methyltransferase activity is based on the use of a single-stranded DNA capable of forming an intramolecular triplex structure through hydrogen bonds (Hoogsteen interactions) between a hairpin duplex domain and a single-strand triplex-forming portion.[30] More specifically, we have designed three nanoswitches displaying the same triplex-forming domains (i.e., 4 cytosines + 6 thymines) but differing in the content of O6-MeGs in the hairpin duplex domain (0, 1, and 2). The idea underlying the molecular design of the triplex nanoswitches is that the methyl group at the guanine O6 position should affect the formation of the Hoogsteen hydrogen bond with the cytosine on the single-strand DNA (Figure 1a), thus influencing the pH-dependent triplex folding/unfolding behaviour of the nanoswitch. The nanoswitches are also labelled with a pH-insensitive FRET pair to follow pH-dependent folding/unfolding (Figure 1b). To demonstrate the effect of O6-MeG on the triplex formation, we have first tested the pH-dependent folding/unfolding behaviour of our three nanoswitches by measuring the fluorescence signal of our FRET pair at a fixed concentration of the nanoswitch and different pH values. As expected, our control triplex nanoswitch (Triplex switch, Figure 1b, top) that lacks any O6-MeG in its sequence, shows signals that are consistent with the formation of a folded triplex at acidic pH (pH 5.0) and suggest the unfolding of the triplex structure at basic pH values (pH 8.5). The pH of semi-protonation (here defined as pKa, the average pKa due to several interacting protonation sites) for this triplex nanoswitch is 7.5 (Figure 1c,d top, and S1). The presence of one O6-MeG (1-Me Triplex switch, Figure 1b, center) in the hairpin duplex (at position 7) strongly destabilizes triplex formation; as a result, we observe at pH 5.0 a FRET signal that is consistent with a partially unfolded triplex structure and we obtain a pKa of 7.0 (Figures 1c,d center, and S1). Finally, the nanoswitch containing two O6-MeG (2-Me Triplex switch, at position 4 and 7, Figure 1b, bottom) is even more destabilized and the pH titration curve with this switch does not reach a plateau at lower pH values suggesting a pKa lower than 6.0 (Figures 1c,d bottom, and S1). The formation of a folded triplex structure of the Triplex switch was confirmed by melting and urea denaturation experiments performed at different pHs (Figures S2, S3). Also, CD spectra of the three nanoswitches further support the formation of pH-dependent Hoogsteen interactions (Figure S4).
Figure 1.
Folding-upon-repair DNA nanoswitches. a) DNA parallel triplets formed between two cytosines and one guanine and involving Watson–Crick (three hydrogen bonds) and Hoogsteen interactions (two hydrogen bonds) require the protonation of the N3 of cytosine in the third strand (top, left) and thus are only stable at acidic pH values (average pKa of protonated cytosines in C-G:C triplet is ≈6.5). Because it affects Hoogsteen base pairing efficiency, methylation of the guanine in the O6-position destabilizes the triplex conformation. b) Programmable DNA-based triplex nanoswitches designed to form an intramolecular triplex structure with 0 (Triplex), 1 (1-Me Triplex) or 2 (2-Me Triplex) O6-MeG in the sequence. c) Fluorescence spectra were obtained for each nanoswitch at pH 5.0 and pH 8.5. d) pH-titration curves of the triplex nanoswitches. Triplex-to-duplex transition is monitored through a pH-insensitive FRET pair at the 3’-end (Cy5) and internally located (Cy3). The pH titration experiments were performed at 25°C, [nanoswitch]=50 nM by measuring the fluorescence signal at different pH values in 50 mM Na2HPO4, 150 mM NaCl buffer. Spectra were obtained by excitation at 530(±5) nm and acquisition between 545 and 700 nm (±10) nm.
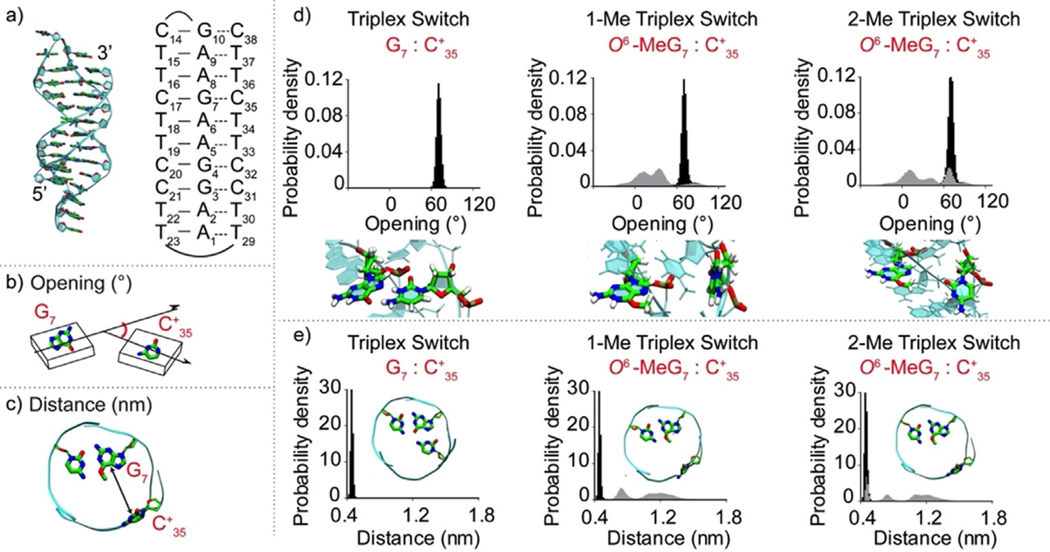
To better understand the effect that guanine methylation has on the stability of our triplex nanoswitches we performed overall molecular dynamics (MD) simulations. We first considered the non-methylated nanoswitch (Triplex switch) where all cytosines involved in Hoogsteen base pairing are protonated (Figure 2a). Then, we introduced the O6-MeG at position 7 (creating the 1-Me Triplex switch), and at both positions 4 and 7 (2-Me Triplex switch). The obtained trajectories encompassed >4.5 μs of sampling, including multiple replicas and providing solid statistics for the analysis of the DNA triplex structural changes. We focused on the C17-G7:C+35 triplet, as this triplet share the methylated guanine in both the 1-Me and 2-Me Triplex switches. Our analysis took into consideration two parameters: 1) the base-pair opening angle, which describes the in-plane opening between G7:C+35 paired Hoogsteen-forming bases, as defined by Lu and Olson[31] (Figure 2b; see Tables S1–S4 for all DNA base-pair parameters); 2) the distance between the centers of mass of the atoms of the G7:C+35 Hoogsteen-forming bases at the interface with each other (Figure 2c).
Figure 2.
Molecular dynamics simulations of triplex nanoswitches. a) Structure (left) and schematic representation (right) of the Triplex switch with numeration of DNA nucleotides. Graphical representation of b) opening angle and c) distance parameters for the G7:C+35 pair. d) Opening angle distribution for the Triplex switch (left), 1-Me Triplex switch (center), and 2-Me Triplex switch (right). Below each graph, side-view snapshots of the analysed base pairs are reported. The opening distribution for 1Me- and 2Me- Triplex switches are shown in grey. The distribution of the non-methylated Triplex switch (black) is also reported in each graph as a comparison. e) Distance parameter for the G7:C+35 Hoogsteen interaction for the Triplex switch, 1-Me Triplex switch, and 2-Me Triplex switch. In each graph, a top-view snapshot of the triplet most probable configuration is shown. The distance distribution for 1Me- and 2Me- triplex switches is shown in grey. The distribution of the non-methylated Triplex switch (black) is also shown in each graph as a comparison.
The narrow distributions of both the opening and distance parameters for the C17-G7:C+35 triplet in the Triplex switch (with an average value of 73° and 0.45 nm, respectively) correspond to a well-defined triplex structure (Figure 2d,e, left and Table S1). Conversely, in the case of the 1-Me and 2-Me Triplex switches, distributions of both parameters are broader and have multiple maxima, showing that the same triplet adopts multiple conformational states (Figure 2d,e, center, right). Remarkably, in case of 1-Me and 2-Me Triplex switches the distance distribution shifts towards higher values. This clearly shows that the O6-methylation of guanine hampers the efficient interaction with the Hoogsteen pairing cytosine, leading to a local unwinding of the triplex (see representative structures, Figure 2d, and highlights on Figure 2e, center, right and Figures S5–8). Similar shifts and broadening of opening and distance distributions were also observed in the case of C20-G4:C+32 triplet in 2Me-Triplex switch, whereas in the case of 1Me-Triplex switch, these distributions were virtually the same as in case of non-methylated Triplex switch (Figures S9–11). Although at a lower extent, we also observed the broadening of the opening and distance distributions for the C17-G7:C+35 triplet during additional ≈1.5 μs long simulations of the Triplex switch in the deprotonated form (Figure S12), further supporting the notion that methylation strongly destabilizes the formation of the triplet. Finally, to achieve more meaningful structural information on the triplex destabilization induced by the methylation, we obtained free energy profiles that can be associated with the increase of the distance between the two fluorophores on each switch. As expected, under protonated conditions the control Triplex is the most stable, followed by the 1-Me Triplex and then by the 2-Me Triplex (Figure S13). These results provide an atomic-level understanding of the triplex stability, indicating that the efficient Hoogsteen base pairing is critical for the triplex formation and both cytosine deprotonation at high pH and guanine methylation should reduce the propensity of DNA hairpin to adopt triplex conformation.
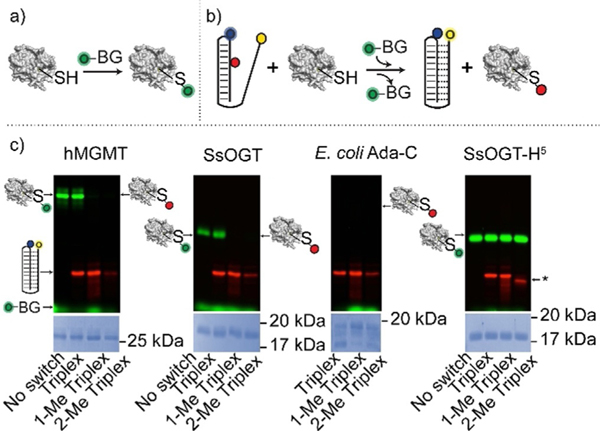
Because the methyl group at the guanine O6 position prevents the efficient formation of Hoogsteen interactions with the respective cytosine in the triplex-forming domain, the O6-MeG-containing triplex nanoswitches characterized above could be used as suitable probes for the monitoring of methyltransferase activity. The enzymatic removal of the methyl group of O6-MeG in the nanoswitches would restore the optimal conditions for Hoogsteen interactions and triplex formation by the nanoswitch. To achieve this goal, we initially performed an indirect assay using a fluorescein derivative of O6-benzyl-guanine (O6-BG)[4,15,32,33] (Figure 3a) to demonstrate that O6-MeG-containing triplex nanoswitches are suitable substrates for methyltransferase enzymes (Figure 3b). We first demonstrated that no interaction occurs between the nanoswitch and the fluorescent-labelled substrate in the absence of the protein. As expected, visible separated bands for both the O6-BG fluorescent derivative and the three DNA nanoswitches tested can be observed (Figure S14). It is worth noting that the nanoswitch containing two O6-MeGs (2-Me Triplex) shows a slightly, but significant, higher mobility (marked by an asterisk in Figure 3c) compared to the single-methylated triplex (1-Me Triplex) and the non-methylated triplex (Triplex) (Figure S14). Although speculative, a possible explanation of the different migration of the 2-Me Triplex could be that this oligonucleotide adopts a partial folding in denaturing conditions. We have pre-incubated the methyltransferase enzymes with our nanoswitches and then added an equimolar amount of the O6-BG fluorescent derivative that interacts with the free enzymes in the solution (Figure 3b). In the case of hMGMT, a fluorescent protein band was observed in the presence of the non-methylated Triplex, suggesting that no repair reaction on this DNA occurred (Figure 3c). On the contrary, 1-Me Triplex nanoswitch and 2-Me Triplex nanoswitch, showed a complete absence of the fluorescent band (Figure 3c). This is likely due to the suicidal nature of methyltransferase enzymes,[3] as the methyl group is irreversibly transferred to the enzyme, which is no longer available for the O6-BG fluorescent derivative, thus demonstrating that methylated nanoswitches are effective substrates for hMGMT. The same results were further confirmed using the thermostable Saccharolobus solfataricus homologue (SsOGT), an enzyme responsible for the direct repair of O6-alkylguanine in double-stranded DNA at high temperatures[4,16,32,33] and the Escherichia coli homologue (Ada-C), which is reported to be insensitive to O6-BG derivatives.[17,18] In the latter case, the absence of the enzyme fluorescent bands despite being correctly loaded as confirmed by the Coomassie staining analysis (Figure 3c, E. coli Ada-c), and the similar migration of all the triplexes, confirm that the DNA-repair occurred. The methylated DNA triplex nanoswitches are also highly specific as they showed no enzymatic activity when incubated with a mutant of SsOGT (SsOGT-H5) that was previously reported to be catalytically active on O6-BG derivatives, but unable to bind and react with ds-DNA.[4,32–35]
Figure 3.
Characterization of triplex nanoswitches as suitable substrates of methyltransferase enzymes. a) Suicide reaction of an AGT enzyme with a fluorescein derivative of O6-BG. b) The alkyl transfer suicide reaction between the enzyme and a DNA methylated triplex hampers the covalent bond between the enzyme and the fluorescein group. c) SDS-PAGE images of reactions in which different enzymes (i.e. hMGMT, SsOGT, E. coli Ada-C, SsOGT-H5) after pre-incubation with the DNA nanoswitches and then reacted with the O6-BG fluorescent derivative. The enzymes that have reacted with the fluorescein derivative appear as green bands on the gel. The absence of any fluorescent bands indicates that the enzyme has not reacted with the fluorescent substrate. Fluorescence-labelled triplex nanoswitches appear as red bands. The asterisk shows the higher mobility of the 2-Me Triplex nanoswitch compared to the other Triplex switches (see also Figure S14). The experiments were performed by pre-incubating for 60 min the different AGT enzymes with the relevant triplex nanoswitches and then adding the O6-BG fluorescent derivative. Samples were then loaded on 15% acrylamide SDS-PAGE. Gels underwent Coomassie staining for the determination and correction of the protein amount loaded.
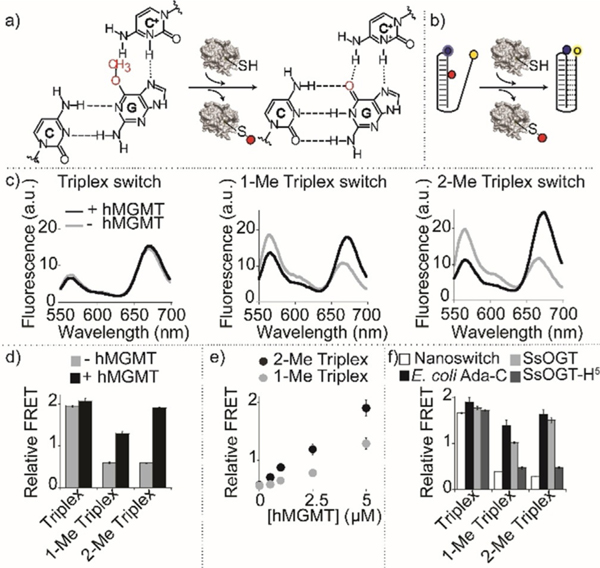
Prompted by the results described above, we tested whether the methyltransferase repair activity on our nanoswitches could result in a conformational switch, and a consequent measurable FRET signal change, providing a means of direct detection of enzymatic activity (Figure 4a,b). Initially, we focused on the detection of the activity of hMGMT; as expected, no significant difference in the FRET signal of the control nanoswitch (non-methylated Triplex) before (1.9±0.1) and after (2.1±0.1) hMGMT incubation can be observed (Figure 4c, left). Under the experimental conditions used, both the fluorescence spectra suggest that the non-methylated triplex switch is completely folded (at pH 5.0). The same experiment carried out using the methylated triplex nanoswitches shows, instead, a strong difference in FRET signals before and after hMGMT incubation (Figure 4c, center, right). For both nanoswitches, the FRET signals observed before hMGMT incubation confirm a partially unfolded configuration (1-Me Triplex, 0.6±0.1, 2-Me Triplex, 0.6±0.1, Figure 4d). Upon hMGMT incubation, the enzymatic demethylation of O6-MeG in the triplex nanoswitches restores their ability to form a triplex structure. This is particularly clear for 2-Me triplex nanoswitch that shows a FRET signal (1.9±0.1) after hMGMT incubation that is within the error of the control non-methylated nanoswitch (2.1±0.1) (Figure 4d). Of note, the folding dynamics of these switches are extremely rapid (Kfolding and Kunfolding =10 s−1 and 2 s−1, respectively)[36] and thus the rate-determining step in these measurements is given by the enzyme-catalyzed reaction. We found out that with hMGMT a saturation of signal is observed after 15 minutes of enzymatic reaction (Figure S15). Both methylated triplex nanoswitches show a change in the relative FRET values that are linearly dependent on the concentration of hMGMT in the range between 0.5 and 5 μM (R22-Me Triplex = 0.99, R21-Me Triplex = 0.95) (Figure 4e). Similar experiments were also performed with other methyltransferase enzymes, both methylated triplex nanoswitches upon incubation with SsOGT and E. coli Ada-C enzymes gave FRET signal changes consistent with triplex folding suggesting efficient enzymatic activity (Figure 4 f). The same experiment performed with SsOGT-H5 (a DNA repair defective mutant, but catalytically active) produced no effect on the FRET signal of both nanoswitches (Figure 4 f) once again confirming the specificity of the folding-upon-repair mechanism. To demonstrate the possibility of using this platform in more complex media, we have also performed the measurement of the enzymatic activity of hMGMT in 10% serum and observed FRET signal changes well distinguishable from the control experiment in absence of enzyme (Figure S16).
Figure 4.
Detection of methyltransferase activity with triplex nanoswitches. a) Methyltransferase enzymatic activity on DNA methylated nanoswitches leads to folding-upon-repair of the triplex DNA structure. b) Enzymatic activity detection can be achieved by monitoring folding/unfolding of the Triplex nanoswitch by fluorescence FRET signaling. c) Spectra and d) relative FRET signals obtained with the Triplex nanoswitches before and after incubation with 5 μM (0.1 μgμL−1) hMGMT. e) Relative FRET signals observed at different hMGMT concentrations for 1-Me and 2-Me Triplex nanoswitches. f) Relative FRET signals with different methyltransferase enzymes. Spectra were obtained by excitation at 530(±5) nm and acquisition between 545 and 700 nm (±10) nm. Relative FRET signals were obtained by first incubating the DNA nanoswitches (0.5 μM) in the absence or presence of AGTs (0.1 μgμL−1) in 10 μL solution of 50 mM Na2HPO4 buffer, 150 mM NaCl, pH 7.5 at 30°C for 60 minutes. The reaction mixtures were then diluted to 100 μL using 50 mM Na2HPO4 buffer, 250 mM NaCl at pH 5.0, and heat-inactivated for 2 minutes at 70°C before performing the fluorescence spectra at 25°C.
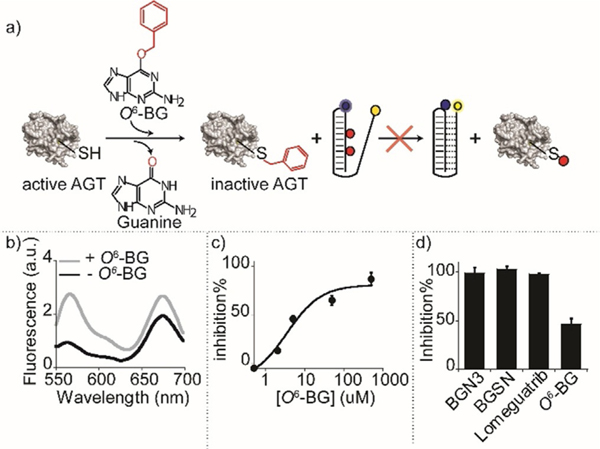
To demonstrate the utility of our platform for the study and characterization of new hMGMT inhibitors as possible drug candidates,[37] we used our nanoswitches to measure the activity of the human enzyme in the presence of inhibitors, inactivators, or pseudo-substrates. The addition of an hMGMT inhibitor in the reaction mixture should prevent the enzyme from repairing the methylated DNA, and no significant change of the nanoswitch FRET signal should be observed (Figure 5a). We initially tested O6-BG, a widely characterized methyltransferase inactivator.[38,39] The relative FRET signal of the nanoswitch incubated with hMGMT and O6-BG (Figure 5b, grey line) is, at 0.8±0.1, similar to the values observed in the absence of enzyme (Figure 5b, black line) an effect consistent with the inactivation of hMGMT activity by O6-BG. The measured inactivation, as expected, follows a concentration-dependent behaviour with a measured IC50 = 3.5±2 μM (Figure 5c). Moreover, our platform is able to measure the inactivation efficiency of different molecules including 4-azido-N-(4-(hydroxymethyl) benzyl) butanamide (BGN3), (N-(4-(((2-amino-9H-purin-6-yl)oxy)-methyl)benzyl)-4-azidobutanamide (BGSN), Lomeguatrib,[40,41] O6-Benzylguanine (O6-BG),[42] demonstrating once again its versatility (Figure 5d).
Figure 5.
Detection of methyltransferase inhibitors. a) Pre-incubation of the enzyme with an inhibitor (here O6-BG) leads to irreversible inhibition of the enzyme that is thus not able to demethylate the DNA nanoswitch. b) Spectra obtained with the 2-Me Triplex nanoswitch in the presence of hMGMT (5 μM) before and after incubation with the inhibitor O6-BG (5 μM). c) % Inhibition plot obtained at different concentrations of O6-BG inhibitor. d) % Inhibition obtained with the 2-Me Triplex nanoswitch with an equimolar concentration of different enzymatic inhibitors and hMGMT (5 μM). Spectra were obtained by excitation at 530(±5) nm and acquisition between 545 and 700 nm-(±10) nm. Relative FRET signals were obtained by enzymatic incubation of 5 μM hMGMT and inhibitor at 30°C for 60 minutes in 10 μL solution of 50 mM Na2HPO4 buffer, 150 mM NaCl at pH 7.5. 2-Me Triplex nanoswitch (0.5 μM) was then added to the reaction mixtures and incubated for another 60 min at 30°C. The reaction mixtures were diluted to 100 μL using 50 mM Na2HPO4 buffer, 250 mM NaCl at pH 5.0, and heat-inactivated for 2 minutes at 70°C before performing the fluorescence spectra at 25°C.
Conclusion
Here, we have designed folding-upon-repair DNA nanoswitches containing O6-MeG nucleobases that undergo a conformational switch from a duplex to a triplex conformation upon enzymatic demethylation. We have first demonstrated that the presence of a methyl group at the guanine O6 position in the duplex portion of the nanoswitch strongly affects the triplex formation. Molecular simulations were used to determine how the O6-methylation of guanine bases prevents the efficient formation of Hoogsteen interactions with the cytosine in the triplex-forming domain, resulting in a local unwinding of the triplex structure. Such methylated triplex-based nanoswitches are versatile tools for the direct measurement of methyltransferase activity as they form the natural enzymatic substrate duplex conformation when methylated and, upon enzymatic demethylation, they can fold into a measurable triplex structure. We showed that the methylated nanoswitches were efficiently recognized by these enzymes leading to measurable FRET signal changes upon repair activity. Finally, we have also measured hMGMT activity in the presence of several enzyme inactivators, demonstrating the possibility of using in a high-throughput system our nanoswitches for the screening of novel potential inactivators/pseudo-substrates/inhibitors of hMGMT.[37]
Our folding-upon-repair DNA nanoswitches are reagentless, highly specific, and versatile. These features make our proposed approach convenient and easy to perform. Although the current experimental protocol requires a change in the pH of the solution after the enzymatic reaction making it less amenable to automatization, we believe that the principle we have described together with the ease with which these probes are designed make the approach suitable to develop a suite of activity-based DNA nanoswitches for other DNA repair enzymes.
Supplementary Material
Acknowledgements
This work was supported by the European Research Council, ERC (project n.336493) (FR), by Associazione Italiana per la Ricerca sul Cancro, AIRC (project n. 14420) (FR), by the Italian Ministry of Health (project n. GR-2010-2317212) and by the Italian Ministry of University and Research (Project of National Interest, PRIN, 2017YER72K. Computational work has been supported by the National Science Foundation under Grant No. CHE-1905374 and by the National Institute of Health under Grant No. R01-EY027440 (GP). Computer time for MD simulations has been awarded by XSEDE via the Grant No. TG-MCB160059 (GP). We would like to thank Mateusz Kogut for valuable suggestions regarding DNA triplex folding simulations. F.R., G.P., and A.P. would like to thank all the authors for their efforts in writing this work, technical assistance, but mainly for human support during the difficult and delicate period of stay-at-home following the COVID-19 outbreak.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Nada Farag, Department of Chemistry, University of Rome Tor Vergata Via della Ricerca Scientifica, 00133 Rome (Italy).
Rosanna Mattossovich, Institute of Biosciences and BioResources National Research Council of Italy Via Pietro Castellino 111, 80131 Naples (Italy).
Rosa Merlo, Institute of Biosciences and BioResources National Research Council of Italy Via Pietro Castellino 111, 80131 Naples (Italy).
Dr. Łukasz Nierzwicki, Department of Bioengineering, University of California Riverside 900 University Avenue, Riverside, CA 52512 (USA).
Giulia Palermo, Department of Bioengineering, University of California Riverside 900 University Avenue, Riverside, CA 52512 (USA); Department of Chemistry, University of California Riverside 900 University Avenue, Riverside, CA 52512 (USA).
Dr. Alessandro Porchetta, Department of Chemistry, University of Rome Tor Vergata Via della Ricerca Scientifica, 00133 Rome (Italy).
Dr. Giuseppe Perugino, Institute of Biosciences and BioResources National Research Council of Italy Via Pietro Castellino 111, 80131 Naples (Italy).
Francesco Ricci, Department of Chemistry, University of Rome Tor Vergata Via della Ricerca Scientifica, 00133 Rome (Italy).
References
- [1].Madders ECET, Parsons JL in Advances in Experimental Medicine and Biology, Vol. 1241 (Eds.: Crusio WE, Lambris JD, Radeke HH, Rezaei N), Springer, Cham, 2020, pp. 59–75. [DOI] [PubMed] [Google Scholar]
- [2].Gerson SL, Nat. Rev. Cancer 2004, 4, 296–307. [DOI] [PubMed] [Google Scholar]
- [3].Pegg AE, Chem. Res. Toxicol 2011, 24, 618–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mattossovich R, Merlo R, Miggiano R, Valenti A, Perugino G, Int. J. Mol. Sci 2020, 21, 2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fu D, Calvo JA, Samson LD, Nat. Rev. Cancer 2012, 12, 104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gerson SL, J. Clin. Oncol 2002, 20, 2388–2399. [DOI] [PubMed] [Google Scholar]
- [7].Nagel ZD, Beharry AA, Mazzucato P, Kitange GJ, Sarkaria JN, Kool ET, Samson LD, PLoS One 2019, 14, e0208341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hurst-Calderone S, Kohn KW, Cancer Res. 1987, 47, 6229–6235. [PubMed] [Google Scholar]
- [9].Klein S, Oesch F, Anal. Biochem 1992, 205, 294–299. [DOI] [PubMed] [Google Scholar]
- [10].Luu KX, Kanugula S, Pegg AE, Pauly GT, Moschel RC, Biochemistry 2002, 41, 8689–8697. [DOI] [PubMed] [Google Scholar]
- [11].Tintoré M, Aviñó A, Ruiz FM, Eritja R, Fàbrega C, Nucleic Acids J. 2010, 2010, 632041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Beharry AA, Nagel ZD, Samson LD, Kool ET, PLoS One 2016, 11, e0152684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wilson DL, Beharry AA, Srivastava A, O’Connor TR, Kool ET, Angew. Chem. Int. Ed 2018, 57, 12896–12900; Angew. Chem. 2018, 130, 13078–13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen S, Su J, Zhao Z, Shao Y, Dou Y, Li F, Deng W, Shi J, Li Q, Zuo X, Song S, Fan C, Nano Lett. 2020, 20, 7028–7035. [DOI] [PubMed] [Google Scholar]
- [15].Vettone A, Serpe M, Hidalgo A, Berenguer J, del Monaco G, Valenti A, Rossi M, Ciaramella M, Perugino G, Extremophiles 2016, 20, 1–13. [DOI] [PubMed] [Google Scholar]
- [16].Morrone C, Miggiano R, Serpe M, Massarotti A, Valenti A, del Monaco G, Rossi M, Rossi F, Rizzi M, Perugino G, Ciaramella M, Biochim. Biophys. Acta 2017, 1861, 86–96. [DOI] [PubMed] [Google Scholar]
- [17].Elder RH, Margison GP, Rafferty JA, Biochem. J 1994, 298, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Goodtzova K, Kanugula S, Edara S, Pauly GT, Moschel RC, Pegg AE, J. Biol. Chem 1997, 272, 8332–8339. [DOI] [PubMed] [Google Scholar]
- [19].Harroun SG, Prévost-Tremblay C, Lauzon D, Desrosiers A, Wang X, Pedro L, Vallée-Bélisle A, Nanoscale 2018, 10, 4607–4641. [DOI] [PubMed] [Google Scholar]
- [20].Modi S, Swetha MG, Goswami D, Gupta GD, Mayor S, Krishnan Y, Nat. Nanotechnol 2009, 4, 325–330. [DOI] [PubMed] [Google Scholar]
- [21].Patino T, Porchetta A, Jannasch A, Lladó A, Stumpp T, Schäffer E, Ricci F, Sánchez S, Nano Lett. 2019, 19, 3440–3447. [DOI] [PubMed] [Google Scholar]
- [22].Porchetta A, Vallée-Bélisle A, Plaxco KW, Ricci F, J. Am. Chem. Soc 2013, 135, 13238–13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hansen-Bruhn M, Nielsen LDF, Gothelf KV, ACS Sens. 2018, 3, 1706–1711. [DOI] [PubMed] [Google Scholar]
- [24].Rossetti M, Ippodrino R, Marini B, Palleschi G, Porchetta A, Anal. Chem 2018, 90, 8196–8201. [DOI] [PubMed] [Google Scholar]
- [25].Liu L, Wu H-C, Angew. Chem. Int. Ed 2016, 55, 15216–15222; Angew. Chem. 2016, 128, 15440–15446. [DOI] [PubMed] [Google Scholar]
- [26].Bell NAW, Keyser UF, J. Am. Chem. Soc 2015, 137, 2035–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ranallo S, Rossetti M, Plaxco KW, A. Vall8e-B8lisle, F. Ricci, Angew. Chem. Int. Ed 2015, 54, 13214–13218; Angew. Chem. 2015, 127, 13412–13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Arts R, Ludwig SKJ, Van Gerven BCB, Estirado EM, Milroy LG, Merkx M, ACS Sens. 2017, 2, 1730–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Vallée-Bélisle A, Ricci F, Plaxco KW, Proc. Natl. Acad. Sci. USA 2009, 106, 13802–13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Idili A, Vallée-Bélisle A, Ricci F, J. Am. Chem. Soc 2014, 136, 5836–5839. [DOI] [PubMed] [Google Scholar]
- [31].Lu XJ, Olson WK, Nucleic Acids Res. 2003, 31, 5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Perugino G, Vettone A, Illiano G, Valenti A, Ferrara MC, Rossi M, Ciaramella M, J. Biol. Chem 2012, 287, 4222–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Perugino G, Miggiano R, Serpe M, Vettone A, Valenti A, Lahiri S, Rossi F, Rossi M, Rizzi M, Ciaramella M, Nucleic Acids Res. 2015, 43, 8801–8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rossi F, Morrone C, Massarotti A, Ferraris DM, Valenti A, Perugino G, Miggiano R, Biochem. Biophys. Res. Commun 2018, 500, 698–703. [DOI] [PubMed] [Google Scholar]
- [35].Merlo R, Del Prete S, Valenti A, Mattossovich R, Carginale V, Supuran CT, Capasso C, Perugino G, Enzyme Inhib J. Med. Chem 2019, 34, 490–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mariottini D, Idili A, Nijenhuis MAD, Ercolani G, Ricci F, J. Am. Chem. Soc 2019, 141, 11367–11371. [DOI] [PubMed] [Google Scholar]
- [37].Kaina B, Christmann M, DNA Repair 2019, 78, 128–141. [DOI] [PubMed] [Google Scholar]
- [38].Quinn JA, Jiang SX, Reardon DA, Desjardins A, Vredenburgh JJ, Rich JN, Gururangan S, Friedman AH, Signer DD, Sampson JH, McLendon RE, Herndon JE, Walker A, Friedman HS, J. Clin. Oncol 2009, 27, 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Reinhard J, Eichhorn U, Wiessler M, Kaina B, Int.J.Cancer 2001,93, 373–379. [DOI] [PubMed] [Google Scholar]
- [40].Middleton MR, Kelly J, Thatcher N, Donnelly DJ, McElhinney RS, McMurry TBH, McCormick JE, Margison GP, Int. J. Cancer 2000, 85, 248–252. [DOI] [PubMed] [Google Scholar]
- [41].Ranson M, Middleton MR, Bridgewater J, Lee SM, Dawson M, Jowle D, Halbert G, Waller S, McGrath H, Gumbrell L, McElhinney RS, Donnelly D, McMurry TBH, Margison GP, Clin. Cancer Res 2006, 12, 1577–1584. [DOI] [PubMed] [Google Scholar]
- [42].Merlo R, Caprioglio D, Cillo M, Valenti A, Mattossovich R, Morrone C, Massarotti A, Rossi F, Miggiano R, Leonardi A, Minassi A, Perugino G, Enzyme Inhib J. Med. Chem 2021, 36, 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.