Abstract
The spread of mupirocin-resistant Staphylococcus aureus strains in hospitals and communities is a universal challenge. Limited data is available on the genetic features of high-level mupirocin resistant- (HLMUPR-) S. aureus isolates in Tehran. In the present research, we investigated 48 high-level mupirocin resistance S. aureus by antimicrobial activity, virulence analysis, biofilm formation, multilocus sequence typing (MLST), and staphylocoagulase (SC) typing. All the HLMUPR strains were positive for mupA gene. The frequency of multidrug resistance was 97.9%. Twenty-one (43.8%) were toxinogenic with 14 producing pvl (29.2%), 5 tst (10.4%), and two eta (4.2%). Among the HLMUPR isolates, biofilm production was detected in 45 (89.6%) isolates with complete dominance clfB, clfA genes, and a noticeably high frequency fnbA (95.8%), followed by fnbB (93.8%), eno and icaD (each 83.3%), sdrC (81.3%), ebps (79.2%), icaA (75%), sdrD (66.7%), fib (60.4%), sdrE (50%), cna (41.7%), and bap (4.2%). Coagulase typing distinguished isolates into four genotypic patterns including III (50%), II (27.1%), and type IVa (22.9%). A total of three clonal complexes (CCs) and 4 sequence types (STs) including CC/ST22 as the most prevalent (52.1%), CC8/ST239 (20.8%), CC/ST8 (16.7%), and CC/ST5 (10.4%) were identified in current work. According to our analysis, nonbiofilm producer isolates belonged to CC8/ST239 (6.3%) and CC/ST8 (4.2%). Fusidic acid-resistant isolates belonged to CC/ST45 (n = 3) and CC8/ST239 (n = 1). Observations highlighted the circulation of the CC/ST22 HLMUPR S. aureus strains with strong biofilm-production ability in our hospitals, indicating the possibility of transmission of this type between community and hospital.
1. Introduction
As the most frequent pathogen, Staphylococcus aureus causes various diseases ranging from skin and soft tissue infections (SSTIs) to bacteremia, endocarditis, and osteomyelitis in hospitals and communities. Recent evidence has highlighted a dramatic increase in hospital-acquired S. aureus infections worldwide with significant morbidity and mortality [1–3]. The high prevalence of antibiotic resistance due to inappropriate antibiotic use and the remarkable ability of this bacterium to acquire resistance to multiple antimicrobials, particularly mupirocin-resistant S. aureus (mupR-S. aureus), complicated the treatment of related infections. Moreover, the scenario worsens with resistance to commonly used antibiotics, biofilm-forming ability, and various virulence factors for S. aureus [4–6].
Mupirocin is a topical antibiotic with excellent activity in treating SSTIs and eradicating MRSA carriage in nursing homes, healthcare workers, patients, and control of MRSA outbreaks. However, an increase in mupirocin resistance has been reported widely in many areas of the world following its extensive and widespread use [7–10]. Mupirocin, an analog of isoleucyl, interferes with protein synthesis by competitive inhibition of the bacterial isoleucyl-tRNA synthetase (IRS). Resistance to mupirocin has been described in two categories: (i) high level that occurs via plasmid-borne gene mupA and or the newly described mupB gene, coding for an alternate isoleucyl-tRNA synthetase (IleS), and (ii) low level, which results from point mutations in the chromosomal encoding isoleucyl-tRNA synthetase (ileS) gene. Data worldwide indicate that lack of proper and timely identification, distribution of molecular types, and managing of high-level mupirocin resistance S. aureus infections (HLMUPR) could lead to decolonization failure, increase in carriage rate, and subsequently wide range of staphylococcal infections [10, 11]. Diagnosis of HLMUPR in S. aureus clinical isolates is a great challenge due to the distribution of different types related to the type of infection, their antimicrobial profile, and biofilm formability [10]. On the other hand, timely diagnosis of HLMUPR in S. aureus clinical isolates is of great significance for formulating appropriate treatment plans, shortening hospitalization time, and reducing the disease burden [11]. Since extensive studies have not been performed in connection with HLMUPR S. aureus infection and the high importance of its timely diagnosis and treatment, therefore, it has become urgently needed to figure out the antibacterial activity, carriage of virulence determinants, biofilm-forming ability, and specific genes that may be involved in biofilm formation and molecular types in HLMUPR strains isolated from various clinical samples. In order to produce a more comprehensive description of the molecular epidemiology and phenotypic properties of HLMUPR S. aureus in Iran, we characterized 390 S. aureus isolates collected from several geographically dispersed Tehran hospitals over the past two years. The frequency of fourteen selected genes involved in biofilm production was assessed to identify genes involved in biofilm production. Coagulase (coa) typing and multilocus sequence typing (MLST) were applied to determine the molecular types of the HLMUPR strains.
2. Materials and Methods
2.1. Isolation S. aureus, Genomic DNA Extraction, and Identification of HLMUPR Isolates
In the present study, 390 S. aureus strains from clinical samples of patients referred to the hospitals affiliated to Shahid Beheshti University of Medical Sciences from January 2018 to December 2019 were screened for HLMUPR. Identification of S. aureus was performed by microbiological and biochemical tests and the presence of the nuc gene using polymerase chain reaction (PCR) assay [12]. The DNA extraction was performed by the phenol-chloroform extraction method previously described by Goudarzi et al. [12]. The purity of the extracted DNA was evaluated by a NanoDrop-2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) at 260/280, respectively. An A260/A280 purity ratio of ~1.8 was considered for the PCR assay.
The study was ethically approved by the Ethics Committee of the Shahid Beheshti University of Medical Sciences in Tehran, Iran (IR. SBMU. MSP.REC. 1398. 353), and consent was obtained from consent participants. HLMUPR strains were detected by the broth microdilution method (MIC value was ≥512 μg/ml) according to the clinical and the laboratory standards institute (CLSI) guideline (CLSI 2019) and PCR of mupA gene [4]. HLMUPR strains that were obtained on or after 96 hours of admission to a hospital were considered as hospital onset (HO) while a positive culture prior to four days of hospitalization along with one or more of the following criteria: (i) a history of hospitalization, surgery, dialysis, or residence in a long-term care facility in 12 months prior to culture date or (ii) the presence of a central vascular catheter (CVC) within two days before S. aureus culture was considered as community onset (CO).
2.2. In Vitro Antimicrobial Susceptibility of HLMUPR Isolates
The antibiotic resistance pattern of isolates was evaluated using the Kirby Bauer method according to the CLSI instructions against amikacin (AMK), clindamycin (CLI), ciprofloxacin (CIP), erythromycin (ERY), gentamicin (GEN), kanamycin (KAN), linezolid (LIN), penicillin (PEN), quinupristin-dalfopristin (SYN), rifampicin (RIF), tobramycin (TOB), tetracycline (TET), teicoplanin (TEC), and trimethoprim-sulfamethoxazole (SXT) (Mast Co., UK). MRSA strains were identified phenotypically using the Cefoxitin disk diffusion method (30 μg) according to the CLSI guidelines and detection of the mecA gene as previously described [4].
The microdilution was performed to determine minimum inhibitory concentration (MIC) titer for antibiotics vancomycin (VAN), tigecycline (TIG), and fusidic acid (FUS) according to the procedure detailed in our previous report [4]. Results for fusidic acid and tigecycline were interpreted based on the European Committee for antimicrobial susceptibility testing (EUCAST) recommendations (http://www.eucast.org). The inducible and constitutive macrolide-lincosamide-streptogramin group B (iMLSB and cMLSB) resistance phenotypes were identified by erythromycin and clindamycin disks by the CLSI guideline (CLSI 2019). S. aureus ATCC 25923, ATCC 43300, and ATCC 29213 strains were used for antibiotic susceptibility testing quality control.
2.3. Microtiter Plate (MtP) Assay to Determine Biofilm Formability
The ability of bacterial biofilm formation was assessed using MtP assay as Yousefi et al. [9] described. All tests were run in quadruplicate and rerun three separate times to confirm reproducibility. Staphylococcus epidermidis ATCC 35984 strain was used as a positive control strain for biofilm formation. TSB supplemented with 1% glucose was used as a negative control. Considering an optical density cut-off (ODc) to be represented by average OD of negative control + 3 × standard deviation (SD) of negative control, strains were classified into the following categories: without biofilm (OD ≤ 0.059), weak (ODc < OD ≤ 2 × ODc), moderate (2 × ODc < OD ≤ 4 × ODc), and strong (OD > 4 × ODc) [9].
2.4. Screening for Virulence-Related Genes
For determination of the prevalence of Panton-Valentine leukotoxin (pvl), toxic shock syndrome toxin (tsst-1), and exfoliative toxin (eta and etb) genes, PCR reaction was carried out with specific oligonucleotide primers [12]. For definitive confirmation of USA300 strains, the existence of the arginine catabolic mobile (ACME) elements in all the isolates was investigated employing PCR assay as previously described by Boswihi et al. [13].
2.5. Detection of Adhesion and Biofilm Genes
The existence of the clumping factors A and B (clfA and clfB), laminin-binding protein (eno), elastin binding (ebpS), collagen-binding protein (cna), fibrinogen-binding protein (fib), fibronectin-binding protein (fnbA and fnbB), biofilm-associated protein (bap), serine-aspartate repeat (sdrC, sdrD, and sdrE), and intercellular adhesion (icaD and icaA) was determined employing PCR assay [14].
2.6. Molecular Typing Methods
2.6.1. coa Typing
Multiplex PCR was carried out to determine coagulase (coa) types (I-X) based on the method provided by Hirose and coworkers. In this assay, four sets were applied, including (i) set A that identified coa types I, II, III, IVa, IVb, Va, and VI; (ii) set B that identified coa types VII, VIII, and X; (iii) set C that identified coa types IX and Vb; and (iv) set D that differentiated IVa and IVb [15].
2.6.2. MLST
We used the procedure described by the MLST database (https://pubmlst.org/) and the Enright et al. method to characterize HLMUPR isolates; PCR assay was carried out using recommended primers of housekeeping genes (pta, arcC, tpi, aroE, gmk, yqiL, and glp) on the website. Sequence types (STs) were determined by submitting the allelic profile to the online MLST database website.
2.7. Statistical Analysis
SPSS Statistics version 22.0 for Windows was used for statistical analysis. In addition, data were analyzed using the chi-square or Fisher's exact tests. P values less than 0.05 were deemed statistically significant.
3. Results
3.1. Antimicrobial Activities
Of the total S. aureus isolates, 48 (12.3%) isolates were confirmed as HLMUPR. In our study, HLMUPR isolates were collected from hospitalized patients, 32 (66.7%) patients were male, and 16 patients were female (33.3%) with a median age of 38.6 years, ranging from 15 to 63 years. In terms of sample type, the isolates were obtained from wound (20; 41.7%), abscess (10; 20.8%), blood (9; 18.8%), urine (6; 12.5%), and respiratory tract secretions (3; 6.2%). The HLMUPR isolates accounted for 62.5% and 37.5% of HO (30/48) and CO (18/48) cases. All HLMUPR isolates were resistant to methicillin (MRSA). The distribution of resistance among HO and CO HLMUPR strains to drugs is provided in Table 1.
Table 1.
Distribution of resistance among HO and CO HLMUPR MRSA isolated from clinical samples.
| Antibiotic | Mupirocin-resistant S. aureusn (%) | Total n (%) | |
|---|---|---|---|
| Hospital onset | Community onset | ||
| Penicillin | 30 (62.5) | 18 (37.5) | 48 (100) |
| Gentamicin | 24 (60) | 16 (40) | 40 (83.3) |
| Kanamycin | 13 (61.9) | 8 (38.1) | 21 (43.8) |
| Amikacin | 14 (70) | 6 (30) | 20 (41.7) |
| Tobramycin | 8 (53.3) | 7 (46.7) | 15 (31.3) |
| Tetracycline | 19 (59.4) | 13 (40.6) | 32 (66.7) |
| Erythromycin | 22 (64.7) | 12 (35.3) | 34 (70.8) |
| Clindamycin | 13 (65) | 7 (35) | 20 (41.7) |
| Ciprofloxacin | 22 (59.5) | 15 (40.5) | 37 (77.1) |
| Rifampin | 7 (63.6) | 4 (36.4) | 11 (22.9) |
| Trimethoprim-sulfamethoxazole | 9 (64.3) | 5 (35.7) | 14 (29.2) |
| Quinupristin-dalfopristin | 7 (63.6) | 4 (36.4) | 11 (22.9) |
| Fusidic acid | 4 (100) | 0 (0) | 4 (8.3) |
Present results exhibited resistance of 100% of isolates for PEN, followed by 83.3% for GEN, 77.1% for CIP, 70.8% for ERY, 66.7% for TET, 43.8% for KAN, 41.7% for CLI, 41.7% for AMK, 31.3% for TOB, 29.2% for SXT, 22.9% for RIF, 22.9% for SYN, and 8.3% for FUS. Ten resistance patterns were identified, wherein PEN, GEN, KAN, AMK, TET, ERY, CLI, CIP (31.3%, 15/48), PEN, GEN, TET, TOB, ERY, CIP, SXT (20.8%, 10/48), and PEN, GEN, CIP, RIF, SYN (16.7%, 8/48) were the top three frequently identified patterns. We did not identify any HLMUPR isolates with resistance to tigecycline, linezolid, and vancomycin. According to the data, there is a statistically significant relationship between HLMUPR MRSA and resistance to PEN (P value 0.022), GEN (P value 0.003), TET (P value 0.002), ERY (P value 0.006), and CIP (P value 0.047). Five isolates (10.4%) were inhibited by 0.5 μg/mL of vancomycin, 12 isolates (25%) by 1 μg/mL, and 31 isolates (64.6%) by 2 μg/mL. Of the 48 HLMUPR strains susceptible to tigecycline, 29 had MIC values of 0.25 μg/ml (60.4%) and 19 had MIC of 0.5 μg/ml (39.6%). A total of 4 HLMUPR isolates (8.3%) were resistant to fusidic acid, of which two had MIC 16 μg/mL and two exhibited MIC titer of 8 μg/mL. Thoroughly, all isolates except one isolate showed resistance to ≥3 classes of antibiotics and considered multidrug resistance (MDR) isolates. Of the total isolates, 20, 12, and 2 isolates showed cMLSB, iMLSB, and MS phenotypes accounting for 41.7%, 25%, and 4.2%, respectively. Inducible and constitutive clindamycin resistance rate among HO-HLMUPR strains (14.6% and 27.1%) was higher than CO HLMUPR strains (10.4% and 14.6%). All the HLMUPR strains with MS phenotype were isolated from HO cases (Table 2).
Table 2.
A summary of MDR among samples under study.
| Simultaneous resistance to antibiotics | Resistance profile | Resistance patterna | Type of samplesb (n; % indicated when not 100%) | Number of isolates (%) |
|---|---|---|---|---|
| Eight | A | PEN, GEN, KAN, AMK, TET, ERY, CLI, CIP | W (6; 40), B (3; 20), A (6; 40) | 15 (31.3) |
|
| ||||
| Seven | B | PEN, GEN, TOB, TET, ERY, CIP, SXT | W (7; 70), A (2; 20), U (1; 10) | 10 (20.8) |
| C | PEN, GEN, KAN, TOB, TET, ERY, CLI | R (1; 20), A (2; 40), U (2; 40) | 5 (10.4) | |
|
| ||||
| Five | D | PEN, CIP, RIF, SYN, FUS | W (1; 50), B (1; 50) | 2 (4.2) |
| E | PEN, AMK, ERY, SXT, FUS | B (1) | 1 (2.1) | |
| F | PEN, GEN, CIP, RIF, SYN | W (3; 37.5), U (1; 12.5), B (2; 25), R (2; 25) | 8 (16.6) | |
| G | PEN, GEN, AMK, CIP, RIF | U (1) | 1 (2.1) | |
| H | PEN, KAN, CIP, TET, SYN | U (1) | 1 (2.1) | |
|
| ||||
| Four | I | PEN, GEN, TET, FUS | B (1) | 1 (2.1) |
| J | PEN, AMK, ERY, SXT | W (2; 66.7), B (1; 33.3) | 3 (6.2) | |
|
| ||||
| Without | K | - | W (1) | 1 (2.1) |
aAMK: amikacin; CLI: clindamycin; CIP: ciprofloxacin; ERY: erythromycin; FA: fusidic cid; GEN: gentamicin; KAN: kanamycin; PEN: penicillin; RIF: rifampicin; SYN: quinupristin-dalfopristin; SXT: trimethoprim-sulfamethoxazole; TET: tetracycline; TOB: tobramycin. bW: wound; B: blood; A: abscess; U: urine; R: respiratory tract secretions.
3.2. Phenotypic Biofilm Formation
Of the 48 strains of HLMUPR studied, 43 (89.6%) isolates were biofilm producers in varying degrees. Twenty-five (52.1%) isolates were identified as strong biofilm producers, 10 (20.8%) were moderate producers, and eight (16.7%) were found to be weak producers. A statistically significant relationship between resistance to mupirocin at a high level and strong biofilm production (P value 0.015) among HLMUPR MRSA isolates was seen.
Five (10.4%) were confirmed as nonbiofilm producer isolates which all belonged to CO cases. Figure 1 represents the analysis of biofilm formability of isolates based on HO and CO cases.
Figure 1.
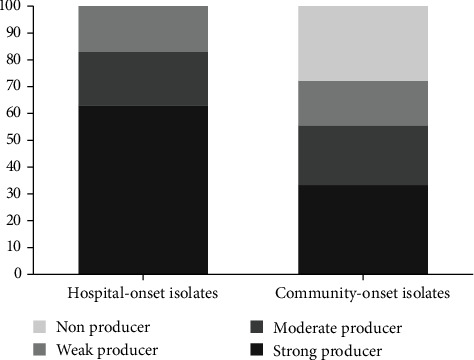
Biofilm formability of isolates based on HO and CO cases.
3.3. Toxin Detection
Of 48 HLMUPR MRSA strains, 21 (43.8%) were toxinogenic, with 14 producing pvl (29.2%), five tst (10.4%), and two eta (4.2%). Our analysis indicated a statistically significant relationship between HLMUPR MRSA and toxin carriage (P value 0.012).
3.4. Genotypic Biofilm Formation
The findings showed that adhesion and biofilm-related genes varied among our isolates. According to our results, among adhesion encoding genes, the clfA and clfB genes were all detected (100%), fnbA was present in 46 strains (95.8%), fnbB in 45 (93.8%), eno in 40 (83.3%), sdrC in 39 (81.3%), ebps in 38 (79.2%), sdrD in 32 (66.7%), fib in 29 (60.4%), sdrE in 24 (50%), and cna in 20 (41.7%). Regarding biofilm-related genes, icaD was present in 10 (20.8%) isolates, icaA in 8 (16.7%) isolates, icaA+icaD in 28 (58.3%) isolates, and icaD+bap in two (4.2%) isolates. Seven different biofilm genetic patterns were identified, wherein clfA, clfB, fnbA, fnbB, eno, ebps, sdrC, icaD, icaA (31.3%, 15/48), clfA, clfB, fnbA, fnbB, eno, ebps, fib, icaA, icaD, sdrD, sdrC, sdrE (25%, 12/48), and clfA, clfB, fnbA, fnbB, ebps, eno, cna, fib, icaD, sdrD, sdrE (18.8%, 9/48) were the top three frequently identified patterns. Biofilm and adhesion-associated genes and their distribution among different STs are provided in Figure 2.
Figure 2.
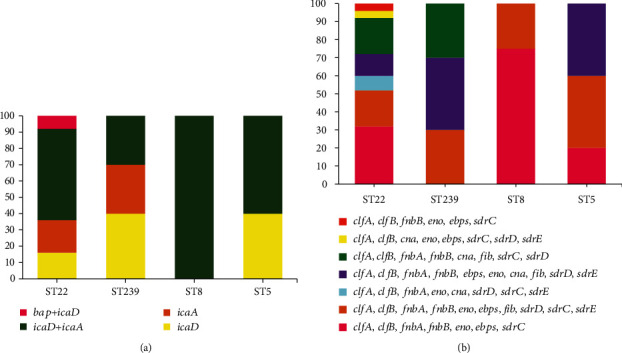
(a) Distribution of biofilm-associated genes in HLMUPR isolates. (b) Distribution of adhesion-associated genes in different molecular types of HLMUPR isolates.
3.5. Molecular Typing
In the present work, HLMUPR was classified into three classes, according to the coa typing. The predominant coa type was III which included 24 isolates (50%), followed by type II (27.1%, 13/48) and type IVa (22.9%, 11/48). According to the MLST, HLMUPR MRSA isolates were assigned to 4 sequences types (STs), including ST22 as the most prevalent (52.1%), followed by ST239 (20.8%), ST8 (16.7%), and ST5 (10.4%). Our analysis clustered tested strains into 3 clonal complexes (CCs), namely, CC22 (52.1%, 25/48), CC8 (37.5%, 18/48), and CC5 (10.4%, 5/48). According to our analysis, nonbiofilm producer isolates belonged to CC8/ST239 (6.3%) and CC/ST8 (4.2%). Regarding biofilm-related genes, the bap gene was detected in CC/ST22 isolates. Among the 21-toxinogenic isolates, pvl was observed in isolates with CC/ST22 (20.8%, 10/48) and CC/ST8 (8.3%, 4/48). A statistically significant relationship between CC/ST22 and pvl carriage (P value 0.007), coa type III (P value 0.041), and strong biofilm formability (P value 0.002) was seen. There is also a significant relationship between CC8 and coa type III (P value 0.047) and strong biofilm formability (P value 0.022). All the tst-positive isolates belonged to CC8/ST239 (10.4%, 5/48). We found two CC/ST45 strains (4.2%) that carried the eta gene. Fusidic acid-resistant isolates belonged to CC/ST45 (n = 3) and CC8/ST239 (n = 1). Twelve isolates with iMLSB phenotype belonged to CC/ST22 (41.7% (5/12)), CC8/ST239 (33.3% (4/12)), CC/ST8 (16.7%, (2/12)), and CC/ST5 (8.3% (1/12)). Out of 20 strains with cMLSB phenotype, 12 isolates belonged to CC/ST22 (60%), 7 isolates to CC8/ST239 (35%), and 1 isolate to CC/ST8 (5%). Out of two isolates with MS phenotype, one isolate belonged to CC/ST8 and another to CC/ST5. Table 3 provides data related to the characterization of HLMUPR MRSA strains under study.
Table 3.
Characteristics of the HLMUPR MRSA strains isolated from patients.
| Clonal complex (CC) (n) | Sequence type (n) | coa type (%) | Antibiotic resistance profilea (%) | Toxin genes (%) | Biofilm statusb (%) | Adhesion/biofilm profilec (%) | Type of infection (%) |
|---|---|---|---|---|---|---|---|
| 22 | ST22 (25) | III (48), IVa (32), II (20) | A (36), B (36), C (20), F (8) | pvl (40) | S (72), M (28) | I (8), II (40), III (20), IV (16), VI (16) | HO (64), CO (36) |
|
| |||||||
| 8 | ST239 (10) | III (70), II (30) | A (50), F (20), I (10), H (10), G (10) | tst (50) | S (50), W (20), N (30) | II (50), IV (20), VI (20), VII (10) | HO (60), CO (40) |
| ST8 (8) | III (62.5), IVa (37.5) | F (50), J (37.5), A (12.5) | pvl (50) | S (25), W (50), N (25) | III (37.5), VI (37.5), V (25) | HO (62.5), CO (37.5) | |
|
| |||||||
| 5 | ST5 (5) | II (100) | D (40), E (20), B (20), K (20) | eta (40) | M (60), W (40) | V (60), VII (40) | HO (60), CO (40) |
4. Discussion
The findings showed that 12.3% of S. aureus isolates were confirmed as HLMUPR and all carried mupA gene. Dadashi et al.'s systematic review and meta-analysis study reported an 8.1% prevalence of HLMUPR MRSA clinical isolates lower than our reported rate. They also reported the prevalence rate of HLMUPR MRSA isolates in Europe, Asia, and the USA, accounting for 8.0%, 12.1%, and 5.9%, respectively [11]. Moreover, Shittu et al. reported that the prevalence of HLMUPR MRSA isolates in Africa ranged between 0.5 and 38% [16]. However, much higher rates were also reported by studies conducted from India (26.1%) [5], the USA (19.3%) [17], and Korea (5.7%) [6]. Reasons for the high resistance rate in Asian countries and our research could be irrational use, various policies in the prescription of these antibiotics, easy access to antibiotics, low cost of the drugs, and spreading of dominant types in these areas. Recent available data has shown there is an increase in the prevalence of mupA-positive MRSA isolates. In this connection, Monecke et al. noted a sharp increase in the prevalence of these isolates during the study period from 1.1% in 2000-2015 to 15.9% in 2016 and 17.6% in 2017 [18]. This high prevalence of mupA gene is probably because this gene, linked to mupirocin resistance at a high level, is contained within a plasmid whose acquisition was reasonably easy through horizontal gene transfer [10, 11]. In the study, the prevalence of iMLSB was found to be 25% which was near to reported rate from Nepal (21%) [19], higher than those reported in Brazil (7.9%) [20], and lower than the rates reported from Jordan (76.7%) [21]. We observed that 41.7% of isolates had cMLSB phenotype. This was lower than the similar studies carried out in Iran (82.9%) [22] and higher than the rate stated in Nepal [19] (38%) and Greece (20.1%) [23]. It seems that the acquisition of erythromycin resistance in HLMUPR MRSA maybe dynamic and associated with injudicious use of macrolides in patients infected with HLMUPR MRSA isolates [10, 11].
According to the earlier published data, mupirocin-resistant strains are more likely to be resistant to fusidic acid. In our survey, of the forty-eight strains, four isolates were fusidic acid-resistant (8.3%). These results differ from those found in Canada (7%), Ireland (19.9%), Greece (62.4%), and Australia (7%) [24, 25], as well as a study carried out in Iran during four years on 726 tested S. aureus isolates, and a low prevalence of resistance to fusidic acid was reported (3%) [26].
In the current research, 29.2% of isolates carried pvl gene. Gambian-based research of the prevalence of PVL S. aureus in the 11 years reported a rate of 61.4% (180/293) of PVL among S. aureus isolates [27]. Different frequency of PVL-positive HLMUPR strains was reported in researches conducted from Iran (27.9%) [28], Ireland (17%) [25], and China (15.1%) [29]. Possible factors involved in the observed high prevalence of pvl gene include study population, design, and circulating clones in hospitals and communities.
We detected the tst-encoding gene in 10.4% of tested isolates. Nevertheless, Shahini Shams-Abadi et al. showed a relatively high prevalence of tst gene among S. aureus strains tested (21.3%) [30]. Differences are probably attributed to the origin of isolates, dissemination of specific clones, and sample type.
As mentioned earlier, three different coa types were detected in our study. Our data indicated low variability and heterogeneity of coa gene. Similarly, a previous study conducted by Omar et al. indicated three coa types and ten subtypes [31]. Based on published data by Afrough et al. in Iran [32], six coa types with the majority of the pattern I C 1 (21.7%) were detected among S. aureus isolates. Consistent with our finding, the results of coa typing of 100 S. aureus clinical isolates by Salehzadeh et al. noted two genotypes and three subtypes [33]. Contrary to Hirose et al.'s study [15], which displayed a high prevalence of coa type II (91.9%), VII (3.9%), and I (1.7%), in our study, coa type III was the most predominant coa type among S. aureus isolates (50%), followed by type II (27.1%) and type IVa (22.9%). Similar results in some studies indicate the predominance of coa type III in MRSA isolates [31, 34]. The presence of coa type III in the present research and other recent studies suggests that this coa type is actively circulating in Tehran province's healthcare setting.
According to the evidence, there is high variability in biofilm-related genes among the MRSA strains and specific genotypes [35]. Earlier studies reported different prevalence rates of biofilm formation among S. aureus with data ranging from 43 to 88% [2]. In the present study, the prevalence of biofilm producers was high (89.6%) which was more significant reported in China (66%) [36] and Iran (62.9%) [9]. It is well established that biofilm formation depends on different factors, but it seems that resistance to mupirocin at a high level could affect the accessory gene regulator system involved in the expression of adhesion factors and biofilm induction.
Many studies have noted the critical role of adhesion-related genes in adherence to the host components and producing biofilm. The clfA, clfB, fnbA, and fnbB genes are essential for microorganism adhesion and might be related to biofilm formability. In the present study, the complete dominance was observed for clfA and clfB genes followed by fnbA (95.8%) and fnbB (93.8%). Similarly, the published results by Yousefi and coworkers in Iran revealed a 69.2% prevalence of biofilm production and the presence of fnbA, clfA, and icaA genes in all examined strains [9]. These rates were comparable to those in other researches [36–40].
Other adhesion-related genes were eno and icaD (each 83.3%), sdrC (81.3%), ebps (79.2%), icaA (75%), sdrD (66.7%), fib (60.4%), cna (41.7%), and bap (4.2%). Nourbakhsh et al. noticed a moderate prevalence of fnbB (46.6%), clfA (41.4%), clfB (44.1%), fnbA (38.1%), ebps (26.5%), and cna (18.3%) genes among their tested isolates [37]. Conversely, in Ghasemian et al.'s study from Iran, the most common biofilm-related genes were cna and ebps accounted for 78% and 7% of isolates, respectively [38]. In multicenter research performed from 2015 to 2018 from major hospitals in the West Bank-Palestine, out of 248 tested S. aureus isolates, 207 (83.5%) possessed the icaD gene. In addition, icaA fnbB, cna, fib, ebps, fnbA, clfA/clfB, and eno genes were found in 16.5, 29, 39.9, 62.2, 76.2, 78.2, 80.2, and 94.8% of isolates, respectively. Azmi and colleagues also showed that none of the strains possessed the bap gene [35].
Serine-aspartate repeat (sdr) family proteins have the primary role in the development of ica-independent biofilms. Regarding sdr genes, sdrC and sdrD were present in 81.3% and 66.7% of isolates. These values agree with the findings of Uribe-García et al. from Mexico who indicated the presence of sdrC, sdrD, and sdrE in the majority of isolates [8]. The percentages obtained in this study were also similar to those reported in Chen et al.'s study which demonstrated the presence of sdrC in the majority of isolates (94.3%) [14]. Earlier data plus the findings of the present study have revealed discrepancies in genetic characteristics of biofilm producers that could likely attribute to the distribution of variants of the genotype of S. aureus in a different geographic area.
Some published data documented that the ability of strains of S. aureus to produce biofilm has related to the synthesis of polysaccharide intercellular adhesion (PIA), which its production is under the control of the icaADBC operon. In this study, a high prevalence of icaA (75%) and icaD (83.3%) was found, which is in line with the findings of Azmi and colleagues [35] and support those of Sharma and coworkers [39] who reported that icaA/D found abundantly among biofilm-producing isolates. Contrarily, Azmi et al. [35] indicated that icaA and icaD genes were present in 16.5% and 83.5% of strains. These findings suggest that the ica-dependent system may not be an absolute mechanism in biofilm production, and these strains perhaps use other systems to produce biofilm.
According to our findings, CC/ST22 was a predominate genotype (52.1%). Forty percent of CC22 isolates were PVL-positive. This finding supports previous results reported by Shore et al. [25], Goudarzi et al. [12], and Monecke et al. [3]. Although biofilm formability and antibiotic resistance profiles in CC/ST22 isolates were found to be varied, resistance to mupirocin and biofilm formability in these strains have been described by several investigators [4, 41].
CC8/ST239 was the second prevailing lineage detected accounting for 20.8% of tested isolates. In line with the present study, ST239 was confirmed as the most predominant genotype in China from 2005 to 2013 in a study conducted by Li et al. [42]. One of the four fusidic acid-resistant MRSA strains belonged to CC8/ST239; this agreed with the findings of Shore et al. from Ireland [25]. Concordant with our study, in a 13-year study, Boswihi et al. in Kuwait displayed fusidic acid-resistant isolates belonged to ST239-MRSA-III that none of them carried fusB or fusC genes [13]. Inducible clindamycin resistance was noted in 40% of the ST239 isolates. In support of this, Abimanyu and coworkers recently reported the emergence of ST239 MRSA strains with both mupirocin and inducible clindamycin resistance in India [43].
Our analysis confirmed CC/ST8 (ACME+ and PVL+) isolates that resembled the USA300. Earlier researches have noted the resistance to mupirocin and trimethoprim-sulfamethoxazole among USA300 isolates [3, 4, 44, 45], in conformity of results obtained from McDougal et al. in the USA, which identified USA300 clone MRSA isolates harboring mupA [44]. We found mupA gene in 12.3% of examined isolates. Contrary to a prior study from Iran, which described ST8 S. aureus isolates with resistance to vancomycin [45], none of CC/ST8 isolates described in this study was vancomycin-resistant. The emergence of the USA300 in the present study reflects its growing importance as an epidemic clone, which may be due to an import of this clone from Asian or European countries.
In this study, we noted a low prevalence of ST5 (10.4%) with 100% biofilm formability. González-Domínguez and colleagues analyzed 147 MRSA strains isolated from different clinical samples and found that HLMUPR-MRSA isolates belonged to ST125 (97.5%) and ST5 (2.5%) [46]. Present work showed that half of the isolates had a high resistance rate to trimethoprim and fusidic acid. These findings confirmed a study conducted by Shore et al. which indicated resistances to trimethoprim and tetracycline among CC5 isolates at a high level [25].
5. Conclusion
Altogether, our study provides evidence for the occurrence of four different lineages of HLMUPR-MRSA in Iran. The high prevalence of CC/ST22 with strong biofilm formation in our hospitals highlights special attention for implementing efficient control protocols and stricter precautions to stop the dissemination of these isolates in both communities to hospitals. There is a need for continuous monitoring of the genotypes of HLMUPR-MRSA isolates to prevent nosocomial outbreaks of these isolates.
Acknowledgments
This work is financially supported by a research grant from the Research Deputy of Shahid Beheshti University of Medical Sciences, Tehran, Iran (Grant No. 25586). We are indebted to Dr. Nobumichi Kobayashi (Sapporo Medical University School of Medicine, Japan) for providing Coagulase-type reference strains of the S. aureus.
Data Availability
All data generated or analyzed during this study are included in this published article.
Disclosure
The funding agency has no role in the design of the project, work execution, analyses, interpretation of the data, and manuscript writing and submission.
Conflicts of Interest
All authors declare that they have no competing interests.
Authors' Contributions
MG, MH, and SZ conceived and designed the study. SZ, AM, BH, PA, MF, MJN, MD, and MH contributed in comprehensive research. MG, MH, and MF wrote the paper. MG, MH, and MF participated in manuscript editing.
References
- 1.Hetem D. J., Bonten M. J. Clinical relevance of mupirocin resistance in _Staphylococcus aureus_. The Journal of Hospital Infection . 2013;85(4):249–256. doi: 10.1016/j.jhin.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Luther M. K., Parente D. M., Caffrey A. R., et al. Clinical and genetic risk factors for biofilm-forming Staphylococcus aureus. Antimicrobial Agents and Chemotherapy . 2018;62(5, article e02252) doi: 10.1128/AAC.02252-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monecke S., Coombs G., Shore A. C., et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One . 2011;6(4):p. e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tayebi Z., Fazeli M., Hashemi A., et al. Molecular characterization of invasive Staphylococcus aureus strains isolated from patients with diabetes in Iran: USA300 emerges as the major type. Infection, Genetics and Evolution . 2021;87, article 104679 doi: 10.1016/j.meegid.2020.104679. [DOI] [PubMed] [Google Scholar]
- 5.Orrett F. A. The emergence of mupirocin resistance among clinical isolates of methicillin-resistant Staphylococcus aureus in Trinidad: a first report. Japanese Journal of Infectious Diseases . 2008;61:p. 107. [PubMed] [Google Scholar]
- 6.Park S. Y., Kim S. M., Park S. D. The prevalence, genotype and antimicrobial susceptibility of High- and low-level mupirocin resistant Methicillin-ResistantStaphylococcus aureus. Annals of Dermatology . 2012;24(1):32–38. doi: 10.5021/ad.2012.24.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosseini M., Shapouri Moghaddam A., Derakhshan S., et al. Correlation between biofilm formation and antibiotic resistance in MRSA and MSSA isolated from clinical samples in Iran: a systematic review and meta-analysis. Microbial Drug Resistance . 2020;26(9):1071–1080. doi: 10.1089/mdr.2020.0001. [DOI] [PubMed] [Google Scholar]
- 8.Uribe-García A., Paniagua-Contreras G. L., Monroy-Pérez E., et al. Frequency and expression of genes involved in adhesion and biofilm formation in Staphylococcus aureus strains isolated from periodontal lesions. Journal of Microbiology, Immunology, and Infection . 2021;54(2):267–275. doi: 10.1016/j.jmii.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Yousefi M., Pourmand M. R., Fallah F., Hashemi A., Mashhadi R., Nazari-Alam A. Characterization of Staphylococcus aureus biofilm formation in urinary tract infection. Iranian Journal of Public Health . 2016;45(4):485–493. [PMC free article] [PubMed] [Google Scholar]
- 10.Khoshnood S., Heidary M., Asadi A., et al. A review on mechanism of action, resistance, synergism, and clinical implications of mupirocin against Staphylococcus aureus. Biomedicine & Pharmacotherapy . 2019;109:1809–1818. doi: 10.1016/j.biopha.2018.10.131. [DOI] [PubMed] [Google Scholar]
- 11.Dadashi M., Hajikhani B., Darban-Sarokhalil D., van Belkum A., Goudarzi M. Mupirocin resistance in _Staphylococcus aureus_ : A systematic review and meta-analysis. Journal of Global Antimicrobial Resistance . 2020;20:238–247. doi: 10.1016/j.jgar.2019.07.032. [DOI] [PubMed] [Google Scholar]
- 12.Goudarzi M., Goudarzi H., Sá Figueiredo A. M., et al. Molecular characterization of methicillin resistant Staphylococcus aureus strains isolated from intensive care units in Iran: ST22-SCCmec IV/t790 emerges as the major clone. PLoS One . 2016;11(5, article e0155529) doi: 10.1371/journal.pone.0155529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boswihi S. S., Udo E. E., Al-Sweih N. Shifts in the clonal distribution of methicillin-resistant Staphylococcus aureus in Kuwait hospitals: 1992-2010. PLoS One . 2016;11(9, article e0162744) doi: 10.1371/journal.pone.0162744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Q., Xie S., Lou X., et al. Biofilm formation and prevalence of adhesion genes amongStaphylococcus aureusisolates from different food sources. Microbiology . 2020;9(1, article e00946) doi: 10.1002/mbo3.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirose M., Kobayashi N., Ghosh S., et al. Identification of staphylocoagulase genotypes I-X and discrimination of type IV and V subtypes by multiplex PCR assay for clinical isolates of Staphylococcus aureus. Japanese Journal of Infectious Diseases . 2010;63(4):257–263. [PubMed] [Google Scholar]
- 16.Shittu A. O., Kaba M., Abdulgader S. M., Ajao Y. O., Abiola M. O., Olatimehin A. O. Mupirocin-resistant Staphylococcus aureus in Africa: a systematic review and meta-analysis. Antimicrobial Resistance and Infection Control . 2018;7(1):p. 101. doi: 10.1186/s13756-018-0382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antonov N. K., Garzon M. C., Morel K. D., Whittier S., Planet P. J., Lauren C. T. High prevalence of mupirocin resistance in Staphylococcus aureus isolates from a pediatric population. Antimicrobial Agents and Chemotherapy . 2015;59(6):3350–3356. doi: 10.1128/AAC.00079-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monecke S., Ruppelt-Lorz A., Müller E., et al. Dissemination of high-level mupirocin-resistant CC22-MRSA-IV in Saxony. Infection Control . 2017;12 doi: 10.3205/dgkh000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adhikari R., Shrestha S., Barakoti A., Amatya R. Inducible clindamycin and methicillin resistant Staphylococcus aureus in a tertiary care hospital, Kathmandu, Nepal. BMC Infectious Diseases . 2017;17(1):p. 483. doi: 10.1186/s12879-017-2584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bottega A., Rodrigues M. A., Carvalho F. A., et al. Evaluation of constitutive and inducible resistance to clindamycin in clinical samples of Staphylococcus aureus from a tertiary hospital. Revista da Sociedade Brasileira de Medicina Tropical . 2014;47(5):589–592. doi: 10.1590/0037-8682-0140-2014. [DOI] [PubMed] [Google Scholar]
- 21.Jarajreh D.’a., Aqel A., Alzoubi H., al-Zereini W. Prevalence of inducible clindamycin resistance in methicillin-resistant Staphylococcus aureus: the first study in Jordan. Journal of Infection in Developing Countries . 2017;11(4):350–354. doi: 10.3855/jidc.8316. [DOI] [PubMed] [Google Scholar]
- 22.Khashei R., Malekzadegan Y., Sedigh Ebrahim-Saraie H., Razavi Z. Phenotypic and genotypic characterization of macrolide, lincosamide and streptogramin B resistance among clinical isolates of staphylococci in southwest of Iran. BMC Research Notes . 2018;11(1):1–6. doi: 10.1186/s13104-018-3817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doudoulakakis A., Spiliopoulou I., Spyridis N., et al. Emergence of a Staphylococcus aureus clone resistant to mupirocin and fusidic acid carrying exotoxin genes and causing mainly skin infections. Journal of Clinical Microbiology . 2017;55(8):2529–2537. doi: 10.1128/JCM.00406-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castanheira M., Watters A. A., Mendes R. E., Farrell D. J., Jones R. N. Occurrence and molecular characterization of fusidic acid resistance mechanisms among Staphylococcus spp. from European countries (2008) The Journal of Antimicrobial Chemotherapy . 2010;65(7):1353–1358. doi: 10.1093/jac/dkq094. [DOI] [PubMed] [Google Scholar]
- 25.Shore A. C., Tecklenborg S. C., Brennan G. I., Ehricht R., Monecke S., Coleman D. C. Panton-Valentine leukocidin-positive Staphylococcus aureus in Ireland from 2002 to 2011: 21 clones, frequent importation of clones, temporal shifts of predominant methicillin-resistant S. aureus clones, and increasing multiresistance. Journal of Clinical Microbiology . 2014;52(3):859–870. doi: 10.1128/JCM.02799-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahimi F., Bouzari M., Katouli M., Pourshafie M. R. Antibiotic resistance pattern of methicillin resistant and methicillin sensitive Staphylococcus aureus isolates in Tehran, Iran. Jundishapur Journal of Microbiology . 2013;6(2):144–149. doi: 10.5812/jjm.4896. [DOI] [Google Scholar]
- 27.Darboe S., Dobreniecki S., Jarju S., et al. Prevalence of Panton-Valentine leukocidin (PVL) and antimicrobial resistance in community-acquired clinical Staphylococcus aureus in an urban Gambian hospital: a 11-year period retrospective pilot study. Frontiers in Cellular and Infection Microbiology . 2019;9:p. 170. doi: 10.3389/fcimb.2019.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abadi M. S. S., Nikokar I., Alfatemi S. M. H., Malekzadegan Y., Azizi A., Ebrahim-Saraie H. S. Epidemiology of Panton-Valentine leukocidin harbouring Staphylococcus aureus in cutaneous infections from Iran: a systematic review and meta-analysis. Wounds . 2017;7:p. 8. [PubMed] [Google Scholar]
- 29.Liu Q., Han L., Li B., Sun J., Ni Y. Virulence characteristic and MLST-agr genetic background of high-level mupirocin-resistant, MRSA isolates from Shanghai and Wenzhou, China. PLoS One . 2012;7(5) doi: 10.1371/journal.pone.0037005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shahini Shams-Abadi M., Halaji M., Hoseini-Alfatemi S., Gholipour A., Mojtahedi A., Sedigh E.-S. H. Epidemiology of toxic shock syndrome toxin-1 harboring Staphylococcus aureus obtained from clinical samples in Iran: a systematic review and meta-analysis. Annali di Igiene . 2018;30 doi: 10.7416/ai.2018.2239. [DOI] [PubMed] [Google Scholar]
- 31.Omar N. Y., Ali H. A. S., Harfoush R. A. H., El Khayat E. H. Molecular typing of methicillin resistant Staphylococcus aureus clinical isolates on the basis of protein A and coagulase gene polymorphisms. International Journal of Microbiology . 2014;2014:11. doi: 10.1155/2014/650328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Afrough P., Pourmand M. R., Sarajian A. A., Saki M., Saremy S. Molecular investigation of Staphylococcus aureus, coa and spa genes in Ahvaz hospitals, staff nose compared with patients clinical samples. Jundishapur Journal of Microbiology . 2013;6:p. 10. doi: 10.5812/jjm.5377. [DOI] [Google Scholar]
- 33.Salehzadeh A., Zamani H., Langeroudi M. K., Mirzaie A. Molecular typing of nosocomial <i>Staphylococcus aureus</i> strains associated to biofilm based on the coagulase and protein A gene polymorphisms. Iranian Journal of Basic Medical Sciences . 2016;19(12):1325–1330. doi: 10.22038/ijbms.2016.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohajeri P., Azizkhani S., Farahani A., Norozi B. Genotyping of coa and aroA genes of methicillin-resistant staphylococcus aureus strains isolated from nasal samples in western Iran. Journal of Microbiology . 2016;9(1) doi: 10.5812/jjm.26460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Azmi K., Qrei W., Abdeen Z. Screening of genes encoding adhesion factors and biofilm production in methicillin resistant strains of Staphylococcus aureus isolated from Palestinian patients. BMC Genomics . 2019;20(1):p. 578. doi: 10.1186/s12864-019-5929-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L., Yu F., Yang L., Li Q., Zeng X. Z., Xu Y. Prevalence of virulence genes and biofilm formation among Staphylococcus aureus clinical isolates associated with lower respiratory infection. African Journal of Microbiology Research . 2010;4:2566–2569. [Google Scholar]
- 37.Nourbakhsh F., Namvar A. E. Detection of genes involved in biofilm formation in Staphylococcus aureus isolates. GMS Hygiene and Infection Control . 2016;11 doi: 10.3205/dgkh000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghasemian A., Najar Peerayeh S., Bakhshi B., Mirzaee M. Several virulence factors of multidrug-resistant Staphylococcus aureus isolates from hospitalized patients in Tehran. International Journal of Enteric Pathogens . 2015;3(2):1–6. doi: 10.17795/ijep25196. [DOI] [Google Scholar]
- 39.Sharma V., Sharma S., Dahiya D. K., Khan A., Mathur M., Sharma A. Coagulase gene polymorphism, enterotoxigenecity, biofilm production, and antibiotic resistance in Staphylococcus aureus isolated from bovine raw milk in North West India. Annals of Clinical Microbiology and Antimicrobials . 2017;16(1):p. 65. doi: 10.1186/s12941-017-0242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serray B., Oufrid S., Hannaoui I., et al. Genes encoding adhesion factors and biofilm formation in methicillin-resistant Staphylococcus aureus in Morocco. Journal of Infection in Developing Countries . 2016;10(8):863–869. doi: 10.3855/jidc.8361. [DOI] [PubMed] [Google Scholar]
- 41.Yuan W., Liu J., Zhan Y., et al. Molecular typing revealed the emergence of pvl-positive sequence type 22 methicillin-susceptible Staphylococcus aureus in Urumqi, Northwestern China. Infection and Drug Resistance . 2019;Volume 12:1719–1728. doi: 10.2147/IDR.S202906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li S., Sun S., Yang C., et al. The changing pattern of population structure of Staphylococcus aureus from bacteremia in China from 2013 to 2016: ST239-030-MRSA replaced by ST59-t437. Frontiers in Microbiology . 2018;9:p. 332. doi: 10.3389/fmicb.2018.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abimanyu N., Murugesan S., Krishnan P. Emergence of methicillin-resistant Staphylococcus aureus ST239 with high-level mupirocin and inducible clindamycin resistance in a tertiary care center in Chennai, South India. Journal of Clinical Microbiology . 2012;50(10):3412–3413. doi: 10.1128/JCM.01663-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDougal L. K., Fosheim G. E., Nicholson A., et al. Emergence of resistance among USA300 methicillin-resistant Staphylococcus aureus isolates causing invasive disease in the United States. Antimicrobial Agents and Chemotherapy . 2010;54(9):3804–3811. doi: 10.1128/AAC.00351-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Havaei S. A., Azimian A., Fazeli H., et al. Genetic characterization of methicillin resistant and sensitive, vancomycin intermediate Staphylococcus aureus strains isolated from different Iranian hospitals. ISRN Microbiology . 2012;2012:6. doi: 10.5402/2012/215275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.González-Domínguez M., Seral C., Potel C., et al. Genotypic and phenotypic characterization of methicillin-resistant Staphylococcus aureus (MRSA) clones with high-level mupirocin resistance. Diagnostic Microbiology and Infectious Disease . 2016;85(2):213–217. doi: 10.1016/j.diagmicrobio.2016.02.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


