Abstract
Introduction
The advent of immune checkpoint blockade (ICB) has led to significantly improved disease outcome in lung adenocarcinoma (ADC), but response of ALK/EGFR-positive tumors to immune therapy is limited. The underlying immune biology is incompletely understood.
Methods
We performed comparative mRNA expression profiling of 31 ALK-positive, 40 EGFR-positive and 43 ALK/EGFR-negative lung ADC focused on immune gene expression. The presence and levels of tumor infiltration lymphocytes (TILs) as well as fourteen specific immune cell populations were estimated from the gene expression profiles.
Results
While total TILs were not lower in ALK-positive and EGFR-positive tumors compared to ALK/EGFR-negative tumors, specific immunosuppressive characteristics were detected in both subgroups: In ALK-positive tumors, regulatory T cells were significantly higher compared to EGFR-positive (fold change: FC = 1.9, p = 0.0013) and ALK/EGFR-negative tumors (FC = 2.1, p = 0.00047). In EGFR-positive tumors, cytotoxic cells were significantly lower compared to ALK-positive (FC = − 1.7, p = 0.016) and to ALK/EGFR-negative tumors (FC = − 2.1, p = 2.0E-05). A total number of 289 genes, 40 part of cytokine–cytokine receptor signaling, were differentially expressed between the three subgroups. Among the latter, five genes were differently expressed in both ALK-positive and EGFR-positive tumors, while twelve genes showed differential expression solely in ALK-positive tumors and eleven genes solely in EGFR-positive tumors.
Conclusion
Targeted gene expression profiling is a promising tool to read out tumor microenvironment characteristics from routine diagnostic lung cancer biopsies. Significant immune reactivity including specific immunosuppressive characteristics in ALK- and EGFR-positive lung ADC, but not a total absence of immune infiltration supports further clinical evaluation of immune-modulators as partners of ICB in such tumors.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-021-02981-w.
Keywords: Lung adenocarcinoma, ALK fusion, EGFR mutation, Immunotherapy, Immune checkpoint blockade, Immunosuppression
Introduction
Non-small cell lung cancer (NSCLC) is the leading cause of cancer-related mortality worldwide [1]. Despite major advances in therapeutic options, the majority of patients are diagnosed at advanced stage with a median survival rate below 2 years. Genetic profiling is of key importance for treatment decision in NSCLC, since targeted therapies, mainly tyrosine kinase inhibition (TKI), have shown dramatically superior efficacy as comparted to standard of care chemotherapy in oncogene-addicted tumors [2]. EGFR mutations and ALK fusions represent the most frequent targetable alterations with a prevalence of, respectively, 10–20% and 3–5% in a Caucasian population with lung adenocarcinomas (ADC) [3].
While the advent of immune checkpoint blockade (ICB) led to significant advances in disease control and survival of driver-negative NSCLC, it quickly became apparent that ALK- and EGFR-positive tumors show limited response to ICB. For example, even despite encouraging early results from the Keynote-001 trail suggesting potential efficacy of pembrolizumab in TKI-naïve EGFR-mutated NSCLC, these were invalidated in a subsequent phase 2 trial [4, 5]. Similarly, Checkmate 012 [6], a phase 1 trial investigating first-line nivolumab monotherapy or in combination with standard therapies for NSCLC demonstrated no meaningful activity in EGFR-mutated tumors. The same trial also investigated the combination of nivolumab with platinum-based chemotherapy and showed reduced activity in EGFR-mutant compared to EGFR-wildtype tumors [7]. Furthermore, several clinical trials combining ICB and TKI resulted in significant toxicity without a signal of improved activity above TKI [7–9]. The only exception so far has been the IMpower150 study, which demonstrated a survival benefit for TKI-pretreated patients with ALK/EGFR-positive NSCLC when the PD-L1 inhibitor atezolizumab was combined with both the antiangiogenic agent bevacizumab and chemotherapy [10, 11].
The biological underpinnings for ICB primary resistance in patients with EGFR-/ALK-positive NSCLC are incompletely understood. Lower tumor mutational burden (TMB), for example, in tumors of never smokers [12, 13], a different nature of PD-L1 expression (intrinsic, induced by oncogenic signaling rather than tumor-infiltrating lymphocytes) [14, 15] and features of the tumor microenvironment (TME) [16–19], possibly influenced by MAPK-signaling [20], have all been suggested as potential causes. The TME is increasingly recognized as crucial parameter for the efficacy of immunotherapies in general, but biological data, especially for the less frequent ALK-positive tumors, are limited [21].
We employed the NanoString nCounter technology and the PanCancer Human IO 360 Panel to investigate the TME in 114 formalin-fixed and paraffin-embedded (FFPE) biopsies of clinically annotated ALK-positive, EGFR-positive and ALK/EGFR-negative advanced lung ADC. We also analyzed gene expression data of earlier stage, resectable tumors from the TCGA lung ADC cohort.
Material and methods
Study cohort
The retrospective study cohort included 31 ALK-positive, 40 EGFR-positive and 43 ALK/EGFR-negative lung ADC patients diagnosed and treated at the Heidelberg University Hospital between 2007 and 2020 (Table 1). ALK and EGFR status were determined at the Heidelberg Institute of Pathology using our routine diagnostic workflow of combined DNA and RNA sequencing starting from formalin-fixed and paraffin-embedded (FFPE) lung biopsies [22]. Tumors harboring activating EGFR mutations were classified as EGFR-positive, and tumors harboring oncogenic ALK fusions were classified as ALK-positive (Suppl. 1).
Table 1.
Clinicopathological characteristics of the study cohort comprising 114 lung adenocarcinomas
| Mutation subtype | ALK-positive | EGFR-positive | ALK/EGFR-negative |
|---|---|---|---|
| Number | 31 | 40 | 43 |
| Age: median (min.- max.) | 58 (33–90) | 69.5 (46–83) | 64 (41–89) |
| Sex: | |||
| Male | 15 (48%) | 7 (18%) | 21 (49%) |
| Female | 16 (52%) | 33 (83%) | 22 (51%) |
| Smoking history: | |||
| Smoker | 9 (29%) | 15 (38%) | 41 (95%) |
| Non-smoker | 17 (55%) | 25 (63%) | 2 (5%) |
| Unknown | 5 (16%) | 0 (0%) | 0 (0%) |
| Tumor stage: | |||
| I | 0 (0%) | 0 (0%) | 0 (0%) |
| II | 1 (3%) | 4 (10%) | 0 (0%) |
| III | 7 (23%) | 10 (25%) | 0 (0%) |
| IV | 23 (74%) | 26 (65%) | 43 (100%) |
| Prior therapy: | |||
| Naïve | 31 (100%) | 40 (100%) | 26 (60%) |
| Chemotherapy | 0 (0%) | 0 (0%) | 17 (40%) |
| Response to ICB | no ICB | no ICB | 16 LTR*, 6 IR†, 21 RP‡ |
*Long-term responders
†Intermediate progressors
‡Rapid progressors
EGFR/ALK-positive patients were therapy-naïve, i.e., received neither TKI nor chemo- or immunotherapy prior to biopsy. ALK/EGFR-negative patients underwent biopsy immediately before start of ICB treatment and were further subdivided according to the subsequent ICB response in 16 long-term responders (LTR; durable response to ICB of 12 months or more), 21 rapid progressors (RP; disease progression within two months of ICB start) and 6 patients with an intermediate duration of response to ICB (IR). For all patients, only biopsies from the primary (lung) tumor with sufficient available mRNA for expression profiling were analyzed. The study was approved by the ethics committee of Heidelberg University (S-145/2017). The subcohort of ALK/EGFR-negative tumors was also analyzed in a study comparing the gene expression differences between LTR and RP [23].
TCGA lung adenocarcinoma cohort
Gene expression and clinical data of the TCGA lung ADC cohort (LUAD) were downloaded from the PanCanAtlas webpage of the Genomic Data Commons repository (https://gdc.cancer.gov/about-data/publications/pancanatlas). Only samples diagnosed as lung adenocarcinoma and of the type “primary solid tumor” (code: 01) were included. Information on ALK fusion status was obtained from a gene fusion analysis based on RNA-Seq data [24]. The TCGA cohort included 5 ALK-positive, 57 EGFR-positive and 439 ALK/EGFR-negative lung ADC (Suppl. 2).
Targeted gene expression profiling
Targeted mRNA expression profiling was conducted on the NanoString nCounter gene expression platform (NanoString Technologies, Seattle, WA) using a 770-gene panel (PanCancer Human IO 360 Panel) focused on genes connected with the interplay between tumor, tumor microenvironment and immune response in cancer. Per sample, 100 ng of total RNA in a final volume of 5 μl was mixed with a 3′ biotinylated capture probe and a 5′ reporter probe tagged with a fluorescent barcode sequence from the PanCancer IO 360 gene expression code set. Probes and target transcripts were hybridized at 65 °C for 18 h according to the manufacturer’s recommendations. Hybridized samples were run on the NanoString nCounter preparation station using the high-sensitivity protocol, in which excess capture and reporter probes are removed and transcript-specific complexes are immobilized on a streptavidin-coated cartridge. The samples were scanned at maximum resolution on the nCounter Digital Analyzer.
Immunohistochemistry
For detection of FOXP3 and CD8 protein expression, 3 μm thick paraffin sections were prepared. Deparaffinization and tissue staining were performed using a Benchmark Ultra IHC Staining module according to standard protocols (FOXP3 Monoclonal Antibody, clone 236A/E7, eBioscience, Invitrogen, Thermo Fisher Scientific Inc., Waltham, MA and CONFIRM anti-CD8 Rabbit Monoclonal Primary Antibody, clone SP57, Roche, Mannheim, Germany). Staining was visualized using the Vectastain elite ABC detection system (Vector, Burlingame, CA, USA) and using 3,3′-Diaminobenzidine (Optiview DAB IHC Detection Kit or ultraView Universal DAB Detection Kit, Ventana, Roche, Mannheim, Germany) as chromogen. Hematoxylin was used for counterstaining of cell nuclei. Evaluation and scoring were performed by an expert pathologist using the image analysis software QuPath (Open Source Digital Pathology, https://github.com/qupath).
Data analysis
Statistical analysis was performed using the programming language R. Sample normalization of the gene expression data was performed by fitting a linear model to negative and positive controls and subsequent housekeeping gene normalization. For the latter, the 20 panel genes with the lowest coefficient of variation and an expression level of at least 100 in the TCGA lung ADC dataset were used as housekeepers (AKT1, API5, DNAJC14, EIF2B4, ELA, ERCC3, GLUD1, HDAC3, HMGB1, IFNAR1, MLH1, OAZ1, PUM1, RIPK1, SF3A1, STAT3, TBC1D10B, TLK2, TMUB2 and UBB). The gene expression profile of each sample was scaled by the median expression level of the housekeeping genes. Gene expression data were log2-transformed prior to statistical analysis and visualization.
The abundance of 14 immune cell populations (B cells, CD45+ cells, CD56dim NK cells, CD8+ T cells, cytotoxic cells, dendritic cells, exhausted CD8+ T cells, macrophages, mast cells, neutrophils, NK cells, T cells, Th1 cells and Treg cells) was estimated from the mRNA expression of marker genes as described and validated before [25]. The abundance of cell populations was reported on log2 scale. The “immunological distance” between two tumors was defined as the Manhattan distance between in the space of the 14 immune cell populations. Analyses were carried out on the following two levels: (1) on the level of the abundance of 14 immune cell populations and (2) on the level of the mRNA expression of 770 genes.
For heatmap displays, each of the cell populations (or genes) was centered (but not scaled) with respect to the mean abundance (or mean mRNA expression) over the samples. Hierarchical clustering was performed using Pearson correlations as similarity measure and the average linkage as measure of distances between clusters. Correlations between clusters and genetic subgroups were assessed using Fisher's exact test.
Differences between ALK-positive, EGFR-positive and ALK/EGFR-negative tumors were assessed for significance using the Kruskal–Wallis as omnibus test and the Wilcoxon test as post hoc test. The Benjamini–Hochberg procedure was used for p value correction, and lists of cell populations or genes were compiled controlling the false discovery rate (FDR) at 5%. KEGG Mapper was used to visualize the cytokine–cytokine receptor network (pathway hsa04060) [26].
Results
The study cohort comprised 31 ALK-positive, 40 EGFR-positive and 43 ALK/EGFR-negative lung ADC patients (Table 1). Biopsies of each of the 114 primary tumors underwent gene expression profiling with an assay of 770 genes focused on immune-related genes. The abundance of 14 immune cell populations in the TME was estimated using an already established method [25].
Overall level of immune cell infiltration
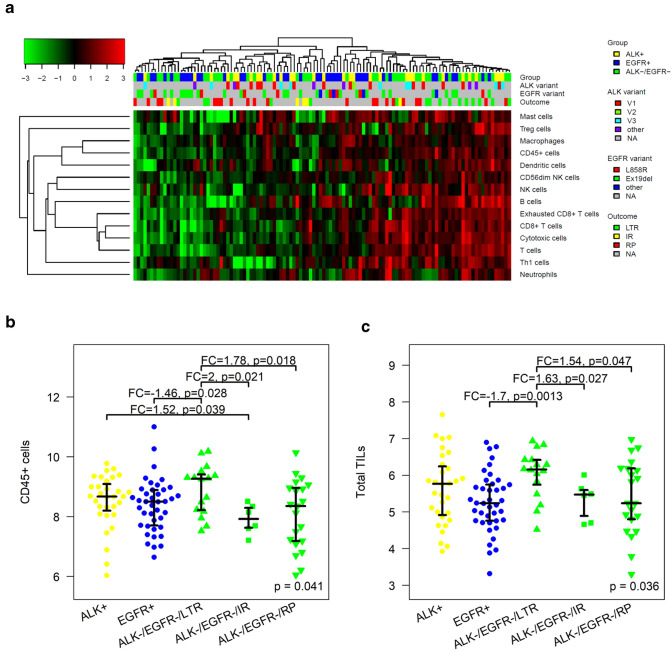
The levels of the immune cell populations were grouped by hierarchical clustering and visualized in a heatmap (Fig. 1a). The markers of cytotoxic cells, T cells, CD8+ T cells and exhausted CD8+ T cells clustered tightly together (all pairwise Spearman correlations ρ > 0.77). Moreover, macrophages and CD45+ cells showed a strong positive correlation (ρ = 0.67). The tumors clustered together in two main immunological groups, “cold” tumors (n = 58) and “hot” tumors (n = 56). The immunological groups did not correlate with the mutation type (ALK-positive, EGFR-positive or ALK/EGFR-negative) of the tumors (p = 0.82). Furthermore, the immunological groups did neither correlate with the type of ALK fusion (V1 vs. V3, p = 1) nor with the type of EGFR mutation (Ex19del vs. L858R, p = 0.69).
Fig. 1.
Immunological analysis of 114 lung adenocarcinomas by targeted gene expression profiling. a Clustering of the tumors by the abundance of 14 immune cell populations. b CD45+ cells were higher in EGFR/ALK-negative tumors that showed durable ICB response (long-term responders, LTR) compared to EGFR/ALK-negative tumors that progressed rapidly (rapid progressors, RP) and compared to EGFR-positive tumors. Levels of CD45+ cells between ALK-positive, EGFR-positive and rapid progressing ALK/EGFR-negative tumors did not differ significantly. IR = intermediate responders. c Similar as in B but for total TILs. Distributions are shown with median, lower and upper quartile
In line with the cluster analysis, neither the levels of CD45+ cells nor of total TILs (calculated as in [25]) correlated with the mutation type (p = 0.47 and p = 0.11). Additionally, we split the ALK/EGFR-negative tumors in long-term responders (LTR) and in rapid progressors (RP) with respect to ICB and carried out a four-group-analysis of ALK-positive tumors, EGFR-positive tumors, LTR and RP. Both, CD45+ cells and total TILs were significantly higher in LTR, but not in RP compared to EGFR-positive tumors (Fig. 1b, c). Neither CD45+ cells nor total TILs showed significant differences when comparing LTR or RP with ALK-positive tumors.
To test whether ALK/EGFR-positive tumors are “immunologically closer” to RP than to LTR, we operationalized the concept of immunological similarity by the introduction of an immunological distance based on the abundance of the 14 immune cell populations. It turned out that both ALK-positive tumors and EGFR-positive tumors were significantly closer to LTR than to RP tumors falsifying the hypothesis (Suppl. 3). This observation suggested distinct rather than common immunosuppressive features of the three subgroups, a hypothesis that we further addressed by comparative analyses of immune cell populations and gene expression profiles between the subgroups.
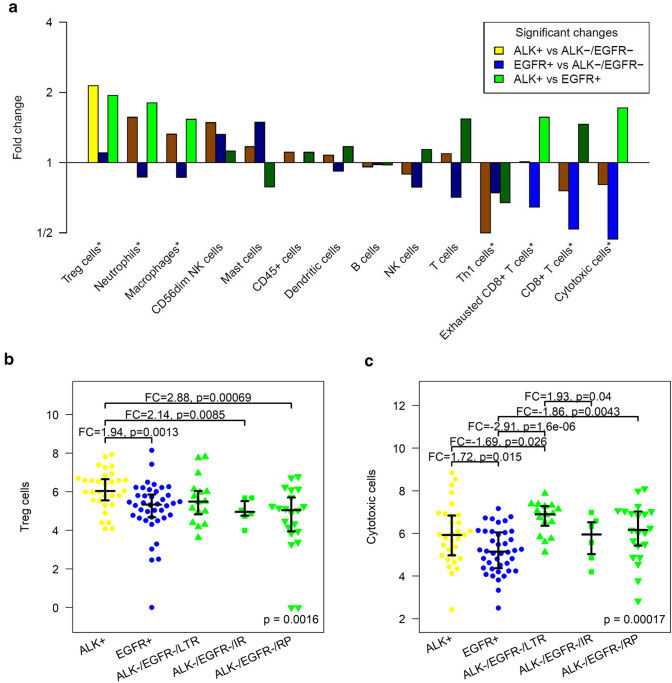
Specific immune cell populations
Seven of 14 immune cell populations were significantly different in ALK-positive, EGFR-positive and ALK/EGFR-negative tumors using omnibus testing (Fig. 2a, marked by*). Regulatory T cells (Tregs) were significantly higher in ALK-positive tumors compared to ALK/EGFR-negative tumors (FC = 2.1, p = 0.00047). Cytotoxic cells, CD8+ T cells and exhausted CD8+ T cells were significantly lower in EGFR-positive tumors compared to ALK/EGFR-negative tumors (FC = − 2.1, p = 1.1E-05; FC = − 1.9, p = 0.0037 and FC = − 1.6, p = 0.0045). Here, ‘cytotoxic cells’ (markers genes: PRF1, GZMA, GZMB, GZMH, GNLY, CTSW, KLRB1, KLRD1, KLRK1 and NKG7) refer to a broader cell population of granzyme releasing cells including cytotoxic T cell and cytotoxic NK cells compared to the more specific population of ‘CD8+ T cells’ (marker genes: CD8A and CD8B). Tregs, neutrophils, cytotoxic cells, exhausted CD8+ T cells and macrophages were significantly higher in ALK-positive compared to EGFR-positive tumors (FC = 1.9, p = 0.0013; FC = 1.8, p = 0.00078; FC = 1.7, p = 0.015; FC = 1.6, p = 0.017 and FC = 1.5, p = 0.00074). Thus, ALK-positive tumors stood out by significantly higher Tregs compared to EGFR-positive and ALK/EGFR-negative tumors, while EGFR-positive tumors stood out by significantly lower cytotoxic cells and exhausted CD8+ T cells compared to the two other subgroups.
Fig. 2.
Levels of specific immune cell populations differ between ALK-positive, EGFR-positive and ALK/EGFR-negative lung adenocarcinomas. a Significantly higher Tregs in ALK-positive tumors compared to ALK/EGFR-negative tumors. Significantly lower cytotoxic cells, CD8+ T cells and exhausted CD8+ T cells in EGFR-positive tumors compared to ALK/EGFR-negative tumors. Significantly higher Tregs cells, neutrophils, macrophages, exhausted CD8+ T cells and cytotoxic cells in ALK-positive compared to EGFR-positive tumors. * = significant in omnibus test. b Significantly higher regulatory T cells in ALK-positive tumors compared to EGFR-positive tumors and to ALK/EGFR-negative tumors that progressed rapidly or intermediately after ICB. c Significantly lower cytotoxic cells in EGFR-positive tumors compared to ALK-positive tumors, to ALK/EGFR-negative tumors that showed durable ICB response and to ALK/EGFR-negative tumors that progressed rapidly under ICB
In two more analyses, we investigated subgroups of the ALK/EGFR-negative tumors that were homogeneous regarding (1) prior treatment, (2) the duration of response to immune therapy, (3) smoking history and (4) TP53 mutation status. Firstly, the results obtained for the complete group of ALK/EGFR-negative tumors stayed correct for the subgroup of treatment-naïve tumors: Tregs were higher in ALK-positive cancer compared to treatment-naïve ALK/EGFR-negative cancer (FC = 2.3, p = 0.00015). Cytotoxic cells, CD8+ T cells and exhausted CD8+ T cells were lower in EGFR-positive cancer compared to treatment-naïve ALK/EGFR-negative cancer (FC = − 2.2, p = 4.1E-05; FC = − 2.0, p = 0.01 and FC = − 1.7, p = 0.0082). Secondly, the group of ALK/EGFR-negative tumors was split in long-term responders (LTR) and rapid progressors (RP) under ICB (Fig. 2b, c, Suppl. 4). Tregs were significantly higher in ALK-positive tumors compared to RP (FC = 2.9, p = 0.00069), while a non-significant trend to higher levels (FC = 1.4, p = 0.094) was observed in comparison to LTR. Cytotoxic cells were significantly lower in EGFR-positive tumors compared to both RP and LTR (FC = − 1.9, p = 0.0043 and FC = − 2.9, p = 1.6E-06). Thirdly, levels of Tregs and cytotoxic cells did not differ significantly between smokers and non-smokers, while Tregs were enhanced in both ALK-positive tumors of smokers and non-smokers and cytotoxic cells were depleted in both EGFR-positive tumors of smokers and non-smokers (Suppl. 5A/B). Likewise, TP53 mutations did not confound the regulation patterns of Tregs and cytotoxic cells detected in the study cohort (Suppl. 5C/D).
Next, we investigated the validity of the findings in a cohort of earlier lung ADC (Suppl. 2). Of the three immune cell populations that were significantly reduced in EGFR-positive tumors compared to ALK/EGFR-negative tumors, the reduction of cytotoxic cells and CD8+ T was also found in the TCGA cohort (FC = − 1.6, p = 0.015 and FC = − 1.6, p = 0.0018). A significantly higher abundance of Tregs in ALK-positive tumors was not detected in the TCGA cohort, but this analysis was heavily underpowered with only five ALK-positive cases available.
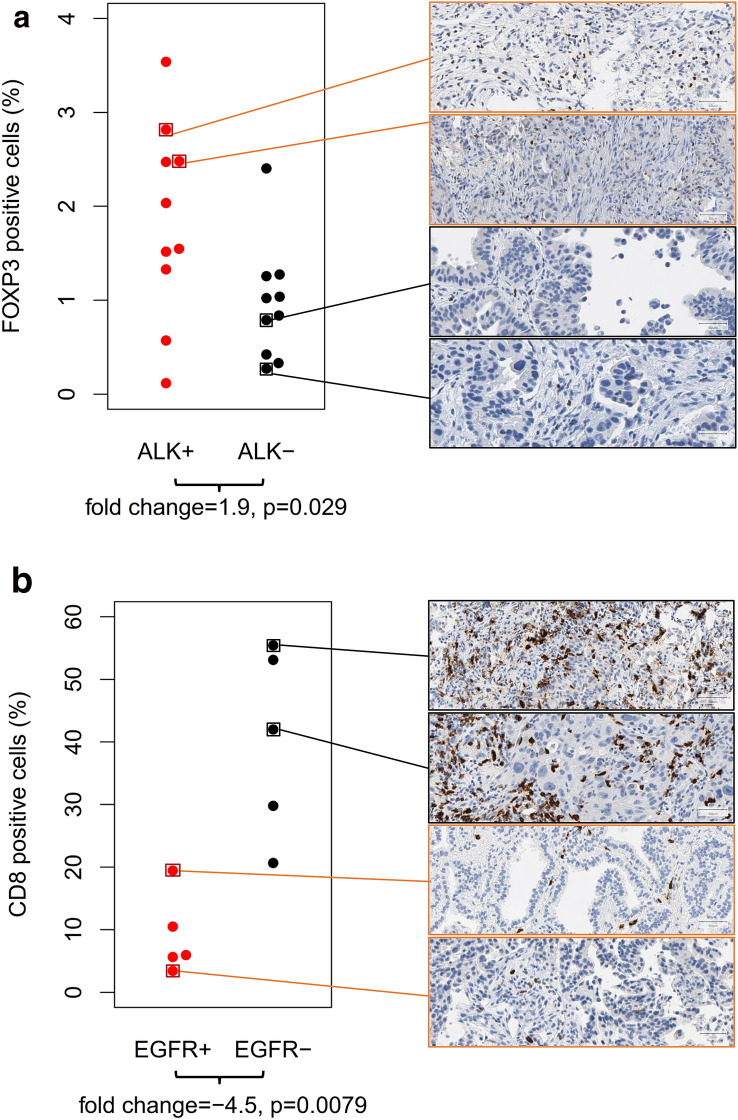
FOXP3 and CD8 protein expression
Representative example cases of the study cohort were analyzed using immunohistochemistry (Fig. 3). FOXP3 and CD8 protein expression were quantified by counting the positive cells in relation to all cells (tumor, immune and other cells) in the tissue section. The percentage of FOXP3-positive cells was lower than 4% in all of the twenty analyzed cases. We observed a diffuse distribution FOXP3-positive immune cells in the intra- and peritumoral stroma, while we did not observe any clusters of two or more adjacent FOXP3-positive cells. The percentage of FOXP3 was significantly higher (p = 0.029) in the ALK-positive cases (mean: 1.84%) compared to the ALK-negative cases (mean: 0.96%). The percentage of CD8-positive cells ranged between virtually 0% and 60% in the ten analyzed example cases. CD8-positive immune cells showed formation of small groups as well as of larger dense clusters. The percentage of CD8-positive cells was significantly lower (p = 0.0065) in the EGFR-positive cases (mean: 9%) compared to the EGFR-negative cases (mean: 40%). Altogether, the protein expression analysis confirmed enrichment of FOXP3-positive cells in ALK-positive cancer and depletion of CD8-postive cells in EGFR-positive cancer.
Fig. 3.
Immunohistochemical analysis of FOXP3 and CD8 protein expression in representative example cases of the study cohort. Positive cells were quantified as percentage of all (tumor and stroma) cells. a Comparison of ALK-positive tumors with high Treg mRNA marker and ALK-negative tumors with low Treg mRNA marker. The percentage of FOXP3-positive cells was significantly higher in the ALK-positive tumors (mean: 1.84%) than in ALK-negative tumors (mean: 0.96%). b Comparison of EGFR-positive tumors with low cytotoxic cell mRNA marker and EGFR-negative tumors with high cytotoxic cell mRNA marker. The percentage of CD8-positive cells was significantly lower in the EGFR-positive tumors (mean: 9%) compared to the EGFR-negative tumors (mean: 40%)
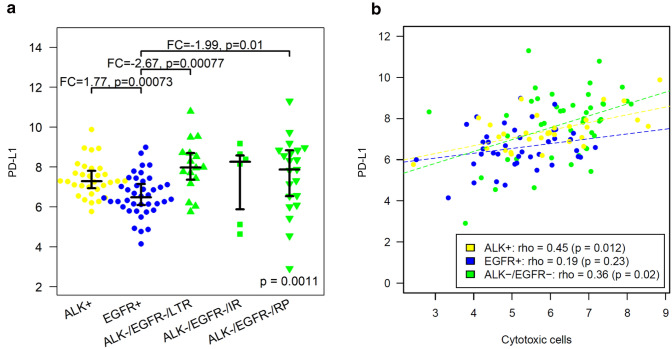
PD-L1 expression
Levels of PD-L1 mRNA were analyzed in a four-group-analysis (Fig. 4a). PD-L1 mRNA expression was significantly lower in EGFR-positive tumors compared to the ALK-positive tumors (FC = − 1.8, p = 0.00073) and compared to both groups of ALK/EGFR-negative tumors (LTR: FC = − 2.7, p = 0.00077; RP: FC = − 2.0, p = 0.01). Significant positive correlations with PD-L1 mRNA expression were observed for eight of the 14 immune cell populations, namely for cytotoxic cells (ρ = 0.45), CD8+ T cells (ρ = 0.38), exhausted CD8+ cells (ρ = 0.34), neutrophils (ρ = 0.33), T cells (ρ = 0.32), Th1 cells (ρ = 0.31), dendritic cells (ρ = 0.3) and Treg cells (ρ = 0.28).
Fig. 4.
PD-L1 expression in ALK-positive, EGFR-positive and ALK/EGFR-negative lung adenocarcinoma. a Significantly lower PD-L1 mRNA expression in EGFR-positive tumors compared to ALK-positive tumors, ALK/EGFR-negative tumors that progressed rapidly after ICB and ALK/EGFR-negative tumors that responded durable to ICB. b PD-L1 mRNA expression and levels of cytotoxic cells correlated positively significantly in ALK-positive and ALK/EGFR-negative tumors, while the correlation was weak and non-significant in EGFR-positive tumors
Thus, by far the strongest correlation was observed between PD-L1 mRNA and cytotoxic cells (Fig. 4b). Partly, this correlation was induced by the incidence of low PD-L1 expression and low abundance of cytotoxic cells in EGFR-positive tumors and at the same time higher levels of both markers in ALK-positive and ALK/EGFR-negative tumors. Additionally, PD-L1 expression and cytotoxic cells correlated positively within some of the subgroups, namely in ALK-positive tumors (ρ = 0.45, p = 0.012) and in ALK/EGFR-negative tumors (ρ = 0.36, p = 0.02), but not EGFR-positive tumors where the correlation was not significant.
Gene expression analysis
In omnibus testing, 289 of 770 investigated genes showed significantly different expression levels in ALK-positive, EGFR-positive and ALK/EGFR-negative tumors (FDR = 5%, Suppl. 6). In detail, 145 genes were differentially expressed between ALK-positive and ALK/EGFR-negative tumors, 122 genes were differentially expressed between EGFR-positive and ALK/EGFR-negative tumors, and 192 genes were differentially expressed between ALK-positive and EGFR-positive tumors. The results could be partially confirmed by analyzing the independent TCGA cohort of early stage lung ADC with a match of 44 genes (30%), 63 genes (52%) and 28 genes (15%) of the respective differentially expressed genes. Confirmation rates were imperfect, but clearly above the random baseline of 5% corresponding to the p value threshold 0.05. A bias was expected, because of the limited sample size (only five tumors with ALK fusions in the TCGA cohort), clinical differences between the in-house and the TCGA cohort (later stage vs. earlier stage tumors) and different analysis platforms (NanoString nCounter vs. RNA-Seq).
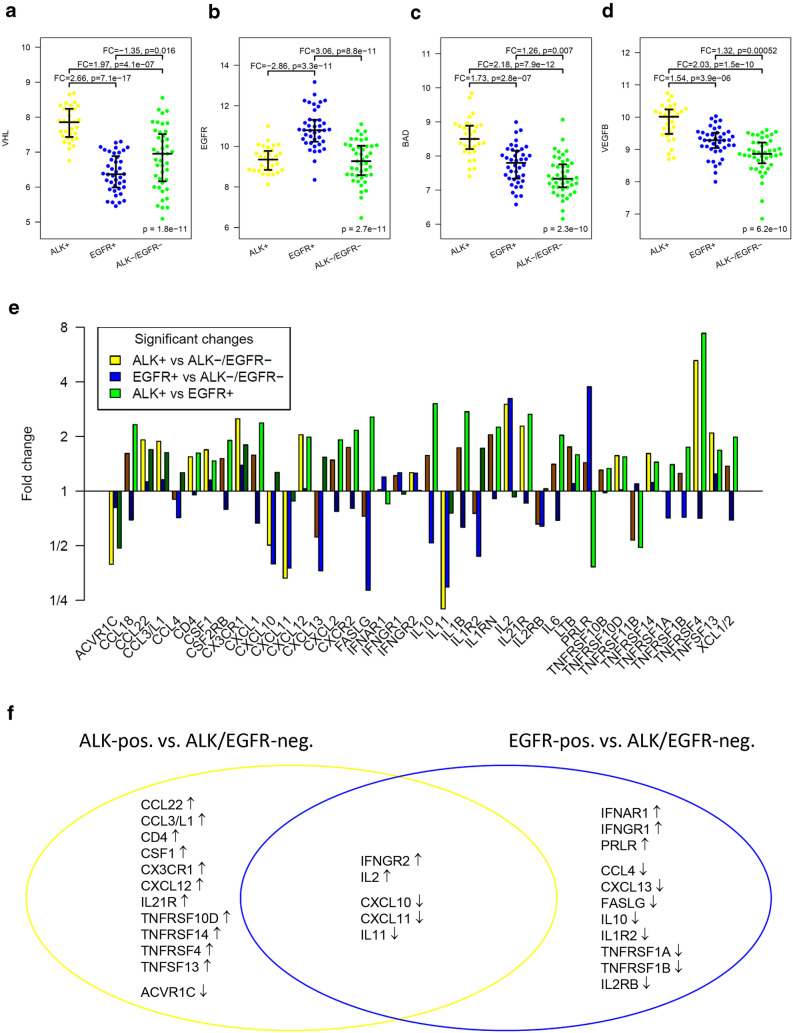
These were the four most significant gene expression changes detected by the omnibus test in the study cohort (Fig. 5a–d): VHL was overexpressed in ALK-positive tumors compared to both EGFR-positive tumors and ALK/EGFR-negative tumors. EGFR was overexpressed in EGFR-positive tumors compared to both ALK-positive tumors and ALK/EGFR-negative tumors. BAD showed the highest expression in ALK-positive tumors, an intermediate expression in EGFR-positive tumors and the lowest expression in ALK/EGFR-negative tumors. VEGFB showed the same decreasing expression pattern as BAD. The overexpression of EGFR in EGFR-positive tumors compared to ALK-positive and ALK/EGFR-negative tumors was also found in the TCGA cohort (FC = 2.2, p = 0.0097 and FC = 3.1, p = 3.1E-16), while we did not detect differential expression of the other three genes.
Fig. 5.
Differential gene expression between ALK-positive, EGFR-positive and ALK/EGFR-negative lung adenocarcinoma. a–d Expression levels of most significantly differentially expressed genes in omnibus testing: a VHL was overexpressed in ALK-positive tumors compared to both EGFR-positive tumors and ALK/EGFR-negative tumors. b EGFR was overexpressed in EGFR-positive tumors compared to both ALK-positive tumors and ALK/EGFR-negative tumors. c BAD showed the highest expression in ALK-positive tumors, an intermediate expression in EGFR-positive tumors and the lowest expression in ALK/EGFR-negative tumors. d VEGFB showed the same decreasing expression pattern as BAD. e–f Analysis of the cytokine–cytokine receptor system: e Fold changes of the 40 differentially expressed of the 121 investigated cytokines and cytokine receptors. f Twelve genes were differentially expressed exclusively between ALK-positive and ALK/EGFR-negative tumors, eleven genes were differentially expressed exclusively between EGFR-positive and ALK/EGFR-negative tumors, while five genes were differentially expressed in both comparisons. ↑ = upregulation, ↓ = downregulation
Analysis of the cytokine–cytokine receptor signaling network
Beyond the estimation of immune cell abundance in the TME, gene expression profiling offers the opportunity to gain insight into the regulation of immune response. Of 294 genes annotated in the KEGG map of cytokines and cytokine receptors, 121 genes (41%) were covered by the used targeted expression assay. Of these, 40 genes (33%) were differentially expressed, 17 between ALK-positive and ALK/EGFR-negative tumors, 16 between EGFR-positive and ALK/EGFR-negative tumors and 26 between ALK-positive and EGFR-positive tumors (Fig. 5e). Compared to ALK/EGFR-negative tumors, five genes were differently expressed in both ALK-positive and EGFR-positive tumors, while twelve genes showed differential expression solely in ALK-positive tumors and eleven genes showed differential expression solely in EGFR-positive tumors (Fig. 5f). We assigned the changes to the map of the interaction of cytokines and receptors (Suppl. 7).
IL2, an interferon that is important for the proliferation of T and B lymphocytes and in particular for the differentiation of CD4+ T cells to T helper cells and Tregs, was overexpressed in ALK-positive and in EGFR-positive tumors compared to ALK/EGFR-negative tumors (FC = 3.0, p = 0.00013 and FC = 3.2, p = 3.0E-05). IL11, an interferon that is known to stimulate the T cell-dependent development of immunoglobulin-producing B cells, was underexpressed in ALK-positive and in EGFR-positive tumors compared to ALK/EGFR-negative tumors (FC = − 4.5, p = 2.7E-05 and FC = − 3.4, p = 0.00024). Tumor necrosis factor TNFRSF4, suggested to play a role in CD4+ T cell response as well as in T cell-dependent B cell proliferation and differentiation, was strongly overexpressed in ALK-positive tumors compared to ALK/EGFR-negative tumors and compared to EGFR-positive tumors (FC = 5.2, p = 4.0E-07 and FC = 7.4, p = 1.0E-07). FASLG, a cytokine that binds to the receptor FAS that transduces apoptotic signals in cells, was underexpressed in EGFR-positive tumors compared to ALK/EGFR-negative tumors and compared to ALK-positive tumors (FC = − 3.5, p = 2.5E-05 and FC = − 2.6, p = 0.0033), an observation possibly related to the lower abundance of cytotoxic cells in EGFR-positive tumors. Prolactin receptor (PRLR), which has been suggested as a therapeutic target in subgroups of breast and of prostate cancer [27], was overexpressed in EGFR-positive tumors compared to ALK/EGFR-negative tumors and compared to ALK-positive tumors (FC = 3.8, p = 0.0004 and FC = 2.6, p = 0.013).
Discussion
About 45% to 50% of all NSCLC show a variety of mutually exclusive genetic driver lesions that can be therapeutically exploited. Collectively, EGFR mutations and ALK fusions represent the largest subgroup comprising approx. 20–25%. While these tumors respond well to TKIs, the efficacy of ICB is generally poor despite tremendous trial efforts in the last few years. These observations led to the hypothesis of a poorly immunogenic “immune desert” that prevents successful application of ICB. Early studies of various sample sizes and disease stages investigating immune cell compositions of NSCLC by either immunohistochemistry or flow cytometry revealed the presence of a broad range of infiltrating immune cells including T cells, NK cells, macrophages, B cells, dendritic cells and granulocytes [28–35].
Complementing and extending these studies, we compiled a retrospective cohort of carefully clinically annotated and confirmed ALK- and EGFR-positive NSCLC, molecularly analyzed at the Heidelberg routine diagnostic laboratory [22]. We profiled routine diagnostic formalin-fixed and paraffin-embedded biopsies after mutational analysis by NGS using the NanoString nCounter technology [36]. This technology, which can be directly applied in a routine diagnostic setting and is tissue sparing, facilitates focused analysis of mRNA profiles of 770 genes with fragmented nucleic acids as input material. It enables a multilayered analysis of specific immune cell types as well as expression levels of cytokines, cytokine receptors and other immune genes.
In the first analysis presented here, we identified two groups of immunologically “cold” and “hot” tumors, which were not restricted to a specific subgroup (ALK-positive, EGFR-positive or ALK/EGFR-negative) or oncogene variant (e.g., ALK V1 vs. V3 or EGFR Ex19del vs. L858R). In the analysis of specific cell populations, different modifications of the TME were detected in ALK-positive and in EGFR-positive tumors compared to ALK/EGFR-negative tumors, respectively: While ALK-positive cases displayed a Treg enriched TME, EGFR-positive cases showed a cytotoxic cell-depleted TME suggesting different mechanisms that abrogate a strong and sustained immune cell response. These two different TMEs appear to be governed by specific immunomodulatory networks, which might be clinically exploitable [37]. Our results confirm other studies analyzing genetic subtypes of NSCLC that found low infiltrates of T cells and CD8+ cells in EGFR-mutant tumors [17, 18]. By contrast, data for ALK-positive NSCLC are highly limited, a few groups identified low CD8 populations in these tumors as well [21, 38]. A more recent study presented reported changes in the immune TME associated with TKI inhibition [39].
For ALK-positive disease, we observed a significant downregulation of CXCL10 and CXCL11 negatively influencing general T cell recruitment [40], while at the same time upregulating CXCL12 [41–43] as well as CCL22 [44–46] which attract CXCR4+ Tregs. Consistent with the suggested regulation mechanism, T cell levels in the study cohort correlated significantly with CXCL10 and CXCL11 (ρ = 0.44, p = 1.6E-06 and ρ = 0.47, p = 1.1E-07). Tregs correlated significantly with CCL22 (ρ = 0.64, p = 2.8E-14), while the correlation of Tregs and CXCL12 was not significant. Downregulation of CXCL10 and CXCL11 was also observed in EGFR-positive disease.
Comparing ALK-positive and EGFR-positive disease, we noted strong upregulation of IL10 in ALK-positive tumors. IL10 is involved in negative feedback mechanisms which decrease the antigen-presenting activity of dendritic cells and inhibit cytotoxic functions and cytokine-release of T and NK lymphocytes [47]. Similarly, pleiotropic IL6 was upregulated, which was reported to negatively influence T cell response and innate immunity [48, 49]. Notably, the upregulation of CSF1 and CCL18—as identified in the TME of ALK-positive tumors —increases type 2 (M2) tumor-associated macrophages (TAM) which have tumorigenic functions and are known to contribute to immune evasion [50, 51]. VEGFB was also upregulated in ALK-positive NSCLC. This might be explained by angiogenic effects but could also be due to potential immunosuppressive effects recently described for VEGF signaling [52, 53]. Interestingly, we identified upregulation of some costimulatory molecules influencing T cell response, including TNFRSF4 (aka OX40 [54]). We also observed upregulation of TNFSF13 (aka APRIL), a molecule influencing B cell development. The latter two phenomena may represent an unfruitful attempt to overcome a generally immune response deprived TME.
The proinflammatory cytokine IL2 was found to be upregulated in both ALK- and EGFR-positive tumors, while IL11, a member of the IL6 family, was found to be downregulated in both subgroups. IL2 is known to modulate the development and expansion of Tregs exerting immunosuppressive effects [55]. High IL11 has been linked to cell proliferation and tumorigenesis in in vitro and in vivo models of NSCLC as well as to unfavorable prognosis [56].
EGFR-positive tumors feature a CD8+ deprived environment that is modulated by downregulation of CCL4, a molecule that supports recruitment of CD103+ dendritic cells which active CD8+ cells [57] as well as downregulated CXCL10, the ligand for CXCR3 on cytotoxic T cells and downregulated CXCL11 negatively modulating CD8+ T cell migration [58, 59]. IFNGR1 and IFNGR2 were upregulated, possibly as a compensatory yet non-successful mechanism to overcome impaired CD8+ T cell-mediated immune response.
At the Heidelberg Institute of Pathology, mRNA extracts of lung cancer biopsies are acquired within the routine workflow of lung cancer biopsies and used for gene fusion analysis [22]. For most of the tumors in the study cohort, leftovers of such extract were available and suitable for gene expression profiling. Thus, targeted gene expression represents a tissue-saving method to read out multi-dimensional information on the immune TME from diagnostic lung cancer biopsies.
It is a limitation of gene expression analysis of bulk tissues that expression levels are summarized over all cells in the tissue, while most of the human genes are expressed in various cell types, e.g., in cancer cells and in different other cells in the TME. Thus, in general, the cells types that are causative for the detected gene expression changes cannot be identified from a bulk tissue analysis. Nevertheless, there are transcripts that are expressed by a only a single cell type and conversely there are cell types that can be characterized by the expression of specific marker genes, a methodology that has been exploited to distinguish between 14 immune cell populations in current study.
An advantage of nucleic acid-based technologies is the parallel monitoring of a high number of genes, while spatial resolved technologies such as immunohistochemistry and multispectral imaging usually allow the monitoring of only one or a few genes in parallel and are more difficult to generalize. Thus, nucleic acid-based technologies can be very helpful for screening and knowledge discovery, while spatial resolved technologies should be employed for validation and in-depth cell type-specific analysis. In the current study, we validated an accumulation of Tregs in ALK-positive tumors and a depletion of CD8+ T cells in EGFR-positive tumors using IHC.
Spatial tumor heterogeneity represents an additional potential limitation for studies in metastatic NSCLC based on a single biopsy of each patient, but this method of sample collection is currently used in the routine molecular pathology.
Collectively, our data revealed two different types of TME modification in ALK- and EGFR-positive NSCLC, respectively, which we term Treg-modulated and CD8-modulated immune TME, each accompanied by a specific characteristic of cytokine signaling. Therefore, tailored therapy strategies will probably be required for oncogene-addicted tumors and we expect detailed profiling of the TME to be instrumental for guiding the selection of appropriate immunomodulatory partners to ICB. Furthermore, our results support the need for exploration of new therapeutic approaches to transform these TME into “hot” states [60]. This may be achievable by opportunistic combinations of ICB with radio- and chemotherapy, or by more subtle, specific approaches that either inhibit specific immunosuppressive agents enriched in “cold” tumors or supplement and boost proinflammatory molecules depleted in “cold” tumors. The detection of significant immune reactivity in oncogene-addicted NSCLC, in contrast the conception of an absolute “immune desert,” suggests that well-aimed use of immunomodulators currently explored in various pre-clinical studies and clinical trials [37, 61] may also hold promise in EGFR- and ALK-positive NSCLC.
Supplementary Information
Below is the link to the electronic supplementary material.
Abbreviations
- ADC
Adenocarcinoma
- ALK-positive tumor
Tumor with activating ALK gene fusion
- ALK/EGFR-negative tumor
Tumor that is neither ALK-positive nor EGFR-positive
- EGFR-positive tumor
Tumor with activating somatic mutation in EGFR
- FC
Fold change
- ICB
Immune checkpoint blockade
- LTR
Long-term responders (at least one year after start of ICB)
- NSCLC
Non-small cell lung cancer
- RP
Rapid progressors (within two months after start of ICB)
- rho
Spearman correlation coefficient
- TILs
Tumor-infiltrating lymphocytes
- TKI
Tyrosine kinase inhibitor
- TMB
Tumor mutational burden
- TME
Tumor microenvironment
- Tregs
Regulatory T cells
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was funded by The German Center for Lung Research (Deutsches Zentrum für Lungenforschung, DZL) and by Roche Pharma.
Declarations
Conflict of interest
Dr. Budczies reports grants from German Cancer Aid, outside the submitted work. Dr. Kirchner, Mr. Kluck, Ms. Glade, Dr. Allgäuer, Dr. Kriegsmann, Dr. Winter, Dr. Goldmann, Dr. Wermke have nothing to disclose. Dr. Kazdal reports personal fees from Pfizer Pharma GmbH, outside the submitted work. Dr. Herth reports personal fees from Schering-Plough, personal fees from Pfizer, personal fees from Basilea, personal fees from Boehringer Ingelheim, personal fees from Novartis, personal fees from Roche, personal fees from Astellas, personal fees from Gilead, personal fees from MSD, personal fees from Lilly, personal fees from Intermune, personal fees from Fresenius, personal fees from Essex, personal fees from AstraZeneca, personal fees from Bracco, personal fees from MEDA Pharma, personal fees from Chiesi, personal fees from Siemens, personal fees from Covidien, personal fees from Pierre Fabre, personal fees from Grifols, personal fees from Bayer, personal fees from Siemens, personal fees from Pfizer, personal fees from MeVis, personal fees from Boehringer Ingelheim, personal fees from German Cancer for Lung Research, outside the submitted work; In addition, Dr. Herth has a patent Method and Device For Representing the Microstructure of the Lungs. IPC8 Class: AA61B5055FI, PAN: 20080208038 pending. Dr. Herth reports personal fees from Schering-Plough, personal fees from Pfizer, personal fees from Basilea, personal fees from Boehringer Ingelheim, personal fees from Novartis, personal fees from Roche, personal fees from Astellas, personal fees from Gilead, personal fees from MSD, personal fees from Lilly, personal fees from Intermune, personal fees from Fresenius, personal fees from Essex, personal fees from AstraZeneca, personal fees from Bracco, personal fees from MEDA Pharma, personal fees from Chiesi, personal fees from Siemens, personal fees from Covidien, personal fees from Pierre Fabre, personal fees from Grifols, personal fees from Bayer, personal fees from Siemens, personal fees from Pfizer, personal fees from MeVis, personal fees from Boehringer Ingelheim, personal fees from German Cancer for Lung Research, outside the submitted work. Dr. Meister reports grants from German Center for Lung Research (DZL), during the conduct of the study. Dr. Muley reports grants and non-financial support from Roche Diagnostics GmbH, Penzberg, Germany, outside the submitted work; In addition, Dr. Muley has a patent WO2019158460 pending, a patent WO2019211418 pending, a patent WO2019215223 pending, a patent EP3391053 issued and a patent EP3365679 pending. Dr. Fröhling reports personal fees from Amgen, grants from AstraZeneca, personal fees from Bayer, personal fees from Eli Lilly, grants from Pfizer, grants and personal fees from PharmaMar, personal fees from Roche, outside the submitted work. Dr. Waller is a consultant/advisory board member for Mylan Inc. Dr. Reck reports personal fees from Amgen, personal fees from AstraZeneca, personal fees from BMS, personal fees from Boehringer Ingelheim, personal fees from Lilly, personal fees from Merck, personal fees from MSD, personal fees from Novartis, personal fees from Pfizer, personal fees from Roche, personal fees from Samsung, outside the submitted work. Dr. Peters reports personal fees from Abbvie, personal fees from Amgen, personal fees from AstraZeneca, personal fees from Bayer, personal fees from Biocartis, personal fees from Boehringer Ingelheim, personal fees from Bistrol-Myers Squibb, personal fees from Clovis, personal fees from Daiichi Sankyo, personal fees from Debiopharm, personal fees from Eli Lilly, personal fees from F. Hoffmann-La Roche, personal fees from Foundation Medicine, personal fees from Illumina, personal fees from Janssen, personal fees from Merck Sharp and Dohme, personal fees from Merck Serono, personal fees from Merrimack, personal fees from Novartis, personal fees from Pharma Mar, personal fees from Pfizer, personal fees from Regeneron, personal fees from Sanofi, personal fees from Seattle Genetics and Takeda, personal fees from AstraZeneca, personal fees from Boehringer Ingelheim, personal fees from Bristol-Myers Squibb, personal fees from Eli Lilly, personal fees from F. Hoffmann-La Roche, personal fees from Merck Sharp and Dohme, personal fees from Novartis, personal fees from Pfizer, personal fees from Takeda, non-financial support from Sponsored by Amgen, non-financial support from AstraZeneca, non-financial support from Boehringer Ingelheim, non-financial support from Bristol-Meyers Squibb, non-financial support from Clovis, non-financial support from F. Hoffmann-La Roche, non-financial support from Illumina, non-financial support from Merck Sharp and Dohme, non-financial support from Merck Serono, non-financial support from Novartis, non-financial support from Pfizer, non-financial support from Sanofi, personal fees from Bioinvent, outside the submitted work. All fees to Institution. Dr. Schirmacher reports grants from QuIP, during the conduct of the study; grants and personal fees from BMS, grants and personal fees from MSD, grants and personal fees from Roche, grants and personal fees from AstraZeneca, grants and personal fees from Novartis, personal fees from Chugai, personal fees from AbbVie, grants from Sanofi-Aventis, personal fees from Ipsen, grants and personal fees from Pfizer, grants from Illumina, grants from Thermo Fisher, outside the submitted work. Dr. Thomas reports personal fees from AbbVie, grants, personal fees and non-financial support from BMS, personal fees and non-financial support from Boehringer, grants and personal fees from Celgene, personal fees from Lilly, personal fees and non-financial support from MSD, personal fees and non-financial support from Novartis, grants and personal fees from Roche, personal fees from Takeda, grants from AstraZeneca, outside the submitted work. Dr. Christopoulos reports grants and personal fees from Novartis, grants and personal fees from Roche, grants and personal fees from AstraZeneca, personal fees from Pfizer, grants and personal fees from Takeda, personal fees from Chugai, personal fees from Boehringer, outside the submitted work. Dr. Stenzinger reports personal fees from Astra Zeneca, grants and personal fees from Bayer, grants and personal fees from BMS, personal fees from MSD, personal fees from Takeda, personal fees from Seattle Genetics, grants from Chugai, personal fees from Novartis, personal fees from Illumina, personal fees from Thermo Fisher, personal fees from Eli Lily, personal fees from Takeda, outside the submitted work.
Footnotes
Co-first authors: Jan Budczies, Martina Kirchner, Klaus Kluck
Co-last authors: Petros Christopoulos, Albrecht Stenzinger
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Howlader N, Noone AM, Krapcho M, et al (2020) SEER cancer statistics review (CSR) 1975–2017. National Cancer Institute
- 2.Reck M, Rabe KF. Precision diagnosis and treatment for advanced non-small-cell lung cancer. N Engl J Med. 2017;377:849–861. doi: 10.1056/NEJMra1703413. [DOI] [PubMed] [Google Scholar]
- 3.Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv192–iv237. doi: 10.1093/annonc/mdy275. [DOI] [PubMed] [Google Scholar]
- 4.Garon EB, Wolf B, Lisberg A, et al. Prior TKI Therapy in NSCLC EGFR mutant patients associates with lack of response to Anti-PD-1 treatment. J Thorac Oncol. 2015;10:269. [Google Scholar]
- 5.Lisberg A, Cummings A, Goldman JW, et al. A phase II study of pembrolizumab in EGFR-mutant, PD-L1+, tyrosine kinase inhibitor naive patients with advanced NSCLC. J Thorac Oncol. 2018;13:1138–1145. doi: 10.1016/j.jtho.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gettinger S, Rizvi NA, Chow LQ, et al. Nivolumab monotherapy for first-line treatment of advanced non-small-cell lung cancer. J Clin Oncol. 2016;34:2980–2987. doi: 10.1200/JCO.2016.66.9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizvi NA, Hellmann MD, Brahmer JR, et al. Nivolumab in combination with platinum-based doublet chemotherapy for first-line treatment of advanced non-small-cell lung cancer. J Clin Oncol. 2016;34:2969–2979. doi: 10.1200/JCO.2016.66.9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Creelan BC, Yeh T, Kim S-W, et al. Phase I study of gefitinib (G) plus durvalumab (D) for locally advanced/metastatic non-small cell lung cancer (NSCLC) harbouring epidermal growth factor receptor (EGFR) sensitising mutations. Ann Oncol. 2019;30:ii31–ii37. [Google Scholar]
- 9.Yang JC, Gadgeel SM, Sequist LV, et al. Pembrolizumab in combination with erlotinib or gefitinib as first-line therapy for advanced NSCLC with sensitizing EGFR mutation. J Thorac Oncol. 2019;14:553–559. doi: 10.1016/j.jtho.2018.11.028. [DOI] [PubMed] [Google Scholar]
- 10.Mok TSK, Socinski MA, Reck M, et al. IMpower150: An exploratory analysis of efficacy outcomes in patients with EGFR mutations. Ann Oncol. 2019;29:ix177–ix178. [Google Scholar]
- 11.Socinski MA, Jotte RM, Cappuzzo F, et al. Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC. N Engl J Med. 2018;378:2288–2301. doi: 10.1056/NEJMoa1716948. [DOI] [PubMed] [Google Scholar]
- 12.Offin M, Rizvi H, Tenet M, et al. Tumor mutation burden and efficacy of EGFR-tyrosine kinase inhibitors in patients with EGFR-mutant lung cancers. Clin Cancer Res. 2019;25:1063–1069. doi: 10.1158/1078-0432.CCR-18-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singal G, Miller PG, Agarwala V, et al. Association of patient characteristics and tumor genomics with clinical outcomes among patients with non-small cell lung cancer using a clinicogenomic database. JAMA. 2019;321:1391–1399. doi: 10.1001/jama.2019.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang N, Zeng Y, Du W, et al. The EGFR pathway is involved in the regulation of PD-L1 expression via the IL-6/JAK/STAT3 signaling pathway in EGFR-mutated non-small cell lung cancer. Int J Oncol. 2016;49:1360–1368. doi: 10.3892/ijo.2016.3632. [DOI] [PubMed] [Google Scholar]
- 15.Chen N, Fang W, Zhan J, et al. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-driven NSCLC: implication for optional immune targeted therapy for NSCLC patients with EGFR mutation. J Thorac Oncol. 2015;10:910–923. doi: 10.1097/JTO.0000000000000500. [DOI] [PubMed] [Google Scholar]
- 16.Mazzaschi G, Madeddu D, Falco A, et al. Low PD-1 expression in cytotoxic CD8+ tumor-infiltrating lymphocytes confers an immune-privileged tissue microenvironment in NSCLC with a prognostic and predictive value. Clin Cancer Res. 2018;24:407–419. doi: 10.1158/1078-0432.CCR-17-2156. [DOI] [PubMed] [Google Scholar]
- 17.Toki MI, Mani N, Smithy JW, et al. Immune marker profiling and programmed death ligand 1 expression across NSCLC mutations. J Thorac Oncol. 2018;13:1884–1896. doi: 10.1016/j.jtho.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong ZY, Zhang JT, Liu SY, et al. EGFR mutation correlates with uninflamed phenotype and weak immunogenicity, causing impaired response to PD-1 blockade in non-small cell lung cancer. Oncoimmunology. 2017;6:e1356145. doi: 10.1080/2162402X.2017.1356145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haratani K, Hayashi H, Tanaka T, et al. Tumor immune microenvironment and nivolumab efficacy in EGFR mutation-positive non-small-cell lung cancer based on T790M status after disease progression during EGFR-TKI treatment. Ann Oncol. 2017;28:1532–1539. doi: 10.1093/annonc/mdx183. [DOI] [PubMed] [Google Scholar]
- 20.Liu L, Mayes PA, Eastman S, et al. The BRAF and MEK Inhibitors dabrafenib and trametinib: effects on immune function and in combination with immunomodulatory antibodies targeting PD-1, PD-L1, and CTLA-4. Clin Cancer Res. 2015;21:1639–1651. doi: 10.1158/1078-0432.CCR-14-2339. [DOI] [PubMed] [Google Scholar]
- 21.Gainor JF, Shaw AT, Sequist LV, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin Cancer Res. 2016;22:4585–4593. doi: 10.1158/1078-0432.CCR-15-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volckmar AL, Leichsenring J, Kirchner M, et al. Combined targeted DNA and RNA sequencing of advanced NSCLC in routine molecular diagnostics: analysis of the first 3000 Heidelberg cases. Int J Cancer. 2019;145:649–661. doi: 10.1002/ijc.32133. [DOI] [PubMed] [Google Scholar]
- 23.Budczies J, Kirchner M, Kluck K, Kazdal D, Glade J, Allgäuer M, Kriegsmann M, Heußel CP, Herth FJ, Winter H, Meister M, Muley T, Fröhling S, Peters S, Seliger B, Schirmacher P, Thomas M, Christopoulos P, Stenzinger A. A gene expression signature associated with B cells predicts benefit from immune checkpoint blockade in lung adenocarcinoma. Oncoimmunology. 2021;10(1):1860586. doi: 10.1080/2162402X.2020.1860586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu X, Wang Q, Tang M, et al. TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res. 2018;46:D1144–D1149. doi: 10.1093/nar/gkx1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danaher P, Warren S, Dennis L, et al. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer. 2017;5:18. doi: 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Sato Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020;29:28–35. doi: 10.1002/pro.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Sullivan CC, Bates SE. Targeting prolactin receptor (PRLR) signaling in PRLR-positive breast and prostate cancer. Oncologist. 2016;21:523–526. doi: 10.1634/theoncologist.2016-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banat GA, Tretyn A, Pullamsetti SS, et al. Immune and inflammatory cell composition of human lung cancer stroma. PLoS ONE. 2015;10:e0139073. doi: 10.1371/journal.pone.0139073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Shibli KI, Donnem T, Al-Saad S, et al. Prognostic effect of epithelial and stromal lymphocyte infiltration in non-small cell lung cancer. Clin Cancer Res. 2008;14:5220–5227. doi: 10.1158/1078-0432.CCR-08-0133. [DOI] [PubMed] [Google Scholar]
- 30.Ohri CM, Shikotra A, Green RH, et al. Macrophages within NSCLC tumour islets are predominantly of a cytotoxic M1 phenotype associated with extended survival. Eur Respir J. 2009;33:118–126. doi: 10.1183/09031936.00065708. [DOI] [PubMed] [Google Scholar]
- 31.Al-Shibli K, Al-Saad S, Donnem T, et al. The prognostic value of intraepithelial and stromal innate immune system cells in non-small cell lung carcinoma. Histopathology. 2009;55:301–312. doi: 10.1111/j.1365-2559.2009.03379.x. [DOI] [PubMed] [Google Scholar]
- 32.Schalper KA, Brown J, Carvajal-Hausdorf D, et al. Objective measurement and clinical significance of TILs in non-small cell lung cancer. J Natl Cancer Inst. 2015 doi: 10.1093/jnci/dju435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganesan AP, Johansson M, Ruffell B, et al. Tumor-infiltrating regulatory T cells inhibit endogenous cytotoxic T cell responses to lung adenocarcinoma. J Immunol. 2013;191:2009–2017. doi: 10.4049/jimmunol.1301317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lizotte PH, Ivanova EV, Awad MM, et al. Multiparametric profiling of non-small-cell lung cancers reveals distinct immunophenotypes. JCI Insight. 2016;1:e89014. doi: 10.1172/jci.insight.89014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kargl J, Busch SE, Yang GH, et al. Neutrophils dominate the immune cell composition in non-small cell lung cancer. Nat Commun. 2017;8:14381. doi: 10.1038/ncomms14381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talla SB, Rempel E, Endris V, et al. Immuno-oncology gene expression profiling of formalin-fixed and paraffin-embedded clear cell renal cell carcinoma: performance comparison of the nanostring ncounter technology with targeted RNA sequencing. Genes Chromosomes Cancer. 2020;59:406–416. doi: 10.1002/gcc.22843. [DOI] [PubMed] [Google Scholar]
- 37.Berraondo P, Sanmamed MF, Ochoa MC, et al. Cytokines in clinical cancer immunotherapy. Br J Cancer. 2019;120:6–15. doi: 10.1038/s41416-018-0328-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu SY, Dong ZY, Wu SP, et al. Clinical relevance of PD-L1 expression and CD8+ T cells infiltration in patients with EGFR-mutated and ALK-rearranged lung cancer. Lung Cancer. 2018;125:86–92. doi: 10.1016/j.lungcan.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 39.Isomoto K, Haratani K, Hayashi H, et al. Impact of EGFR-TKI treatment on the tumor immune microenvironment in EGFR mutation-positive non-small cell lung cancer. Clin Cancer Res. 2020;26:2037–2046. doi: 10.1158/1078-0432.CCR-19-2027. [DOI] [PubMed] [Google Scholar]
- 40.Vilgelm AE, Richmond A. Chemokines modulate immune surveillance in tumorigenesis, metastasis, and response to immunotherapy. Front Immunol. 2019;10:333. doi: 10.3389/fimmu.2019.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- 42.Zou W, Machelon V, Coulomb-L'Hermin A, et al. Stromal-derived factor-1 in human tumors recruits and alters the function of plasmacytoid precursor dendritic cells. Nat Med. 2001;7:1339–1346. doi: 10.1038/nm1201-1339. [DOI] [PubMed] [Google Scholar]
- 43.Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
- 44.Martinenaite E, Munir Ahmad S, Hansen M, et al. CCL22-specific T Cells: Modulating the immunosuppressive tumor microenvironment. Oncoimmunology. 2016;5:e1238541. doi: 10.1080/2162402X.2016.1238541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gobert M, Treilleux I, Bendriss-Vermare N, et al. Regulatory T cells recruited through CCL22/CCR4 are selectively activated in lymphoid infiltrates surrounding primary breast tumors and lead to an adverse clinical outcome. Cancer Res. 2009;69:2000–2009. doi: 10.1158/0008-5472.CAN-08-2360. [DOI] [PubMed] [Google Scholar]
- 46.Rapp M, Wintergerst MWM, Kunz WG, et al. CCL22 controls immunity by promoting regulatory T cell communication with dendritic cells in lymph nodes. J Exp Med. 2019;216:1170–1181. doi: 10.1084/jem.20170277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Garra A, Vieira P. T(H)1 cells control themselves by producing interleukin-10. Nat Rev Immunol. 2007;7:425–428. doi: 10.1038/nri2097. [DOI] [PubMed] [Google Scholar]
- 48.Tsukamoto H, Fujieda K, Senju S, et al. Immune-suppressive effects of interleukin-6 on T-cell-mediated anti-tumor immunity. Cancer Sci. 2018;109:523–530. doi: 10.1111/cas.13433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Somasundaram A, Cillo A, Oliveri L, et al. IL-6, IL-8 drive LAG3/PD1 immune suppression on effector and naïve, peripheral blood CD8+ T cells in cancer patients. J Immunol. 2019;202(1 Supplement):195.9. [Google Scholar]
- 50.Cannarile MA, Weisser M, Jacob W, et al. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J Immunother Cancer. 2017;5:53. doi: 10.1186/s40425-017-0257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J, Yao Y, Gong C, et al. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell. 2011;19:541–555. doi: 10.1016/j.ccr.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voron T, Colussi O, Marcheteau E, et al. VEGF-A modulates expression of inhibitory checkpoints on CD8+ T cells in tumors. J Exp Med. 2015;212:139–148. doi: 10.1084/jem.20140559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lapeyre-Prost A, Terme M, Pernot S, et al. Immunomodulatory activity of VEGF in cancer. Int Rev Cell Mol Biol. 2017;330:295–342. doi: 10.1016/bs.ircmb.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 54.Gough MJ, Ruby CE, Redmond WL, et al. OX40 agonist therapy enhances CD8 infiltration and decreases immune suppression in the tumor. Cancer Res. 2008;68:5206–5215. doi: 10.1158/0008-5472.CAN-07-6484. [DOI] [PubMed] [Google Scholar]
- 55.Nelson BH. IL-2, regulatory T cells, and tolerance. J Immunol. 2004;172:3983–3988. doi: 10.4049/jimmunol.172.7.3983. [DOI] [PubMed] [Google Scholar]
- 56.Zhao M, Liu Y, Liu R, et al. Upregulation of IL-11, an IL-6 family cytokine, promotes tumor progression and correlates with poor prognosis in non-small cell lung cancer. Cell Physiol Biochem. 2018;45:2213–2224. doi: 10.1159/000488166. [DOI] [PubMed] [Google Scholar]
- 57.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 58.Christensen JE, de Lemos C, Moos T, et al. CXCL10 is the key ligand for CXCR3 on CD8+ effector T cells involved in immune surveillance of the lymphocytic choriomeningitis virus-infected central nervous system. J Immunol. 2006;176:4235–4243. doi: 10.4049/jimmunol.176.7.4235. [DOI] [PubMed] [Google Scholar]
- 59.Gao Q, Wang S, Chen X, et al. Cancer-cell-secreted CXCL11 promoted CD8+ T cells infiltration through docetaxel-induced-release of HMGB1 in NSCLC. J Immunother Cancer. 2019;7:42. doi: 10.1186/s40425-019-0511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seliger B. Combinatorial approaches with checkpoint inhibitors to enhance anti-tumor immunity. Front Immunol. 2019;10:999. doi: 10.3389/fimmu.2019.00999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang J, Shalabi A, Hubbard-Lucey VM. Comprehensive analysis of the clinical immuno-oncology landscape. Ann Oncol. 2018;29:84–91. doi: 10.1093/annonc/mdx755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.