Abstract
Aims
To compare specific T-cell responses between laboratory employees in South Africa with and without previously diagnosed SARS-CoV-2 infection.
Methods
Employees at a private pathology laboratory in South Africa were invited to participate in a nationwide cross-sectional study. T-cell proliferation to SARS-CoV-2 nucleocapsid (N)-proteins and spike (S)-proteins was measured by flow cytometry and compared between participants.
Results
Based on classification according to SARS-CoV-2 reverse transcription (RT)-PCR results, a total of 81% (42/52) of positive participants demonstrated T-cell proliferation to SARS-CoV-2 N-proteins or S-proteins (95% CI 67.5% to 90.4%), while 62% (68/110) of negative participants also had detectable T-cell responses to SARS-CoV-2 proteins (95% CI 52.1% to 70.9%). When classified according to SARS-CoV-2 serology results, 92.6% (50/54) of positive participants demonstrated T-cell proliferation to SARS-CoV-2 proteins (95% CI 82.1 to 97,9 %), while 56% (60/108) of negative participants demonstrated T-cell proliferation (95% CI 45.7% to 65.1%). The magnitude of the T-cell responses as determined by a stimulation index, was significantly higher in the group previously infected by SARS-CoV-2 than in the negative group. A statistically significant difference in T-cell proliferation was noted between high risk and low risk groups for exposure to SARS-CoV-2 within the negative group, but no significant difference in magnitude of the response.
Conclusions
A significant proportion of South African laboratory employees who were not previously diagnosed with COVID-19 demonstrated T-cell reactivity to SARS-CoV-2 N-proteins and S-proteins. The pre-existing T-cell proliferation responses may be attributable to cross-reactive immune responses to other human coronaviruses, or possibly asymptomatic infection.
Keywords: COVID-19, immunology, diagnostic techniques and procedures, virology
Introduction
The novel coronavirus, SARS-CoV-2, emerged from China in December 2019, and has since spread globally to infect millions in an ongoing pandemic.1 At the inception of this pandemic many assumptions were made regarding the immune response to the virus, herd immunity and the plausibility of an effective vaccine that could be produced successfully and implemented in pandemic control.2
As Europe spiked into a second wave during their winter months, closely followed by South Africa, many unanswered questions remained with regard to immune response to SARS-CoV-2, correlates of protection and immune response to vaccination. South Africa, as a developing country with a very high HIV, TB and poverty burden appears to have experienced a less severe first wave of COVID-19 than expected.3 4 The reasons for this may be multifactorial, but it is reasonable to pose the question whether there may be some level of pre-existing immunity to SARS-CoV-2 in the South African community.4 An emerging body of evidence worldwide has described T-cell reactivity to SARS-CoV-2 in individuals with no known prior exposure to SARS-CoV-2.5 Literature suggests that memory T cells may prove critical for long-term immune protection against COVID-19, and that cross-reactive memory T cells which could arise from prior exposure to other circulating coronaviruses may provide a form of background immunity to COVID-19, even when no antibodies are present.6 7 This may well have implications for vaccine development and determining vaccine-derived immunity in future, as well as contribute to current knowledge of herd immunity and the transmission dynamics of COVID-19.8 Testing for memory T cells against SARS-CoV-2 may also provide evidence of immunity in seronegative patients who had mild or asymptomatic COVID-19.8
The aim of this study was to compare laboratory employees previously diagnosed with SARS-CoV-2 and laboratory employees not deemed to have been infected by SARS-CoV-2 with respect to their T-cell responses to SARS-CoV-2.
Individuals not previously infected by SARS-CoV-2 should not produce an antibody response to SARS-CoV-2 as measured in highly specific commercially available antibody assays, but a subpopulation may produce a cross-reactive T-cell response to SARS-CoV-2, due to prior exposure to other circulating human coronaviruses.5 Most individuals previously infected by SARS-CoV-2 should produce a T-cell and antibody response to SARS-CoV-2, however literature suggests that a small subset may not produce or sustain detectable SARS-CoV-2 specific antibodies.2
The majority of published T-cell reactivity studies to SARS-CoV-2 have been limited to research laboratories, due to the high level of technical skill involved in T-cell proliferation testing. Results in this study were determined by an in-house, validated, flow-cytometric T-cell proliferation assay to SARS CoV-2 S-protein and N-protein suitable to scalability and routine diagnostic use. Access to T-cell proliferation tests to SARS-CoV-2 in a routine diagnostic setting could contribute to data to fill current knowledge gaps in the immune response to SARS-CoV-2.
Methods
Study design
The study design is a cross-sectional study of two groups of a population consisting of Ampath private pathology laboratory employees, one group previously diagnosed with SARS-CoV-2 and one group not deemed to have been infected by SARS-CoV-2.
Study participants
Ampath pathology employs approximately 5500 employees in various phlebotomy, pathology and administrative positions nationally in South Africa. All staff members were eligible to participate in this study.
Recruitment of participants
Ampath staff members were invited by means of a confidential internal email sent from a customised, private study email address to participate in this study. A total of 459 staff members responded to this email to voluntarily participate in this study. Volunteers were selected sequentially and invited to answer a questionnaire and provide blood specimens for this study. Enrolment continued until sufficient participants from the two predetermined study groups, that is, participants with and without previously confirmed COVID-19, submitted a completed questionnaire and blood specimens. Volunteers who were diagnosed with SARS-CoV-2 infection by reverse transcription-polymerase chain reaction (RT-PCR) less than 2 weeks prior to enrolment were asked to delay blood sampling by 2 weeks postdiagnosis.
Informed consent was obtained from all volunteers prior to enrolment. Participants were randomised in a 1:2 ratio of employees diagnosed with COVID-19 and employees not diagnosed previously. In order to detect at least a 25% difference between the groups, assuming a maximum of 50% positivity (in the COVID-19 negative group) and a minimum of 75% positivity (in the COVID-19 positive group), and with a minimum statistical power of 90%, a minimum sample size of 47 COVID-19 positive and 94 COVID-19 negative participants would be required. The actual number of participants recruited were 52 COVID-19 positive participants and 110 presumed COVID-19 negative participants.
Study participants received a questionnaire to complete electronically, which included information on travel history, questions related to risk level of potential exposure at work, episodes of self-quarantine or contact with a known COVID-19 patient, COVID-19 related symptoms experienced since February 2020, treatment received if diagnosed with COVID-19, underlying medical conditions and chronic medication, previous SARS-CoV-2 RT-PCR testing performed, as well as the date and results of tests.
A blood collection form which was linked to each participant’s unique study number, was sent via email. Participants were required to have a venepuncture at any of Ampath’s 350 national blood collection depots to collect four citrate and one clotted blood specimens.
Recruitment occurred from 22 September 2020 to 9 November 2020.
Research was conducted in line with the stipulations of the Declaration of Helsinki (2013). Volunteer confidentiality was maintained and all participants were awarded a deidentified study number on enrolment. Laboratory staff and investigators did not have access to any participant information and were blinded on analysis and interpretation of test results.
Laboratory protocol
All testing was performed in the Ampath National Immunology Reference Laboratory in Centurion, South Africa, accredited by the South African National Accreditation System in accordance with ISO 15912 standards. Antibody responses to both the SARS-CoV-2 S-protein and the N-protein were measured with commercial kits, namely Anti-SARS-CoV-2 IgG S-protein ELISA (EUROIMMUN, Lubeck, Germany) and Elecsys Anti-SARS-CoV-2 IgM//IgG N-protein chemiluminescence assay (Roche Diagnostics, Penzberg, Germany). The respective sensitivities and specificities of these assays as determined by internal validation studies were 96.67% sensitivity with 98.7% specificity for the EUROIMMUN Spike IgG assay and 92.31% sensitivity with 100% specificity for the Roche Elecsys IgM/IgG to N-protein.
All clotted blood samples for antibody testing were centrifuged and frozen at −20°C until all study specimens were collected. Specimens were processed in concordance with routine laboratory practice for serology testing and run as a batch once all samples had been received.
T-cell proliferation tests were performed to SARS-CoV-2 S-protein and N-proteins. The antigens that were used for the T-cell stimulation were the following:
SARS-CoV-2 N-protein-Miltenyi Biotec B.V. & Co. KG, Bergisch Gladbach, Germany (www.miltenyi biotec.com): Nucleocapsid phosphoprotein of SARS-CoV-2, which is peptides consisting of 15 amino acid lengths, which stimulates CD4 + and CD8+T cells, eliciting secretion of effector cytokines and thus upregulation of activation surface markers, measured by flow cytometry. The peptides were lyophilised, stored at −20°C. The average purity of the peptides were >70% using HPLC methods. The product contains no preservatives.
SARS-CoV-2 S protein—Cape BioPharms, Cape Town, South Africa (www.capebiopharms.com): Spike glycoprotein of SARS-CoV-2, obtained from DNA encoding the S1 portion of the Spike protein, fused to polyhistidine and SEKDEL tag at the C-terminus, recombinantly expressed in Nicotiana benthamiana (relative of the tobacco plant, indiginous to Australia), purified using immobilised metal chromatography and determined by SDS-PAGE.
A standardised, in-house flow cytometric lymphocyte proliferation test method currently in use at Ampath to detect T-cell responses to other recall antigens, for example, Varicella-Zoster-Virus (VZV), Candida albicans and Tetanus toxoid was modified for the purpose of this study. The sensitivity of this in-house SARS-CoV-2 T-cell proliferation assay as determined by an in-house validation in patients with known positive SARS-CoV-2 PCR tests (n=40) was 90% and the specificity in a known SARS-CoV-2 negative population (n=102) was 34%. The specificity was expected to be low, due to the anticipated presence of cross-reactive T-cells.
T-cell proliferation testing was performed in accordance with our in-house validation protocol. We collected four citrate tubes per study participant, to ensure adequate numbers of viable cells as determined on in-house validation studies for cellular tests. Fresh peripheral blood mononuclear cells (PBMCs) were harvested by Ficoll density gradient centrifugation. Viability studies were performed for each sample, using a vital stain (Tryphan blue). Visualisation was performed by an experienced scientist under a light microscope before stimulation and testing could be conducted. Samples in which the viability of cells could not be confirmed, were recollected. The maximum validated cut-off time from collection to processing for this assay is 24 hours. However, sample collection appointments were arranged to ensure the shortest turnaround time possible and samples were processed immediately when they reached the laboratory.
PBMCs were stimulated with SARS-CoV-2 S- and N- proteins for a duration of 6 days. A negative control and a positive mitogen control was included for each patient. Lymphocyte proliferation was detected by flow-cytometric (BD FACSCanto II) measurement of intracellular expression of Ki67 in T-cells. The results were reported as a Stimulation Index (SI), which was calculated by dividing the T-cell proliferation to the N-protein and S-protein antigens by the T-cell proliferation to the negative control. Our cut-off used for positivity was a SI greater than 3. This is similar to criteria used for the cut-off of T-cell proliferation to other specific antigens (tetanus, varicella-zoster-virus and candida) used in the laboratory, and supported by previously published reference ranges.9
An explanation of the flow cytometry analysis is detailed in figure 1.
Figure 1.
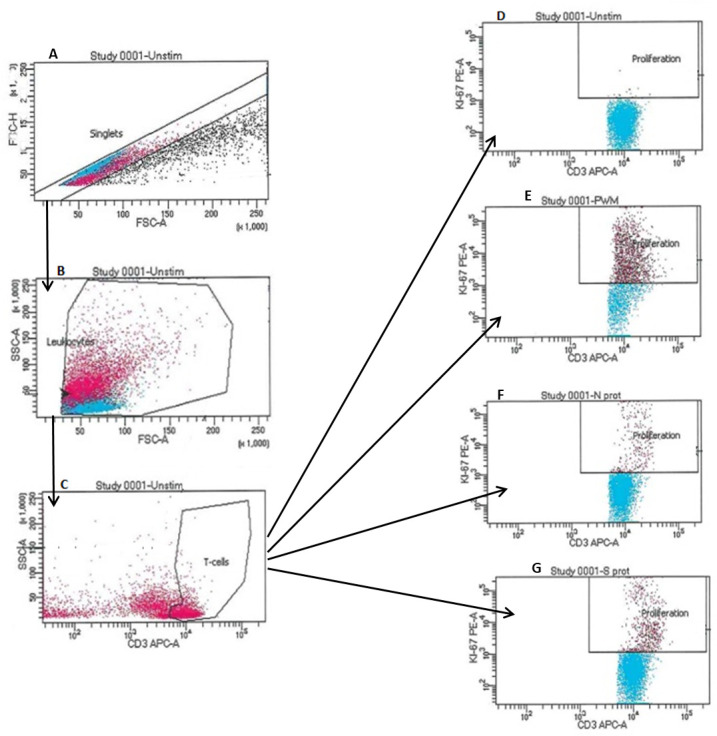
Flow cytometric gating strategies used to identify the response of T-cells to the N-protein and S-protein of SARS-CoV2. Gating was first set on singlet events (A), followed by gating on target populations, leucocytes (B) and then CD3 expressing T-cells (C). The stimulation of the T-cells were measured using intracellular—PE Ki-67 (D). Buffer was added to the negative control (unstimulated) in (D). The positive control in (E) was pokeweed mitogen. The stimulation of the T-cells can be seen in the proliferation gate (E). T-cell activation seen in the proliferation gate (F) when exposed to nucleocapsid (N) protein of SARS-CoV2. T-cell activation seen in the proliferation gate (G) when exposed to spike (S) protein of SARS-CoV2. SSC, side scatter.
Statistical analysis
Data analysis was performed by comparing T-cell proliferation and antibody responses to SARS-CoV-2 S- and N-protein obtained from the SARS-CoV-2 RT-PCR positive group, with the group deemed not to have been infected by SARS-CoV-2 previously. The primary outcome was the proportion of participants with T-cell proliferation responses to SARS-CoV-2 S- and N-protein in the SARS-CoV-2 groups (positive vs negative). This was performed using a Fisher’s exact test.
Data analysis was also performed comparing T-cell proliferation to N- and S-protein in a SARS-CoV-2 positive and negative group classified by serology results. These results were also compared by a Fischer’s exact test.
In a secondary analysis, the group not deemed to have been infected by SARS-CoV-2 previously, was compared in two subgroups, one with a high risk of exposure (answered yes to any of the questions relating to previous international travel in 2020, previous quarantine, previous SARS-CoV-2 PCR performed or reporting any two of the three symptoms of fever, dry cough or loss of smell or taste during the same month) and one with low risk of exposure (the remainder of the negative group). Laboratory results of these two subgroups were compared and analysed by a Fischer’s exact test.
The magnitude of the T-cell responses to the N- and S-proteins of SARS-CoV-2, as indicated by the SI, were compared between the positive and the negative group categorised according to RT-PCR results using a Mann Whitney test. The magnitude of the T-cell responses to the N-and S-proteins of SARS-CoV-2 were similarly compared between the high and low risk negative groups. Statistical analysis was conducted using STATA Release V.16 (StataCorp). Statistical significance was set at a p <0.05 (95% CI).
Results
Characteristics of COVID-19 positive and negative participants
A total of 52 COVID-19 participants with previous positive SARS-CoV-2 RT-PCR results (positive group) and 110 participants who never were confirmed or suspected to have COVID-19 (negative group) were recruited. The majority of the participants in the positive group had mild illness at the time of infection with SARS-CoV-2, as only 7.7% (4/52) of the participants required admission to hospital. Among the hospital admissions, only three participants were given supplemental oxygen, and none required ventilation. The answers provided in the questionnaire were used to subdivide the negative group into a high-risk group for exposure (38 participants) and a low-risk group for exposure (72 participants). Indications for classifying participants as negative, high risk, were those who travelled internationally, quarantined, experienced COVID-19 symptoms (at least two of; fever, dry cough and loss of taste/smell) and those who had a negative SARS-CoV-2 RT-PCR test performed within the duration of the study.
Serological classification of participants in positive and negative groups was based on the presence of antibodies to SARS-CoV-2 S-protein or N- protein. A total of 54 participants were assigned to the positive group and 108 participants to the negative group. All of the participants in the positive group demonstrated the presence of SARS-CoV-2 N-protein antibodies and (53/54) participants demonstrated antibodies to S-protein.
SARS-CoV-2-specific T-cell and antibody responses in positive and negative groups by RT-PCR classification
A positive T-cell response to SARS-CoV-2 was demonstrated in 81% (42/52; 95% CI 67.5% to 90.4%) of the participants previously diagnosed with COVID-19 by SARS-CoV-2 RT-PCR in this study, while 62% (68/110; 95% CI 52.1% to 70.9%) of participants who had not been diagnosed with COVID-19 previously had a T-cell response to SARS-CoV-2 N-protein or S-protein (p=0.011). A total of 79% (41/52) of the participants in the positive group diagnosed by SARS-CoV-2 PCR, had also demonstrated a positive antibody response to SARS-CoV-2. In 40 of the 41 patients with positive antibody responses a positive result was demonstrated on both antibody assays (N-protein and S-protein assays). One patient had a positive antibody response on the SARS-CoV-2 N-protein IgM/G only, but had positive T-cell responses to both SARS-CoV-2 N-protein and S-protein. In the presumed negative group, a total of 7% (8/110) of the participants had positive antibody results on both antibody assays (N-protein and S-protein assays) as well as positive T-cell proliferation to SARS-CoV-2. The results of the T-cell and antibody responses are summarised in table 1.
Table 1.
Detection of SARS-CoV-2 T-cell responses to S-protein and N-protein and antibody responses to S-proteins (EUROIMMUN) and N-proteins (Roche Elecsys)
| SARS-CoV-2 antibody assay | Negative group (n=110) | Positive group (n=52) | P value Antibody results |
P value T-cell results |
||
| Positive antibody results | Positive T-cell response | Positive antibody results | Positive T-cell response | |||
| SARS-CoV-2 N-protein target | 8 (7.27%) | 61 (55.45%) | 41 (78.84%) | 42 (80.77%) | <0.001 | 0.001 |
| SARS-CoV-2 S-protein target | 8 (7.27%) | 53 (48.18%) | 40 (76.92%) | 38 (73.08%) | <0.001 | 0.002 |
| Any SARS-CoV-2 protein target | 8 (7.27%) | 68 (61.82%) | 41 (78.84%) | 42 (80.77%) | <0.001 | 0.011 |
SARS-CoV-2 T-cell responses in positive and negative groups according to serology classification
When the data were reanalysed according to antibody test results to the two different SARS-CoV-2 antigens on two different serology platforms for classification, a positive T-cell response to SARS-CoV-2 was demonstrated in 93% (50/54; 95% CI 82.11% to 97.9%) of the participants in the reclassified positive group, while 56% (60/108; 95% CI 45.7% to 65.1%) of the participants in the negative group had a T-cell response to SARS-CoV-2 N- or S-protein (p<0.001). The data are summarised in table 2.
Table 2.
Detection of SARS-CoV-2 T-cell responses to S-protein and N-protein in positive and negative groups according to serology classification
| SARS-CoV-2 T-cell response | Negative group (n=108) | Positive group (n=54) | P value T-cell results |
| SARS-CoV-2 N-protein target | 54 (50%) | 49 (90.74%) | <0.001 |
| SARS-CoV-2 S-protein target | 45 (41.67%) | 46 (85.19%) | <0.001 |
| Any SARS-CoV-2 protein target | 60 (55.56%) | 50 (92.59%) | <0.001 |
Magnitude of T-cell responses in positive and negative groups
The SI of T-cell proliferation tests to the N- and S-proteins of SARS-CoV-2 were compared between the positive group and negative group using a Mann-Whitney test. The positive group demonstrated a higher magnitude of T-cell proliferation to SARS-CoV-2 N-protein and S-protein than the negative group, which was statistically significant (p<0.001). The data are summarised in table 3 and figure 2.
Table 3.
Magnitude of T-cell responses to SARS-CoV-2 S-protein and N-protein expressed as a Stimulation Index (SI) in the SARS-CoV-2 positive and negative group
| N-protein SI | S-protein SI | |||
| Negative group | Positive group | Negative group | Positive group | |
| Median | 6 | 18.6 | 5.66 | 13 |
| IQR | 5–12 | 8.66–26 | 4–8 | 7–25 |
| Range | 3–202 | 3.33–83 | 3–96 | 3.4–65 |
Figure 2.
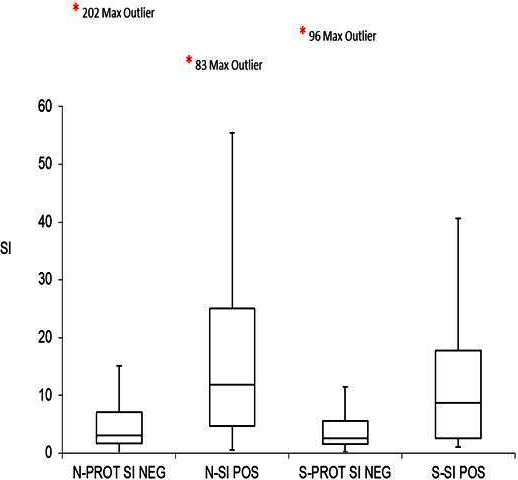
Comparison of T-cell Stimulation Index (SI) to SARS-CoV-2 S-protein and N-protein between the SARS-CoV-2 positive and negative groups. *indicates the SI value of the maximum outlier in each group. NEG, negative; N-PROT, nucleocapsid protein; POS, positive, S-PROT, spike protein.
SARS-CoV-2 T-cell responses in negative group with high and low risk of exposure
Results for both T-cell proliferation and antibody responses to SARS-CoV-2 N-protein and S-protein antigens were compared within the RT-PCR negative group reflecting classification in a high-risk and low-risk group based on questionnaire responses. Of the SARS-CoV-2 negative group, 71% (27/38) of participants with high risk of exposure had a positive T-cell response to SARS-CoV-2 N- or S-protein, while 57% (41/72) of participants with a low risk of exposure had a positive T-cell response to SARS-CoV-2 (p=0.018). The data table can be viewed online as online supplemental table 1.
jclinpath-2021-207556supp001.pdf (163.2KB, pdf)
Magnitude of T-cell responses in negative group with high and low risk of exposure
The magnitude of the T-cell responses were compared between the high-risk and low-risk exposure groups within the negative group, expressed as a SI, with no statistical significance found between these groups. The data is summarised in figure 3. A complete data table can be viewed online as online supplemental table 2.
Figure 3.
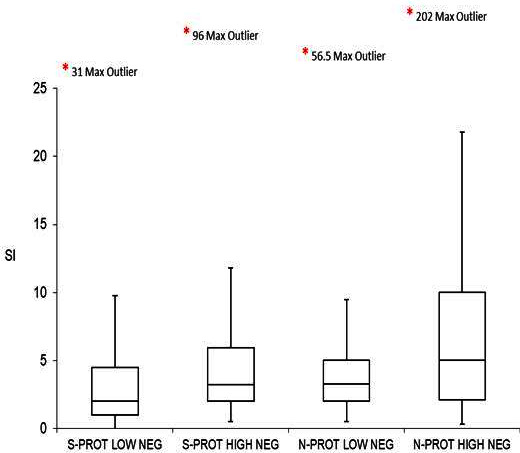
Comparison of T-cell Stimulation Index (SI) to SARS-CoV-2 S-protein and N-protein between the SARS-CoV-2 negative high-risk and low-risk groups. *indicates the SI value of the maximum outlier within each group. NEG, negative; N-PROT, nucleocapsid protein; S-PROT, spike protein.
jclinpath-2021-207556supp002.pdf (35.1KB, pdf)
Discussion
The findings of the current study indicate that while SARS-CoV-2 specific T-cell responses were detectable in the majority of study participants with previously diagnosed SARS-CoV-2 infection, a significant proportion of participants who were not previously diagnosed with COVID-19 also demonstrated T-cell reactivity to SARS-CoV-2 N- and S-proteins. The positive T-cell responses in the negative group may represent cross reactivity with other human coronaviruses associated with the common cold, or alternatively could be due to asymptomatic SARS-CoV-2 infection. The SI of both the N- and S-protein was significantly higher (p<0.001) in the positive group than the negative group, indicating that SARS-CoV-2 infection elicits a greater T-cell response to N-protein and S-protein than cross-reactive T-cells.
The negative group, divided in a high risk and low risk group for previous exposure to SARS-CoV-2 based on questionnaire responses, showed a statistically significant difference in T-cell proliferation to SARS-CoV-2 N- and S-proteins. 71% (27/38) of the high-risk negative group and 57% (41/72) of the low-risk negative group demonstrated T-cell proliferation to one or both of the tested SARS-CoV-2 antigens (p=0.018). However, the difference in the magnitude of T-cell responses between the high and low risk negative groups was not statistically significant.
In the presumed negative group, a total of 7% (8/110) of the participants had positive antibody results on both antibody assays (N-protein and S-protein assays) as well as positive T-cell proliferation to SARS-CoV-2, which strongly suggests that these individuals had been infected by SARS-CoV-2, but were undiagnosed by RT-PCR. It was anticipated from the outset of this study that the correct classification of groups as previously infected with SARS-CoV-2 or not, might prove to be a challenge. PCR testing has been shown to be a less than perfect gold standard for the diagnosis of SARS-CoV-2 infection with some false positive, and in particular false negative results to be anticipated.10 11 Asymptomatic SARS-CoV-2 infections are well described and such individuals may not be tested for SARS-CoV-2 infection.12 It is notable that 50% (4/8) of these participants were quarantined previously due to close contact with a confirmed COVID-19 case, increasing the likelihood that they had undiagnosed asymptomatic SARS-CoV-2 infections.
Six participants in the positive group had a complete lack of antibody and T-cell immune response to SARS-CoV-2, which may indicate that their SARS-CoV-2 PCR tests could have been false positive results. These individuals were furthermore asymptomatic and did not reveal a history of an immunodeficiency or the use of immunosuppressive medication on their study questionnaire.
Given the limitations of RT-PCR for classification of prior COVID-19 infection, reanalysis of T-cell responses according to classification by means of serology was performed. The T-cell responses demonstrated a better correlation when analysed according to serology classification than classification by RT-PCR.
Positioning of this study in context of existing literature
Emerging literature has shifted focus from the role of antibodies to the potential role of T-cells in immunity to SARS-CoV-2.5 Studies suggest that memory T cells may prove critical for long-term immune protection against COVID-19, and that cross-reactive memory T-cells which could arise from prior exposure to other circulating coronaviruses may provide a form of background immunity to COVID-19, or affect the disease severity, even when no antibodies are present.13 14
A study by Le Bert et al demonstrated SARS-CoV-2 N-protein specific T cells in individuals with a history of SARS-CoV-1 infection, and that this population of T cells expanded after an encounter with SARS-CoV-2 N-protein peptides. They also frequently detected SARS-CoV-2 specific T cells in individuals with no history of either contact or infection with SARS-CoV-1 or SARS-CoV-2.6 Grifoni et al also detected SARS-CoV-2-reactive CD4 +T cells in as many as 60% of unexposed donors recruited between 2015 and 2018, which further supports cross reactive T cell recognition between circulating ‘‘common cold’’ coronaviruses and SARS-CoV-2.15 Similar findings by Sekine et al demonstrated possible cross-reactive T cell responses directed against either the S or membrane proteins in 28% of the healthy individuals who donated blood before SARS-CoV-2 emerged.16 Finally, Braun et al detected SARS-CoV-2 S-reactive CD4 +T cells in 83% of patients with COVID-19 but also in 35% of unexposed healthy donors. S-protein reactive CD4 +T cells from SARS-CoV-2-naive donors were found to have a similar interaction with human endemic coronaviruses 229E and OC43 and SARS-CoV-2. This suggests that prior endemic coronavirus infections may generate S-cross-reactive T cells.7
A recent study measuring T-cell responses to COVID-19 in 2826 healthcare workers and firefighters performed by Oxford Immunotek in collaboration with Public health England using a research version of a COVID-19 TB-Spot test, demonstrated higher T-SPOT responses to spike, membrane and nucleoprotein in persons with either confirmed previous SARS-CoV-2 infection or who were seropositive as detected with anti-SARS-CoV-2 immunoassays, in comparison with persons deemed to have been uninfected.17 The investigators found that participants with higher T-SPOT responses were less likely to test positive for COVID-19 by RT-PCR during a median follow-up period of 118 days.
The immune response to COVID-19 is complex and highly variable between individuals. While the vast majority of persons infected with SARS-CoV-2 will develop IgG antibodies to the virus, a subset will not, which limits the usefulness of serology tests to determine prior exposure to SARS-CoV-2.18 In addition, it has been well described that SARS-CoV-2-specific antibodies may start to wane after 3–4 months following infection.19 Other factors such as the age of the patient and disease severity may also impact antibody responses, with higher antibody levels recorded in older and hospitalised cases of COVID-19 in previous studies.20 In contrast, CD4 +T cell responses have been shown to be lower in those who require hospitalisation due to more severe COVID-19 than individuals with mild illness, and the CD4 +memory T cells remain detectable for a period beyond 6 months in most individuals.18 In essence, the immune response (including the immune memory) is heterogeneous, and the option of testing SARS-CoV-2-specific T cell responses is an important tool which can greatly assist in routine clinical practice. The majority of participants who were included in the positive group of our own study population reportedly had mild COVID-19 illness, and provides further insight into the complexities of the immune response to infection with SARS-CoV-2 in a population where lower antibody levels were to be expected.
In summary, emerging literature suggests that memory T cells may prove critical for long-term immune protection against COVID-19, and that cross-reactive memory T cells which could arise from prior exposure to other circulating coronaviruses may provide a form of background immunity to COVID-19, or affect the disease severity, even when no antibodies are present.
Contribution of this study
Our study contributes to the emerging international data pool and was designed to determine the level of T-cell reactivity to SARS-CoV-2 S-protein and N-protein in our study population, consisting of South African laboratory workers who were either previously diagnosed with COVID-19 or not. The results were generated using an in-house flow cytometric T-cell proliferation assay within a diagnostic laboratory, which is scalable and suitable for routine use. T-cell proliferation tests usually require a high level of technical skill, which often limits its application to research settings. Immunological status against SARS-CoV-2 was studied in an integrated manner within a routine diagnostic setting, which may become important for evaluating durability of the immune response, of particular concern considering viral mutations, new strains and vaccination rollout in South Africa. The ability to measure and compare the magnitude of T-cell response to SARS-CoV-2 S-protein creates opportunities to measure T-cell as well as antibody responses prevaccination and postvaccination. This could become relevant to inform decisions regarding choice of vaccine, dosing schedules as well as monitoring vaccine response in immunocompromised individuals.
Limitations of this study
This study was performed on a relatively small number of volunteers from various sectors within a pathology practice, recruited from multiple sites throughout South Africa. Larger studies, including volunteers from multiple sectors and population groups would be needed to extrapolate data to encompass the greater South African population. The participants included in this study are all employees within a healthcare setting, with a potentially higher rate of exposure to SARS-CoV-2 and other respiratory viral infections than the general public. This may have introduced spectrum bias in this particular study.
One of the main limitations of this study is the lack of a reliable gold standard test to assess previous SARS-CoV-2 infection. PCR testing has limitations and false positive results as well as false-negative results occur. Similarly, serology results have limitations, as antibody levels may wane over time, and not all individuals will necessarily produce a detectable antibody response.
A further limitation of the study is the low specificity of the T-cell proliferation assay, which may be attributed to cross-reactivity following prior infections with other human coronaviruses in subjects who were not exposed to SARS-CoV-2, as the objective was to measure SARS-CoV-2 specific responses. It would have been ideal to have included peptides to other human coronaviruses, but these peptides were not commercially available in South Africa at the time of this study.
Future research
Correlates of protection and post-vaccination immunity are important future research topics. A follow-up prospective cohort study of participants and their immune response to SARS-CoV-2 over time, as well as future COVID-19 infections, vaccination and subsequent T-cell immune responses should be studied in an attempt to determine correlates of protection. Future research measuring T-cell and antibody responses in vaccinated participants as well as participants infected by new SARS-CoV-2 variants, should also be considered.
Conclusion
The findings from the study provides evidence of significant pre-existing T-cell immunity to SARS-CoV-2 in South African individuals not previously diagnosed with COVID-19, which may be attributable to pre-existing cross-reactive immune responses to other human coronaviruses or asymptomatic infection. The magnitude of T-cell responses to both SARS-CoV-2 S-proteins and N-proteins were greater in participants previously diagnosed with COVID-19, indicating that a significant T-cell response was elicited post-SARS-CoV-2 infection in comparison to the group considered to have had pre-existing cross-reactive T-cell immunity. This study demonstrates that T-cell proliferation assays can be used in a routine diagnostic setting, which could be of particular importance for measuring postvaccination T-cell responses.
Take home messages.
A significant number of participants in this study not previously diagnosed with COVID-19 were shown to have pre-existing T-cell immunity to SARS-CoV-2, which may be attributable to pre-existing cross-reactive immune responses to other human coronaviruses or asymptomatic infection.
The magnitude of T-cell responses to both SARS-CoV-2 S-proteins and N-proteins were significantly greater in participants previously diagnosed with COVID-19 in comparison to the group considered to have had pre-existing cross-reactive T-cell immunity.
This study measured SARS-CoV-2 specific T-cell responses by means of an in-house T-cell proliferation assay within a routine diagnostic laboratory setting, which could be of particular importance for measuring postvaccination T-cell responses.
Acknowledgments
We would like to thank the following people for their contribution to data entry and administrative support: Marlene Buitendach and Hester Pieters. We would like to acknowledge Dr Mark da Silva and Dr Louise Murray for proofreading our manuscript before submission.
Footnotes
Handling editor: Stephen R A Jolles.
Contributors: CvR acts as guarantor. CvR and MB were responsible for the writing, literature review, concept and interpretation of the study. PS did the planning and specimen collection together with the administrative processes of the study and reviewing. SvdB assisted with the data interpretation, reviewing and conception and design. CvdM and MvdM were responsible for laboratory assays and data collection. PB helped with statistical analysis and RG was a supervisor and assisted with conception.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as online supplemental information. Deidentified participant data available as online supplemental information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by Research Ethics Committee (Faculty of Health Sciences) of the University of Pretoria, approval number: 631/2020.
References
- 1. Dhama K, Khan S, Tiwari R, et al. Coronavirus disease 2019-COVID-19. Clin Microbiol Rev 2020;33:e00028–20. 10.1128/CMR.00028-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altmann DM, Douek DC, Boyton RJ. What policy makers need to know about COVID-19 protective immunity. Lancet 2020;395:1527–9. 10.1016/S0140-6736(20)30985-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization . Global tuberculosis report 2019, 2019. Available: https://www.who.int/teams/global-tuberculosis-programme/tb-reports
- 4. Musa HH, Musa TH, Musa IH. Addressing Africa’s pandemic puzzle: perspectives on COVID-19 infection and mortality in sub-Saharan Africa. Int J Infect Dis 2020;S1201-9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Doshi P. Covid-19: do many people have pre-existing immunity? BMJ 2020;370:m3563. 10.1136/bmj.m3563 [DOI] [PubMed] [Google Scholar]
- 6. Le Bert N, Tan AT, Kunasegaran K, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020;584:457–62. 10.1038/s41586-020-2550-z [DOI] [PubMed] [Google Scholar]
- 7. Braun J, Loyal L, Frentsch M, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 2020;587:270–4. 10.1038/s41586-020-2598-9 [DOI] [PubMed] [Google Scholar]
- 8. Poland GA, Ovsyannikova IG, Kennedy RB. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet 2020;396:1595–606. 10.1016/S0140-6736(20)32137-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Spickett GP, ed. Oxford Handbook of Clinical Immunology and Allergy. 3 ed. Oxford: Oxford Medical publications, 2013. [Google Scholar]
- 10. Sethuraman N, Jeremiah SS, Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA 2020;323:2249–51. 10.1001/jama.2020.8259 [DOI] [PubMed] [Google Scholar]
- 11. Watson J, Whiting PF, Brush JE. Interpreting a covid-19 test result. BMJ 2020;369:m1808. 10.1136/bmj.m1808 [DOI] [PubMed] [Google Scholar]
- 12. He D, Zhao S, Lin Q, et al. The relative transmissibility of asymptomatic COVID-19 infections among close contacts. Int J Infect Dis 2020;94:145–7. 10.1016/j.ijid.2020.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bozkus CC. SARS-CoV-2-specific T cells without antibodies. Nat Rev Immunol 2020;20:463. 10.1038/s41577-020-0393-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lipsitch M, Grad YH, Sette A, et al. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat Rev Immunol 2020;20:709–13. 10.1038/s41577-020-00460-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020;181:e15:1489–501. 10.1016/j.cell.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sekine T, Perez-Potti A, Rivera-Ballesteros O, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 2020;183:158–68. 10.1016/j.cell.2020.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wyllie D, Mulchandani R, Jones HE. SARS-CoV-2 responsive T cell numbers are associated with protection from COVID-19: a prospective cohort study in key workers. medRxiv 2020. 10.1101/2020.11.02.20222778 [DOI] [Google Scholar]
- 18. Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021;371. doi: 10.1126/science.abf4063. [Epub ahead of print: 05 02 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Muecksch F, Wise H, Batchelor B, et al. Longitudinal serological analysis and neutralizing antibody levels in coronavirus disease 2019 convalescent patients. J Infect Dis 2021;223:389–98. 10.1093/infdis/jiaa659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kowitdamrong E, Puthanakit T, Jantarabenjakul W, et al. Antibody responses to SARS-CoV-2 in patients with differing severities of coronavirus disease 2019. PLoS One 2020;15:e0240502. 10.1371/journal.pone.0240502 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jclinpath-2021-207556supp001.pdf (163.2KB, pdf)
jclinpath-2021-207556supp002.pdf (35.1KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as online supplemental information. Deidentified participant data available as online supplemental information.


