Abstract
Objective
To systematically assess the diagnostic accuracy of rapid point-of-care tests for diagnosis of current SARS-CoV-2 infections in children under real-life conditions.
Design
Systematic review and meta-analysis.
Data sources
MEDLINE, Embase, Cochrane Database for Systematic Reviews, INAHTA HTA database, preprint servers (via Europe PMC), ClinicalTrials.gov, WHO ICTRP from 1 January 2020 to 7 May 2021; NICE Evidence Search, NICE Guidance, FIND Website from 1 January 2020 to 24 May 2021.
Review methods
Diagnostic cross-sectional or cohort studies were eligible for inclusion if they had paediatric study participants and compared rapid point-of care tests for diagnosing current SARS-CoV-2 infections with reverse transcription polymerase chain reaction (RT-PCR) as the reference standard. The Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) tool was used to assess the risk of bias and the applicability of the included studies. Bivariate meta-analyses with random effects were performed. Variability was assessed by subgroup analyses.
Results
17 studies with a total of 6355 paediatric study participants were included. All studies compared antigen tests against RT-PCR. Overall, studies evaluated eight antigen tests from six different brands. Only one study was at low risk of bias. The pooled overall diagnostic sensitivity and specificity in paediatric populations was 64.2% (95% CI 57.4% to 70.5%) and 99.1% (95% CI 98.2% to 99.5%), respectively. In symptomatic children, the pooled diagnostic sensitivity was 71.8% (95% CI 63.6% to 78.8%) and the pooled diagnostic specificity was 98.7% (95% CI 96.6% to 99.5%). The pooled diagnostic sensitivity in asymptomatic children was 56.2% (95% CI 47.6% to 64.4%) and the pooled diagnostic specificity was 98.6% (95% CI 97.3% to 99.3%).
Conclusions
The performance of current antigen tests in paediatric populations under real-life conditions varies broadly. Relevant data were only identified for very few antigen tests on the market, and the risk of bias was mostly unclear due to poor reporting. Additionally, the most common uses of these tests in children (eg, self-testing in schools or parents testing their toddlers before kindergarten) have not been addressed in clinical performance studies yet. The observed low diagnostic sensitivity may impact the planned purpose of the broad implementation of testing programmes.
PROSPERO registration number
CRD42021236313.
Keywords: diagnosis, pediatrics, COVID-19
Summary box.
What is already known on this topic?
Antigen tests are widely used to detect children with current SARS-CoV-2 infection in schools and kindergarten despite an ongoing debate on potential benefits and harms.
Sensitivity estimates of antigen tests in adult populations vary broadly and are substantially lower than reported by manufacturers; however, test performance in paediatric populations remained unknown.
What this study adds?
A systematic literature search and comprehensive author queries allowed the inclusion of 17 studies evaluating the diagnostic accuracy of antigen tests in children.
Real-life performance of current antigen tests for professional use in paediatric populations is below the minimum performance criteria set by WHO, the United States Food and Drug Administration, or the Medicines and Healthcare products Regulatory Agency (UK).
Performance of antigen tests for professional use in paediatric populations is simillar to what has been reported previously for adult populations. No evidence on the performance of self-tests in children was identified.
Summary box.
How might it impact clinical practice in the foreseeable future?
The observed low diagnostic sensitivity may impact the intended purpose of antigen tests in children.
Evidence gaps identified in this systematic review demonstrate current research needs to support evidence-based decision making. In particular, evidence is needed on the real-life performance of tests in schools (self-testing performed by children) and kindergarten (sample collection in toddlers by laypersons).
Introduction
Since the beginning of the COVID-19 pandemic caused by SARS-CoV-2, accurate, fast and early detection of people infected with SARS-CoV-2 followed by effective isolation measures of infected individuals has been considered a cornerstone in the global fight against the spread of SARS-CoV-2. Laboratory-based reverse transcription polymerase chain reaction (RT-PCR) testing is the standard for diagnosing current infections with SARS-CoV-2. However, limited testing capacities at many laboratories worldwide and limited availability of laboratories in developing countries has shown the urgent need for novel diagnostic tests that are easy to use, less expensive, widely available and suitable for point-of-care use. Today, such tests—and in particular antigen tests—are increasingly used to complement testing with RT-PCR to extend testing capacities or when a short turnaround time is essential.1 However, the advantages of antigen tests come at the price of lower diagnostic accuracy, most notably a lower diagnostic sensitivity, which increases the risk of missing cases, including those with pre-symptomatic infection who have yet to enter the most infectious period.2
Whether a lower sensitivity can be compensated by frequent testing remains a topic of controversial discussions.3–6 Additionally, the fact that sensitivity and specificity are not inherent test characteristics but are affected by various factors, including population characteristics, sample quality and study design, needs consideration.7 Data on diagnostic accuracy provided by antigen test manufacturers at market access are often overly optimistic and do not necessarily reflect the test’s performance in practice. Sensitivity of antigen tests in adult populations varies considerably across brands,8 with only a few tests meeting the minimum acceptable sensitivity of 80% or higher as defined by WHO or the United States Food and Drug Administration (US FDA).9 10
Because many countries are implementing public health safety measures that involve the use of antigen tests in adults and also in children, such as mass (self-)testing in schools,11 knowledge about how these tests perform in children is of high importance. However, to our knowledge, systematic reviews analysing the diagnostic test accuracy (DTA) of rapid tests in children are lacking. Therefore, in this systematic review and meta-analysis, we aimed to identify, assess and summarise the best available evidence on the real-life performance of rapid tests for diagnosing current SARS-CoV-2 infections in paediatric populations at the point of care.
Methods
The protocol for this systematic review was registered with PROSPERO (ID: CRD42021236313).12 The reporting adhered to the Preferred Reporting Items for Systematic reviews and Meta-Analyses of DTA studies (PRISMA-DTA) guideline13 and two relevant extensions ‘PRISMA-DTA for Abstracts’14 and ‘PRISMA-S for Reporting Literatures Searches in Systematic Reviews’.15
Eligibility criteria
We included diagnostic cross-sectional and cohort studies that evaluated the clinical performance of rapid point-of-care tests for detecting current SARS-CoV-2 infections against the reference standard in paediatric or mixed-age populations. Assessing analytical performance parameters such as the analytical sensitivity (limit of detection) or the analytical specificity (cross-reactivity) was not covered by the current review. Diagnostic case-control studies were excluded because they reflect the test’s performance under ideal conditions and, therefore, often overestimate the diagnostic accuracy.16 Moreover, studies evaluating serological tests were excluded because such tests are not suitable for the initial diagnosis of current SARS-CoV-2 infection.17 We considered a study as eligible if the study population comprised at least 10 paediatric study participants, each identified as positive or negative by the reference standard. In the absence of a true gold standard, laboratory-based RT-PCR alone or in combination with clinical findings or clinical follow-up was defined as the reference standard because it reflects the best available method for diagnosing individuals currently infected with SARS-CoV-2.7 Furthermore, we required reporting of data that allowed constructing a complete 2×2 contingency table. The full set of eligibility criteria is shown in online supplemental table S1 of Appendix 1. The decision rule for author queries is described in online supplemental appendix 2.
bmjebm-2021-111828supp001.pdf (7.5MB, pdf)
bmjebm-2021-111828supp002.pdf (111KB, pdf)
Information sources
We performed a comprehensive search for primary studies and secondary publications (systematic reviews and Health Technology Assessment (HTA) reports) in the following electronic bibliographic databases: MEDLINE (Ovid), Embase (Ovid), the Cochrane Library (Wiley), and preprint servers (Europe PMC) including medRxiv and bioRxiv (see Hamelers and Parkin18 for a full list of included preprint servers). Here, secondary publications were solely used as sources for potentially relevant studies. In addition, we searched two study registries (ClinicalTrials.gov and the WHO International Clinical Trials Registry Platform (ICTRP)) for relevant clinical studies. Other information sources comprised the International HTA Database, the Foundation of Innovative Diagnostics (FIND) COVID-19 website, and the Evidence Search and Guidance websites of Britain’s National Institute for Health and Care Excellence (NICE).
Search strategy
In accordance with the Cochrane Handbook for DTA Reviews,19 the search strategy included concepts addressing the index test and the target condition. The development of the search strategy followed an objective approach that involved text-analytic procedures to identify candidate search terms based on the method described by Hausner and colleagues.20 Further details are available in online supplemental appendix 2. The last search in bibliographical databases and study registries was conducted on 7 May 2021. Other information sources were last searched on 24 May 2021. All search strategies are provided in online supplemental appendix 3.
bmjebm-2021-111828supp003.pdf (114.3KB, pdf)
Study selection
The screening of literature retrieved from bibliographical databases involved a two-step screening procedure and was performed independently by two researchers using the web-based Trial Selection Database (webTSDB).21 In a first step, potentially eligible primary studies and secondary publications were identified from screening titles and abstracts of retrieved citations. In a second step, the full texts of these articles were obtained and evaluated. Publications that met the eligibility criteria were included. Any discrepancies were resolved by consensus between the two researchers before finalising each screening step. Reference lists of relevant systematic reviews and HTAs (independent of mentioning paediatric study participants) were manually screened to identify further relevant studies. For the screening of records from study registries, both screening steps were combined. Furthermore, documents identified through searching other information sources were screened for eligibility or information about potentially relevant studies.
Data collection
The individual steps of data collection and data extraction were performed by one researcher. All output was checked by a second researcher to ensure its validity and completeness. Any disagreements were resolved by consensus. See online supplemental appendix 2 for further details.
Quality assessment
We used the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) tool22 to evaluate the methodological quality and applicability of the included studies at the study level. The tool was tailored to our review by adding one signalling question and review-specific guidance was provided to facilitate judgments; see online supplemental appendix 4. The quality assessment of each included study was performed by one researcher. A second researcher verified all judgments. Any disagreements were resolved by consensus. The results were summarised in the text and visualised as a table and figure.
bmjebm-2021-111828supp004.pdf (131.9KB, pdf)
Diagnostic accuracy measures and data synthesis
For each included study, diagnostic sensitivity, diagnostic specificity, positive predictive value (PPV) and negative predictive value (NPV) with corresponding 95% CIs were calculated based on the extracted 2×2 tables. Individual study participants were used as the unit of analysis throughout this work. If a study reported repeat testing of individuals only the initial test was included in our analyses. If a study evaluated more than one test in the same study population, we reported all test evaluations, but only one randomly chosen test was included in the meta-analyses to avoid the necessity to adjust for multiplicity.
The meta-analyses were based on recommendations provided in the methodological guideline ‘Meta-analysis of diagnostic test accuracy studies’ by the European Network for Health Technology Assessment (EUnetHTA)23; see online supplemental appendix 2 for further details.
Results
Study selection
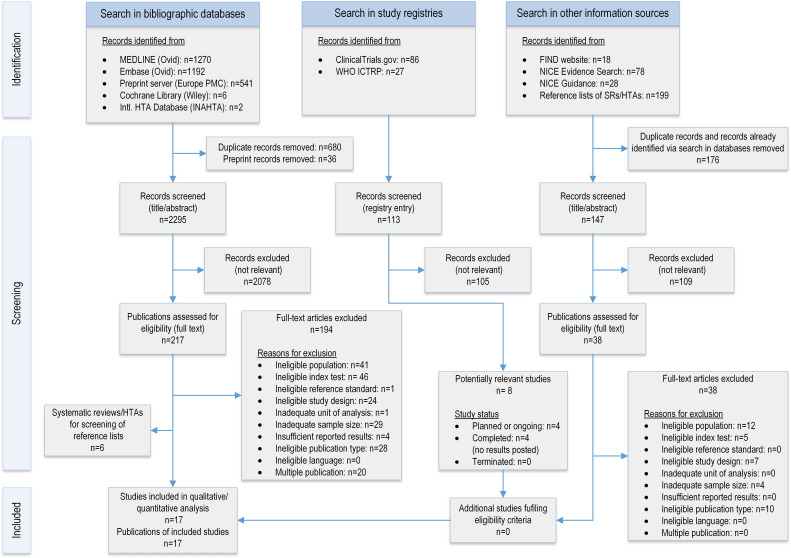
Overall, 3011 records were retrieved from five bibliographical databases. The PRISMA flow diagram is shown in figure 1 and outlines the process of identifying relevant studies from different information sources. References that were excluded at the full-text level can be found in online supplemental appendix 5 with the reason for their exclusion. After removing 36 preprint records identified via MEDLINE and 680 duplicate records, 2295 records were screened for eligibility; 2078 records were excluded at the title/abstract level. Full-text publications of 217 records were retrieved for further assessment. Nine studies24–32 met all eligibility criteria for inclusion. Furthermore, 21 studies33–53 were identified as eligible for author queries to obtain study data on paediatric subgroups. The authors of nine studies did not respond to our request for data.33 34 39 40 42 44–46 49 In four cases,36 47 52 53 the authors reported that the required number of individuals who tested positive or negative by the reference standard was not reached, and in one case43 no data on age were recorded. Eventually, author queries led to the inclusion of eight further studies,35 37 38 41 48 50 51 54 resulting in a total of 17 relevant studies for this review (12 peer-reviewed journal articles and five preprints). The full list of included studies is reported in online supplemental table S3 of Appendix 1.
Figure 1.
PRISMA (Preferred Reporting Items for Systematic reviews and Meta-Analyses) flow diagram showing the selection process of primary studies included in this systematic review and meta-analysis.
bmjebm-2021-111828supp005.pdf (260.7KB, pdf)
Furthermore, we screened 113 records identified from study registries and 323 records identified from other information sources. The search for studies in study registries allowed us to identify four planned or ongoing and four completed studies with no results posted, see online supplemental table S4 of Appendix 1 for further details. Information retrieval from other information sources included screening 18 records retrieved from the FIND website, 78 records from NICE Evidence Search, 28 records from NICE Guidance, and 23 records from reference lists of six systematic reviews8 55–59 identified via searching bibliographical databases. As a result, no additional study that met the inclusion criteria was identified.
Study characteristics
All 17 included studies (6355 paediatric study participants) evaluated the performance of antigen tests against the reference standard RT-PCR. The main study characteristics for each individual study are summarised in table 1, further details are reported in table 2 and online supplemental table S5 of Appendix 1. Fourteen studies evaluated the test performance in mixed-age populations (adults and children), including 24 to 928 paediatric study participants. Three studies with a sample size between 440 and 1620 individuals exclusively recruited children. In eight studies, the purpose of testing included diagnostic testing of individuals with symptoms suggestive of SARS-CoV-2 infection. Six studies reported the inclusion of individuals who were asymptomatic but were at increased risk of infection due to previous exposure to SARS-CoV-2. Here, ‘asymptomatic’ refers to any individual who is healthy, infected but pre-symptomatic or infected but without symptoms at the time of testing. Symptom status definitions were reported in nine of 16 studies that included individuals who were symptomatic. Individuals with at least one symptom (mostly self-reported) were considered symptomatic. Evaluating the performance of antigen tests in a screening setting (eg, community mass testing) was the main objective of six studies. Eight antigen tests (six lateral flow immunochromatographic assays and two fluorescent immunoassays) from six different brands were used in 18 test evaluations, whereas antigen tests by Abbott were most investigated (Panbio COVID-19 Ag Rapid Test n=6, BinaxNOW COVID-19 Ag Card n=5). In more than half of the test evaluations (n=11), nasopharyngeal samples were collected for the index test. Six test evaluations used anterior nasal specimens for the index test. In all studies, the reference standard was RT-PCR performed in a laboratory setting.
Table 1.
Characteristics of included studies. All studies aimed to identify individuals currently infected with SARS-CoV-2. The reference standard was RT-PCR performed in the laboratory setting (created by the authors)
| Study identifier | Publication status (year) |
Study design | Setting | Purpose of testing | Location (recruitment period) | Name of index test | Total number of study participants | Paediatric study participants | Funding/potential COI | |||
| (n) | Male (%) |
Symptomatic (%) |
Age (years) |
|||||||||
| Akingba*et al 35 | Preprint (2021) | Cross-sectional | Community testing site (mobile clinic) | D | South Africa (Nov 20) | Panbio COVID-19 Ag Rapid Test | 667 | 41 | 39.0 | 100† | Median: 13,‡ IQR: 10–16,‡ range: 3–17‡ | None/none |
| Bianco*et al 37 | Published (2021) | Cross-sectional | Hospital ED, hospital unit | nr | Italy (Oct–Dec 20) |
LumiraDx SARS-CoV-2 Ag Test | 907§ | 165 | nr | 9.7 | Mean: 7.2, range: 0–18 | None/none |
| Dřevinek* et al 38 | Preprint (2020) | Cross-sectional | Hospital TC | D, A, S | Czech Republic (Oct 20) |
1: Panbio COVID-19 Ag Rapid Test 2: Standard F COVID-19 FIA Ag |
591 | 31 | 38.7‡ | 32.3‡ | Median: 15,‡ IQR: 13.5–16,‡ range: 11–17‡ |
Public/none |
| González-Donapetry et al 24 | published (2021) | Cross-sectional | Hospital ED | D | Spain (Sept–Oct 20) |
Panbio COVID-19 Ag Rapid Test | 440 | 440 | 59.1 | 100† | Median: 3, IQR: 1–7, range:0–15 | None/none |
| Homza* et al 41 | Published (2021) | Cross-sectional | Hospital TC | D, A | Czech Republic (nr) |
ECOTEST COVID-19 Antigen Rapid Test | 494 | 24 | 58.3 | 45.8 | Mean: 13.17, SD: 2.79, range: 7–17 | Public/none |
| Kiyasu* †† et al 54 | Preprint (2021) | Cohort | Hospital TC | nr | Japan (Oct 20–Jan 21) |
QuickNavi COVID-19 Ag | 1881 | 90 | 68.9 | 4.4 | Median: 12,‡ IRQ: 6–15,‡ range: 0–17‡ | Private/yes |
| L’Huillier et al 25 | Preprint (2021) | Cross-sectional | Hospital TC | D, A, S | Switzerland (Nov 20–Mar 21) | Panbio COVID-19 Ag Rapid Test | 885 | 885 | 50.1 | 64.8 | Median: 11.8, IQR: 9.0–14.3, range: 0–16 | Public/none |
| Möckel et al 26 | Published (2021) | Cross-sectional | Hospital ED | D | Germany (Oct–Nov 20) |
Roche SARS-CoV-2 Rapid Antigen Test | 483 | 196‡‡ | 55 | 87.1 | Median: 3, IQR: 1–9 | Public/none |
| Pilarowski et al 27 | Published (2020) | Cross-sectional | Community testing site | S | USA (Nov–Dec 20) |
BinaxNOW COVID-19 Ag Card | 3320 | 209 | nr | nr 30.9¶ |
≤18 (47%<13) |
Public, private/yes |
| Pollock et al 28a | Published (2021) | Cross-sectional | Community testing site | S | USA (Oct–Dec 20) |
BinaxNOW COVID-19 Ag Card | 2482 | 928 | 47.0‡ | 10.7 | ≤18 (69%‡ ≤ 13) |
Public/none |
| Pollock et al 29b | Preprint (2021) | Cross-sectional | Community testing site | S | USA (Jan 21) |
CareStart COVID-19 Antigen test | 1603 | 253 | 46.2‡ | 12.6 | ≤18 (62%‡ ≤ 13) |
Public/none |
| Prince-Guerra et al 30 | Published (2021) | Cross-sectional | Community testing site | S | USA (Nov 20) |
BinaxNOW COVID-19 Ag Card | 3419 | 236 | nr | nr 24.2¶ |
Range: 10–17 | nr/none |
| Shah* et al 48 61 | Published (2021) | Cohort | Community testing site | S | USA (Nov–Dec 20) |
BinaxNOW COVID-19 Ag Card | 2024 | 217 | nr | 53.4‡ | Range: 5–17 | Public/none |
| Sood et al 31 | Published (2021) | Cross-sectional | Community testing site | S, A | USA (Nov–Dec 20) |
BinaxNOW COVID-19 Ag Card | 1429 | 783 | 49.5 | 23.5 | Range: 5–17, 65%: 5–12, 35%: 13–17 | Public, private/yes |
| Takeuchi*†† et al 50 | Published (2021) | Cohort | Hospital TC | D, A | Japan (Oct–Dec 20) |
QuickNavi COVID-19 Ag | 1208 | 164 | 61.6 | 54.9 | Median: 10,‡ IQR: 5-14, ‡ range: 0-17‡ | Private/yes |
| Torres* et al 51 | Published (2021) | Cross-sectional | Hospital TC | A | Spain (Oct–Nov 20) |
Panbio COVID-19 Ag Rapid Test | 634 | 73 | 47.9 | 0** | Median: 13, range: 9–17 | None/none |
| Villaverde et al 32 | Published (2021) | Cohort | Hospital ED | D | Spain (Sept–Oct 20) |
Panbio COVID-19 Ag Rapid Test | 1620 | 1620 | nr | 100† | Range: 0–16 | Public/none |
A: testing of asymptomatic individuals and at increased risk of infection due to previous exposure to SARS-CoV-2; D: diagnostic testing of symptomatic individuals; S: screening of individuals irrespective of symptoms (e.g. mass testing, pretravel testing).
*Study included due to unpublished paediatric study data obtained from author via author queries.
†Not explicitly stated, but as per inclusion criteria only study participants who are symptomatic.
‡Own calculation.
§Analysis population (total number of study participants not reported).
¶Value reported for whole study population.
**Not explicitly stated, but as per inclusion criteria only study participants who are asymptomatic.
††The study reported by Kiyasu et al 54 is specified as ‘extension study’ of a previous study reported by Takeuchi et al.50 Both studies were included in this systematic review and considered as two separate studies due to different but overlapping recruitment periods, substantial differences in the proportion of paediatric study participants who were symptomatic and differences in how discordant RT-PCR test results were re-evaluated.
‡‡The overall paediatric study population as defined by the authors consisted of n=202 individuals and included n=6 adult chaperones.
COI, conflicts of interest; Hospital ED, hospital emergency department; Hospital TC, hospital test centre; nr, not reported; RT-PCR, reverse transcription polymerase chain reaction.
Table 2.
Characteristics of the studies’ index test and reference standard. (created by the authors)
| Study identifier | Index test | Reference standard | ||||||
| Name (manufacturer) | Test method*/readout* | Target analyte* | Specimen type used in study | RT-PCR assay | Viral target | Positivity threshold | Specimen type used in study | |
| Akingba | Panbio COVID-19 Ag Rapid Test (Abbott) | LFA/visual | N protein | NP | Seegene nCoV assay | Three targets (not specified) | At least one target Ct value <38; inconclusive: two targets negative and one target positive with Ct value≥38 |
NP (same swab used for antigen test) |
| Bianco | LumiraDx SARS-CoV-2 Ag Test (LumiraDx) | Microfluidic FIA/automated | N protein | Nasal | XpertXpress SARS-CoV-2 assay (Cepheid)† | nr | nr | NP |
| Dřevinek | 1: Panbio COVID-19 Ag Rapid Test (Abbott) 2: Standard F COVID-19 FIA Ag (SD Biosensor) |
1: LFA/visual 2: FIA/automated |
1: N protein 2: N protein |
1: NP 2: NP |
Allplex SARS-nCoV-2 (Seegene) | N, E and RdRP/S genes | At least one target with Ct value≤40 | NP+OP |
| González-Donapetry | Panbio COVID-19 Ag Rapid Test (Abbott) | LFA/visual | N protein | NP | Vircell SARS-CoV-2 real-time PCR kit (Vircell) | N and E gene | Both targets with Ct value≤40 | NP |
| Homza | ECOTEST COVID-19 Antigen Rapid Test (Assure Tech) | LFA/visual | N protein | NP (nostril 1) | COVID-19 Multiplex RT-PCR Kit (Diana Biotechnologies) | S gene and gene coding the EndoRNAse | nr | NP (nostril 2) |
| Kiyasu | QuickNavi COVID-19 Ag (Denka) | LFA/visual | N protein | NP | RT-PCR (National Institute of Infectious Diseases, Japan)‡ | nr | nr | NP |
| L’Huillier | Panbio COVID-19 Ag Rapid Test (Abbott) | LFA/visual | N protein | NP | 1: Cobas SARS-CoV-2 assay (Roche) or 2: Nimbus RT-PCR assay | 1: nr 2: nr |
1: unclear 2: unclear |
NP |
| Möckel | Roche SARS-CoV-2 Rapid Antigen Test (SD Biosensors) | LFA/visual | N protein | ONP | 1: Cobas SARS-CoV-2 assay (Roche) or 2: SARS-CoV-2 E-gene assay (TibMolbiol) | 1: nr 2: E gene |
1: unclear 2: unclear |
ONP |
| Pilarowski | BinaxNOW COVID-19 Ag Card (Abbott) | LFA/visual | N protein | AN (bilateral) | RenegadeXP§ (RenegadeBio) | N gene | ‘No Ct cut-off’, Ct cut-off value=30 and 35 | AN (bilateral) |
| Pollock a | BinaxNOW COVID-19 Ag Card (Abbott) | LFA/visual | N protein | AN (bilateral) | CRSP SARS-CoV-2 Real-time RT-PCR Diagnostic Assay (MIT/Harvard) | N2 gene | Ct cut-off value=40 (in addition: 25, 30, 35) | AN (bilateral) |
| Pollock b | CareStart COVID-19 Antigen test (Access Bio) | LFA/visual | N protein | AN (bilateral) | CRSP SARS-CoV-2 Real-time RT-PCR Diagnostic Assay (MIT/Harvard) | N2 gene | Ct cut-off value=40 (in addition: 25, 30, 35) | AN (bilateral) |
| Prince-Guerra | BinaxNOW COVID-19 Ag Card (Abbott) | LFA/visual | N protein | AN (bilateral) | 1: CDC 2019-nCoV Real-time RT-PCR Diagnostic Panel or 2: Fosun COVID-19 RT-PCR Detection Kit | 1: nr 2: nr |
1: nr 2: nr |
NP (bilateral) |
| Shah | BinaxNOW COVID-19 Ag Card (Abbott) | LFA/visual | N protein | AN (nostril 1) | TaqPath SARS-CoV-2 Combo Kit (Thermo Fisher Scientific) | S, N and Orf1Ab genes | Ct value≤37 for at least two targets; inconclusive: one target positive | AN (nostril 2) |
| Sood | BinaxNOW COVID-19 Ag Card (Abbott) | LFA/visual | N protein | AN | Curative SARS-Cov-2 Assay (EUA for testing at KorvaLabs) | nr | Ct value≤40 | Oral fluid¶ |
| Takeuchi | QuickNavi COVID-19 Ag (Denka) | LFA/visual | N protein | NP | RT-PCR (National Institute of Infectious Diseases, Japan)** | nr | nr | NP |
| Torres | Panbio COVID-19 Ag Rapid Test (Abbott) | LFA/visual | N protein | NP (left nostril) | TaqPath SARS-CoV-2 Combo Kit (Thermo Fisher Scientific) | N gene | Ct value≤35 (in addition: ≤30, ≤25, ≤20) | NP (right nostril) |
| Villaverde | Panbio COVID-19 Ag Rapid Test (Abbott) | LFA/visual | N protein | NP | RT-PCR (not further specified) | E and RdRp genes | nr | NP |
*Technical specification taken from FIND Test Directory/manufacturer’s instructions for use if not reported in paper.
†XpertXpress SARS-CoV-2 assay has FDA EUA for both point-of-care and laboratory use (https://www.fda.gov/media/136316/download—accessed online: 20 June 2021). The authors reported that RT-PCR was performed ‘at the Microbiology and Virology unit’ of the ‘University Hospital Citta della Salute e della Scienza di Torino, Turin (Italy)’ which is, according to the authors, the ‘largest tertiary care facility in Europe’,37 therefore we considered the reference standard as eligible.
‡Classified as ‘reference real-time RT-PCR’ which was performed at the manufacturer’s site. RT-PCR was performed in an in-house microbiology laboratory. In case of discordant RT-PCR test results, a re-evaluation was performed ‘with a published for SARS-CoV-2’54. The final decision was based on the result of the latter RT-PCR test.
§PCR-positive results were confirmed by the "standard US Centers for Disease Control and Prevention methodology using Qiagen viral RNA purification kits and singleplex RT-PCR detection of the nucleoprotein gene".27
¶The FDA’s Accelerated Emergency Use Authorisation (EUA) Summary (https://www.fda.gov/media/137089/download—accessed online: 30 May 2021) states that the ‘collection of … oral fluid specimens is limited to symptomatic individuals within 14 days of COVID-19 symptom onset … Negative results for SARS-CoV-2 RNA from oral fluid specimens should be confirmed by testing of another specimen type authorised for use with this test if clinically indicated’.
**Classified as ‘reference real-time RT-PCR’50 which was performed at the manufacturer’s site. RT-PCR was performed in an in-house microbiology laboratory. In case of discordant PCR test results, a re-evaluation was performed using a BioFire Respiratory Panel 2.1 on the BioFire FilmArray system.
AN, anterior nasal (nares); Ct, cycle threshold; FIA, fluorescent immunoassay; LFA, lateral flow immunochromatographic assay; NP, nasopharyngeal; nr, not reported; ONP, oro-nasopharyngeal; OP, oropharyngeal; RT-PCR, reverse transcription polymerase chain reaction.
Risk of bias and applicability
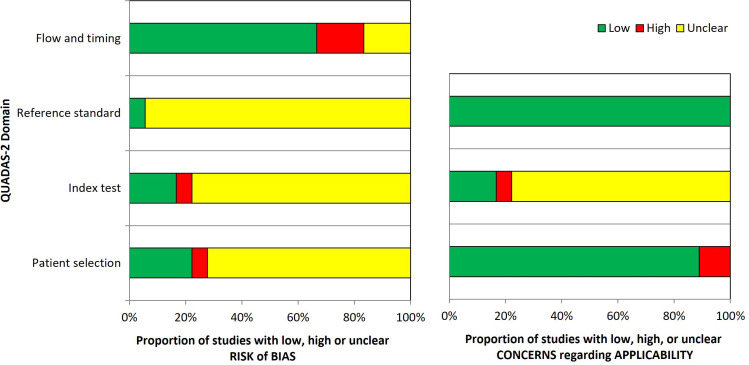
The results of the quality assessment are summarised in online supplemental table S6 of Appendix 1 and figure 2. Quality among studies varied. Only one study was at low risk of bias in all four domains of the QUADAS-2 tool. For patient selection, more than half of the studies were at high (n=1) or unclear (n=12) risk of bias because inadequate exclusion of participants occurred, or it was not clear whether a consecutive or random sample was enrolled into the study. All but one study was judged as having an unclear risk of bias for the reference standard due to insufficient reporting of blinding. Risk of bias in the flow and timing domain was high in three studies due to more than 5% of missing outcome data.
Figure 2.
QUADAS-2 (Quality Assessment of Diagnostic Accuracy Studies 2) risk of bias and applicability concerns. Graphical summary showing the review authors’ judgment about each domain as percentages across 18 test evaluations reported in 17 included studies.
Overall applicability concerns were high in three studies due to high concerns in either the patient selection or index test domain. Three studies were of low concern and the remaining 11 were rated unclear due to insufficient reporting in at least one domain.
Results of individual studies
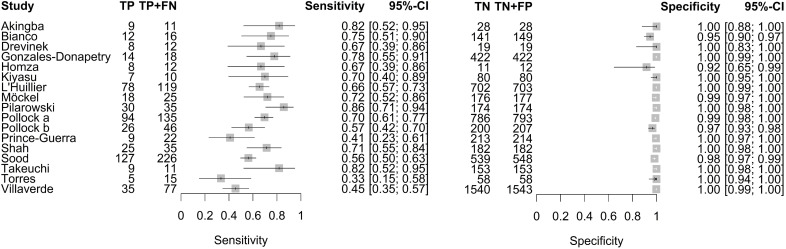
RT-PCR positivity rate, diagnostic sensitivity, diagnostic specificity as well as PPV and NPV (and their 95% CIs) of individual studies based on data from 2×2 contingency tables for paediatric populations are reported in online supplemental table S7 and figure 3. The RT-PCR positivity rate, which corresponds to the SARS-CoV-2 prevalence in the sample population, varied between 4.1% and 50%, with a median of 14,5% over n=17 studies. The sensitivity and specificity ranged from 33.3% to 85.7% and 91.7% to 100%, respectively. PPV and NPV ranged from 60.0% to 98.7% and 73.3% to 98.9%, respectively.
Figure 3.
Forest plot of sensitivity and specificity of antigen tests in entire paediatric study populations irrespective of symptoms. The point estimates of sensitivity and specificity from each study (identified by name of first author) are shown as squares; the corresponding 95% CIs are represented as horizontal lines. TP, true positive; FN, false negative; TN, true negative; FP, false positive.
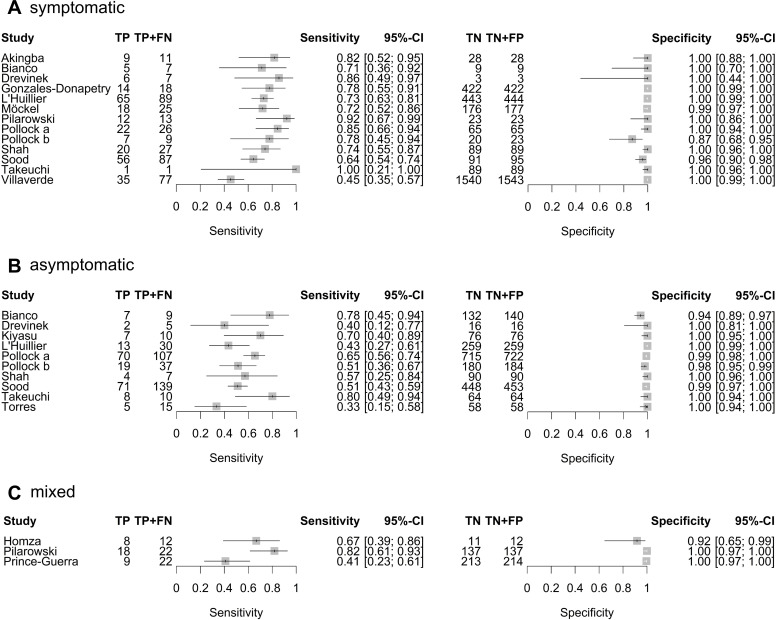
For individual studies, separate analyses for subgroups based on symptom status are reported in online supplemental tables S8-S10 of Appendix 1 and figure 4. Here, populations were defined as symptomatic or asymptomatic if at least 80% of paediatric study participants were reported as being symptomatic or asymptomatic at the time of testing, respectively. Mixed populations refer to populations with no predominant symptom status. RT-PCR positivity rates of the primary analysis population and the different subgroups based on symptom status are presented in online supplemental figure S1 of Appendix 1. Two studies38 41 were performed in high-prevalence populations with RT-PCR positivity rates of 38.7% and 50.0%, respectively. The median RT-PCR positivity rate was 13.2% (n=10 test evaluations) in asymptomatic populations, 13.8% in mixed populations (n=3 test evaluations) and 25.7% in symptomatic populations (n=13 test evaluations). Thus, the median RT-PCR positivity rate in symptomatic study populations was about 12 percentage points higher than in asymptomatic study populations, indicating a slight trend in the RT-PCR positivity rate with respect to the proportion of symptomatic subjects.
Figure 4.
Forest plot of sensitivity and specificity of antigen tests in (A) symptomatic, (B) asymptomatic and (C) mixed paediatric study populations. The point estimates of sensitivity and specificity from each study (identified by name of first author) are shown as squares; the corresponding 95% CIs are represented as horizontal lines. TP, true positive; FN, false negative; TN, true negative; FP, false positive.
Synthesis of results
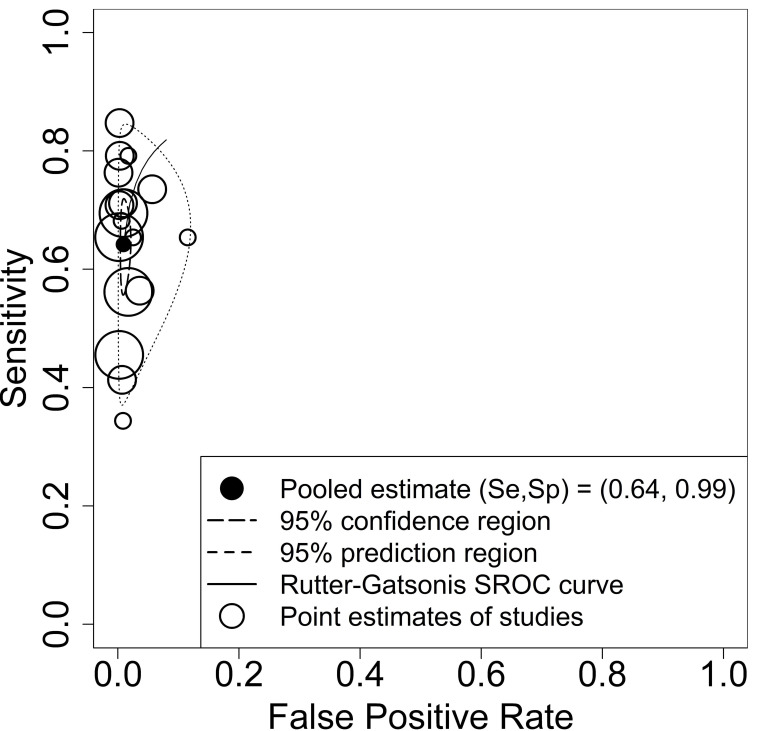
In our primary meta-analyses, we used data from 17 studies evaluating the diagnostic accuracy of antigen tests in paediatric participants. Estimated pooled sensitivity and specificity were 64.2% (95% CI 57.4% to 70.5%) and 99.1% (95% CI 98.2% to 99.5%), respectively. While the estimates for the sensitivity revealed high heterogeneity and thus justified the application of the bivariate model with random effects, the estimates for the specificity were limited to a small range, as shown in figure 5. Consequently, the estimated summary receiver operating characteristic (SROC) curve cannot be meaningfully interpreted. As prespecified in the protocol, we performed subgroup analysis evaluating the diagnostic accuracy according to symptom status. Estimated pooled sensitivity and specificity in asymptomatic children was 56.2% (95% CI 47.6% to 64.4%) and 98.6% (95% CI 97.3% to 99.3%), respectively, based on data from 2439 asymptomatic children in 10 studies. Estimated pooled sensitivity and specificity in symptomatic children was 71.8% (95% CI 63.6% to 78.8%) and 98.7% (95% CI 96.6% to 99.5%), respectively, based on data from 3413 symptomatic children in 13 studies. Estimated pooled sensitivity and specificity in the mixed population of symptomatic and asymptomatic children from three studies including 419 children was 63.4% (95% CI 37.3% to 83.5%) and 98.7% (95% CI 90.8% to 99.8%), respectively. The corresponding SROC curves are shown in online supplemental figure S2 of Appendix 1. The likelihood ratio test (LRT) for differences between the three groups revealed a p-value of pLRT=0.066. Since the bivariate meta-analysis might be influenced by the differences in the prevalence,60 we performed a bivariate meta-regression taking the prevalences within the studies directly into account. The LRT between the models without and with prevalence revealed a statistically significant p-value of pLRT=0.003. Results for the other subgroup analyses did not show relevant differences in the pooled estimates (setting pLRT=0.400; index test sample type pLRT=0.303; reference standard sample type pLRT=0.723; RT-PCR cycle threshold (Ct) cut-off value pLRT=0.105; publication status pLRT=0.551). The prediction regions of these analyses also showed a higher heterogeneity for sensitivity compared with specificity, see online supplemental figures S3-S6 of Appendix 1. Due to insufficient data, we did not perform subgroup analysis with respect to test type (antigen vs molecular) and end-user (layperson (self-testing) vs trained staff/healthcare worker). Except for one study48 61 where the testing procedure involved supervised self-collection of samples by study participants, in all other studies, testing was conducted by trained staff and/or healthcare workers (if reported). Univariate meta-analysis with random effects for sensitivity and specificity in cases where only a few studies were included (mixed population of symptomatic and asymptomatic children) did not show remarkable differences to the bivariate analysis. The results of all bivariate meta-analyses are summarised in table 3.
Figure 5.
Summary receiver operating characteristic (SROC) plot of sensitivity and specificity of antigen tests for diagnosis of current SARS-CoV-2 infections in entire paediatric study populations irrespective of symptoms. Each circle represents the point estimate of an individual study, whereas the size of the circle correlates with the number of paediatric study participants (small circle: 500 participants). The pooled estimate (black dot) of the pair of sensitivity (Se) and specificity (Sp) is surrounded by its 95% confidence region (closed curve with short dashes) and prediction region (closed curve with long dashes). The estimation of the SROC curve is based on the bivariate approach by Rutter and Gatsonis.77
Table 3.
Results of the bivariate meta-analyses: pooled sensitivity and specificity with 95% CI (created by the authors)
| Test evaluations included in analysis (n) | Paediatric study participants included in analysis (n) | Sensitivity (95% CI) |
Specificity (95% CI) |
|
| All studies | 17 | 6287 | 64.2 (57.4 to 70.5) | 99.1 (98.2 to 99.5) |
| Subgroup analysis | ||||
| (a) Symptom status | ||||
| - symptomatic population | 13 | 3407 | 71.8 (63.6 to 78.8) | 98.7 (96.6 to 99.5) |
| - asymptomatic population | 10 | 2431 | 56.2 (47.6 to 64.4) | 98.6 (97.3 to 99.3) |
| - mixed population | 3 | 419 | 63.4 (37.3 to 83.5) | 98.7 (90.8 to 99.8) |
| (b) Setting | ||||
| - community testing site | 8 | 2680 | 64.1 (54.7 to 72.6) | 98.7 (97.6 to 99.3) |
| - hospital test centre/emergency department | 9 | 3607 | 64.1 (53.8 to 73.2) | 99.4 (98.2 to 99.8) |
| (c) Sample type (index test) | ||||
| - nasopharyngeal | 10 | 3505 | 64.3 (54.7 to 73.0) | 99.4 (98.5 to 99.8) |
| - not nasopharyngeal | 7 | 2782 | 64.6 (54.4 to 73.7) | 98.5 (96.7 to 99.3) |
| (d) Sample type (reference standard) | ||||
| - nasopharyngeal | 11 | 3670 | 65.4 (56.3 to 73.5) | 99.1 (97.7 to 99.7) |
| - not nasopharyngeal | 6 | 2617 | 64.2 (53.1 to 74.0) | 98.9 (97.6 to 99.5) |
| (e) RT-PCR positivity threshold | ||||
| - Ct cut-off value=25 | 5 | 2062 | 92.4 (72.7 to 98.2) | 92.7 (85.4 to 96.5) |
| - Ct cut-off value=30 | 6 | 2271 | 83.3 (63.9 to 93.4) | 96.1 (91.8 to 98.2) |
| (f) Publication status | ||||
| - preprint | 5 | 1235 | 63.2 (55.6 to 70.3) | 98.9 (95.9 to 99.7) |
| - peer reviewed | 12 | 5052 | 64.3 (54.8 to 72.7) | 99.1 (98.1 to 99.6) |
Ct, cycle threshold; RT-PCR, reverse transcription-polymerase chain reaction.
Discussion
To our knowledge, this is the first systematic review that focused on evaluating the diagnostic accuracy of rapid point-of-care tests for current SARS-CoV-2 infections in paediatric populations. Our review comprises 17 studies with 18 evaluations of eight different antigen tests in children, whereas comprehensive author queries allowed us to include eight studies that did not provide sufficient data on paediatric study participants in their original study publication. We did not identify any evaluations of molecular-based tests that met our inclusion criteria confirming the current dominant role of antigen tests for rapid point-of-care usage.
Sensitivity estimates of antigen tests varied broadly among studies and were substantially lower than reported by manufacturers. However, one should note that the intended use of most tests is limited to symptomatic individuals. Thus, performance data reported by manufacturers usually refer to symptomatic individuals only. Less variation and only minor discrepancies to performance claims by manufacturers were observed for specificity estimates across studies. Taking into account test-specific pooled results, no test included in this review fully satisfied the minimum performance requirements as recommended by WHO9 (minimum sensitivity ≥80% and minimum specificity ≥97%), the US FDA10 (minimum sensitivity ≥80%, whereas a lower bound of the two-sided 95% CI above 70% is required for over-the-counter use self-tests62) or the Medicines and Healthcare products Regulatory Agency (MHRA) in the UK63 (minimum acceptable sensitivity ≥80% with two-sided 95% CI entirely above 70% and minimum acceptable specificity of 95% with two-sided 95% CI entirely above 90%). Limited performance was also observed in a recent laboratory-based study that evaluated the sensitivity of 122 of these antigen tests using common SARS-CoV-2 specimens with varying viral concentrations.64 Even under such ideal conditions, a wide range of sensitivities was observed, whereas 26 tests missed the study’s sensitivity criteria of 75% for specimens with high SARS-CoV-2 concentrations of around 106 SARS-CoV-2 RNA/ml and higher corresponding to a Ct value less than 25.
The bivariate meta-regression with respect to prevalence was statistically significant. This result is mirrored in the results of the subgroup analysis with respect to symptom status. While specificities were similarly high in symptomatic (98.7% with 95% CI 96.6% to 99.5%) and asymptomatic (98.6% with 95% CI 97.3% to 99.3%) populations, we observed a drop in sensitivity by about 15 percentage points in asymptomatic populations (56.2% with 95% CI 47.6% to 64.4%) compared with symptomatic populations (71.8% with 95% CI 63.6% to 78.8%). The better performance in symptomatic populations might be explained by changes in the viral load over the course of infection and the timing of the test: most symptomatic individuals were tested within 7 days of symptom onset in contrast to individuals who were asymptomatic at the time of testing with more variable disease onset, including individuals in the early (pre-symptomatic) or late stages of infection when viral loads are relatively low.65
As expected, the sensitivity increased when the positivity threshold of the reference standard was set to a lower Ct cut-off value of 30 or 25. However, such analyses should not be over-interpreted because Ct values are not standardised across systems or laboratories, making it difficult to directly compare results between different studies. Furthermore, while the Ct value from RT-PCR is a strong indicator of viral load, there is no specific cut-off viral load which allows distinguishing individuals as being infectious or not. As shown in online supplemental tables S11 and S12 of Appendix 1, an increase in sensitivity comes at the cost of a decrease in specificity as antigen tests also identify some individuals with moderate or low viral loads, who would then be considered as false positives. Despite some methodological differences (such as the stringency of inclusion criteria) and neglecting differences between included studies (eg, settings), the findings of our review are similar to those in the recent Cochrane Review by Dinnes et al 8 or the now published living systematic review by Brümmer et al.66 These similarities between paediatric and adult populations might be explained by the findings by Jones et al,67 who only identified minor differences in viral loads across age groups in a comprehensive analysis of more than 25 000 individuals who tested positive for SARS-CoV-2 by RT-PCR.
It is widely recognised that RT-PCR is an imperfect reference standard for identifying current SARS-CoV-2 infection. However, based on the guidance provided by Reitsma et al,68 we assume that this does not play a pivotal part in the context of DTA studies of SARS-CoV-2 antigen tests because antigen-based testing does not outperform RT-PCR-based molecular testing in terms of diagnostic accuracy.2 The observed analytical variability between RT-PCR assays that may affect the false-negative rate is considered negligible as the analytical sensitivity (limit of detection) of RT-PCR assays is several magnitudes higher than the analytical sensitivity of antigen tests. Furthermore, RT-PCR-based testing in low prevalence settings confirms the very high specificity of the method in practice. Any pre-analytic issues such as the quality of specimen collection, which may affect the diagnostic accuracy of RT-PCR, also apply to antigen tests.
For the current version of our review, publication bias is not considered relevant due to the novelty of the topic. No study included in our review was published before November 2020. All four completed studies that were identified through searching study registries were completed within the last 9 months. Of note is that for all but one study26 included in our review no entry in a study registry was reported.
Despite the roll-out of vaccines, testing continues to be a key to pandemic control. Particularly in populations with low vaccination rates or waning immunity, early identification of outbreaks will remain vital for controlling the spread of SARS-CoV-2. Consequently, multi-layered mitigation strategies will continue to involve screening tests of children in schools and kindergarten to avoid further closures. Whether this would still apply in populations with high childhood vaccination rates remains an open point for discussion.
The high specificity of antigen tests and the corresponding PPVs calculated for the paediatric study populations suggest that antigen testing might be a valuable tool to rapidly identify children with SARS-CoV-2 infection in moderate to high prevalence settings. However, at the same time, it is important to raise awareness that antigen tests should not be used to rule out SARS-CoV-2 infection (or infectiousness) because of their limited sensitivity. Whether increasing the frequency of antigen-based testing leads to an improved overall diagnostic accuracy that allows to effectively reduce transmission of SARS-CoV-2 has yet to be demonstrated in practice.69 The latter two aspects and the urgent need for high-quality screening tests probably led to the recent publication of a new target product profile by the MHRA in the UK,70 which includes increased performance requirements for self-tests to be used in national testing programmes that aim at detecting current SARS-CoV-2 infections in individuals without symptoms. Here, the minimum acceptable sensitivity for tests to ‘rule out’ a current infection is at least 97% with two-sided 95% CI entirely above 95%. The minimum acceptable specificity is 99.5% or higher with two-sided 95% CI entirely above 97%. Furthermore, it is stated that performance claims of repeated testing strategies require adequate clinical evidence rather than evidence from modelling studies only.
Other screening testing methods such as molecular-based pool testing, which involves RT-PCR testing of pooled samples and so-called deconvolution testing of individuals belonging to pools tested positive, are currently under investigation71–73 and may complement mere antigen testing, in particular in low prevalence settings. Additionally, novel tests, for example, lateral flow tests based on clustered regularly interspaced short palindromic repeat (CRISPR), hold great promise for a highly sensitive direct detection of SARS-CoV-274 but have yet to gain market access and demonstrate their value.
Limitations
Because we limited our search to studies published in English or German, it is possible that relevant studies were missed. The chosen implementation of the bivariate approach required a continuity correction for studies with zero cells in 2×2 tables. This approach may introduce bias into the results if multiple small studies with zero cells are included. With regard to bias, one should keep in mind that most studies were at unclear risk of bias in at least three of the four domains of the QUADAS-2 tool because of poor reporting. We acknowledge that infectiousness as a target condition is of higher practical relevance than current SARS-CoV-2 infection, which was chosen as the target condition of this review. While RT-PCR as the corresponding reference standard is a highly sensitive method that is used to detect the presence of viral RNA in a specimen, this does not necessarily indicate that infectious virus is present. Therefore, the actual transmission risk from individuals who tested RT-PCR positive remains unknown. Testing for infectiousness would allow to identify (and isolate) exclusively individuals who could pass the virus to others. However, while there have been attempts to use viral load (estimated from Ct values) or virus viability in cell culture as a proxy to determine individuals who are infectious, up to now, there is no adequate reference standard for infectiousness.69 We included only eight different antigen tests in our review. Thus, with more than 500 antigen tests for professional use that gained market access in the EU,75 the performance of most antigen tests under real-life conditions remains unknown. Sample collection in toddlers by laypersons or self-testing in schools performed by children are likely to influence the real-life test performance but were not addressed in any of the studies included in our review. Furthermore, one should keep in mind that diagnostic accuracy is only one factor affecting the effectiveness of testing programmes.76 We emphasise that all included studies were performed before market authorisation of COVID-19 vaccines for paediatric populations and most individuals identified as RT-PCR positive were likely infected with the wild-type of SARS-CoV-2. Diagnostic accuracy estimates reported in this review may not apply to future variants of SARS-CoV-2 or children who are vaccinated.
Conclusion
The performance of current antigen tests in paediatric populations under real-life conditions varies broadly. Relevant data were only identified for very few antigen tests on the market, and the risk of bias was mostly unclear due to poor reporting. Estimates of sensitivity and specificity and their 95% CIs from the bivariate meta-analyses indicate a subpar real-life performance of current antigen tests in children below the minimum performance criteria set by WHO, the US FDA or the MHRA in the UK. This may affect the planned purpose of the broad implementation of testing programmes. Up to now, the most common uses of these tests in children (eg, self-testing in schools or parents testing their toddlers before kindergarten) have not been addressed in clinical performance studies. Thus, it is of high relevance that these use cases are promptly investigated in independent studies. Moreover, the implementation of routine audits of testing programmes may allow monitoring of test performance in practice outside of studies.
Acknowledgments
We thank Dr Siw Waffenschmidt (IQWiG, Germany) for her valuable guidance during the development of the search strategies, the peer review of the final search strategy, and the literature screening technical support. Further, we are grateful to all study authors who replied to our author queries and who provided us with separate data on the paediatric study participants included in their studies. Finally, we thank the two anonymous peer reviewers and the editor for their critical reading of our manuscript and their valuable feedback.
Footnotes
Contributors: NFR: idea/conceptualisation, protocol, literature search (search strategies, screening), data extraction, quality assessment, data analysis, manuscript draft [lead author], manuscript review; LB: protocol, data extraction, quality assessment, data analysis, manuscript draft (contributor), manuscript review; YZ: protocol, literature search (screening), manuscript review; AV: supervision, protocol, manuscript review. All authors read and approved the final manuscript. The lead author (NFR) is the manuscript’s guarantor and affirms that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned and registered have been explained. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information. The data supporting the findings of this review are available within the article or its appendices The R code used for this review is available from the corresponding author (NFR) on request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study does not involve human participants.
References
- 1. Crozier A, Rajan S, Buchan I, et al. Put to the test: use of rapid testing technologies for covid-19. BMJ 2021;372:n208. 10.1136/bmj.n208 [DOI] [PubMed] [Google Scholar]
- 2. Guglielmi G. Fast coronavirus tests: what they can and can't do. Nature 2020;585:496–8. 10.1038/d41586-020-02661-2 [DOI] [PubMed] [Google Scholar]
- 3. Mina MJ, Parker R, Larremore DB. Rethinking Covid-19 Test Sensitivity - a strategy for containment. N Engl J Med 2020;383:e120. 10.1056/NEJMp2025631 [DOI] [PubMed] [Google Scholar]
- 4. Kmietowicz Z. Covid-19: controversial rapid test policy divides doctors and scientists. BMJ 2021;372:n81. 10.1136/bmj.n81 [DOI] [PubMed] [Google Scholar]
- 5. Larremore DB, Wilder B, Lester E, et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci Adv 2021;7. 10.1126/sciadv.abd5393. [Epub ahead of print: 01 01 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moreno GK, Braun KM, Pray IW. SARS-CoV-2 transmission in intercollegiate athletics not fully mitigated with daily antigen testing. Clin Infect Dis Off Publ Infect Dis Soc Am 2021. 10.1101/2021.03.03.21252838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. CDC . Interim guidance for antigen testing for SARS-CoV-2. Cent. Dis. Control Prev 2021. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html [Google Scholar]
- 8. Dinnes J, Deeks JJ, Adriano A, et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev 2020;8:CD013705. 10.1002/14651858.CD013705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Emergency Prepardness, WHO Headquarters . Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays. interim guidance, 2020. Available: https://www.who.int/publications-detail-redirect/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays [Accessed 12 Jun 2021].
- 10. U.S. food and drug administration, antigen template for test developers, 2020. Available: https://www.fda.gov/media/137907/download [Accessed 12 Jun 2021].
- 11. Torjesen I. What do we know about lateral flow tests and mass testing in schools? BMJ 2021;372:n706. 10.1136/bmj.n706 [DOI] [PubMed] [Google Scholar]
- 12. Fujita-Rohwerder N, Zens Y, Beckmann L. Diagnostic accuracy of rapid point-of-care tests for diagnosis of current SARS-CoV-2 infections in children: a systematic review. PROSPERO 2021;2021:CRD42021236313. https://www.crd.york.ac.uk/prospero/display_record.php?RecordID=236313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Salameh J-P, Bossuyt PM, McGrath TA, et al. Preferred reporting items for systematic review and meta-analysis of diagnostic test accuracy studies (PRISMA-DTA): explanation, elaboration, and checklist. BMJ 2020;370:m2632. 10.1136/bmj.m2632 [DOI] [PubMed] [Google Scholar]
- 14. Cohen JF, Deeks JJ, Hooft L, et al. Preferred reporting items for Journal and conference Abstracts of systematic reviews and meta-analyses of diagnostic test accuracy studies (PRISMA-DTA for Abstracts): checklist, explanation, and elaboration. BMJ 2021;372:n265. 10.1136/bmj.n265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rethlefsen ML, Kirtley S, Waffenschmidt S, et al. PRISMA-S: an extension to the PRISMA statement for reporting literature searches in systematic reviews. Syst Rev 2021;10:39. 10.1186/s13643-020-01542-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rutjes AWS, Reitsma JB, Di Nisio M, et al. Evidence of bias and variation in diagnostic accuracy studies. Can Med Assoc J 2006;174:469–76. 10.1503/cmaj.050090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deeks JJ, Dinnes J, Takwoingi Y, et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev 2020;6:CD013652. 10.1002/14651858.CD013652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hamelers A, Parkin M. A full text collection of COVID-19 preprints in Europe PMC using JATS XML, 2021. https://www.ncbi.nlm.nih.gov/books/NBK569517/ [Google Scholar]
- 19. de Vet H, Eisinga A, Riphagen I. Chapter 7: Searching for Studies. In: Cochrane Handbook for systematic reviews of diagnostic test accuracy. The Cochrane Collaboration, 2008. [Google Scholar]
- 20. Hausner E, Waffenschmidt S, Kaiser T, et al. Routine development of objectively derived search strategies. Syst Rev 2012;1:19. 10.1186/2046-4053-1-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hausner E, Ebrahim S, Herrmann-Frank A. Study selection by means of a web-based Trial Selection DataBase (webTSDB). In: Abstracts of the 19th Cochrane Colloquium. Madrid, Spain: John Wiley & Sons, 2011. [Google Scholar]
- 22. Whiting PF, Rutjes AWS, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529–36. 10.7326/0003-4819-155-8-201110180-00009 [DOI] [PubMed] [Google Scholar]
- 23. European Network for Health Technology Assessment (EUnetHTA) . Meta-Analysis of diagnostic test accuracy studies, 2014. Available: https://eunethta.eu/wp-content/uploads/2018/01/Meta-analysis-of-Diagnostic-Test-Accuracy-Studies_Guideline_Final-Nov-2014.pdf [Accessed 10 Jan 2021].
- 24. González-Donapetry P, García-Clemente P, Bloise I, et al. Think of the children: evaluation of SARS-CoV-2 rapid antigen test in pediatric population. Pediatr Infect Dis J 2021;40:385–8. 10.1097/INF.0000000000003101 [DOI] [PubMed] [Google Scholar]
- 25. L'Huillier AG, Lacour M, Sadiku D, et al. Diagnostic accuracy of SARS-CoV-2 rapid antigen detection testing in symptomatic and asymptomatic children in the clinical setting. J Clin Microbiol 2021;59:e0099121. 10.1128/JCM.00991-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Möckel M, Corman VM, Stegemann MS, et al. SARS-CoV-2 antigen rapid immunoassay for diagnosis of COVID-19 in the emergency department. Biomarkers 2021;26:213–20. 10.1080/1354750X.2021.1876769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pilarowski G, Marquez C, Rubio L. Field performance and public health response using the BinaxNOW TM rapid SARS-CoV-2 antigen detection assay during community-based testing. Clin Infect Dis 2021;73:e3098. 10.1093/cid/ciaa1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pollock NR, Jacobs JR, Tran K, et al. Performance and implementation evaluation of the Abbott BinaxNOW rapid antigen test in a high-throughput Drive-through community testing site in Massachusetts. J Clin Microbiol 2021;59. 10.1128/JCM.00083-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pollock NR, Tran K, Jacobs JR, et al. Performance and operational evaluation of the access bio CareStart rapid antigen test in a high-throughput Drive-through community testing site in Massachusetts. Open Forum Infect Dis 2021;8. 10.1093/ofid/ofab243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Prince-Guerra JL, Almendares O, Nolen LD, et al. Evaluation of Abbott BinaxNOW Rapid Antigen Test for SARS-CoV-2 Infection at Two Community-Based Testing Sites - Pima County, Arizona, November 3-17, 2020. MMWR Morb Mortal Wkly Rep 2021;70:100–5. 10.15585/mmwr.mm7003e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sood N, Shetgiri R, Rodriguez A, et al. Evaluation of the Abbott BinaxNOW rapid antigen test for SARS-CoV-2 infection in children: implications for screening in a school setting. PLoS One 2021;16:e0249710. 10.1371/journal.pone.0249710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Villaverde S, Domínguez-Rodríguez S, Sabrido G, et al. Diagnostic accuracy of the Panbio severe acute respiratory syndrome coronavirus 2 antigen rapid test compared with reverse-transcriptase polymerase chain reaction testing of nasopharyngeal samples in the pediatric population. J Pediatr 2021;232:287–9. 10.1016/j.jpeds.2021.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abdulrahman A, Mustafa F, AlAwadhi AI. Comparison of SARS-COV-2 nasal antigen test to nasopharyngeal RT-PCR in mildly symptomatic patients. medRxiv 2020. 10.1101/2020.11.10.20228973v2 [DOI] [Google Scholar]
- 34. Agulló V, Fernández-González M, Ortiz de la Tabla V, Tabla Odela, et al. Evaluation of the rapid antigen test Panbio COVID-19 in saliva and nasal swabs in a population-based point-of-care study. J Infect 2021;82:186–230. 10.1016/j.jinf.2020.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Akingba OL, Sprong K, Hardie DR. Field performance evaluation of the PanBio rapid SARS-CoV-2 antigen assay in an epidemic driven by 501Y.v2 (lineage B.1.351) in the eastern Cape, South Africa. medRxiv 2021. 10.1101/2021.02.03.21251057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berger A, Nsoga MTN, Perez-Rodriguez FJ, et al. Diagnostic accuracy of two commercial SARS-CoV-2 antigen-detecting rapid tests at the point of care in community-based testing centers. PLoS One 2021;16:e0248921. 10.1371/journal.pone.0248921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bianco G, Boattini M, Barbui AM, et al. Evaluation of an antigen-based test for hospital point-of-care diagnosis of SARS-CoV-2 infection. J Clin Virol 2021;139:104838. 10.1016/j.jcv.2021.104838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dřevínek P, Hurych J, Kepka Z, et al. The sensitivity of SARS-CoV-2 antigen tests in the view of large-scale testing. Epidemiol Mikrobiol Imunol 2021;70:156–60. 10.1101/2020.11.23.20237198v1 [DOI] [PubMed] [Google Scholar]
- 39. Favresse J, Gillot C, Oliveira M, et al. Head-To-Head comparison of rapid and automated antigen detection tests for the diagnosis of SARS-CoV-2 infection. J Clin Med 2021;10:265. 10.3390/jcm10020265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fournier P-E, Zandotti C, Ninove L, et al. Contribution of VitaPCR SARS-CoV-2 to the emergency diagnosis of COVID-19. Journal of Clinical Virology 2020;133:104682. 10.1016/j.jcv.2020.104682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Homza M, Zelena H, Janosek J, et al. Covid-19 antigen testing: better than we know? A test accuracy study. Infect Dis 2021;53:661–8. 10.1080/23744235.2021.1914857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kilic A, Hiestand B, Palavecino E. Evaluation of performance of the BD Veritor SARS-CoV-2 chromatographic immunoassay test in patients with symptoms of COVID-19. J Clin Microbiol 2021;59. 10.1128/JCM.00260-21. [Epub ahead of print: 20 04 2021]. 10.1128/JCM.00260-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Landaas ET, Storm ML, Tollånes MC, et al. Diagnostic performance of a SARS-CoV-2 rapid antigen test in a large, Norwegian cohort. Journal of Clinical Virology 2021;137:104789. 10.1016/j.jcv.2021.104789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marti JLG, Gribschaw J, McCullough M. Differences in detected viral loads guide use of SARS-CoV-2 antigen-detection assays towards symptomatic college students and children. medRxiv 2021. 10.1101/2021.01.28.21250365 [DOI] [Google Scholar]
- 45. Masiá M, Fernández-González M, Sánchez M, et al. Nasopharyngeal Panbio COVID-19 antigen performed at point-of-care has a high sensitivity in symptomatic and asymptomatic patients with higher risk for transmission and older age. Open Forum Infect Dis 2021;8:ofab059. 10.1093/ofid/ofab059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Merino P, Guinea J, Muñoz-Gallego I, et al. Multicenter evaluation of the Panbio™ COVID-19 rapid antigen-detection test for the diagnosis of SARS-CoV-2 infection. Clin Microbiol Infect 2021;27:758–61. 10.1016/j.cmi.2021.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nsoga MTN, Kronig I, Rodriguez FJP. Diagnostic accuracy of PanbioTM rapid antigen tests on oropharyngeal swabs for detection of SARS-CoV-2. medRxiv 2021. 10.1101/2021.01.30.21250314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shah MM, Salvatore PP, Ford L. Performance of repeat BinaxNOW SARS-CoV-2 antigen testing in a community setting, Wisconsin, November-December 2020. Clin Infect Dis 2021. 10.1093/cid/ciab309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shrestha B, Neupane AK, Pant S, et al. Sensitivity and specificity of lateral flow antigen test kits for COVID-19 in asymptomatic population of quarantine centre of Province 3. Kathmandu Univ. Med. J. 2020;18:36–9. 10.3126/kumj.v18i2.32942 [DOI] [PubMed] [Google Scholar]
- 50. Takeuchi Y, Akashi Y, Kato D, et al. The evaluation of a newly developed antigen test (QuickNavi™-COVID19 Ag) for SARS-CoV-2: a prospective observational study in Japan. J Infect Chemother 2021;27:890–4. 10.1016/j.jiac.2021.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Torres I, Poujois S, Albert E, et al. Evaluation of a rapid antigen test (Panbio™ COVID-19 Ag rapid test device) for SARS-CoV-2 detection in asymptomatic close contacts of COVID-19 patients. Clin Microbiol Infect 2021;27:636.e1–636.e4. 10.1016/j.cmi.2020.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Torres I, Poujois S, Albert E, et al. Point-Of-Care evaluation of a rapid antigen test (CLINITESTⓇ rapid COVID-19 antigen test) for diagnosis of SARS-CoV-2 infection in symptomatic and asymptomatic individuals. J Infect 2021;82:e11–12. 10.1016/j.jinf.2021.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Turcato G, Zaboli A, Pfeifer N, et al. Clinical application of a rapid antigen test for the detection of SARS-CoV-2 infection in symptomatic and asymptomatic patients evaluated in the emergency department: a preliminary report. J Infect 2021;82:e14–16. 10.1016/j.jinf.2020.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kiyasu Y, Takeuchi Y, Akashi Y, et al. Prospective analytical performance evaluation of the QuickNavi™-COVID19 Ag for asymptomatic individuals. J Infect Chemother 2021;27:1489–92. 10.1016/j.jiac.2021.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brümmer LE, Katzenschlager S, Gaeddert M, et al. Accuracy of novel antigen rapid diagnostics for SARS-CoV-2: a living systematic review and meta-analysis. PLoS Med 2021;18:e1003735. 10.1371/journal.pmed.1003735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee J, Song Jae‐Uk. Diagnostic accuracy of the Cepheid Xpert Xpress and the Abbott ID now assay for rapid detection of SARS‐CoV‐2: a systematic review and meta‐analysis. J Med Virol 2021;93:4523–31. 10.1002/jmv.26994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mustafa Hellou M, Górska A, Mazzaferri F, et al. Nucleic acid amplification tests on respiratory samples for the diagnosis of coronavirus infections: a systematic review and meta-analysis. Clin Microbiol Infect 2021;27:341–51. 10.1016/j.cmi.2020.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yong Chua PE, Gwee SXW, Wang MX, et al. SARS-CoV-2 diagnostic tests for reopening of borders: a systematic review and meta-analysis. SSRN Journal (Published Online First: 27 January 2021). 10.2139/ssrn.3774144 [DOI] [Google Scholar]
- 59. Yoon SH, Yang S, Cho H, et al. Point-Of-Care testing for the detection of SARS-CoV-2: a systematic review and meta-analysis. Eur Rev Med Pharmacol Sci 2021;25:503–17. 10.26355/eurrev_202101_24422 [DOI] [PubMed] [Google Scholar]
- 60. Hoyer A, Kuss O. Meta-Analysis of diagnostic tests accounting for disease prevalence: a new model using trivariate copulas. Stat Med 2015;34:1912–24. 10.1002/sim.6463 [DOI] [PubMed] [Google Scholar]
- 61. Ford L, Whaley MJ, Shah MM. Characteristics of children and antigen test performance at a SARS-CoV-2 community testing site. medRxiv 2021. 10.1101/2021.07.06.21259792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Food and Drug Administration (FDA) . Supplemental template for developers of molecular and antigen diagnostic COVID-19 tests for screening with serial testing, 2021. Available: https://www.fda.gov/media/146695/download [Accessed 26 Jun 2021].
- 63. Healthcare products regulatory agency (MHRA) . Target product profile: point of care SARS-CoV-2 detection tests, 2021. Available: https://www.gov.uk/government/publications/how-tests-and-testing-kits-for-coronavirus-covid-19-work/target-product-profile-point-of-care-sars-cov-2-detection-tests [Accessed 27 Jun 2021].
- 64. Scheiblauer H, Filomena A, Nitsche A, et al. Comparative sensitivity evaluation for 122 CE-marked rapid diagnostic tests for SARS-CoV-2 antigen, Germany, September 2020 to April 2021. Euro Surveill 2021;26:2100441. 10.2807/1560-7917.ES.2021.26.44.2100441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Walsh KA, Jordan K, Clyne B, et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect 2020;81:357–71. 10.1016/j.jinf.2020.06.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brümmer LE, Katzenschlager S, Gaeddert M, et al. Accuracy of novel antigen rapid diagnostics for SARS-CoV-2: a living systematic review and meta-analysis. PLoS Med 2021;18 10.1371/journal.pmed.1003735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jones TC, Biele G, Mühlemann B, et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science 2021;373:eabi5273. 10.1126/science.abi5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Reitsma JB, Rutjes AWS, Khan KS, et al. A review of solutions for diagnostic accuracy studies with an imperfect or missing reference standard. J Clin Epidemiol 2009;62:797–806. 10.1016/j.jclinepi.2009.02.005 [DOI] [PubMed] [Google Scholar]
- 69. Royal Statistical Society . Diagnostic tests Working Group report, 2021. Available: https://rss.org.uk/RSS/media/File-library/Policy/2021/RSS-Diagnostic-tests-report-FINAL.pdf [Accessed 10 Jun 2021].
- 70. Medicines & Healthcare products Regulatory Agency (MHRA) . Target product profile: in vitro diagnostic (IVD) self-tests for the detection of SARS-CoV-2 in people without symptoms. target Prod. profile vitro Diagn. IVD Self-Tests detect. SARS-CoV-2 people symptoms, 2021. Available: https://www.gov.uk/government/publications/how-tests-and-testing-kits-for-coronavirus-covid-19-work/target-product-profile-in-vitro-diagnostic-ivd-self-tests-for-the-detection-of-sars-cov-2-in-people-without-symptoms [Accessed 10 Jul 2021].
- 71. Joachim A, Dewald F, Suárez I, et al. Pooled RT-qPCR testing for SARS-CoV-2 surveillance in schools - a cluster randomised trial. EClinicalMedicine 2021;39:101082. 10.1016/j.eclinm.2021.101082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pollock NR, Berlin D, Smole SC, et al. Implementation of SARS-CoV2 screening in K–12 schools using In-School pooled molecular testing and deconvolution by rapid antigen test. J Clin Microbiol 2021;59:JCM.:01123–21. 10.1128/JCM.01123-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tsang NNY, So HC, Ng KY, et al. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: a systematic review and meta-analysis. Lancet Infect Dis 2021;21:1233–45. 10.1016/S1473-3099(21)00146-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Broughton JP, Deng X, Yu G, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol 2020;38:870–4. 10.1038/s41587-020-0513-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Federal Institute for drugs and medical devices (BfArM) . List of antigen tests for professional use for direct pathogen detection of the coronavirus SARS-CoV-2. Available: https://antigentest.bfarm.de/ords/f?p=ANTIGENTESTS-AUF-SARS-COV-2 [Accessed 06 Jul 2021].
- 76. Ferrante di Ruffano L, Hyde CJ, McCaffery KJ, et al. Assessing the value of diagnostic tests: a framework for designing and evaluating trials. BMJ 2012;344:e686. 10.1136/bmj.e686 [DOI] [PubMed] [Google Scholar]
- 77. Rutter CM, Gatsonis CA. A hierarchical regression approach to meta-analysis of diagnostic test accuracy evaluations. Stat Med 2001;20:2865–84. 10.1002/sim.942 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjebm-2021-111828supp001.pdf (7.5MB, pdf)
bmjebm-2021-111828supp002.pdf (111KB, pdf)
bmjebm-2021-111828supp003.pdf (114.3KB, pdf)
bmjebm-2021-111828supp004.pdf (131.9KB, pdf)
bmjebm-2021-111828supp005.pdf (260.7KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information. The data supporting the findings of this review are available within the article or its appendices The R code used for this review is available from the corresponding author (NFR) on request.