Abstract
We have developed a PCR assay that is capable of amplifying kinetoplast DNA (kDNA) of Leishmania donovani in a species-specific manner among Old World leishmanias. With Indian strains and isolates of L. donovani the assay was sensitive enough to detect kDNA in an amount equivalent to a single parasite or less. The extreme sensitivity of the assay was reflected in its ability to detect parasite DNA from small volumes of peripheral blood of patients with kala-azar (KA) and from skin lesions from patients with post-KA dermal leishmaniasis (PKDL). A total of 107 clinical leishmaniasis samples were analyzed. Of these 102 (95.3%) were positive by PCR. The test provided a diagnosis of KA with 96% sensitivity using patient whole-blood samples instead of bone marrow or spleen aspirates that are obtained by invasive procedures. The assay was also successful in the diagnosis of 45 of 48 PKDL cases (93.8%). Cross-reactions with pathogens prevalent in the area of endemicity, viz., Mycobacterium tuberculosis, Mycobacterium leprae, and Plasmodium spp., could be ruled out. Eighty-one control samples, including dermal scrapings from healthy portions of skin from patients with PKDL were all negative. Two of twenty controls from the area of endemicity were found positive by PCR assay; however, there was a good possibility that these two were asymptomatic carriers since they were serologically positive for KA. Thus, this PCR assay represents a tool for the diagnosis of KA and PKDL in Indian patients in a noninvasive manner, with simultaneous species identification of parasites in clinical samples.
The protozoan parasites of the genus Leishmania are the causative agents of a group of diseases called leishmaniases, endemic in more than 80 countries worldwide. Considerable morbidity and mortality occur in the visceral infections termed visceral leishmaniasis (VL) or kala-azar (KA), which is a significant infectious disease in the developing world and of late in the developed world because of increased international travel and human immunodeficiency virus (HIV) infection. KA is a symptomatic infection of the liver, spleen, and bone marrow caused by organisms of Leishmania donovani complex. The annual incidence and prevalence of VL cases worldwide are 0.5 million and 2.5 million respectively. Of these 90% of cases occur in India, Nepal, Bangladesh, and Sudan (9). The official estimate of 430,000 VL cases in Bihar State of India over the past 11 years may represent only a fraction of the real numbers. The actual number is believed to be at least five times as great (8). The causative organism in the Indian subcontinent and Africa is L. donovani donovani, while in the Mediterranean basin and South America it is L. donovani infantum.
Post-KA dermal leishmaniasis (PKDL) is an unusual dermatosis that develops as a sequel of KA, producing gross cutaneous lesions in the form of hypopigmented macules, erythema, and nodules. The disease is relatively common in the Indian subcontinent and less frequent in East Africa but exceptional in the American and European continents (25). The need to search for cases of PKDL and treat them as a part of KA control programs has been emphasized since patients with PKDL provide the only known reservoir for the parasite in India (37). The cost and toxicity of current treatment regimens emphasize the importance of establishing control strategies and make diagnosis and typing of leishmaniasis critical (20). Detection and characterization of Leishmania from patients with KA or PKDL are important for deciding treatment regimens as well as for understanding the disease epidemiology. Current diagnostic methods based on parasite detection (stained smears, culture, and histopathology) and immunological methods (direct agglutination test, enzyme-linked immunosorbent assay [ELISA], etc.) have several limitations, including low sensitivity and specificity. Procedures for demonstration of the parasite in spleen or bone marrow in KA and in skin lesions in PKDL are invasive and often not sensitive enough. Immunological methods fail to distinguish between past and present infections and are not reliable in the case of immunocompromised patients (6, 11, 39). Furthermore, neither of these methods addresses the problem of species identification, which is important for determining appropriate treatment regimens and designing control measures. Procedures involving the use of monoclonal antibodies, isoenzyme and schizodeme analysis, and DNA hybridization have to be resorted to (12, 17, 29). Most of these procedures are tedious and require massive cultures of parasites. There is, therefore, an urgent need to develop diagnostic procedures that are simple, sensitive, and specific.
In recent years PCR-based diagnostic methods have been described for leishmaniasis, with a wide range of sensitivity and specificity. An excellent target for a sensitive and rapid detection method is the kinetoplast mini-circle DNA, which is present at thousands of copies per cell. The mini-circles have been used as targets for selective amplification of parasite DNA in various studies (5, 7, 21, 28, 35). The identification of conserved sequence elements represented within the kinetoplast DNA (kDNA) of a given species of Leishmania would allow the design of oligonucleotide primers to be used for species-specific identification of parasites in clinical samples. We have analyzed kDNA sequences from Old World leishmanias and designed primers specific for L. donovani species to detect kDNA from a single parasite in the presence of huge excesses of human DNA. The utility of the primers designed for L. donovani has been examined in clinical samples from patients with KA and PKDL in India. The PCR test was found to be sensitive enough to detect parasite DNA from peripheral blood of patients with KA and from skin lesions of patients with PKDL. Furthermore, the test was specific for L. donovani species of the parasite, leading to simultaneous species identification of the parasite.
MATERIALS AND METHODS
Patients.
Fifty-one KA patients hailing from Bihar and reporting to Safdarjung Hospital (SJH), New Delhi, India, were included in the study at the pretreatment stage. The patients presented with characteristic symptoms of KA such as fever, hepatosplenomegaly, anemia, and leukopenia. Only those patients for whom the diagnosis of KA was confirmed by demonstration of parasites in bone marrow aspirates were taken for the study. Blood was taken from all 51 patients. In addition bone marrow samples were obtained from 8 of these patients. Clinical samples were also taken from a total of 48 patients with PKDL that were originally from Bihar and reported to the Dermatology Department of SJH during the period from 1996 to 2000. Forty-five of these reported history of KA, while the remaining three were not aware of it. The time elapsed after cure from KA in the 45 patients ranged from 1 to 15 years. Clinical diagnosis in 36 patients was based on condition characterized by erythematous indurate areas and papulonodular and hypochromic macules in a bilateral distribution. The remaining 12 patients had a predominantly macular presentation, most of them being the subject of a recent study (26). Slit skin smears stained with Giemsa stain were positive in only 10 cases. Histopathological findings upon skin biopsy were similar to those reported earlier (19, 31). The dermis showed a diffuse infiltration by lymphocytes, histiocytes, and plasma cells. All patients responded well to therapy with sodium antimony gluconate. The control group of patients was comprised of individuals with confirmed cases of malaria (n = 15), pulmonary tuberculosis (n = 15), and lepromatous leprosy (n = 32) from SJH. Twenty healthy volunteers living in an area of endemicity (Muzaffarpur, Bihar) were also included in the control group.
Parasites.
Ten World Health Organization reference strains of Leishmania originating from distinct geographic locations were used in the study. These included L. donovani DD8 (MHOM/IN/80/DD8) from India, L. donovani AG83 (MHOM/IN/83/AG83) from India, L. donovani 1S (MHOM/SD/00/1S-C12D) from Sudan, L. donovani WR 684 (MHOM/ET/67/82) from Ethiopia, L. donovani infantum (MCAN/SP/00/XXX) from Spain, L. tropica WR 683 (MHOM/SU/58/OD) from the Soviet Union, L. tropica WR 664 (MHOM/SU/74/K27) from the Soviet Union, L. major WR662 (MHOM/IL/67/Zericho II/WR662) from Israel, L. major LV39 (MRHO/SU/59/P/LV39) from the Soviet Union, and L. major ASKH (MHOM/SU/73/5ASKH) from the Soviet Union. Three isolates of L. donovani (MHOM/IN/94/IICB6, MHOM/IN/94/IICB7, and MHOM/IN/94/IICB8) were kindly provided by D. Sarkar, Indian Institute of Chemical Biology, Calcutta, India. These were isolated from patients with VL (IICB6 and IICB8) and PKDL (IICB7) originating from Bihar and characterized as L. donovani (10). Ten parasite isolates were set up in culture in our laboratory over the last 2 years from patients with VL and PKDL reporting to SJH. All parasite cultures were set up and propagated in medium 199 supplemented with 25 mM HEPES, pH 7.5, and 10% fetal calf serum (30). Parasites were harvested in late log phase and washed in phosphate-buffered saline prior to DNA isolation.
Sample collection and DNA isolation.
Bone marrow and skin scrapings were collected in NET buffer (150 mM NaCl, 15 mM Tris-HCl [pH 8.30], 1 mM EDTA). Blood was collected in heparinized tubes. Samples were transported to the laboratory at ambient temperature, except for blood collected in areas of endemicity, in which case they were brought on ice. Samples were transferred to 4°C and generally processed on the same day. Blood (0.2 to 1 ml) was treated with RBC lysis buffer (114 mM sodium phosphate [pH 8.0], 1 mM NH4Cl), and the buffycoat was isolated. DNA from parasite cultures as well as from clinical samples (skin scrapings, bone marrow, or blood) was isolated by overnight lysis in NET buffer with proteinase-K (100 μg/ml) and 1% sodium dodecyl sulfate. DNA was extracted by phenol-chloroform extraction and ethanol precipitation. In a few samples DNA was isolated from 0.2 ml of blood using a QIAamp DNA blood minikit (Qiagen) in order to determine if this method provided any advantage over the phenol-chloroform method for DNA extraction.
Oligonucleotide primers.
The 792-bp L. donovani kinetoplast mini-circle sequence (accession no. Y11401) was analyzed using PC-Gene software, and appropriate primers were identified. The two primers used were 5′-AAATCGGC TCCGAGGCGGGAAAC-3′ and 5′-GGTACACTCTATCAGTAGCAC-3′, together designated the LdI primers. These were synthesized using an Applied Biosystems DNA-RNA synthesizer (model 394). The LdI primers amplify a fragment of approximately 600 bp that is seen on the gels.
PCR amplification.
DNA from cultured parasites (1 ng) and from clinical samples (100 ng) was taken for amplification using the LdI primers described above. The reaction mixture (50 μl) contained 10 mM Tris-HCl (pH 8.3) 50 mM KCl, 1.5 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, 50 ng of each primer, and 1.25 U of Taq DNA polymerase (Gibco BRL). Each reaction mixture was overlaid with mineral oil, and amplification was performed in a thermal cycler (Perkin-Elmer, Warrington, Great Britain) programmed for 40 cycles of denaturation at 94°C for 1 min, annealing at 45°C for 1 min, and extension at 72°C for 2 min, preceded by an initial denaturation of 2 min at 94°C. Final extension was for 3 min at 72°C. Products were analyzed by electrophoresis in 1% agarose gel containing ethidium bromide (0.5 μg/ml) in TAE buffer (0.04 M Tris acetate, 0.001 M EDTA) and photographed under UV illumination.
Southern blot analysis.
PCR products were analyzed in 1% agarose gel, and Southern blot analysis was done as described (14). Southern blots were hybridized with 32P-labeled cloned L. donovani kDNA fragments using the conditions described (14).
Sequencing reaction.
The PCR amplification products from culture isolates and clinical samples from patients with KA and PKDL were cloned into the pGEMT-Easy vector system (Promega). DNA sequencing was performed with the ABI PRISM Dye Terminator Cycle sequencing kit and an ABI PRISM automated sequencer (Model 377; Perkin-Elmer). Briefly, the sequencing reaction mixture contained terminator ready reaction mix, DNA template, primer and 5% dimethyl sulfoxide. Dimethyl sulfoxide was added to keep the DNA template denatured since leishmania DNA has a high GC content. The PCR was carried out in a DNA thermal cycler (model 480). The PCR conditions in the Perkin-Elmer analytical manual were followed. Sequences were assembled and edited using Sequencher software (Gene Codes Corporation, Ann Arbor, Mich.) and analyzed with Mac Vector DNA and protein sequence analysis software (Genetics Computer Group Inc., Madison, Wis.).
RESULTS
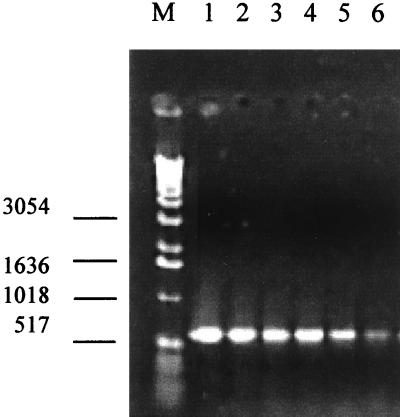
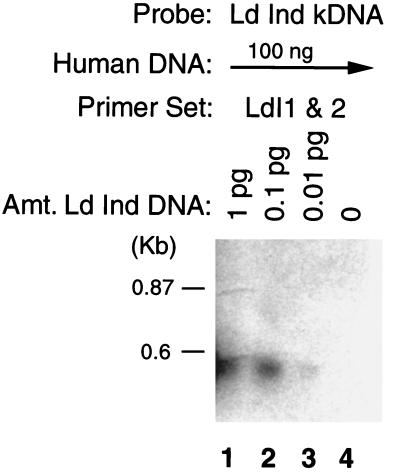
The initial aim of the study was to define a set of PCR primers based on kDNA sequences which would allow sensitive and specific detection of L. donovani. For this purpose, we analyzed kDNA mini-circle sequences from an L. donovani DD8 strain of Indian origin and designed oligonucleotide primers that showed lack of cross-reactivity with organisms phylogenetically or geographically related (G. P. Pogue and H. L. Nakhasi, unpublished data). The sensitivity and effectiveness of the PCR-based detection system was seen in its ability to amplify kDNA fragments from as little as 1 fg of DNA of L. donovani (Fig. 1). When the amplification properties of PCR were combined with the specificity and sensitivity of Southern blot-based DNA hybridization, kDNA fragments could be detected by probes generated from the parasite kDNA sequences in PCRs containing as little as 10 fg of Leishmania DNA diluted in a 10 million-fold excess of human DNA (Fig. 2).
FIG. 1.
Sensitivity of the PCR assay. Shown are the results of PCR amplification of the serially diluted L. donovani (DD8) DNA analyzed on agarose gels. DNA was extracted from parasite cultures and amplified as described in Materials and Methods. Lane M, 1 kb Ladder (Gibco BRL); lane 1, 10 ng of DNA; lane 2, 1 ng of DNA; lane 3, 10 pg of DNA; lane 4, 1 pg of DNA; lane 5, 10 fg of DNA; lane 6, 1 fg of DNA.
FIG. 2.
Sensitivity of PCR amplification of Leishmania kDNA followed by Southern blot analysis. The PCR contained 100 ng of human genomic DNA and the indicated amount of total DNA from L. donovani DD8. The PCR product was probed with parasite kDNA and exposed for about 1 h. Lane 4 represents a PCR containing only human DNA as a control.
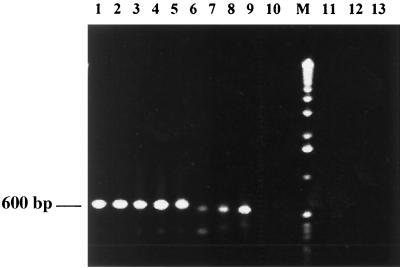
Initially the primers were evaluated with various strains of Old World Leishmania as described under Materials and Methods. Both strains of L. donovani of Indian origin gave positive results by PCR, as did the three isolates from Indian patients (Fig. 3, lanes 1 to 5). These three isolates of L. donovani were isolated 6 years prior to the present study from patients with KA and PKDL and were preserved in the Parasite Bank at the IICB. Strains of L. donovani from Sudan and Ethiopia as well as L. donovani infantum from Spain were positive by PCR, though the bands were of significantly lower intensity (Fig. 3, lanes 6 to 8). DNA from L. major and L. tropica was not amplified, indicating the species specificity of primers (Fig. 3, lanes 9 and 10). Species specificity for L. donovani was further established, since the use of DNA up to 10 ng from three different strains of L. major and two strains of L. tropica as described in Materials and Methods did not give any amplification. Specificity of the primers was also evaluated using DNA (10 ng) from microorganisms causative of the common infectious diseases prevalent in India, such as Plasmodium spp., Mycobacterium leprae, Mycobacterium tuberculosis; there was no amplification with DNA from any of these organisms using our primers (Fig. 3, lanes 11 to 13).
FIG. 3.
Amplification of parasite DNA from various strains and isolates of Leishmania. DNA (1 ng) isolated from parasite cultures was subjected to PCR and analyzed. Lane 1, L. donovani AG83; lane 2, L. donovani DD8; lane 3, L. donovani IICB8; lane 4, L. donovani IICB6; lane 5, L. donovani IICB 7 (PKDL origin); lane 6, L. donovani 1S; lane 7, L. donovani WR684; lane 8 L. donovani infantum; lane 9, L. tropica WR683; lane 10, L. major LV 39, lane M, 1-kb ladder, lane 11, Plasmodium; lane 12, M. leprae; lane 13, M. tuberculosis.
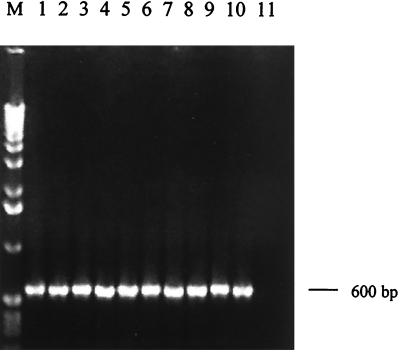
In order to establish the clinical utility of the assay, PCR amplification was evaluated with DNA from several recent isolates of the parasite. Parasite cultures were set up from bone marrow aspirates of five patients with KA that reported to SJH over the last 2 years (designated KA1 to KA5). DNA isolated from each of these cultures was observed to be amplified by PCR (Fig. 4, lanes 1 to 5). The assay was also positive with a number of cultures isolated from dermal lesions of patients with PKDL (PK1 to PK5) (Fig. 4, lanes 6 to 10), while the parasite culture isolated from a patient with cutaneous leishmaniasis hailing from Afghanistan gave no amplification in the PCR test (Fig. 4, lane 11). The sensitivity of the assay with the isolates of KA and PKDL was found to be 1 fg of total DNA.
FIG. 4.
DNA amplification from recent field isolates of KA and PKDL. DNA (1 ng) extracted from cultures of parasite isolates was used for PCR amplification. Lanes: M, 1-kb ladder; 1, KA-1; 2, KA-2; 3, KA-3; 4, KA-4; 5, KA-5; 6, PK-1; 7, PK-2; 8, PK-3; 9, PK-4; 10, PK-5; 11, isolate from a patient with cutaneous leishmaniasis.
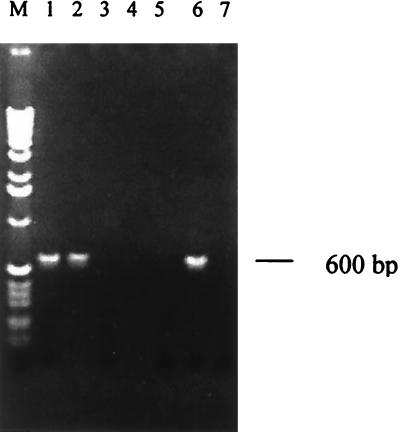
A clinical study was undertaken with Indian patients with KA and PKDL using PCR based on LdI primers. The PCR assay was evaluated with clinical samples from patients with KA and PKDL along with samples from suitable controls. PCR analysis of representative sample from each of test materials—i.e., bone marrow and whole blood from patients with KA, blood from patients with malaria and tuberculosis, blood from control subjects from areas of endemicity, and skin lesion samples from patients with PKDL and leprosy—is shown in Fig. 5. Only samples of bone marrow and blood from patients with KA and from PKDL skin lesion specimens were PCR positive (Fig. 5, lanes 1, 2, and 6). The rest of the samples were negative (Fig. 5, lanes 3 to 5 and 7). All eight samples of bone marrow aspirates of patients with KA gave positive results when subjected to PCR amplification (Table 1). Our results showed that the primers could specifically amplify DNA from peripheral blood of 49 of 51 patients with KA. Identical results were obtained by PCR using DNA extracted by the phenol-chloroform method or with a QIAamp DNA blood minikit, indicating that either method could be employed. DNA from just 0.2 ml of patient's blood was found to be sufficient for the PCR test, indicating the tremendous clinical usefulness of the test. All malaria (n = 15) and tuberculosis (n = 15) blood samples were negative, while two of the 20 samples from control subjects from areas of endemicity were positive by PCR (Table 1). A large majority of specimens from patients with PKDL (45 of 48) gave positive results, while all the specimens from patients with leprosy (32 of 32) were negative. Samples of normal dermal tissue from unaffected parts of skin of patients with PKDL (n = 19) were also negative (Table 1). Sequence analysis of the PCR product obtained with DNA from clinical samples (for KA, blood; for PKDL, tissue) as well as from parasite isolates from patients with KA and PKDL revealed that the sequence of the products was identical to that obtained with the DD8 strain of L. donovani (Fig. 6).
FIG. 5.
PCR assay with clinical samples of KA and PKDL. DNA (100 ng) isolated from clinical samples was used for PCR amplification. Lane M, 1-kb ladder, lane 1, KA (bone marrow); lane 2, KA (blood); lane 3, malaria (blood); lane 4, tuberculosis (blood); lane 5, control from the area of endemicity (blood); lane 6, PKDL (skin lesion); lane 7, leprosy (lesion).
TABLE 1.
Results of PCR assay with clinical samples from patients with KA and PKDL and controls
| Source of DNAa | Samples
|
||
|---|---|---|---|
| Total no. | No. PCR positive | % PCR positive | |
| KA (bone marrow) | 8 | 8 | 100 |
| KA (blood) | 51 | 49 | 96 |
| Malaria (blood) | 15 | 0 | 0 |
| Tuberculosis (blood) | 15 | 0 | 0 |
| Controls (blood) | 20 | 2 | 10 |
| PKDL (lesions) | 48 | 45 | 93.8 |
| Leprosy (lesions) | 32 | 0 | 0 |
| PKDL (Healthy tissue) | 19 | 0 | 0 |
The disease and sample type (in parentheses) are given. Controls, control subjects from areas of endemicity.
FIG. 6.
Sequence of PCR product with DNA isolated from L. donovani DD8 strain and isolates and clinical samples from patients with KA and PKDL. PCR products obtained with DNA isolated from L. donovani DD8 strain, parasite isolates from patients with KA and PKDL (two each) and clinical samples (two each of KA blood and PKDL tissue) were subjected to sequence analysis. Identical sequences were obtained for the PCR products in each case, which matched exactly with the published sequence of a 792-bp kDNA mini-circle segment of the DD8 strain of L. donovani (GenBank accession no. Y11401). The positions of primers are indicated in boldface type.
DISCUSSION
We have developed a PCR assay that is species specific for L. donovani kDNA among the Old World leishmanias and could detect the parasite in a highly sensitive manner in clinical samples from Indian patients with KA and PKDL. The assay could detect as little as 1 fg of parasite DNA from Indian strains of L. donovani, an amount that represents the equivalent of approximately 0.1 parasite (13). DNAs from several parasite isolates obtained from patients with KA as well as PKDL originating from the region of endemicity in India were found to be amplified with equal sensitivity. Therefore, the assay is theoretically capable of detecting a single parasite in a biological sample. The extreme sensitivity of our detection system was evident by its ability to amplify parasite DNA from peripheral blood of patients with KA and dermal lesions of patients with PKDL in a large majority of cases. A total of 107 clinical samples from patients with leishmaniasis were examined, and 95% tested positive by PCR. Our PCR described in this work yielded a unique product of approximately 600 bp, and no nonspecific side product or artifacts appeared on the gel. It had the advantage that results were easily and unequivocally interpreted upon analysis of agarose gels. The high level of sensitivity was reflected by the ability of the assay to detect parasite DNA in peripheral blood of patients with KA with 96% sensitivity in the 51 cases examined. Use of peripheral blood is advantageous because the collection procedure is less invasive and safer than the splenic or bone marrow biopsy specimen collection. In earlier studies for diagnosis of VL due to L. donovani, the sensitivity of PCR for blood samples has been found to be in the range of 45 to 94% based on smaller sample sizes ranging from 17 to 42 (1, 4, 15, 21, 22, 32, 35). For detection of VL due to L. donovani infantum, which may have a different pathogenesis, sensitivities between 64 to 97% have been reported with blood samples (16, 18, 21). The sensitivity of detection was 100% in the limited number of bone marrow samples that we examined. Bone marrow is known to have a high load of parasites, while in peripheral blood the parasites are relatively scarce. Studies reporting PCRs with detection sensitivities comparable to ours (less than a single parasite) did not obtain sensitivities as high as that of our assay when using blood samples of patients with KA (15, 35). With clinical samples the sensitivity in practice may be affected by factors such as accessibility of the DNA in parasite-containing biopsy samples and the conditions used in the PCR amplification.
DNA isolated from the pathogens causative of common coendemic diseases (M. leprae, M. tuberculosis, and Plasmodium) was not amplified. Blood from patients with malaria and tuberculosis were PCR negative in all cases (30 of 30), while two of the blood samples from control subjects from areas of endemicity were PCR positive, giving an overall specificity of 96% in the control blood samples examined. The two positive controls from areas of endemicity were relatives of patients with KA and possibly asymptomatic carriers since both samples were positive by ELISA with recombinant antigen k39 and in a dipstick test using immunochromatographic strips coated with rk39 antigen (P. Salotra and G. Sreenivas, unpublished data), tests reported to be specific for KA (34, 36). A recent study has reported a PCR assay that could often detect parasitemia a few weeks before the appearance of any clinical signs or symptoms (16).
In India, 10 to 20% percent of patients apparently cured of KA develop PKDL. As there is no known animal reservoir in India, patients with PKDL are considered an important source of transmission in recent epidemics of KA in India (37). The disease is easily confused with a number of skin disorders, primarily leprosy, due to similarities in the clinical presentation, therefore, a high level of clinical expertise is needed to diagnose PKDL. Detection of L. donovani bodies in skin lesions by microscopy gives a positive result in only about 58% of cases, as parasites are scanty (33). Early recognition and treatment of PKDL would contribute significantly to the control of KA, as patients with PKDL constitute a reservoir for the Leishmania parasite (37, 38). Our assay, validated in a large number of cases, provided a highly sensitive method for diagnosis of PKDL. The sensitivity of the assay was 93.8% for PKDL, which is significantly higher than that reported (82.7%) in a recent study with 32 patients with PKDL in Sudan (23). The specificity of the test was 100%, as all of the control tissues examined (32 leprosy lesions and 19 dermal samples from healthy regions of skin of patients with PKDL were negative.
Species specificity of the assay was carefully evaluated by testing DNA from different strains and species of Old World Leishmania. The assay was found to be positive with several World Health Organization reference strains of L. donovani originating from distinct geographical regions. L. donovani from Ethiopia and Sudan and L. donovani infantum from Spain gave PCR products of identical size but of comparatively lower intensity, probably due to lower copy numbers of the target kDNA sequence. Variations among L. donovani strains from different geographic regions have also been detected by random amplified polymorphic DNA PCR (3) and arbitrary primed PCR analysis (24). The primers were found to be species specific for L. donovani, as DNA from two other Leishmania species examined (L. major and L. tropica) was not amplified. One clinical isolate of L. tropica from a patient with cutaneous leishmaniasis was also negative, while several clinical isolates from patients with KA and PKDL were all positive. The PCR products amplified from clinical samples of patients with KA and PKDL showed nucleotide sequences identical to those of the cultured parasites.
Our PCR provides a useful tool for the simultaneous typing of parasites while diagnosis is being performed on clinical samples. Such a tool is necessary to complement diagnostic assays since most of them do not furnish the taxonomic information about the parasite required to determine the appropriate therapeutic regimens and control measures. Early detection and simultaneous typing would enable implementation of specific treatment. Leishmania is increasingly recognized as an opportunistic pathogen during coinfection with HIV (2, 27). Since incidence of HIV infection is on the increase in India, cases of coinfection with Leishmania are likely to present in future. In such cases immunological tests have particularly low sensitivity, and our assay would provide a rapid detection as well as species identification of Leishmania. Since this method is rapid and reproducible, we believe that it can be used for the reliable identification and characterization of cultured parasites. The test has other potential values in detecting and typing parasites in vectors for epidemiological surveys and in retrospective studies of archival material.
ACKNOWLEDGMENTS
We thank Shyam Sundar, Banaras Hindu University, Varanasi, India, for helping us procure blood samples from controls in areas of endemicity and Dwijen Sarkar, IICB, for providing strains and isolates of Leishmania. We acknowledge the expert technical assistance of P. D. Sharma.
Part of this work was supported by a grant from the Indo-U.S. Vaccine Action Program.
REFERENCES
- 1.Adhya S, Chatterjee M, Hassan M Q, Mukherjee S, Sen S. Detection of Leishmania in the blood of early kala-azar patients with the aid of polymerase chain reaction. Trans R Soc Trop Med Hyg. 1995;89:622–624. doi: 10.1016/0035-9203(95)90416-6. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht H, Sobottka I, Emminger C, Jablonowski H, Just G, Stoehr A, Kubin T, Salzberger B, Lutz T, Lunzen J V. Leishmaniasis and HIV. Arch Pathol Lab Med. 1996;120:189–198. [PubMed] [Google Scholar]
- 3.Andresen K, Gaafar A, El Hassan A M, Ismail A, Dafalla M, Theander T G, Kharazmi A. Evaluation of the polymerase chain reaction in the diagnosis of cutaneous leishmaniasis due to Leishmania major. A comparison with direct microscopy of smears and sections from lesions. Trans R Soc Trop Med Hyg. 1996;90:133–135. doi: 10.1016/s0035-9203(96)90112-1. [DOI] [PubMed] [Google Scholar]
- 4.Andresen K, Gasim S, El Hassan A M, Khalil E A, Barker D C, Theander T G, Kharazmi A. Diagnosis of visceral leishmaniasis by polymerase chain reaction using blood, bone marrow and lymph node samples from patients from the Sudan. Trop Med Int Health. 1997;2:440–444. [PubMed] [Google Scholar]
- 5.Aviles H, Belli A, Armijos R, Monroy F P, Harris E. PCR detection and identification of Leishmania parasites in clinical specimens in Ecuador: a comparison with classical diagnostic methods. J Parasitol. 1999;85:181–187. [PubMed] [Google Scholar]
- 6.Badaro R, Benson D, Eulalio M C, Freire M, Cunha S, Netto E M, Sampaio D P, Madureira C, Burns J M, Houghton R L, David J R, Reed S G. rK39: A cloned antigen of Leishmania chagasi that predicts active visceral leishmaniasis. J Infect Dis. 1996;173:758–761. doi: 10.1093/infdis/173.3.758. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharyya R, Das K, Sen S, Roy S, Majumder H K. Development of a genus specific primer set for detection of Leishmania parasites by polymerase chain reaction. FEMS Microbiol Lett. 1996;135:195–200. doi: 10.1111/j.1574-6968.1996.tb07989.x. [DOI] [PubMed] [Google Scholar]
- 8.Bihar Directorate of Health Service. Monograph on Kala-azar incidence in Bihar (1985–1991; 1991–1996). Patna, Bihar, India: Office of the Chief Malaria Officer, Bihar Directorate of Health Services; 1998. [Google Scholar]
- 9.Bora D. Epidemiology of visceral leishmaniasis in India. Natl Med J India. 1999;12:62–68. [PubMed] [Google Scholar]
- 10.Chatterjee M, Manna M, Bhaduri A N, Sarkar D. Recent kala-azar cases in India: isozyme profiles of Leishmania parasites. Indian J Med Res. 1995;102:165–172. [PubMed] [Google Scholar]
- 11.Gradoni L, Scalone A, Gramiccia M. HIV-Leishmania co-infections in Italy; serological data as an indication of the sequence of acquisition of the two infections. Trans R Soc Trop Med Hyg. 1993;87:94–96. doi: 10.1016/0035-9203(93)90441-r. [DOI] [PubMed] [Google Scholar]
- 12.Grimaldi G, Jr, David J R, MacMohan-Pratt D. Identification and distribution of New World Leishmania species characterized by serodeme analysis using monoclonal antibodies. Am J Trop Med Hyg. 1987;36:280–287. doi: 10.4269/ajtmh.1987.36.270. [DOI] [PubMed] [Google Scholar]
- 13.Grimaldi G, Jr, Tesh R B. Leishmaniasis of the New World: current concepts and implications for future research. Clin Microbiol Rev. 1993;6:230–250. doi: 10.1128/cmr.6.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi, M., D. M. Dwyer, and H. L. Nakhasi. Cloning and characterization of differentially expressed genes from in vitro grown amastigotes of Leishmania donovani. Mol. Biochem. Parasitol. 58:345–354. [DOI] [PubMed]
- 15.Katakura K, Kawazu S-I, Naya T, Nagakura K, Ito M, Aikawa M, Qu J-Q, Guan L-R, Zuo X-P, Chai J-J, Chang K-P, Matsumoto Y. Diagnosis of kala-azar by nested PCR based on amplification of the Leishmania mini-exon gene. J Clin Microbiol. 1998;36:2173–2177. doi: 10.1128/jcm.36.8.2173-2177.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lachaud L, Dereure J, Chabbert E, Reynes J, Mauboussin J M, Oziol E, Dedet J P, Bastien P. Optimized PCR using patient blood samples for diagnosis and follow-up of visceral leishmaniasis, with special reference to AIDS patients. J Clin Microbiol. 2000;38:236–240. doi: 10.1128/jcm.38.1.236-240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopes U, Mohen H, Grimaldi G, Jr, Marzachi M, Pacheco R, Morel C. Schizodeme and zymodeme characterization of Leishmania in the investigation of foci of visceral and cutaneous leishmaniasis. J Parasitol. 1984;70:89–98. [PubMed] [Google Scholar]
- 18.Mathis A, Deplazes P. PCR and in vitro cultivation for detection of Leishmania spp. in diagnostic samples from humans and dogs. J Clin Microbiol. 1995;33:1145–1149. doi: 10.1128/jcm.33.5.1145-1149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherjee A, Ramesh V, Misra R S. Post kala-azar dermal leishmaniasis: a light and electron microscopic study of 18 cases. J Cutan Pathol. 1993;20:320–325. doi: 10.1111/j.1600-0560.1993.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 20.Navin T R, Arana B A, Arana F E, De Merida A M, Castillo A L, Pozuelos J L. Placebo controlled clinical trials of meglumine antomoniate (glucantime) vs. localized controlled heat in the treatment of cutaneous leishmaniasis in Guatemala. Am J Trop Med Hyg. 1990;42:43–50. doi: 10.4269/ajtmh.1990.42.43. [DOI] [PubMed] [Google Scholar]
- 21.Nuzum E, White III F, Thakur C, Dietze R, Wages J, Grogl M, Berman J. Diagnosis of symptomatic visceral leishmaniasis by use of the polymerase chain reaction on patient blood. J Infect Dis. 1995;171:751–754. doi: 10.1093/infdis/171.3.751. [DOI] [PubMed] [Google Scholar]
- 22.Osman O F, Oskam L, Zijlstra E E, Kroon N C, Schoone G J, Khalil E A G, Hassan A M, Karger P A. Evaluation of PCR for diagnosis of visceral leishmaniasis. J Clin Microbiol. 1997;35:2454–2457. doi: 10.1128/jcm.35.10.2454-2457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osman O F, Oskam L, Kroon N C M, Schoone G J, Khalil E-T A G, El Hassan A M, Zijlstra E E, Kager P A. Use of PCR for diagnosis of post-kala-azar dermal leishmaniasis. J Clin Microbiol. 1998;36:1621–1624. doi: 10.1128/jcm.36.6.1621-1624.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pogue G P, Koul S, Lee N S, Dwyer D M, Nakhasi H L. Identification of intra- and interspecific Leishmania genetic polymorphisms by arbitrary primed polymerase chain reactions and use of polymorphic DNA to identify differentially regulated genes. Parasitol Res. 1995;81:282–290. doi: 10.1007/BF00931531. [DOI] [PubMed] [Google Scholar]
- 25.Ramesh V, Mukherjee A. Post kala-azar dermal leishmaniasis. Int J Dermatol. 1995;34:85–91. doi: 10.1111/j.1365-4362.1995.tb03584.x. [DOI] [PubMed] [Google Scholar]
- 26.Ramesh V, Singh N. A clinical and histopathological study of macular type of post kala-azar dermal leishmaniasis. Trop Doc. 1999;29:205–207. doi: 10.1177/004947559902900406. [DOI] [PubMed] [Google Scholar]
- 27.Rios-Buceta L, Buezo G F, Penas P F, Tello E D, Montanes M A, Fernandez J F, Diez A G. Post kala-azar dermal leishmaniasis in a HIV-patient. Int J Dermatol. 1996;35:303–304. doi: 10.1111/j.1365-4362.1996.tb03014.x. [DOI] [PubMed] [Google Scholar]
- 28.Rodgers M R, Popper S J, Wirth D F. Amplification of kinetoplast as a tool in the detection and diagnosis of Leishmania. Exp Parasitol. 1990;71:267–275. doi: 10.1016/0014-4894(90)90031-7. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez N, Guzman B, Rodas A, Takiff H, Bloom B, Convit J. Diagnosis of cutaneous leishmaniasis and species discrimination of parasites by PCR and hybridization. J Clin Microbiol. 1994;32:2246–2251. doi: 10.1128/jcm.32.9.2246-2252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salotra P, Raina A, Negi N S. Immunoblot analysis of the antibody response to antigens of Leishmania donovani in Indian Kala-azar. Br J Biomed Sci. 1999;56:263–267. [PubMed] [Google Scholar]
- 31.Salotra P, Raina A, Ramesh V. Western blot analysis of humoral immune response to Leishmania donovani in patients with post kala-azar dermal leishmaniasis. Trans R Soc Trop Med Hyg. 1999;93:98–101. doi: 10.1016/s0035-9203(99)90197-9. [DOI] [PubMed] [Google Scholar]
- 32.Singh N, Curran M D, Rastogi A K, Middleton D, Sundar S. Diagnostic PCR with Leishmania donovani using sequences from the variable region of kinetoplast minicircle DNA. Trop Med Int Health. 1999;4:448–453. doi: 10.1046/j.1365-3156.1999.00416.x. [DOI] [PubMed] [Google Scholar]
- 33.Singh R P. Observations on dermal leishmanoid in Bihar. Indian J Dermatol. 1968;13:59–63. [PubMed] [Google Scholar]
- 34.Singh S, Sachs A G, Chang K P, Reed S G. Diagnostic and prognostic value of k39 recombinant antigen in Indian leishmaniasis. J Parasitol. 1995;81:1000–1003. [PubMed] [Google Scholar]
- 35.Smyth A J, Ghosh A, Hassan Md Q, Basu D, De Bruijn M H L, Adhya S, Mallik K K, Barker D C. Rapid and sensitive detection of Leishmania kinetoplast DNA from spleen and blood samples of kala-azar patients. Parasitology. 1992;105:183–192. doi: 10.1017/s0031182000074096. [DOI] [PubMed] [Google Scholar]
- 36.Sundar S, Reed S G, Singh V P, Kumar P C K, Murray H W. Rapid accurate field diagnosis of Indian visceral leishmaniasis. Lancet. 1998;351:563–565. doi: 10.1016/S0140-6736(97)04350-X. [DOI] [PubMed] [Google Scholar]
- 37.Thakur C P, Kumar K. Post kala-azar dermal leishmaniasis: a neglected aspect of kala-azar control programmes. Ann Trop Med and Parasitol. 1992;86:355–359. doi: 10.1080/00034983.1992.11812678. [DOI] [PubMed] [Google Scholar]
- 38.World Health Organization. The leishmaniases. World Health Organization technical report series, no. 793. Geneva, Switzerland: World Health Organization; 1990. [Google Scholar]
- 39.Zijlstra E F, Ali M S, El-Hassan A M, El-Toum I A, Satti M, Ghalib H W, Kager P A. Kala-azar; a comparative study of parasitological methods and the direct agglutination test in diagnosis. Trans R Soc Trop Med Hyg. 1992;86:505–507. doi: 10.1016/0035-9203(92)90086-r. [DOI] [PubMed] [Google Scholar]