Abstract
Background
Bread wheat (Triticum aestivum) is an important staple cereal grain worldwide. The ever-increasing environmental stress makes it very important to mine stress-resistant genes for wheat breeding programs. Therefore, dehydrin (DHN) genes can be considered primary candidates for such programs, since they respond to multiple stressors.
Results
In this study, we performed a genome-wide analysis of the DHN gene family in the genomes of wheat and its three relatives. We found 55 DHN genes in T. aestivum, 31 in T. dicoccoides, 15 in T. urartu, and 16 in Aegilops tauschii. The phylogenetic, synteny, and sequence analyses showed we can divide the DHN genes into five groups. Genes in the same group shared similar conserved motifs and potential function. The tandem TaDHN genes responded strongly to drought, cold, and high salinity stresses, while the non-tandem genes respond poorly to all stress conditions. According to the interaction network analysis, the cooperation of multiple DHN proteins was vital for plants in combating abiotic stress.
Conclusions
Conserved, duplicated DHN genes may be important for wheat being adaptable to a different stress conditions, thus contributing to its worldwide distribution as a staple food. This study not only highlights the role of DHN genes help the Triticeae species against abiotic stresses, but also provides vital information for the future functional studies in these crops.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-022-08317-x.
Keywords: Bread wheat, DHN gene family, Expression profile, Biotic stress, Abiotic stress
Background
Bread wheat (Triticum aestivum) is an important staple cereal crop providing ~ 20% of the global dietary protein and calories [1, 2]. It comprises three homologous sub-genomes (AABBDD; 2n = 6x = 42) originating from two natural hybridization events [3, 4]. First, tetraploidization from the hybridization between T. urartu (AA; 2n = 2x = 14) and an unknown close relative of Aegilops speltoides (BB; 2n = 2x = 14) generated the tetraploid wild emmer wheat (T. turgidum ssp. dicoccoides, AABB, 2n = 4x = 28). Wild emmer wheat hybridized with Ae. tauschii (DD; 2n = 2x = 14) about 8000 years ago to produce hexaploid bread wheat [5]. Environmental stressors including abiotic stressors (e.g., drought, salinity, and high and low temperatures) [6–8] and biotic stressors like Fusarium graminearum (Fusarium head blight or FHB), Blumeria graminis (powdery mildew), and Puccinia striiformis (stripe rust) challenge bread wheat yield during its growth phase [9]. The key to facing these challenges is mining for stress-resistant genes and utilizing them for breeding. With the release of the genome assembly and annotation for T. aestivum [10], T. dicoccoides [11], T. urartu [12], and Ae. tauschii [13], a genome-wide analysis of all stress-related genes in wheat and its relatives can now be realized. Furthermore, large-scale RNA sequencing (RNA-seq) provides a rich resource for analyzing their related gene expression patterns not only under diverse stress conditions but also at different developmental stages [14].
Dehydrins (DHNs) are a class of highly hydrophilic, stress-responsive proteins rich in charged and polar amino acids [15, 16]. These proteins accumulate during late embryogenesis and are induced in vegetative tissues by several cell-dehydrating environmental stressors like drought, salinity, and cold [17]. Based on their sequence characteristics, DHNs are defined as proteins containing at least one copy of a conserved motif called the K-segment [18, 19]. The K-segment (consensus EKKGIM [E/D]KIKEKLPG) is a lysine-rich amino acid sequence, forming amphiphilic α-helixes at the protein’s C-terminus [20, 21]. DHNs also possess other conserved motifs, like the N-terminal tyrosine-rich Y-segment (consensus [T/V] D [E/Q]YGNP), and the serine-rich S-segment (consensus LHRSGS4–10(E/D)3) containing a stretch of 4–10 serine residues [22, 23]. The diversity of the conserved domains allows the DHN gene to form combinations of different domains, and then produce different groups [24, 25]. Based on the presence of these conserved motifs (K-, S-, and Y-segment), DHNs are classified into different categories of YnSKn, YnKn, SKn, KnS, and Kn [18, 19, 26].
DHNs are stress proteins protecting plants against dehydration by: (a) binding metal ions and scavenging reactive oxygen species, (b) binding DNA or phospholipids to maintain biological activity, (c) binding proteins to prevent denaturation, and (d) holding water molecules [27, 28]. DHN family members are intrinsically unstructured, heat-stable proteins expressed during the late embryogenesis stage [29, 30]. Their characteristic protein conformational changes result in protein functional changes via a phenomenon called ‘moonlighting’, and thus also called IDPs/IUPs (intrinsically unstructured/disordered proteins). They either may help in forming and stabilizing the plant cytoplasmic glassy state during dehydration, or serve as hub proteins coordinating cellular signaling crosstalk involved in the stress response [31, 32]. Previous studies demonstrated that DHNs are crucial in abiotic stress tolerance; overexpressing the Solanum habrochaites DHN gene enhanced transgenic tomato tolerance against multiple abiotic stressors; overexpressing the oleaster DHN gene OesDHN improved drought tolerance in Arabidopsis; overexpression of four Prunus mume DHNs in Escherichia coli and tobacco resulted in increased freezing resistance; HbDHN1 and HbDHN2 from Hevea brasiliensis significantly increased drought, salt, and osmotic stress tolerance when overexpressed in Arabidopsis [33–36]. These studies indicate the extensive involvement of plant DHNs in abiotic stress tolerance. Several studies have shown that DHNs might also play important roles in both plant development and biotic stress response. For example, Medicago truncatula Y2K4-type dehydrin (MtCAS31) interacts with AtICE1, which is essential for stomatal development [37]; expression of several DHNs in drought-tolerant oak species Quercus ilex are induced by a Phytophthora cinnamomic infection [38].
In this study, we identified the DHN genes and its homologs in bread wheat and its relatives and analyzed their phylogenetic, syntenic, and sequence relationships. We analyzed the putative promoter cis-elements of the TaDHN genes. Then, we investigated the expression profiles of the DHN gene family in response to various environmental stressors (including biotic and abiotic stressors) and hormones. Finally, we analyzed the interaction network of DHN genes and experimentally verified their predicted subcellular location. Therefore, this study (a) provides a comprehensive structural and functional analysis of the DHN gene family in bread wheat and its relatives, and (b) clarifies the important role of DHN genes help against various abiotic stresses.
Results
Characterization of DHN genes in bread wheat and its relatives
We used HMMER 3.1 and BLASTP for searching DHN genes in the genomes of bread wheat and its relatives, based on the Pfam database-derived HMM profile of the DHN domain (PF00257) as a query. Then, we verified the predicted sequences using InterPro and CDD. Finally, we identified 117 putative DHN genes. Among them, we detected 55 DHN genes in T. aestivum, 31 in T. dicoccoides, 15 in T. urartu, and 16 in Ae. tauschii. These DHN gene numbers are directly related to the genome ploidy. The DHN gene names, locus IDs, and other features are shown in Table 1.
Table 1.
The details of DHN genes among bread wheat and its relatives
| Gene Name | Locus ID | Type | Genomic Position | BP | GC (%) | AA | MW (kDa) | pI | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| TaDHN1-A | TraesCS3A02G254600 | YSK2 | 476,563,869–476,564,968(−) | 642 | 68.07 | 213 | 21.83 | 6.75 | Cytoplasm Nucleus |
| TaDHN1-B | TraesCS3B02G286600 | YSK2 | 458,398,889–458,399,630(−) | 654 | 67.13 | 217 | 22.3 | 7.5 | Cytoplasm Nucleus |
| TaDHN1-D | TraesCS3D02G255500 | YSK2 | 357,146,923–357,147,959(−) | 648 | 67.75 | 215 | 22.24 | 7.19 | Cytoplasm Nucleus |
| TaDHN2-A | TraesCS3A02G396200 | YSK3 | 643,459,970–643,461,316(−) | 828 | 67.63 | 275 | 27.02 | 10.13 | Cytoplasm |
| TaDHN2-B | TraesCS3B02G428200 | YSK3 | 667,112,076–667,113,352(−) | 825 | 68.12 | 274 | 27.19 | 10.29 | Cytoplasm |
| TaDHN2-D | TraesCS3D02G390200 | YSK3 | 505,318,572–505,319,988(−) | 828 | 67.87 | 275 | 27.16 | 10.26 | Cytoplasm |
| TaDHN3-A | TraesCS4A02G250900 | Y2SK3 | 562,289,788–562,291,566(−) | 1368 | 70.37 | 455 | 43.74 | 9.28 | Cytoplasm |
| TaDHN3-B | TraesCS4B02G064200 | Y2SK3 | 57,136,426–57,138,194(−) | 1374 | 69.92 | 457 | 43.89 | 9.27 | Cytoplasm |
| TaDHN3-D | TraesCS4D02G063100 | Y2SK3 | 39,233,033–39,234,840(−) | 1293 | 70.15 | 430 | 41.22 | 9.47 | Cytoplasm |
| TaDHN4-A1 | TraesCS5A02G369800 | YSK2 | 569,677,389–569,678,193(+) | 432 | 71.53 | 143 | 14.57 | 8.91 | Cytoplasm |
| TaDHN4-A2 | TraesCS5A02G369900 | YSK2 | 569,682,833–569,683,707(+) | 423 | 70.92 | 140 | 14.24 | 8.91 | Cytoplasm |
| TaDHN4-B1 | TraesCS5B02G372100 | YSK2 | 550,320,429–550,321,418(+) | 432 | 71.76 | 143 | 14.43 | 8.91 | Cytoplasm |
| TaDHN4-B2 | TraesCS5B02G372200 | YSK2 | 550,337,855–550,338,611(+) | 417 | 70.5 | 138 | 14.22 | 8.91 | Cytoplasm |
| TaDHN4-D1 | TraesCS5D02G379200 | YSK2 | 450,373,636–450,374,483(+) | 432 | 71.53 | 143 | 14.52 | 8 | Cytoplasm |
| TaDHN4-D2 | TraesCS5D02G379300 | YSK2 | 450,379,533–450,380,460(+) | 402 | 69.65 | 133 | 13.93 | 9.44 | Cytoplasm |
| TaDHN5-A1 a | TraesCS5A02G424700 | YSK1 | 610,078,219–610,079,136(−) | 336 | 67.26 | 111 | 11.48 | 10.15 | Cytoplasm |
| TaDHN5-A2 | TraesCS5A02G424800 | YSK2 | 610,184,778–610,185,696(−) | 450 | 65.56 | 149 | 15.22 | 9.96 | Cytoplasm |
| TaDHN5-B1 | TraesCS5B02G426700 | YSK2 | 602,483,279–602,484,206(−) | 453 | 66 | 150 | 15.18 | 10.16 | Cytoplasm |
| TaDHN5-B2 | TraesCS5B02G426800 | YSK2 | 602,648,556–602,649,390(−) | 453 | 65.78 | 150 | 15.22 | 9.99 | Cytoplasm |
| TaDHN5-D1 | TraesCS5D02G433200 | YSK2 | 489,012,960–489,013,838(−) | 459 | 66.67 | 152 | 15.35 | 10.16 | Cytoplasm |
| TaDHN5-D2 | TraesCS5D02G433300 | YSK2 | 489,166,583–489,167,421(−) | 465 | 66.67 | 154 | 15.59 | 10.16 | Cytoplasm |
| TaDHN6-A | TraesCS6A02G059800 | YSK2 | 31,583,535–31,584,464(−) | 462 | 68.4 | 153 | 15.51 | 9.46 | Cytoplasm |
| TaDHN6-B | TraesCSU02G086200 | YSK2 | 76,960,851–76,961,538(+) | 456 | 67.76 | 151 | 15.29 | 9.68 | Cytoplasm |
| TaDHN6-D a | TraesCSU02G122200 | YSK1 | 104,041,218–104,042,344(−) | 450 | 69.56 | 149 | 14.86 | 7.5 | Cytoplasm |
| TaDHN7-A | TraesCS6A02G253300 | SK3 | 468,473,627–468,475,112(−) | 807 | 64.19 | 268 | 28.82 | 5.05 | Nucleus |
| TaDHN7-B | TraesCS6B02G273400 | SK3 | 493,352,704–493,354,073(−) | 780 | 62.69 | 259 | 27.97 | 4.98 | Nucleus |
| TaDHN7-D | TraesCS6D02G234700 | SK3 | 329,080,938–329,082,415(−) | 789 | 63.88 | 262 | 28.16 | 4.97 | Nucleus |
| TaDHN8-A a | TraesCS6A02G350100 | K3 | 581,982,926–581,983,580(+) | 573 | 65.1 | 190 | 19.24 | 7.74 | Cytoplasm |
| TaDHN8-B | TraesCS6B02G383200 | K6 | 658,177,094–658,178,907(+) | 1218 | 66.56 | 405 | 40.29 | 7.37 | Cell wall Cytoplasm Nucleus |
| TaDHN8-D a | TraesCS6D02G332500 | K3 | 434,811,674–434,812,738(+) | 540 | 66.3 | 179 | 17.8 | 8.23 | Cytoplasm |
| TaDHN9-A | TraesCS6A02G350200 | K2 | 582,086,438–582,087,160(+) | 282 | 62.06 | 93 | 9.66 | 7.43 | Cytoplasm |
| TaDHN9-D | TraesCS6D02G332600 | K2 | 435,012,400–435,012,681(+) | 282 | 63.12 | 93 | 9.66 | 7.43 | Cytoplasm |
| TaDHN10-A | TraesCS6A02G350300 | K14 | 582,092,081–582,096,138(+) | 2976 | 62.96 | 991 | 101.61 | 6.33 | Cytoplasm |
| TaDHN10-B a | TraesCS6B02G695200LC | K8 | 658,234,035–658,241,687(+) | 1671 | 61.88 | 556 | 57.44 | 6.67 | Cytoplasm |
| TaDHN10-D | TraesCS6D02G332700 | K12 | 435,072,895–435,075,633(+) | 2739 | 63.08 | 912 | 93.49 | 6.45 | Cell wall Cytoplasm Nucleus |
| TaDHN11-A | TraesCS6A02G350500 | YSK2 | 582,264,726–582,266,027(+) | 666 | 68.92 | 221 | 22.05 | 9.45 | Cell wall Cytoplasm Nucleus |
| TaDHN11-B | TraesCS6B02G383500 | YSK2 | 658,402,976–658,404,180(+) | 690 | 69.13 | 229 | 23.01 | 9.79 | Cell wall Cytoplasm Nucleus |
| TaDHN11-D | TraesCS6D02G332900 | YSK2 | 435,351,033–435,352,218(+) | 651 | 69.12 | 216 | 21.62 | 9.5 | Cell wall Cytoplasm Nucleus |
| TaDHN12-A1 | TraesCS6A02G350600 | YSK2 | 582,511,276–582,512,221(+) | 489 | 66.87 | 162 | 16.28 | 9.88 | Cytoplasm |
| TaDHN12-A2 | TraesCS6A02G350700 | YSK2 | 582,516,436–582,517,359(+) | 459 | 64.49 | 152 | 15.52 | 8.05 | Cytoplasm |
| TaDHN12-A3 | TraesCS6A02G350800 | SK2 | 582,630,751–582,631,295(−) | 432 | 63.66 | 143 | 14.82 | 9.88 | Cytoplasm |
| TaDHN12-A4 b | TraesCS6A02G350900 | YSK1 | 582,638,141–582,638,862(−) | 573 | 64.92 | 190 | 20.14 | 11.34 | Cytoplasm |
| TaDHN12-B1 | TraesCS6B02G695700LC | YSK2 | 658,477,430–658,478,020(+) | 489 | 63.27 | 162 | 16.13 | 9.79 | Cytoplasm |
| TaDHN12-B2 | TraesCS6B02G695800LC | YSK2 | 658,496,215–658,496,805(+) | 489 | 63.27 | 162 | 16.13 | 9.79 | Cytoplasm |
| TaDHN12-B3 | TraesCS6B02G695900LC | YSK2 | 658,515,366–658,515,956(+) | 489 | 63.27 | 162 | 16.1 | 9.79 | Cytoplasm |
| TaDHN12-B4 | TraesCS6B02G383600 | YSK2 | 658,530,539–658,531,483(+) | 477 | 66.46 | 158 | 15.84 | 9.69 | Cytoplasm |
| TaDHN12-B5 | TraesCS6B02G383800 | YSK2 | 658,577,562–658,578,499(−) | 501 | 65.67 | 166 | 16.7 | 8.89 | Cytoplasm |
| TaDHN12-D1 | TraesCS6D02G333000 | YSK2 | 435,711,668–435,712,623(+) | 489 | 66.87 | 162 | 16.2 | 8.93 | Cytoplasm |
| TaDHN12-D2 | TraesCS6D02G333100 | YSK2 | 435,749,803–435,750,719(+) | 435 | 65.06 | 144 | 14.51 | 9.79 | Cytoplasm |
| TaDHN12-D3 | TraesCS6D02G333200 | YSK2 | 435,763,700–435,764,598(+) | 468 | 65.6 | 155 | 15.73 | 9.41 | Cytoplasm |
| TaDHN12-D4 | TraesCS6D02G333300 | YSK2 | 435,831,506–435,832,386(−) | 483 | 66.87 | 160 | 16.26 | 8.94 | Cytoplasm |
| TaDHN12-D5 | TraesCS6D02G333600 | YSK2 | 435,962,275–435,963,310(−) | 504 | 66.27 | 167 | 16.71 | 8.05 | Cytoplasm |
| TaDHN13-A | TraesCS7A02G560000 | K3 | 731,882,428–731,883,023(+) | 375 | 61.87 | 124 | 12.83 | 8.08 | Cytoplasm |
| TaDHN13-B | TraesCS7B02G484900 | K3 | 741,668,510–741,668,887(+) | 378 | 64.02 | 125 | 12.67 | 7.04 | Cytoplasm |
| TaDHN13-D | TraesCS7D02G549900 | K2 | 634,439,606–634,440,272(−) | 339 | 63.13 | 112 | 11.53 | 6.8 | Cytoplasm |
| TdDHN1-A | TRIDC3AG038190 | YSK2 | 483,446,623–483,451,982(−) | 660 | 67.42 | 219 | 22.53 | 6.93 | Cytoplasm Nucleus |
| TdDHN1-B | TRIDC3BG042940 | YSK2 | 468,417,850–468,427,038(−) | 654 | 66.97 | 217 | 22.37 | 7.19 | Cytoplasm Nucleus |
| TdDHN2-A a | TRIDC3AG056410 | YSK2 | 639,664,417–639,665,447(−) | 390 | 64.36 | 129 | 13.24 | 10.56 | Cytoplasm Nucleus |
| TdDHN2-B a | TRIDC3BG063150 | YSK2 | 677,652,490–677,653,501(−) | 405 | 65.43 | 134 | 13.87 | 10.69 | Cytoplasm |
| TdDHN3-A a | TRIDC4AG039320 | SK3 | 555,522,541–555,523,348(−) | 552 | 66.67 | 183 | 18.37 | 10.19 | Cytoplasm |
| TdDHN3-B a | TRIDC4BG009930 | Y2SK3 | 55,083,350–55,084,815(−) | 543 | 65.38 | 180 | 18.02 | 10.19 | Cytoplasm |
| TdDHN4-A1 | TRIDC5AG054100 | YSK2 | 564,921,473–564,922,000(+) | 432 | 71.76 | 143 | 14.56 | 8.91 | Cytoplasm |
| TdDHN4-A2 | TRIDC5AG054110 | YSK2 | 564,926,944–564,927,492(+) | 423 | 70.92 | 140 | 14.27 | 8.87 | Cytoplasm |
| TdDHN4-B1 | TRIDC5BG058060 | YSK2 | 556,085,190–556,085,719(+) | 432 | 71.76 | 143 | 14.43 | 8.91 | Cytoplasm |
| TdDHN4-B2 | TRIDC5BG058080 | YSK2 | 556,101,420–556,101,927(+) | 417 | 70.26 | 138 | 14.25 | 8.91 | Cytoplasm |
| TdDHN5-A1 a | TRIDC5AG061380 | SK1 | 605,402,094–605,402,742(−) | 219 | 63.01 | 72 | 7.61 | 10.81 | Nucleus |
| TdDHN5-A2 b | TRIDC5AG061420 | YSK2 | 605,513,222–605,513,988(−) | 522 | 63.03 | 173 | 17.79 | 10.35 | Cytoplasm |
| TdDHN5-B | TRIDC5BG065560 | YSK2 | 608,742,508–608,743,380(−) | 456 | 65.79 | 151 | 15.37 | 10.13 | Cytoplasm |
| TdDHN6-A | TRIDC6AG007480 | YSK2 | 31,000,232–31,000,891(−) | 462 | 67.97 | 153 | 15.44 | 8.93 | Cytoplasm |
| TdDHN6-B | TRIDC6BG010780 | YSK2 | 55,591,718–55,592,361(−) | 456 | 67.76 | 151 | 15.29 | 9.68 | Cytoplasm |
| TdDHN7-A | TRIDC6AG039020 | SK3 | 470,140,072–470,141,622(−) | 807 | 64.19 | 268 | 28.82 | 5.05 | Nucleus |
| TdDHN7-B | TRIDC6BG045690 | SK3 | 486,136,098–486,141,800(+) | 780 | 62.56 | 259 | 27.94 | 4.98 | Nucleus |
| TdDHN8-A a | TRIDC6AG052540 | K4 | 582,467,279–582,468,478(+) | 792 | 68.06 | 263 | 26.21 | 7.44 | Cytoplasm |
| TdDHN8-B b | TRIDC6BG061300 | K7 | 642,747,123–642,748,606(+) | 1410 | 66.67 | 470 | 47.07 | 7.63 | Cell wall Cytoplasm Nucleus |
| TdDHN9-A | TRIDC6AG052550 | K2 | 582,534,354–582,534,931(+) | 282 | 62.06 | 93 | 9.66 | 7.43 | Cytoplasm |
| TdDHN9-B b | TRIDC6BG061310 | K2 | 642,748,977–642,749,569(+) | 354 | 65.82 | 117 | 11.67 | 7.54 | Cytoplasm |
| TdDHN10-A a | TRIDC6AG052570 | K8 | 582,541,438–582,543,392(+) | 1698 | 61.37 | 565 | 58.87 | 6.45 | Cytoplasm |
| TdDHN11-A a | TRIDC6AG052590 | YSK2 | 582,698,877–582,699,685(+) | 489 | 67.28 | 162 | 16.66 | 9.6 | Cell wall Cytoplasm Nucleus |
| TdDHN11-B | TRIDC6BG061340 | YSK2 | 643,041,875–643,042,827(+) | 696 | 69.25 | 231 | 23.13 | 9.45 | Cell wall Cytoplasm Nucleus |
| TdDHN12-A1 | TRIDC6AG052630 | YSK2 | 582,893,775–582,898,592(+) | 489 | 66.87 | 162 | 16.22 | 9.13 | Cytoplasm |
| TdDHN12-A2 | TRIDC6AG052640 | YSK2 | 582,992,994–582,993,790(−) | 474 | 64.14 | 157 | 16.14 | 8.08 | Cytoplasm |
| TdDHN12-A3 | TRIDC6AG052650 | YSK2 | 583,001,187–583,001,807(−) | 441 | 65.76 | 146 | 14.75 | 9.79 | Cytoplasm |
| TdDHN12-B1 | TRIDC6BG061350 | YSK2 | 643,116,359–643,132,143(+) | 477 | 66.67 | 158 | 15.87 | 9.71 | Cytoplasm |
| TdDHN12-B2 | TRIDC6BG061380 | YSK2 | 643,190,550–643,191,397(−) | 501 | 65.87 | 166 | 16.65 | 8.89 | Cytoplasm |
| TdDHN13-A | TRIDC7AG077740 | K3 | 723,743,247–723,743,983(−) | 375 | 61.87 | 124 | 12.83 | 8.08 | Cytoplasm |
| TdDHN13-B | TRIDC7BG076250 | K3 | 750,614,280–750,614,913(+) | 381 | 63.78 | 126 | 12.8 | 7.38 | Cytoplasm |
| TuDHN1 | TuG1812G0300003021.01 | YSK2 | 477,045,519–477,046,650(−) | 660 | 67.42 | 219 | 22.57 | 7.14 | Cytoplasm Nucleus |
| TuDHN3 | TuG1812G0400000583.01 | Y2SK3 | 41,901,253–41,903,075(−) | 1392 | 70.55 | 463 | 44.55 | 9.28 | Cytoplasm |
| TuDHN4–1 | TuG1812G0500003981.01 | YSK2 | 535,938,718–535,939,515(+) | 432 | 71.53 | 143 | 14.55 | 8.03 | Cytoplasm |
| TuDHN4–2 | TuG1812G0500003982.01 | YSK2 | 535,944,152–535,945,001(+) | 423 | 71.16 | 140 | 14.24 | 8.91 | Cytoplasm |
| TuDHN5–1 a | TuG1812G0500004492.01 | YSK1 | 574,218,374–574,219,129(−) | 336 | 67.26 | 111 | 11.46 | 9.8 | Cytoplasm |
| TuDHN5–2 a | TuG1812S0001634800.01 | YSK1 | 1–593(−) | 414 | 67.87 | 138 | 13.91 | 7.97 | Cytoplasm |
| TuDHN6 | TuG1812G0600000621.01 | YSK2 | 31,890,359–31,891,122(−) | 462 | 68.18 | 153 | 15.4 | 8.1 | Cytoplasm |
| TuDHN7 | TuG1812G0600002817.01 | SK3 | 439,323,348–439,324,718(−) | 807 | 64.06 | 268 | 28.85 | 5.05 | Nucleus |
| TuDHN8 | TuG1812G0600003768.01 | K6 | 539,540,401–539,541,995(+) | 1176 | 68.54 | 391 | 38.82 | 7.48 | Cell wall Cytoplasm Nucleus |
| TuDHN9 | TuG1812G0600003769.01 | K2 | 539,598,064–539,599,229(+) | 282 | 62.06 | 93 | 9.66 | 7.43 | Cytoplasm |
| TuDHN11 | TuG1812G0600003775.01 | YSK2 | 539,946,480–539,947,767(+) | 756 | 68.65 | 251 | 24.98 | 9.12 | Cell wall Cytoplasm Nucleus |
| TuDHN12–1 b | TuG1812S0003423700.01 | YSK1 | 1979–2944(+) | 537 | 67.6 | 178 | 17.93 | 10.89 | Cytoplasm |
| TuDHN12–2 | TuG1812S0003424100.01 | YSK2 | 5679–6605(+) | 459 | 64.27 | 152 | 15.64 | 8.89 | Cytoplasm |
| TuDHN12–3 | TuG1812G0600003779.01 | YSK2 | 540,160,098–540,161,020(−) | 510 | 67.06 | 169 | 16.94 | 8.89 | Cytoplasm |
| TuDHN13 | TuG1812G0700005988.01 | K3 | 712,117,702–712,118,431(+) | 375 | 61.87 | 124 | 12.74 | 7.47 | Cytoplasm |
| AetDHN1 | AET3Gv20620600 | YSK2 | 364,603,011–364,610,005(−) | 648 | 67.75 | 215 | 22.24 | 7.19 | Cytoplasm Nucleus |
| AetDHN2 a | AET3Gv20881700 | SK2 | 513,201,759–513,203,320(−) | 396 | 66.41 | 131 | 13.27 | 10.87 | Cytoplasm |
| AetDHN3 | AET4Gv20132600 | Y2SK3 | 41,638,766–41,640,754(−) | 1383 | 67.97 | 460 | 44.58 | 9.03 | Cytoplasm |
| AetDHN4–1 | AET5Gv20866700 | YSK2 | 458,683,120–458,684,124(+) | 432 | 71.53 | 143 | 14.52 | 8 | Cytoplasm |
| AetDHN4–2 | AET5Gv20866800 | YSK2 | 458,689,123–458,689,989(+) | 402 | 69.65 | 133 | 13.93 | 9.44 | Cytoplasm |
| AetDHN5 | AET5Gv20990000 | YSK2 | 498,787,518–499,029,693(−) | 465 | 67.1 | 154 | 15.56 | 10.16 | Cytoplasm |
| AetDHN6 a | AET6Gv20153700 | K1 | 34,801,689–34,803,209(−) | 270 | 66.3 | 89 | 8.86 | 9.64 | Cytoplasm |
| AetDHN7 | AET6Gv20653900 | SK3 | 352,066,646–352,068,329(−) | 789 | 63.88 | 262 | 28.16 | 4.97 | Nucleus |
| AetDHN8 | AET6Gv20864100 | K6 | 458,864,577–458,866,768(+) | 1176 | 66.75 | 391 | 38.89 | 7.59 | Cell wall Cytoplasm Nucleus |
| AetDHN9 | AET6Gv20864400 | K2 | 459,070,823–459,071,706(+) | 282 | 63.12 | 93 | 9.66 | 7.43 | Cytoplasm |
| AetDHN10 a | AET6Gv20864500 | K9 | 459,131,190–459,134,316(+) | 2190 | 63.56 | 729 | 75.05 | 6.35 | Cytoplasm |
| AetDHN11 b | AET6Gv20864900 | YSK1 | 459,422,062–459,423,545(+) | 444 | 67.79 | 148 | 16.32 | 10.99 | Cytoplasm Nucleus |
| AetDHN12–1 | AET6Gv20865700 | YSK2 | 459,787,474–459,841,184(+) | 468 | 65.6 | 155 | 15.73 | 9.41 | Cytoplasm |
| AetDHN12–2 | AET6Gv20866000 | YSK2 | 459,889,128–459,890,166(−) | 483 | 66.87 | 160 | 16.26 | 8.94 | Cytoplasm |
| AetDHN12–3 | AET6Gv20866400 | YSK2 | 460,020,381–460,021,566(−) | 504 | 66.27 | 167 | 16.71 | 8.05 | Cytoplasm |
| AetDHN13 | AET7Gv21347200 | K2 | 641,274,370–641,275,124(+) | 339 | 63.13 | 112 | 11.53 | 6.8 | Cytoplasm |
Ta T.aestivum, Td T.dicoccodies, Tu T.urartu, Aet Ae. tauschii, DHN Dehydrin, “a”: truncated genes, “b”: potenial misannotated genes; BP coding sequence length, AA amino sequence length, MW molecular weight, pI isoelectric point
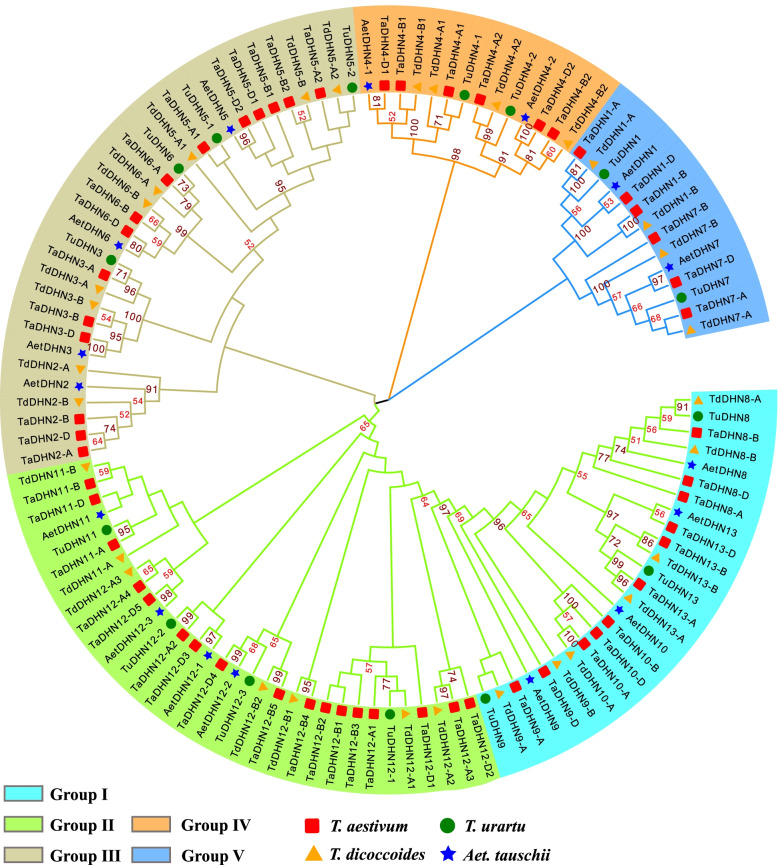
To study the phylogenetic relationships of the DHN family, we constructed an unrooted phylogenetic tree using the 117 DHN protein sequences of bread wheat and its relatives (Fig. 1). The DHN genes were clustered into five major groups. Group I contained DHN8/9/10/13, which encode Kn type DHNs (Table 1). The K-segment copies varied from 2 to 14. Group II contained DHN11/12, which encode YnSKn type DHNs (except TaDHN12-A3 encodes a SK2 type DHN), mainly the YSK2 type (Table 1). Group III contained DHN2/3/5/6, which encode YnSKn type DHNs (except TdDHN3-A, TdDHN5-A1, AetDHN2, and AetDHN6). Group IV contained the DHN4 genes, which encode YSK2 type DHNs. The remaining clade was Group V, containing DHN1 and DHN7, which encode YSK2 and SK3 type DHNs, respectively (Fig. 1 and Table 1).
Fig. 1.
Phylogenetic analysis of the DHN genes in Triticum aestivum and its relatives. The phylogenetic tree was constructed using the neighbor-joining method and MEGA-X software; bootstrap scores > 50% are displayed; Different symbols represent different species, and different background colors represent different groups
We analyzed all DHN protein sequences in wheat and its relatives and identified three types of DHNs (YnSKn, SKn, and Kn). YnSKn was the most common with 81 among 117 DHNs, followed by Kn with 26, and only 10 DHN genes encoded SKn type proteins (Table 1). We also studied the phylogenetic relationships of DHN genes in wheat and its relatives, rice, and A. thaliana. Rice and A. thaliana had far fewer DHN genes than wheat and its relatives, with most being clustered into Group V, while several others belonged to Group I, Group III, and Group IV (Fig. S1). Therefore, the results showed that the DHN gene family conservation is limited to the close relatives of wheat, and very different from non- Triticeae species.
Chromosomal distribution and synteny analysis of the DHN genes
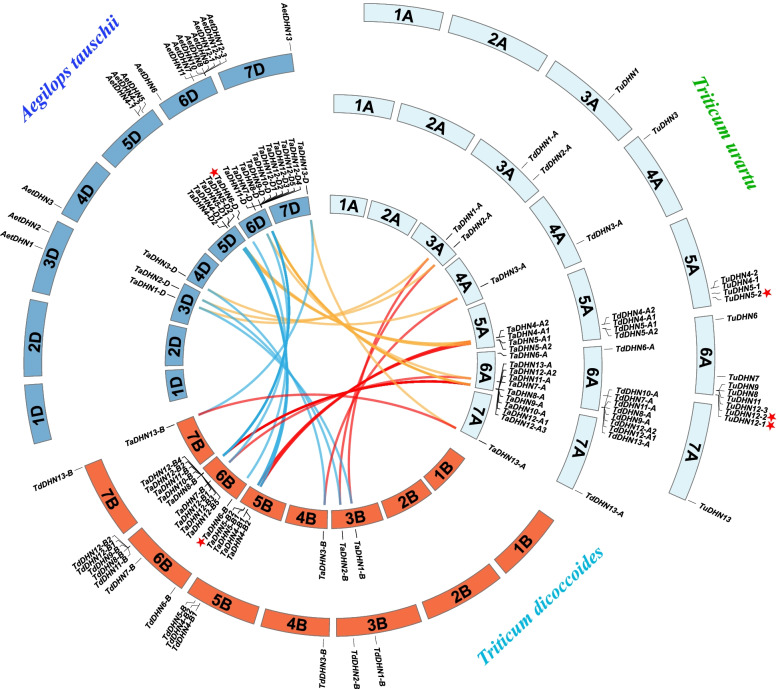
To analyze the DHN gene syntenic relationships between bread wheat and its relatives, we identified orthologous genes among these four released species genomes. There were 18 TaDHNs, 17 TdDHNs, and 15 TuDHNs in the A sub-genome; 18 TaDHNs and 14 TdDHNs in the B sub-genome; and 19 TaDHNs and 16 AetDHNs in the D sub-genome. We identified and mapped the gene pairs of Ta/Td/Tu-A, Ta/Td-B, and Ta/Aet-D to corresponding genomic chromosomes (Fig. 2 and Table S1). Five genes, TaDHN6B/D, TuDHN5–2, and TuDHN12–1/2, were not assigned to chromosomes in the genome annotation file we used. We re-assigned these genes to the corresponding chromosomes based on the homologous and phylogenetic relationships (Table S1 and Fig. 1) and genomic location information (Table 1) of all DHN genes between the different diploid sub-genomes (Fig. 2). The DHN genes were distributed in the third to seventh homologous groups of bread wheat and its relatives, of which the fourth and seventh homologous groups had only one gene copy (except T. urartu-3A, with a missing gene), and the sixth homologous group had the most DHN genes, ranging from 7 to 14.
Fig. 2.
Synteny analysis and chromosomal distribution of DHN genes in bread wheat and its relatives. The hexploid bread wheat (Ta) and tetraploid durum wheat (Td) genomes were split into three and two diploid subgenomes, respectively. The A, B, and D genomes are represented by different colors. The DHN genes were assigned to the corresponding chromosome, according to their genome annotation file, except TaDHN6B/D, TuDHN5–2, and TuDHN12–1/2, which are labeled with a red star, and their positions are predicted by their homologs. Syntenic DHN gene pairs belonging to the same linkage groups between A and B, A and D, and B and D are linked with orange, red, and blue lines, respectively
We also performed gene specific SSRs mining analysis for TaDHN genes, and 31 gene specific SSRs were discovered. These SSRs were distributed in the following four classes: di (dinucleotide), tri (trinucleotide), tetra (tetranucleotide) and penta (pentanucleotide) (FigureS2 and Table S2). Di SSR repeats (~ 67.74%) were far more than other repeats, and the tri SSRs (~ 25.81%) were found to be more than tetra (~ 3.22%) and penta repeats (~ 3.22%) (Fig. S2). After due validation, the predicted genes specific SSRs can be utilized for marker-assisted breeding programs in the future.
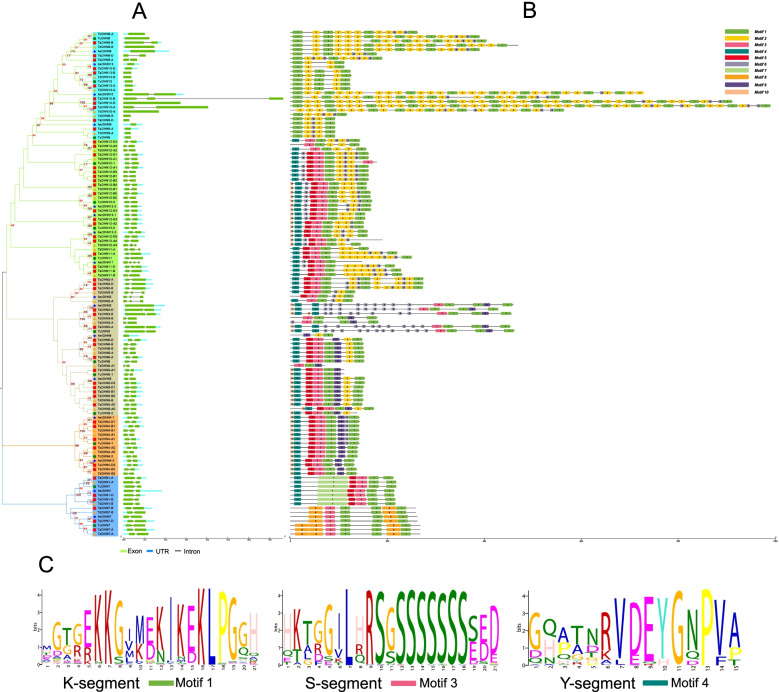
Sequence analysis and re-annotation of the DHN gene family
We collected the structural information of all DHN genes in the annotation file and visualized it using the Gene Structure Display Server (GSDS) web tool. The structural analysis results showed that the exon number varied between one and four. After analyzing the conserved domains of all DHN proteins, we found that all contained one dehydrin core motif (K-segment), but different numbers of Y−/S-segments (Fig. 3B and C). The remaining motifs are shown in Fig. S3. We manually checked the coding sequences and amino sequences of all DHN genes among bread wheat and its relatives, and combined the results with those of the phylogenetic (Fig. 1), synteny (Fig. 2), and sequence structural (Fig. 3) analyses. The truncated genes and potential mis-annotated genes are identified and marked in Table 1, and the identified missing genes are shown in Table S1.
Fig. 3.
Sequence information of the DHN genes among bread wheat and its relatives. A Exon-intron structures. Green boxes, light blue boxes, and black horizontal lines indicate exons, UTRs, and introns, respectively. B Conserved domain composition. Dehydrin core domain (K-segment) and the other two dehydrin-related domains (S-segment and Y-segment) are shown in different colors. C Logos of the dehydrin-conserved domains
According to the ploidy of bread wheat and its relatives, we speculate that the theoretical numbers of DHN genes should be 17 in Ae. tauschii, 17 in T. urartu, 34 in T. dicoccoides, and 52 in T. aestivum. However, TaDHN9-B, TdDHN5-B2/10-B/12-B3, TuDHN2/10, and AetDHN5–2 were missing genes, and with tandem duplication events occurring in TaDHN12-A/B/D, it resulted in the actual gene number deviating slightly from the theoretical gene number (Table S1). In the phylogenetic analysis (Fig. 1), genes belonging to the same group occasionally had individual genes encoding a DHN type that varied from most genes in the group (e.g., Group II genes mostly encoded YSK2 type DHNs, but TaDHN12-A3 encoded SK2 type DHNs). After manually checking the sequence, we found that this occurred due to sequence truncation or potential mis-annotation. The first 28 amino acids of TaDHN12-A3 were mis-annotated, resulting in a loss of the Y-segment.
Analysis of cis-acting elements in the promoter regions of TaDHN genes
DHN genes play important roles in response to various stressors. Cis-acting elements control their target gene expression by interacting with transcription factors [39, 40]. Hence, identifying the cis-acting elements will help understand the potential regulatory mechanism. We analyzed the 1500-bp upstream region from the start codon (ATG) for putative cis-acting elements of all stress-responsive TaDHN genes, and identified eight different types of cis-acting elements (Fig. S4), Among them, the abscisic acid (ABA) responsive element (ABRE), the methyl jasmonate (MeJA) responsive element (MeJA-RE), and the TCA-element are involved in hormone signaling, whereas the drought responsive element (DRE1/DRE core), low temperature responsive (LTR), TC-rich, and MYB binding site (MBS) are involved in the abiotic stress response. The results show varied distribution and abundance of the cis-acting elements among the 55 DHN promoters (Fig. S4). The ABRE elements involved in ABA signaling and osmotic stress [41–43] appeared in all DHN gene promoter regions, and the DRE1/DRE core being abundantly present in 45 DHN promoter regions ensured that DHN gene expression was regulated in response to drought stress [44]. MeJA-RE appeared in the promoter regions of 41 DHN genes, followed by TC-rich repeats and LTR being present in 21. These three types of cis-acting elements also play critical roles in response to abiotic or biotic stress [45]. The MBS cis-acting element appearing in 16 DHN genes is important for the stress (esp. drought) response and ABA signaling [28]. Taken together, the wide distribution of various hormone and stress responsive elements in the promoter regions demonstrates that DHN genes are potentially involved in the environmental stress response in plants.
Expression profile of DHN genes in different tissues and in response to various biotic stressors
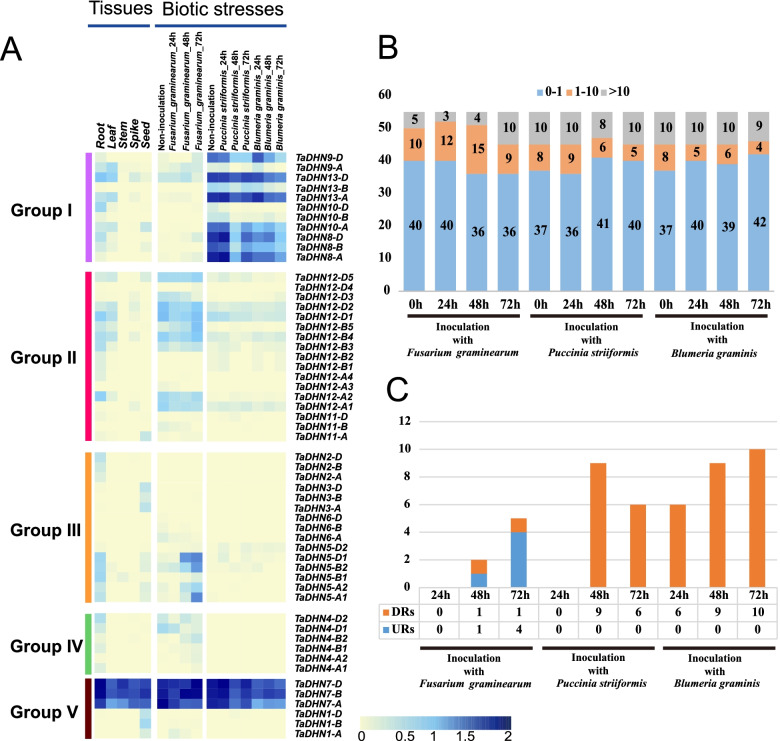
We analyzed the RNA-seq data of five tissues/organs (roots, leaves, stems, spikes, and seeds) to characterize the expression of the bread wheat DHN genes. Of the 55 TaDHN genes, while 65% (n = 36) were expressed in at least one tissue, with a wide expression level range (tpmmax = 1–204) (Fig. 4A and Table S3), the remaining 35% showed no or very low expression (tpmmax < 1), like DHN6 and DHN13 (Fig. 4A and Table S3). About 48% (n = 26) of the DHN genes were expressed in roots (DHN2/4/5 and some DHN8/10/12 genes were expressed specifically in roots). Twelve DHN genes were expressed in seeds and leaves (DHN1/3 genes were expressed specifically in seeds). Few DHN genes were expressed in stems (n = 3) or spikes (n = 4) (Fig. 4A and Table S3).
Fig. 4.
Expression analysis of TaDHN genes in various tissues and stress treatments. A The expression profiles of the TaDHN genes in different tissues under biotic stress. The expression data of 55 TaDHN genes were involved in roots, leaves, stems, spikes, seeds, and pathogen infection (Fusarium graminearum, Fusarium head blight (FHB); Blumeria graminis, powdery mildew; Puccinia striiformis, stripe rust) under different treatment times. B The number of TaDHN genes corresponding to the three expression levels (tpm: 0–1, 1–10, and > 10) under different pathogen infection times. The three expression levels are represented by different colors. (C) The number of up/down-regulated (URs or DRs) genes under different pathogen infection times. The DRs and URs are represented by orange and blue, respectively
We also analyzed the RNA-seq data of bread wheat inoculated with F. graminearum, B. graminis, or P. striiformis to investigate how TaDHN genes function in response to biotic stress. About 55% (n = 30) of the DHN genes (DHN4/5/7/8/9/10/12/13) were expressed with a wide expression level range (tpmmax = 1–671) (Fig. 4A and Table S3). To accurately understand how DHN genes respond to different biotic stressors, we divided them into three categories according to gene expression levels (tpm): 0–1 (no to low), 1–10 (medium), and > 10 (high). Most of the DHN genes showed no or low expression, while many showed medium expression (Fig. 4B). Although the number of highly expressed genes changed with increasing inoculation time, the DHN gene family did not respond strongly when inoculated with F. graminearum, B. graminis, or P. striiformis (Fig. 4B).
We defined DHN genes with tpm fold change > 1 (treatment vs. control) and tpm value change > 10 as up- and down-regulated genes (URs and DRs) to further understand the DHN family gene expression changes under different biotic stressors. No URs or DRs were detected after 24 h in bread wheat inoculated with F. graminearum. There was one DR and one UR at 48 h and one DR and four URs at 72 h (Fig. 4C). TaDHN5-D1 was up-regulated at 48 and 72 h, while TaDHN5-A1/B2 and TaDHN12-B5 were up-regulated only at 72 h (Fig. 4A and Table S3). However, no URs occurred in bread wheat inoculated with B. graminis or P. striiformis, and most genes were either down-regulated or had a very low tpm value (Fig. 4C).
Expression profile of DHN genes in response to various abiotic stressors
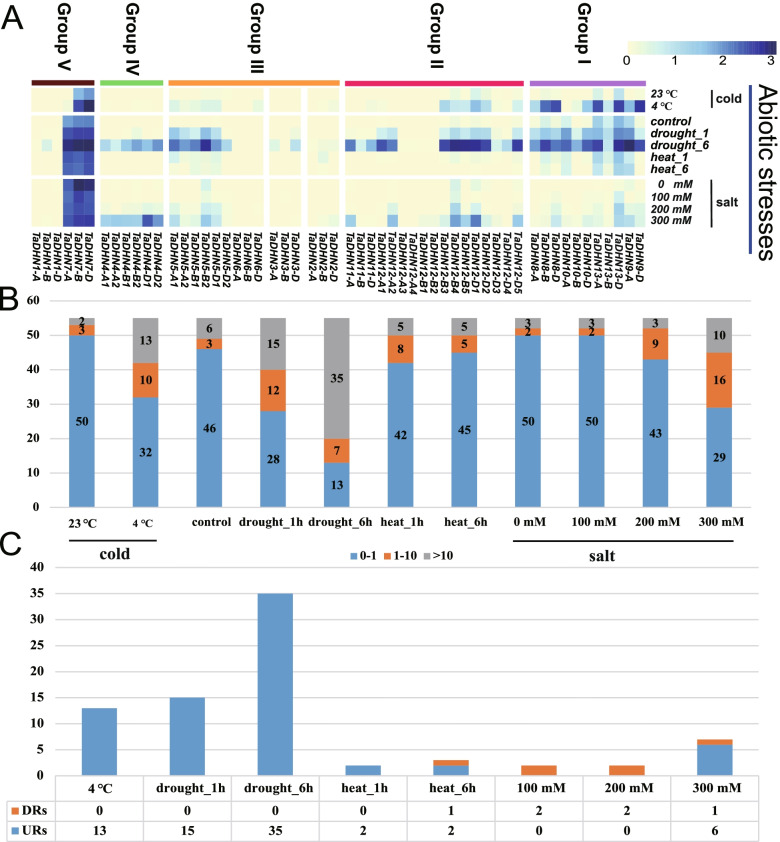
We analyzed the bread wheat RNA-seq data under cold, drought, heat, and salt conditions to understand how TaDHN genes respond to abiotic stress. Of the 55 TaDHN genes, 78% (n = 43) were expressed, and genes from DHN1/2/3 showed no or very low expression (Fig. 5A and Table S3). We also analyzed the DHN family gene expression levels under the four different abiotic stressors. Ten DHN genes showed medium level expression under cold stress (1 < tpm < 10), while fifteen were highly expressed (tpm > 10) (Fig. 5B). Under drought stress, while twelve and fifteen genes had medium and high expression at 1 h, seven had medium and thirty-five had high expression at 6 h. When we subjected bread wheat to heat stress, only five DHN genes were highly expressed at 1 and 6 h, and eight and five were expressed at medium levels at 1 and 6 h, respectively (Fig. 5B). The DHN genes were mostly insensitive to 100 or 200 mM NaCl, as only three genes each was highly expressed at both concentrations, while only two and nine had medium expression levels at 100 and 200 mM NaCl, respectively. However, 10 and 16 DHN genes had high and medium expression levels, respectively, at 300 mM NaCl (Fig. 5B).
Fig. 5.
Expression analysis of the TaDHN genes under various abiotic stressors. A The expression profiles of the TaDHN genes under the abiotic stress treatments. The expression levels of 55 TaDHN genes changed in response to cold, drought, heat, and salt under different treatment times and concentrations. B The numbers of TaDHN genes corresponding to the three expression levels (tpm: 0–1, 1–10, and > 10) under different abiotic treatment times. The three expression levels are represented by different colors. C The number of up/down-regulated (URs or DRs) genes under different abiotic treatment times. The DRs and URs are represented by orange and blue, respectively
Then, we analyzed the DRs and URs of the DHN family in response to the four abiotic stressors. No DRs appeared in response to the cold or drought stressors, but we found 13 URs under cold stress. Most of the 15 and 35 URs responding to drought stress at 1 and 6 h, respectively (Fig. 5C), were highly expressed (Fig. 5A and Table S3). In summary, most DHN genes were insensitive to both heat and 100/200 mM NaCl stressors, as we detected very few DRs or URs under both these conditions. We also detected six URs under the 300 mM NaCl stress, indicating that DHN genes are sensitive to high salinity. Most of the Group I genes expressing Kn type proteins mainly under cold and drought stress were URs (Fig. 5A and C). In contrast, although some Group I DHN genes had high tpm values under the B. graminis and P. striiformis inoculation, surprisingly most were DRs (Fig. 4C).
Response of DHN genes under various hormone treatments
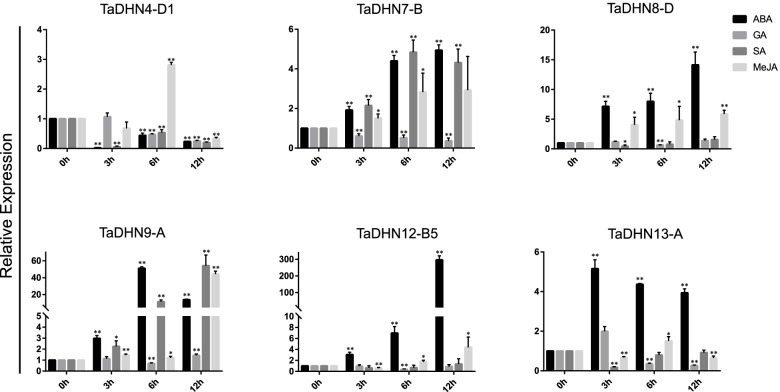
To understand the roles of the TaDHN genes in response to hormones, six TaDHN genes with higher expression levels under various stress conditions were selected to analyze their expression profiles. We found that all responded strongly to the ABA treatment (Fig. 6). Among them, while ABA treatment only down-regulated TaDHN4-D1 significantly (p < 0.01), it up-regulated the others. In contrast, gibberellin (GA) treatment weakly induced or inhibited the expression of DHN genes. While salicylic acid (SA) treatment significantly upregulated TaDHN7-B and TaDHN9-A (p < 0.01), it either down-regulated or did not affect the other genes (Fig. 6). However, MeJA treatment significantly induced all selected genes (p < 0.01 or p < 0.05); TaDHN4-D1 and TaDHN13-A peaked at 6 h, TaDHN9-A and TaDHN12-B5 peaked at 12 h, and TaDHN7-B and TaDHN8-D were up-regulated throughout the entire MeJA treatment period (Fig. 6). In summary, the DHN genes showed various expression patterns under different hormone treatments. All selected DHN genes were highly sensitive to the ABA treatment (particularly TaDHN9-A and TaDHN12-B5 with strikingly high expression). Since ABA signaling is very important in regulating plant stress response [41], the abundance of ABA-related cis-acting elements (Fig. S4) and the strong response of the DHN genes to the ABA treatment reflects the crucial role of the DHN gene family in various stress conditions.
Fig. 6.
Expression analysis of six selected TaDHN genes under different hormone treatments. Expression profiles of six selected TaDHN genes (including TaDHN4-D1, TaDHN7-B, TaDHN8-D, TaDHN9-A, TaDHN12-B5, and TaDHN13-A) were analyzed under ABA (100 μM), GA (50 μM), SA (100 μM), and MeJA (100 μM) treatments. Error bars represent standard deviations (SDs) calculated from three independent biological replicates. P < 0.05 (*) and P < 0.01(**) by Student’s t-test
Interaction network and subcellular localization
In order to further understand how the abiotic stress-induced DHN proteins function, we used the STRING database to annotate the proteins encoded by the wheat DHN genes and their Ae. tauschii homologs (Table S4). Then, using the well-studied AetDHNs we constructed an interaction network (Fig. S5). We found that these DHN proteins were not only closely connected (except EMT32858 and EMT15121, which are annotated as cold shock proteins), but also their functions covered many aspects of wheat abiotic stress response. For example, DHN8/9/10/14 encode cold shock proteins, DHN4/12/13 encode salt-induced proteins (Fig. S5 and Table S4), while DHN5/4–1/12 encode EMT-25371/30993/24840 that interact with a heat shock protein, EMT106830 (Fig. S5 and Table S4). Moreover, we speculate that DHN proteins can cooperate with each other when plants are under abiotic stress.
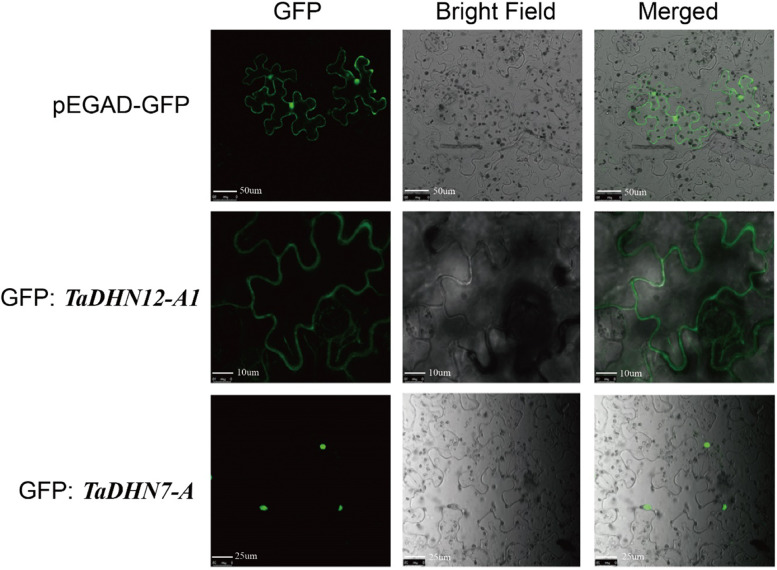
Using bioinformatics, we also predicted the subcellular location of the DHN protein to understand where it might function (Table 1). In order to determine and experimentally verify the accuracy of the prediction, we selected two genes, TaDHN12-A1 and TaDHN7-A1, and found that their encoded proteins were indeed located in the cytoplasm and nucleus, respectively (Fig. 7). The results thus verify and confirm the accuracy of the bioinformatics prediction.
Fig. 7.
Subcellular localization of selected TaDHN-GFP fusion proteins in N. benthamiana leaf epidermal cells. TaDHN12-A1 and TaDHN7-A were localized to the cytoplasm and nucleus, respectively. pEGAD-GFP was used as a positive control
Discussion
Since many DHN genes are key to protecting plants from various environmental stressors, they are potential candidates for crop breeding and improvement. Using a comprehensive approach in this study, we identified 55 DHN genes in hexaploid bread wheat (T. aestivum), 31 in tetraploid durum wheat (T. dicoccoides), 15 in diploid T. urartu, and 16 in diploid Ae. tauschii.
Identification of DHN genes in bread wheat and its relatives
According to the chromosomal distribution and homologous relationship of the DHN gene family (Fig. 2 and Table S1), they were unevenly distributed among different homologous groups, with most being distributed in homologous groups 5 and 6. We also found similar distribution in seven other sub-genomes (Ta-A/B/D, Td−/A/B, Tu-A, and Aet-D), thus providing high confidence for the identification. We also observed the translocation events occurred on T. urartu 4A chromosome; TaDHN3-A/B/D, TdDHN3-A/B, and AetDHN3-D were distributed on the homologous group 4 distal long arm; while TuDHN3 was located on the distal short arm. A previous T. urartu genome study had already reported the translocation event [12]. The close relationships between homologous groups greatly improved the accuracy of identification. For example, based on the annotation file, TuDHN5–2 was initially placed on the unmapped chromosome. But based on its high similarity with DHN5 genes located on seven other sub-genomes, TuDHN5–2 was re-assigned on the 5A chromosome. The missing genes were also identified according to the relationships between homologous groups, like TuDHN2 was identified as a missing gene because although TaDHN2-A/B/D, TdDHN2-A/B, and AetDHN2-D were localized to homologous group 3, their corresponding homoeologs on chromosome 3A of T. urartu genome were absent. Similar gene loss is widespread in other gene families and may occur during the wheat polyploidization process [5, 46, 47]. Although genes were missing from some genomic regions, homologous relationships were clear among the DHN genes located in different diploid sub-genomes, indicating that this gene family is evolutionarily well-conserved. The gene families in different sub-genomes of polyploid plants, such as bread wheat, particularly small or medium-sized families, are conserved in number or sequences. Using the sequence similarity of the gene family between sub-genomes, we can accurately identify the target genes. Furthermore, since automated annotation generates truncated and mis-annotated genes, additional manual checking is necessary for proper identification.
Evolution and expansion of the DHN genes among bread wheat and its relatives
Gene duplication occurs in different ways, including whole-genome duplication, segmental duplication, and single-gene duplication (including tandem and dispersed duplications) events. Duplication events are important in expanding a gene family [48–50]. According to the chromosomal distribution and syntenic relationships between bread wheat and its relatives, allopolyploid events were the main driving force behind expanding the hexaploid wheat DHN family. The DHN4/5/12 genes have undergone tandem duplication events. Three DHN12 genes occur in the diploid genome of wheat ancestors (one gene is missing in Td-B), while the DHN12 gene number has changed in the three wheat sub-genomes (A, B, and D). The A sub-genome of bread wheat has one more copy than the A genome of the ancestors. The B and D sub-genomes have two more copies than the B and D genomes of the ancestors, respectively (Table S1). We analyzed the TaDHN12 genes as tandems. Therefore, we speculate that the DHN gene family in bread wheat has undergone tandem duplication events after polyploidization, leading to more bread wheat DHN genes in the family than in its diploid donors.
The DHN family is a small family present in many plant species, like seven members in Oryza sativa [51], ten in Arabidopsis [52, 53], seven in Pyrus pyrifoli [54], and four each in Vitis vinifera and V. yeshanensis [16]. Bread wheat has the largest DHN gene number (55) among the above-mentioned plants. Even its diploid ancestors T. urartu and Ae. tauschii have greater number of DHN genes than other plant species, i.e., 15 and 17, respectively. Bread wheat has > 7.8 times higher number of DHN genes than rice, and this phenomenon cannot generally be explained by their ploidy. We hypothesize that the expanding DHN gene family may help Triticeae crops rapidly adapt to different stress conditions, particularly water-related stressors, like drought, therefore, contributing to the global distribution of bread wheat and its relatives. Whether we can detect DHN gene copy number variations in different wheat varieties is an interesting issue, with the recent release of the wheat pan-genomic data [55].
In the present study, tandem duplications occurred in linkage groups five and six, and tandem duplication genes (DHN4/5/12) appeared in clusters at corresponding chromosomes. The genes were combined with the expression profile results. Interestingly, while these tandem genes were mostly up-regulated under various abiotic stressors, the non-tandem genes like DHN1/2/3/6 had no or very low expression (low tpm values), thus indicating that non-tandem genes are abiotic stress-insensitive. Notably, drought and cold stress up-regulated the DHN 8/9/10/11 genes with high tpm values (Fig. 5A). These genes and the DHN12 genes existed as gene clusters and were continuous in position (Fig. 2 and Table 1). These findings combined with the sequence characteristics indicate that these genes (DHN8/9/10/11/12) may have originated from tandem duplications of an ancestral gene. The need for ecological adaptability pushed them to subsequently evolve into the Kn and YSK2 groups. Adaptive evolution may have driven these tandem duplication events in the DHN gene family. Thus, tandem duplication events are the main reason for expansion of the DHN gene family in bread wheat diploid donors.
Expression analysis of the TaDHN genes
We analyzed the expression profiles of the bread wheat DHN genes under various biotic and abiotic stress conditions. TaDHN1/2/3/6 were insensitive to all the stress conditions in this study, with no or very low expression in all six tissues (Fig. 4A and Fig. 5A). The TaDHN4 genes had root-specific expression and were mainly drought- and salt-inducible (Fig. 4A). Previous studies demonstrated that the TaDHN5 genes and their homologs contributed towards drought and salt tolerance [56, 57]. This is consistent with our expression profile analysis results, which showed TaDHN5 genes were indeed drought and salt stress-inducible (Fig. 5A). Some TaDHN5 genes were also biotic stress-inducible (inoculation with F. graminearum), indicating that these genes may help in resistance against F. graminearum.
The TaDHN7 genes were generally highly expressed (high tpm values) under all stress conditions (except DHN7-A in cold stress) and were constitutively expressed in all tissues (Fig. 4A and Fig. 5A). Among all DHN genes in bread wheat, TaDHN7-A/B/D are the only three genes that encode SK3-type proteins (Table 1). A previous study identified many SK3-type DHN genes having important functions under various abiotic stress conditions. For example, overexpression of ShDHN in tomato not only improves drought and cold stress tolerance, but also seedling growth under osmotic and salt stress [33]. Overexpression of MusaDHN-1 in banana improves drought and salt stress tolerance [58]. A functional analysis demonstrated that SpDHN1 in Stipa purpurea is important in drought stress resistance [59]. These studies of SK3-type DHN proteins indicate that TaDHN7 genes may also be important in bread wheat facing various abiotic stressors. Taken together, TaDHN genes mainly responded to cold, drought, and high salinity stressors, but were insensitive to heat, low or medium salinity, and most biotic stressors.
Conclusions
We comprehensively analyzed the DHN gene family, using molecular characterization, phylogenetic classification, chromosomal distributions, gene structure, conserved motifs, and missing, truncated, and mis-annotated genes, as well as cis-acting elements. Based on six RNA-seq datasets, the DHN genes exhibited distinct tissue-specific expression patterns, and we identified the induced genes under different stress conditions. Conserved, duplicated DHN genes may be important in helping wheat adapt to various conditions, therefore, contributing to its distribution as a global staple food. The cooperation of multiple DHNs may be important in protecting plants from abiotic stress. Therefore, our study results will not only help in further study of the stress resistance mechanisms of the DHN gene family, but also facilitate wheat breeding by fine-tuning its important traits.
Methods
Plant materials
The bread wheat variety “Chinese Spring” and N. benthamiana were used for RT-PCR and subcellular localization, respectively. And these materials are presented from State Key Laboratory of Crop Biology, College of Agronomy, Shandong Agricultural University (Taian, China).
Sequence search, identification, and naming of the DHN genes
The genome sequences and gene annotations of bread wheat (T. aestivum) and wild emmer wheat (T. dicoccoides) were obtained from the Ensemble Plants website (http://plants.ensembl.org/) [60]. The genome files for T. urartu and Ae. tauschii were obtained from the (http://www.mbkbase.org/Tu/) and (http://aegilops.wheat.ucdavis.edu/ATGSP/annotation/) websites, respectively (Table S5) [13, 61]. To identify the DHN genes in bread wheat and its relatives, HMMER 3.1 (http://www.hmmer.org/) with default parameter settings and the BLAST algorithm for proteins (BLASTP) with the threshold expectation value set to 1E-20 were performed using the hidden Markov model (HMM) (version 3.0) profiles of the dehydrin domain (PF00257) obtained from the Pfam database (http://pfam.xfam.org/) as the query [62, 63]. We merged all hits obtained and removed the redundant hits. All non-redundant protein sequences were further analyzed with the NCBI conserved domain database (CDD, https://www.ncbi.nlm.nih.gov/cdd) and InterPro (http://www.ebi.ac.uk/interpro/) to confirm the conserved domain of the DHN protein in each candidate sequence [64, 65]. The methodology flowchart of the identification of DHN gene family was also provided (Fig. S6). Tandem genes were screened by a custom Perl script, according to the following standards: (i) length of alignable sequence covers > 70% of longer gene; (ii) similarity of aligned regions > 70%; (iii) The physical distance between the align genes on the chromosome < 500 kb.
We suggest a consistent naming pattern for all DHN genes of bread wheat and its relatives, considering the genomic location and phylogenetic and syntenic relationships of the DHN genes between different diploid sub-genomes (Ta/Td/Tu-A, Ta/Td-B, and Ta/Aet-D). (i) Each DHN gene name starts with an abbreviation for the species name. For example, T. aestivum (Ta), followed by the abbreviation of dehydrin gene family: DHN; (ii) the gene names include an A, B, or D, indicating the sub-genome where they are located. For example, TaDHN1-A; (iii) putative homologs between sub-genomes have identical gene names except for the sub-genome identifier or species name (e.g., TaDHN7-A, TaDHN7-B, TaDHN7-D, TdHN7-A, TdDHN7-B, TuDHN7, and AetDHN7); (iv) tandem genes are consecutively numbered (e.g., TaDHN4-A1 and TaDHN4-A2).
Phylogenetic and synteny analysis
All identified DHN protein sequences were aligned using the MUSCLE [66] program with default parameters. Phylogenetic trees were constructed using MEGA X software with the neighbor joining method and the following parameters: bootstrap (1000 replicates) and the Jones-Taylor-Thornton substitution model [67].
All identified DHN genes in wheat and its relatives were located on pseudo-chromosomes based on the physical location information acquired from the genomic database. To understand the relationship between the DHN genes identified in wheat and its relatives at the genomic level, the hexploid bread wheat (Ta) and tetraploid durum wheat (Td) genomes were split into three and two diploid sub-genomes (AA, BB, DD and AA, BB), respectively. A collinear analysis was performed using the five sub-genomes with diploid T. urartu and Ae. tauschii genomes and JCVI software (https://github.com/tanghaibao/jcvi/wiki). The results were visualized by Circos [68].
Analysis of DHN gene characteristics and SSRs mining
Isoelectric points and molecular weights were determined using ExPASy (https://web.expasy.org/protparam/). Subcellular localization of all DHN genes was predicted using the Cell-PLoc (version 2.0) website (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/) [69]. Exon-intron structures of the DHN genes in bread wheat and its relatives were displayed using the Gene Structure Display Server (GSDS, http://gsds.gao-lab.org/index.php) [70]. The promoter sequences (1500-bp upstream of the ATG translation start codon) of the DHN genes were extracted from the bread wheat genome sequence (IWGSC v1.0). Cis-acting elements were predicted in the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [39], and the promoter sequences was provided (Table S6). The SSRs mining analysis was performed by GMATA software [71], and the specific markers were developed by Primer-BLAST [72].
Expression profiles of the DHN genes in RNA-seq
To understand the expression profiles of the DHN genes in different tissues and under different stress conditions, six transcriptome datasets were downloaded from the NCBI (https://www.ncbi.nlm.nih.gov/) with accession numbers SRP043554, SRP045409, SRP300360, SRP041017, ERP013829, and ERP107574.
The RNA-seq data accession numbers SRP043554, SRP300360 and ERP013829 involved cold, salt and FHB infections. The SRP045409 data involved drought and heat stress. The SRP041017 data involved stripe rust and powdery mildew. The ERP107574 data were collected from various bread wheat tissues. The expression levels of the DHN genes were quantified as transcripts per kilobase million (TPM). The tpm value was calculated using Kallisto software [73].
Plant cultivation, RNA isolation and RT-PCR
To investigate the expression patterns of the DHN genes in wheat under different hormone treatments, T. aestivum cv. Chinese Spring was used for the reverse transcription-polymerase chain reaction (RT-PCR) analysis. Bread wheat was planted in a growth chamber at 23 °C under a 16 h/8 h (light/dark) photoperiod. Then, 2-week-old seedlings were transferred to a hormone treatment solution containing 100 μM ABA, 50 μM GA, 100 μM SA, or 100 μM MeJA. The leaf tissues were harvested at 0, 3, 6, and 12 h and stored at − 80 °C after being frozen in liquid nitrogen. Total RNA of all samples was extracted using the RNAprep Pure Plant Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. cDNA was generated with a one-step reverse transcription kit (TIANGEN). The Lightcycler 96 system (Roche, Mannheim, Germany) was used for the RT-PCR assay with the SYBR qPCR Master Mix (Vazyme, Nanjing, China); three technical replicates were carried out. Primer information could be found in supplementary (Table S7).
Interaction network construction and subcellular localization
STRING website (https://string-db.org/) was used to analyze the interaction of DHN proteins with a confidence parameter set at 0.4 threshold [74]. Gene-specific primers were designed to amplify the coding sequences of the two selected TaDHN genes (Table S8). Amplified fragments were ligated in-frame to the 5′-terminus with the expression vector pEGAD-GFP. Then, Constructed plasmids were infiltrated into abaxial air space of six-week-old N. benthamiana leaves using the transformed Agrobacterium strain GV3101. Infiltrated parts of the leaves were marked and fluorescence was observed under the confocal laser scanning microscope (Leica, German) after 48 h of infiltration.
Supplementary Information
Acknowledgements
We would like to thank State Key Laboratory of Crop Biology, Shandong Agricultural University for providing computing resources during data analysis. We would like to thank TopEdit (www.topeditsci.com) for its linguistic assistance during the preparation of this manuscript.
Authors’ contributions
HWW designed the research; YCH and YJC analyzed the data. MH performed the experiments; YCH wrote the manuscript, HWW and LRK revised the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This work were supported by the National Natural Science Foundation of China (31871610 and 32171963) and the Shandong Provincial Scientific Innovation Project for Young Scholars of Universities (2019KJF026).
Availability of data and materials
The datasets used during the current study are available from NCBI (https://www.ncbi.nlm.nih.gov/) with accession numbers SRP043554, SRP045409, SRP300360, SRP041017, ERP013829, and ERP107574. The materials (“Chinese Spring” and N. benthamiana) used to support the findings of this study are available from the corresponding author upon request.
Declarations
Ethics approval and consent to participate
The materials are owned by the corresponding author, no permissions are required. The study is in compliance with relevant institutional, national, and international guidelines and legislation..
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yongchao Hao and Ming Hao contributed equally to this work.
References
- 1.Salamini F, Ozkan H, Brandolini A, Schafer-Pregl R, Martin W. Genetics and geography of wild cereal domestication in the near east. Nat Rev Genet. 2002;3(6):429–441. doi: 10.1038/nrg817. [DOI] [PubMed] [Google Scholar]
- 2.Shiferaw B, Smale M, Braun H-J, Duveiller E, Reynolds M, Muricho G. Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Security. 2013;5(3):291–317. [Google Scholar]
- 3.Pont C, Leroy T, Seidel M, Tondelli A, Duchemin W, Armisen D, Lang D, Bustos-Korts D, Goue N, Balfourier F, et al. Tracing the ancestry of modern bread wheats. Nat Genet. 2019;51(5):905–911. doi: 10.1038/s41588-019-0393-z. [DOI] [PubMed] [Google Scholar]
- 4.Liu X, Liu Z, Niu X, Xu Q, Yang L. Genome-wide identification and analysis of the NPR1-like gene family in bread wheat and its relatives. Int J Mol Sci. 2019;20(23):5974. doi: 10.3390/ijms20235974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hao Y, Xu S, Lyu Z, Wang H, Kong L, Sun S. Comparative analysis of the glutathione S-Transferase gene family of four Triticeae species and Transcriptome analysis of GST genes in common wheat responding to salt stress. Int J Genomics. 2021;2021:6289174. doi: 10.1155/2021/6289174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curtis T, Halford NG. Food security: the challenge of increasing wheat yield and the importance of not compromising food safety. Ann Appl Biol. 2014;164(3):354–372. doi: 10.1111/aab.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rey E, Abrouk M, Keeble-Gagnere G, Karafiatova M, Vrana J, Balzergue S, Soubigou-Taconnat L, Brunaud V, Martin-Magniette ML, Endo TR, et al. Transcriptome reprogramming due to the introduction of a barley telosome into bread wheat affects more barley genes than wheat. Plant Biotechnol J. 2018;16(10):1767–1777. doi: 10.1111/pbi.12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian R, Yang Y, Chen M. Genome-wide survey of the amino acid transporter gene family in wheat (Triticum aestivum L.): identification, expression analysis and response to abiotic stress. Int J Biol Macromol. 2020;162:1372–1387. doi: 10.1016/j.ijbiomac.2020.07.302. [DOI] [PubMed] [Google Scholar]
- 9.Figueroa M, Hammond-Kosack KE, Solomon PS. A review of wheat diseases-a field perspective. Mol Plant Pathol. 2018;19(6):1523–1536. doi: 10.1111/mpp.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.International Wheat Genome Sequencing C, investigators IRp, Appels R, Eversole K, Feuillet C, Keller B, Rogers J, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361(6403):eaar7191. [DOI] [PubMed]
- 11.Avni R, Nave M, Barad O, Baruch K, Twardziok SO, Gundlach H, Hale I, Mascher M, Spannagl M, Wiebe K, et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science. 2017;357:93–97. doi: 10.1126/science.aan0032. [DOI] [PubMed] [Google Scholar]
- 12.Ling HQ, Ma B, Shi X, Liu H, Dong L, Sun H, Cao Y, Gao Q, Zheng S, Li Y, et al. Genome sequence of the progenitor of wheat a subgenome Triticum urartu. Nature. 2018;557(7705):424–428. doi: 10.1038/s41586-018-0108-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo MC, Gu YQ, Puiu D, Wang H, Twardziok SO, Deal KR, Huo N, Zhu T, Wang L, Wang Y, et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature. 2017;551(7681):498–502. doi: 10.1038/nature24486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramirez-Gonzalez RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, et al. The transcriptional landscape of polyploid wheat. Science. 2018;361(6403):eaar6089. [DOI] [PubMed]
- 15.Zhang HF, Liu SY, Ma JH, Wang XK, Haq SU, Meng YC, Zhang YM, Chen RG. CaDHN4, a salt and cold stress-responsive Dehydrin gene from pepper decreases Abscisic acid sensitivity in Arabidopsis. Int J Mol Sci. 2019;21(1):26. doi: 10.3390/ijms21010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, He M, Zhu Z, Li S, Xu Y, Zhang C, Singer SD, Wang Y. Identification of the dehydrin gene family from grapevine species and analysis of their responsiveness to various forms of abiotic and biotic stress. BMC Plant Biol. 2012;12:140. doi: 10.1186/1471-2229-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hand SC, Menze MA, Toner M, Boswell L, Moore D. LEA proteins during water stress: not just for plants anymore. Annu Rev Physiol. 2011;73:115–134. doi: 10.1146/annurev-physiol-012110-142203. [DOI] [PubMed] [Google Scholar]
- 18.Close TJ. Dehydrins: a commonalty in the response of plants to dehydration and low temperature. Physiol Plant. 1997;100:291–296. [Google Scholar]
- 19.Timothy JC. Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol Plant. 1996;97:795–803. [Google Scholar]
- 20.Cuevas-Velazquez CL, Rendon-Luna DF, Covarrubias AA. Dissecting the cryoprotection mechanisms for dehydrins. Front Plant Sci. 2014;5:583. doi: 10.3389/fpls.2014.00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Z, Wang X, Zhang L. Structural and functional dynamics of Dehydrins: a plant protector protein under abiotic stress. Int J Mol Sci. 2018;19(11):3420. doi: 10.3390/ijms19113420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008;148(1):6–24. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo D, Hou X, Zhang Y, Meng Y, Zhang H, Liu S, Wang X, Chen R. CaDHN5, a Dehydrin gene from pepper, plays an important role in salt and osmotic stress responses. Int J Mol Sci. 2019;20(8):1989. doi: 10.3390/ijms20081989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stival Sena J, Giguere I, Rigault P, Bousquet J, Mackay J. Expansion of the dehydrin gene family in the Pinaceae is associated with considerable structural diversity and drought-responsive expression. Tree Physiol. 2018;38(3):442–456. doi: 10.1093/treephys/tpx125. [DOI] [PubMed] [Google Scholar]
- 25.Perdiguero P, Barbero MC, Cervera MT, Soto A, Collada C. Novel conserved segments are associated with differential expression patterns for Pinaceae dehydrins. Planta. 2012;236(6):1863–1874. doi: 10.1007/s00425-012-1737-4. [DOI] [PubMed] [Google Scholar]
- 26.Campbell S, Close TJ. Dehydrins: genes, proteins, and associations with phenotypic traits. New Phytol. 1997;137:61–74. [Google Scholar]
- 27.Liu Y, Song Q, Li D, Yang X, Li D. Multifunctional roles of plant Dehydrins in response to environmental stresses. Front Plant Sci. 2017;8:1018. doi: 10.3389/fpls.2017.01018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Zheng J, Su H, Xia K, Jian S, Zhang M. Molecular cloning and functional characterization of the Dehydrin (IpDHN) gene from Ipomoea pes-caprae. Front Plant Sci. 2018;9:1454. doi: 10.3389/fpls.2018.01454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagaraju M, Kumar SA, Reddy PS, Kumar A, Rao DM, Kavi Kishor PB. Genome-scale identification, classification, and tissue specific expression analysis of late embryogenesis abundant (LEA) genes under abiotic stress conditions in Sorghum bicolor L. PLoS One. 2019;14(1):e0209980. doi: 10.1371/journal.pone.0209980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maskin L, Frankel N, Gudesblat G, Demergasso MJ, Pietrasanta LI, Iusem ND. Dimerization and DNA-binding of ASR1, a small hydrophilic protein abundant in plant tissues suffering from water loss. Biochem Biophys Res Commun. 2007;352(4):831–835. doi: 10.1016/j.bbrc.2006.11.115. [DOI] [PubMed] [Google Scholar]
- 31.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 32.Tompa P, Szasz C, Buday L. Structural disorder throws new light on moonlighting. Trends Biochem Sci. 2005;30(9):484–489. doi: 10.1016/j.tibs.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Liu H, Yu C, Li H, Ouyang B, Wang T, Zhang J, Wang X, Ye Z. Overexpression of ShDHN, a dehydrin gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses in tomato. Plant Sci. 2015;231:198–211. doi: 10.1016/j.plantsci.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 34.Chiappetta A, Muto A, Bruno L, Woloszynska M, Van Lijsebettens M, Bitonti MB. A dehydrin gene isolated from feral olive enhances drought tolerance in Arabidopsis transgenic plants. Front Plant Sci. 2015;6:392. doi: 10.3389/fpls.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bao F, Du D, An Y, Yang W, Wang J, Cheng T, Zhang Q. Overexpression of Prunus mume Dehydrin genes in tobacco enhances tolerance to cold and drought. Front Plant Sci. 2017;8:151. doi: 10.3389/fpls.2017.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao Y, Xiang X, Geng M, You Q, Huang X. Effect of HbDHN1 and HbDHN2 genes on abiotic stress responses in Arabidopsis. Front Plant Sci. 2017;8:470. doi: 10.3389/fpls.2017.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie C, Zhang R, Qu Y, Miao Z, Zhang Y, Shen X, Wang T, Dong J. Overexpression of MtCAS31 enhances drought tolerance in transgenic Arabidopsis by reducing stomatal density. New Phytol. 2012;195(1):124–135. doi: 10.1111/j.1469-8137.2012.04136.x. [DOI] [PubMed] [Google Scholar]
- 38.Robin C, Capron G, Desprez-Loustau ML. Root infection by Phytophthora cinnamomi in seedlings of three oak species. Plant Pathol. 2001;50:708–716. [Google Scholar]
- 39.Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, YVd P, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu S, Wang X, Chen W, Yao J, Li Y, Fang S, Lv Y, Li X, Pan J, Liu C, et al. Cotton DMP gene family: characterization, evolution, and expression profiles during development and stress. Int J Biol Macromol. 2021;183:1257–1269. doi: 10.1016/j.ijbiomac.2021.05.023. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida T, Mogami J, Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol. 2014;21:133–139. doi: 10.1016/j.pbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 42.Trivedi DK, Gill SS, Tuteja N. Abscisic acid (ABA): biosynthesis, regulation, and role in abiotic stress tolerance. Abiotic Stress Response in Plants. 2016;15:311–322. [Google Scholar]
- 43.Bulgakov VP, Avramenko TV. Linking Brassinosteroid and ABA signaling in the context of stress acclimation. Int J Mol Sci. 2020;21(14):5108. [DOI] [PMC free article] [PubMed]
- 44.Yan H, Wang Y, Hu B, Qiu Z, Zeng B, Fan C. Genome-wide characterization, evolution, and expression profiling of VQ gene family in response to Phytohormone treatments and abiotic stress in Eucalyptus grandis. Int J Mol Sci. 2019;20(7):1765. doi: 10.3390/ijms20071765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeda S, Sugimoto K, Otsuki H, Hirochika H. A 13-bp cis-regulatory element in the LTR promoter of the tobacco retrotransposon Tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant J. 1999;4:383–393. doi: 10.1046/j.1365-313x.1999.00460.x. [DOI] [PubMed] [Google Scholar]
- 46.Kumar A, Batra R, Gahlaut V, Gautam T, Kumar S, Sharma M, Tyagi S, Singh KP, Balyan HS, Pandey R, et al. Genome-wide identification and characterization of gene family for RWP-RK transcription factors in wheat (Triticum aestivum L.) PLoS One. 2018;13(12):e0208409. doi: 10.1371/journal.pone.0208409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar A, Sharma M, Gahlaut V, Nagaraju M, Chaudhary S, Kumar A, Tyagi P, Gajula M, Singh KP. Genome-wide identification, characterization, and expression profiling of SPX gene family in wheat. Int J Biol Macromol. 2019;140:17–32. doi: 10.1016/j.ijbiomac.2019.08.105. [DOI] [PubMed] [Google Scholar]
- 48.Maere S, Bodt SD, Raes J, Casneuf T, Montagu MV, Kuiper M, YVd P. Modeling gene and genome duplications in eukaryotes. PNAS. 2005;102(15):5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MLaJS C. The evolutionary fate and consequences of duplicate genes. SCIENCE. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Lu X, Malik WA, Chen X, Wang J, Wang D, Wang S, Chen C, Guo L, Ye W. Differentially expressed bZIP transcription factors confer multi-tolerances in Gossypium hirsutum L. Int J Biol Macromol. 2020;146:569–578. doi: 10.1016/j.ijbiomac.2020.01.013. [DOI] [PubMed] [Google Scholar]
- 51.Verma G, Dhar YV, Srivastava D, Kidwai M, Chauhan PS, Bag SK, Asif MH, Chakrabarty D. Genome-wide analysis of rice dehydrin gene family: its evolutionary conservedness and expression pattern in response to PEG induced dehydration stress. PLoS One. 2017;12(5):e0176399. doi: 10.1371/journal.pone.0176399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hundertmark M, Hincha DK. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9:118. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bies-Etheve N, Gaubier-Comella P, Debures A, Lasserre E, Jobet E, Raynal M, Cooke R, Delseny M. Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol Biol. 2008;67(1–2):107–124. doi: 10.1007/s11103-008-9304-x. [DOI] [PubMed] [Google Scholar]
- 54.Hussain S, Niu Q, Qian M, Bai S, Teng Y. Genome-wide identification, characterization, and expression analysis of the dehydrin gene family in Asian pear (Pyrus pyrifolia) Tree Genet Genomes. 2015;11(5):1–1. [Google Scholar]
- 55.Walkowiak S, Gao L, Monat C, Haberer G, Kassa MT, Brinton J, Ramirez-Gonzalez RH, Kolodziej MC, Delorean E, Thambugala D, et al. Multiple wheat genomes reveal global variation in modern breeding. Nature. 2020;588(7837):277–283. doi: 10.1038/s41586-020-2961-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saibi W, Feki K, Ben Mahmoud R, Brini F. Durum wheat dehydrin (DHN-5) confers salinity tolerance to transgenic Arabidopsis plants through the regulation of proline metabolism and ROS scavenging system. Planta. 2015;242(5):1187–1194. doi: 10.1007/s00425-015-2351-z. [DOI] [PubMed] [Google Scholar]
- 57.Brini F, Hanin M, Lumbreras V, Amara I, Khoudi H, Hassairi A, Pages M, Masmoudi K. Overexpression of wheat dehydrin DHN-5 enhances tolerance to salt and osmotic stress in Arabidopsis thaliana. Plant Cell Rep. 2007;26(11):2017–2026. doi: 10.1007/s00299-007-0412-x. [DOI] [PubMed] [Google Scholar]
- 58.Souran MM, Dorrazehi GM, Hossein MS, Piry H, Ainali A, Dadkani AG, Dehvari M. Study of Musa DHN-1 gene expression from Banana cultivar of dwarf Cavendish under salinity treatment. Pharm Lett. 2016;5:413–418. [Google Scholar]
- 59.Yang Y, Sun X, Yang S, Li X, Yang Y. Molecular cloning and characterization of a novel SK3-type dehydrin gene from Stipa purpurea. Biochem Biophys Res Commun. 2014;448(2):145–150. doi: 10.1016/j.bbrc.2014.04.075. [DOI] [PubMed] [Google Scholar]
- 60.Howe KL, Contreras-Moreira B, De Silva N, Maslen G, Akanni W, Allen J, Alvarez-Jarreta J, Barba M, Bolser DM, Cambell L, et al. Ensembl genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 2020;48(D1):D689–D695. doi: 10.1093/nar/gkz890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng H, Wang K, Chen Z, Cao Y, Gao Q, Li Y, Li X, Lu H, Du H, Lu M, et al. MBKbase for rice: an integrated omics knowledgebase for molecular breeding in rice. Nucleic Acids Res. 2020;48(D1):D1085–D1092. doi: 10.1093/nar/gkz921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021;49(D1):D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wheeler TJ, Eddy SR. Nhmmer: DNA homology search with profile HMMs. Bioinformatics. 2013;29(19):2487–2489. doi: 10.1093/bioinformatics/btt403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48(D1):D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blum M, Chang HY, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021;49(D1):D344–D354. doi: 10.1093/nar/gkaa977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chou K-C, Shen H-B. Cell-PLoc 2.0: an improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat Sci. 2010;02(10):1090–1103. doi: 10.1038/nprot.2007.494. [DOI] [PubMed] [Google Scholar]
- 70.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X, Wang L. GMATA: An integrated software package for genome-scale SSR mining, marker development and viewing. Front Plant Sci. 2016;7:1350. doi: 10.3389/fpls.2016.01350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34(5):525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 74.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the current study are available from NCBI (https://www.ncbi.nlm.nih.gov/) with accession numbers SRP043554, SRP045409, SRP300360, SRP041017, ERP013829, and ERP107574. The materials (“Chinese Spring” and N. benthamiana) used to support the findings of this study are available from the corresponding author upon request.