Significance
Paneth cells produce granules containing antimicrobial peptides, such as α-defensin for host defense. Examination of the production and utilization of fucosyltransferase 2 (Fut2) by Paneth cells revealed two distinct subsets: Fut2+ Paneth cells and Fut2− Paneth cells. Compared with Fut2− Paneth cells, Fut2+ Paneth cells were enriched in granules containing antimicrobial peptides. IL-22 and IL-17a were found to be essential for commensal bacteria-dependent Fut2+ Paneth cell development and function as part of gut defense.
Keywords: Paneth cell, α-defensin, fucosyltransferase 2, IL-22, IL-17a
Abstract
Paneth cells are intestinal epithelial cells that release antimicrobial peptides, such as α-defensin as part of host defense. Together with mesenchymal cells, Paneth cells provide niche factors for epithelial stem cell homeostasis. Here, we report two subtypes of murine Paneth cells, differentiated by their production and utilization of fucosyltransferase 2 (Fut2), which regulates α(1,2)fucosylation to create cohabitation niches for commensal bacteria and prevent invasion of the intestine by pathogenic bacteria. The majority of Fut2− Paneth cells were localized in the duodenum, whereas the majority of Fut2+ Paneth cells were in the ileum. Fut2+ Paneth cells showed higher granularity and structural complexity than did Fut2− Paneth cells, suggesting that Fut2+ Paneth cells are involved in host defense. Signaling by the commensal bacteria, together with interleukin 22 (IL-22), induced the development of Fut2+ Paneth cells. IL-22 was found to affect the α-defensin secretion system via modulation of Fut2 expression, and IL-17a was found to increase the production of α-defensin in the intestinal tract. Thus, these intestinal cytokines regulate the development and function of Fut2+ Paneth cells as part of gut defense.
The intestinal epithelium contains various types of epithelial cell, some of which use α(1,2)fucosylation to create cohabitation niches for commensal bacteria such as Bacteroides and prevent pathogenic bacterial invasion (1–4). α(1,2)fucosylation by intestinal epithelial cells is regulated by the enzymes fucosyltransferase 1 and 2 (Fut1 and Fut2) (5). Fut1 is expressed constitutively, whereas Fut2 expression is induced by external stimuli, such as signals from commensal bacteria (5).
In our previous study, we detected α(1,2)fucose in the columnar epithelial cells of villus epithelium and in crypts of the small intestinal mucosa (2), which is where Paneth cells preferentially localize (6). Adjacent to Paneth cells are epithelial stem cells that express the stem cell–specific marker Lgr5 (7). When Paneth cell–associated mediators such as Wnt3a are supplied in the culture medium, Lgr5+ stem cells form three-dimensional organoids containing intestinal epithelial cells (8). Nonepithelial cells, such as mesenchymal cells, also provide Wnt signals to epithelial stem cells (9).
Paneth cells produce granules containing α-defensin and other antimicrobial peptides as part of host defense (10–12). Defective granule formation or reduced secretion of α-defensin in the gut lumen can lead to dysbiosis or aggravation of gastrointestinal disorders, such as severe terminal ileitis and colitis (13). A recent study profiling the types of epithelial cell in the small intestine has suggested that Paneth cells can be separated into different subsets, reflecting the various environments within the intestinal tract (14). However, little is known about the mechanism of differentiation, regulation, and function of these Paneth cell subsets.
Here, we examined α(1,2)fucosylation in Paneth cells and found two Paneth cell subtypes based on their production and utilization of Fut2. Compared with Fut2− Paneth cells, Fut2+ Paneth cells were found to produce many mature α-defensin–rich granules, from which α-defensin is secreted to the ileum. Also, we found that interleukin 22 (IL-22) and IL-17a regulated Fut2+ Paneth cell development and the production of α-defensin in the gastrointestinal tract.
Results
Existence of Fut2+ and Fut2− Paneth Cell Subsets.
First, we examined α(1,2)fucose localization by staining wild-type (WT) mouse ileum and duodenum with Ulex europaeus agglutinin-1 (UEA-1), a lectin that reacts specifically with α(1,2)fucose (15). As reported previously (2), ileal, but not duodenal, villus epithelium was positive for α(1,2)fucose production (Fig. 1A). However, ileal and duodenal crypt regions were both positive for α(1,2)fucose production (Fig. 1A). Also, as reported previously (2), in Fut2LacZ/LacZ mice [Fut2-deficient mice; α(1,2)fucose produced by Fut1 only (5, 16)], complete loss of α(1,2)fucose production was observed in ileal villus epithelial cells but not in ileal crypt cells (Fig. 1A). In contrast, in Fut1LacZ/LacZ mice [Fut1-deficient mice; α(1,2)fucose produced by Fut2 only (5, 16)], total loss of α(1,2)fucose production was observed in duodenal crypt cells but not in ileal crypt cells (Fig. 1A). These results suggest that α(1,2)fucosylation in duodenal crypt epithelial cells is mediated by Fut1 not by Fut2, whereas that in ileal crypt epithelial cells is mediated by both Fut1 and Fut2.
Fig. 1.
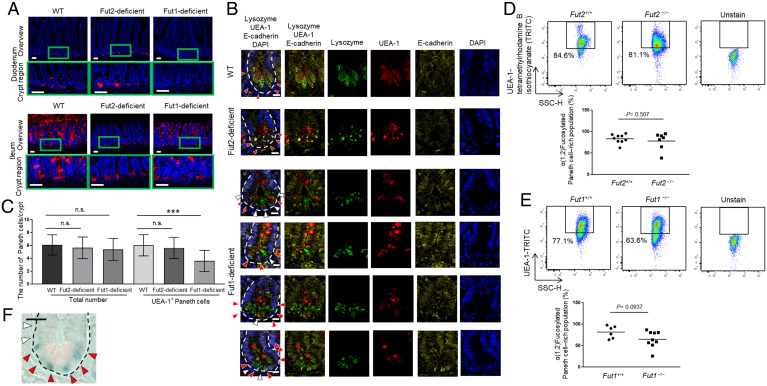
Paneth cells are divided into two subtypes: Fut2+ and Fut2− Paneth cells. (A) Sections of ileum and duodenum of WT, Fut2-deficient, and Fut1-deficient mice were stained with UEA-1 (red) and DAPI (counterstain; blue). Crypt regions are indicated with green boxes. (Scale bars, 50 µm.) Data are representative of three independent experiments. (B) Sections of ileum of WT, Fut2-deficient, and Fut1-deficient mice were stained with UEA-1 (red), anti-lysozyme antibody (green), DAPI (counterstain; blue), and anti–E-cadherin antibody (plasma membrane; yellow). Red arrows, lysozyme+ UEA-1+ cells; white arrows, lysozyme+ UEA-1− cells; and white dotted lines delineate crypts. (Scale bars, 10 μm.) Data are representative of three independent experiments. (C) Numbers of lysozyme+ UEA-1+ cells and lysozyme+ UEA-1− cells per crypt were counted in 49 ileal crypts pooled from nine WT mice, 40 ileal crypts pooled from eight Fut2-deficient mice, and 23 ileal crypts pooled from three Fut1-deficient mice. Data are presented as mean ± SD. n.s., not significant, ***P < 0.001, Student’s t test. (D and E) Flow cytometric analysis of ileal crypt epithelial cells from Fut2-deficient (Fut2−/−) and WT (Fut2+/+) mice (D) and Fut1-deficient (Fut1−/−) and WT (Fut1+/+) mice (E). The UEA-1+ (fucosylated) Paneth cell–rich fraction is indicated by the black boxes. Mean percentage of the fucosylated, Paneth cell–rich population is shown. P values by Student’s t test are shown. (F) Fut2 promoter activity was examined by X-gal staining. Sections of ileum from Fut2LacZ/+ mice were stained with anti-lysozyme antibody (red), and Fut2 promoter activity (i.e., β-gal derived from the LacZ gene) was detected as a blue color. Red arrows, Paneth cells expressing Fut2 and white arrows, Paneth cells not expressing Fut2. The dotted lines delineate crypts. (Scale bar, 20 μm.) Data are representative of three independent experiments.
To examine whether α(1,2)fucosylation in Paneth cells is Fut1 and/or Fut2 dependent, tissue sections of ileal crypt were stained with anti-lysozyme antibody [a Paneth cell marker (10)], UEA-1, and anti–E-cadherin antibody to label the plasma membrane (17). In both WT and Fut2-deficient mice, the whole Paneth cell population was positive for α(1,2)fucose production (Fig. 1B). However, in Fut1-deficient mice, the ileal crypts contained a mixture of α(1,2)fucose-positive and -negative Paneth cells (Fig. 1B).
We then counted the total number of Paneth cells and the number of α(1,2)fucosylated Paneth cells per ileal crypt in WT, Fut2-deficient, and Fut1-deficient mice (Fig. 1C). The total number of Paneth cells per ileal crypt in Fut2-deficient mice and Fut1-deficient mice was comparable with that in WT mice. However, Fut1-deficient mice had a significantly lower number of α(1,2)fucosylated Paneth cells per crypt compared with that in WT mice (P < 0.001). To confirm these findings, we used flow cytometry to examine the CD45− CD24int side scatter (SSC)high cell fraction, which is a known Paneth cell–rich population (18) (Fig. 1 D and E). In the ileal crypts of WT mice (Fut2+/+; Paneth cells producing both Fut1 and Fut2) and Fut2-deficient mice (Fut2−/−; Paneth cells producing only Fut1), this cell fraction contained mostly α(1,2)fucose-positive cells, and no significant differences in population sizes were observed when compared with the WT population (Fig. 1D). However, in Fut1-deficient mice (Fut1−/−; Paneth cells producing only Fut2), there was a tendency for this fraction to contain fewer α(1,2)fucose-positive cells compared with that in WT mice (Fut1+/+) (P = 0.0937) (Fig. 1E).
Together, these results suggest that α(1,2)fucose is produced in all Paneth cells by Fut1 but that in some Paneth cells it is also produced by Fut2. In situ X-gal staining confirmed that Fut2 was expressed in ileal Paneth cells (Fig. 1F). Thus, we concluded that Paneth cells can be divided into Fut2+ and Fut2− subsets.
Distinct Biological Characteristics of Fut2+ and Fut2− Paneth Cells.
To confirm the localization of Fut2+ and Fut2− Paneth cells, we examined the duodenal and ileal crypts of Fut1-deficient mice. Ileal crypts contained significantly more α(1,2)fucosylated Paneth cells than did duodenal crypts (P < 0.001; Fig. 2 A and B), suggesting that Fut2+ Paneth cells are preferentially observed in ileal crypts, and Fut2− Paneth cells are preferentially observed in duodenal crypts.
Fig. 2.
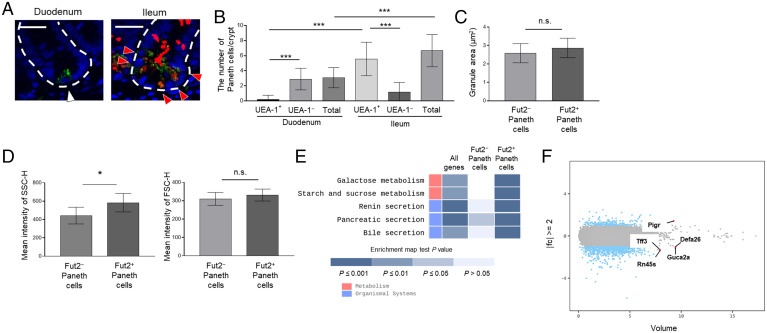
Characterization of Fut2+ and Fut2− Paneth cells. (A) Sections of duodenum and ileum from Fut1-deficient mice were stained with UEA-1 (red), anti-lysozyme antibody (green), and DAPI (counterstain; blue). Red arrows, lysozyme+ UEA-1+ cells; white arrow, lysozyme+ UEA-1− cell; and white dotted lines delineate crypts. (Scale bars, 20 μm.) Data are representative of three independent experiments. (B) The numbers of lysozyme+ UEA-1+ cells and lysozyme+ UEA-1− cells per crypt were counted in 48 duodenal and 39 ileal crypts pooled from four Fut1-deficient mice. Data are presented as mean ± SD ***P < 0.001, Student’s t test. (C) The granule area in ileal Fut2+ and Fut2− Paneth cells was measured in 10 crypts per mouse with the ImageJ software. Data are presented as mean ± SD (n = 8 Fut2LacZ/+ mice per group). n.s., not significant, Student’s t test. (D) SSC-H and FSC-H profiles of ileal Fut2+ and Fut2− Paneth cells detected by using the fluorescent probe Zinpyr-1 and UEA-1 (see SI Appendix, Fig. S1 for the gating strategy). Data are presented as mean ± SD (n = 6 Fut1LacZ/LacZ mice per group). *P < 0.05, n.s., not significant, Student’s t test. (E) Enrichment pathway heatmap obtained from a KEGG analysis. The heatmap shows, from left to right, genes with at least twofold different (|fc| ≥ 2) expression between Fut2+ and Fut2− Paneth cells; genes that are highly expressed in Fut2−, but not Fut2+, Paneth cells; and genes that are highly expressed in Fut2+, but not Fut2−, Paneth cells. P values were obtained using the modified Fisher’s exact test. Blue colored boxes mean P values. Empty boxes mean that there is not matched gene. (F) Volume plot analysis of the genes examined in the transcriptomics analysis. Blue dots indicate the genes with at least twofold different (|fc| ≥ 2) expression between Fut2+ and Fut2− Paneth cells. Red dots indicate the five genes with the highest expression volumes among those with |fc| ≥ 2.
Next, we examined the functional differences between Fut2+ and Fut2− Paneth cells. First, we examined the granule size of these Paneth cell subsets in ileum and found it to be comparable between Fut2+ and Fut2− Paneth cells (Fig. 2C). Then, we conducted a flow cytometric analysis using Fut1-deficient mice and the fluorescent probe Zinpyr-1, which reacts with Paneth cell granules (19); Zinpyr-1+ UEA-1+ cells were classified as Fut2+ Paneth cells, and Zinpyr-1+ UEA-1− cells were classified as Fut2− Paneth cells (SI Appendix, Fig. S1). Although the forward scatter (FSC)-H profile was comparable between the two cell types, Fut2+ Paneth cells had a significantly higher side scatter height (SSC-H) profile than did Fut2− Paneth cells (P < 0.05; Fig. 2D). SSC represents the granularity and structural complexity inside cells, and FSC represents cell size (20), suggesting that although both cell types have comparable cell and granule sizes, Fut2+ cells produce more granules and, therefore, more antimicrobial molecules than Fut2− Paneth cells.
To further examine the differences between Fut2+ and Fut2− Paneth cells, we conducted a transcriptomics analysis. Highly purified Fut2+ and Fut2− Paneth cell populations were isolated from the ileum of Fut1-deficient mice (SI Appendix, Fig. S1). Differentially Expressed Gene analysis (21) revealed 821 genes whose expression differed by at least twofold between the two cell populations. Representative genes are shown in SI Appendix, Table S1. The results of a Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (22) of each gene group are shown in Fig. 2E. Briefly, genes related to the carbohydrate metabolism (e.g., Hkdc1, Lct, Mgam, Sis, and Enpp3) and secretion systems (e.g., Cftr and Clca1) were up-regulated in Fut2+ Paneth cells (Fig. 2E and SI Appendix, Table S1). When we sorted the genes with more than a twofold difference in expression by expression volume (defined as the geometric mean of two groups’ expression level) (23), the top five genes were Pigr, Tff3, Defa26, Rn45s, and Guca2a (Fig. 2F). Pigr was highly expressed in Fut2+ Paneth cells (SI Appendix, Table S1). These results suggest that there are differences in the biological roles of ileal Fut2+ and Fut2− Paneth cells.
Fut2-Mediated α(1,2)Fucosylation Requires Both Commensal Microbial Signals and IL-22.
Based on our finding that Fut2+ Paneth cells were preferentially localized in the ileum rather than the duodenum (Fig. 2 A and B) and the fact that the ileum has a larger bacterial load than the duodenum (23), we examined whether the commensal microbiota influences the differentiation of Fut2+ Paneth cells. We treated Fut1-deficient mice with an antibiotic mixture containing ampicillin, vancomycin, neomycin, and metronidazole and found that this treatment eradicated Fut2-mediated α(1,2)fucosylation in Paneth cells (Fig. 3A). We then repeated the experiment, but after the initial antibiotic treatment, the mice were rehoused to allow bacterial recolonization. We found that bacterial recolonization resulted in recovery of Fut2-mediated α(1,2)fucosylation (Fig. 3A). These results were confirmed by flow cytometric analysis (Fig. 3B). Together, these findings suggest that the commensal microbiota induces Fut2-mediated α(1,2)fucosylation of ileal Paneth cells.
Fig. 3.
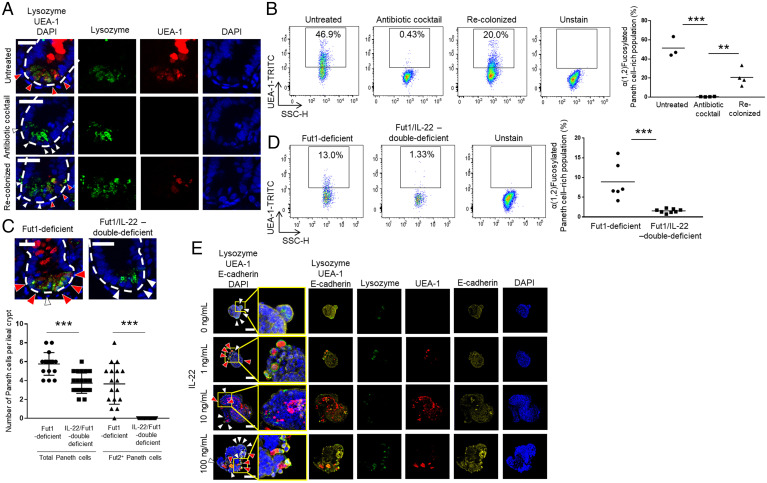
Commensal bacteria and IL-22 induce differentiation of Fut2+ Paneth cells. (A and C) Sections of untreated, antibiotic mixture–treated, and bacterial recolonized ileum isolated from Fut1-deficient mice (A) or ileum from Fut1-deficient mice and Fut1/IL-22-double–deficient mice (C) were stained with UEA-1 (red), anti-lysozyme antibody (green), and DAPI (counterstain; blue). Red arrows indicate lysozyme+ UEA-1+ cells (Fut2+ Paneth cells); white arrows indicate lysozyme+ UEA-1− cells (Fut2− Paneth cells); and white dotted lines delineate crypts. (Scale bars, 20 μm.) Three to five crypts per mouse were observed. One to two mice per group were used. Data are representative of three independent experiments. The numbers of Fut2+ Paneth cells and Fut2− Paneth cells per crypt were counted. The data were pooled from four Fut1-deficient mice and five Fut1/IL-22-double–deficient mice. ***P < 0.001, Student’s t test. (B and D) Flow cytometric analysis of crypt epithelial cells isolated from ileum of untreated, antibiotic mixture–treated, bacterial recolonized, Fut1-deficient mice (B) and whole small intestine from Fut1-deficient mice and Fut1/IL-22-double–deficient mice (D). The UEA-1+ (fucosylated) Paneth cell–rich fraction is indicated with black boxes. Mean percentage of the fucosylated, Paneth cell–rich population is shown. **P < 0.01, ***P < 0.001, Student’s t test. (E) Organoids derived from duodenum of Fut1-deficient mice were treated with the indicated concentrations of recombinant murine IL-22 for 2 d. Samples were stained with UEA-1 (red), anti-lysozyme antibody (green), DAPI (counterstain; blue), and anti–E-cadherin antibody (plasma membrane; yellow). Red arrows, lysozyme+ UEA-1+ cells and white arrows, lysozyme+ UEA-1− cells. (Scale bars, 50 μm.) Organoids were derived from one mouse per experiment. Three to four organoids per experiment were observed. Data are representative of three independent experiments.
We and another group have reported that Fut2-mediated α(1,2)fucosylation is regulated by IL-22 (2–4). IL-22 is also an important cytokine for the growth of organoids derived from intestinal crypts (24). Therefore, we speculated that the IL-22 signaling pathway induces development and maturation of Fut2+ Paneth cells. To examine this hypothesis, we generated mice deficient in both Fut1 and IL-22 and subjected them to microscopic and flow cytometric analyses. These analyses showed that whereas Fut1-deficient mice had at least several α(1,2)fucosylated Paneth cells per crypt, the double-deficient mice had no α(1,2)fucosylated Paneth cells in their crypts (Fig. 3 C and D). These findings suggest that the differentiation of Fut2+ Paneth cells and the production of α(1,2)fucose via Fut2 in Paneth cells are regulated in a IL-22–dependent manner. To examine whether IL-22 directly induces the production of α(1,2)fucose by Fut2 in Paneth cells, we used organoids derived from the duodenum of Fut1-deficient mice, which contain few or no Fut2+ Paneth cells (Figs. 1A and 2 A and B). Organoids were cultured with recombinant mouse IL-22 (mIL-22) for 2 d. Microscopic analysis showed that control organoids did not harbor Paneth cells containing α(1,2)fucose, whereas those treated with any of the concentrations of mIL-22 tested (1, 10, and 100 ng/mL) did (Fig. 3E). mIL-22 also up-regulated the expressions of Fut2 and Lyz1; Lyz1 encodes lysozyme, a marker of Paneth cells (10) (SI Appendix, Fig. S2). Next, to examine whether IL-22 is involved in the regulation of Paneth cell development, we compared the number of Paneth cells per ileal crypt in IL-22–deficient and WT mice. IL-22–deficient mice had fewer Paneth cells in their ileum compared with that in WT mice (SI Appendix, Fig. S3), suggesting that IL-22 is part of the regulatory machinery underlying the development of Paneth cells. Taken together, these findings suggest that IL-22 regulates the development of Fut2+ Paneth cells as well as the induction of Fut2-mediated α(1,2)fucosylation.
Loss of IL-22 Signaling Reduces the Fut2+ Paneth Cell Population, Decreasing α-Defensin in the Intestinal Lumen.
Because our in vivo and in vitro findings suggested that IL-22 plays a critical role in the regulation of Fut2+ Paneth cell development (Fig. 3 C–E and SI Appendix, Figs. S2 and S3), we next examined whether IL-22 is also involved in the regulation of α-defensin secretion by Paneth cells. When we measured α-defensin concentration in the feces of IL-22–deficient mice and WT mice by using an enzyme-linked immunosorbent assay (ELISA) (25), fecal α-defensin concentration was significantly lower in the IL-22–deficient mice compared with that in the WT mice (P < 0.01; Fig. 4A). However, the Paneth cell granules in the IL-22–deficient mice still contained α-defensin (SI Appendix, Fig. S4A), suggesting that IL-22 does not regulate α-defensin production. These results also suggest that the observed reduction in the amount of α-defensin in the feces of IL-22–deficient mice (Fig. 4A) is partially due to the loss of the development of Fut2+ Paneth cells (Fig. 3 C and D and SI Appendix, Fig. S3), which we found to harbor a high density of granules containing antimicrobial peptides, such as α-defensin (Fig. 2D).
Fig. 4.
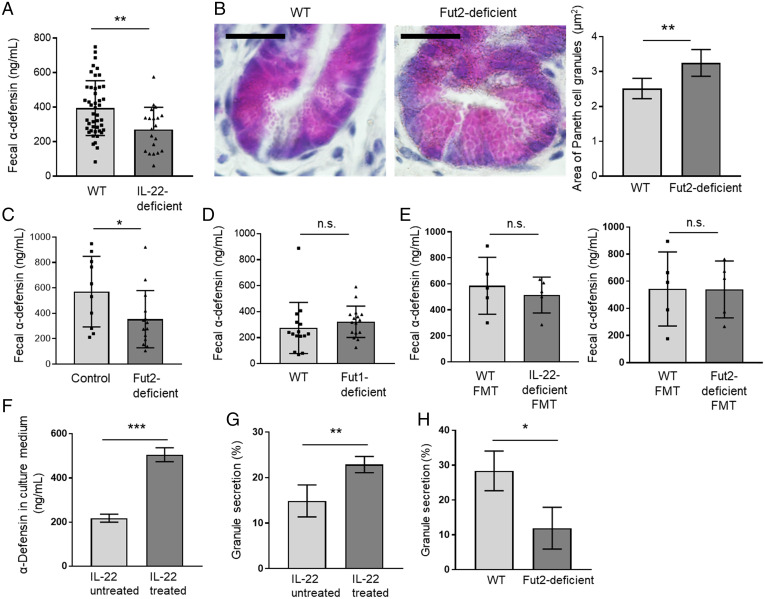
IL-22 regulates the amount of α-defensin in the intestinal lumen. (A, C, D, and E) α-defensin concentration in the feces of WT (n = 45) and IL-22–deficient mice (n = 20) (A), control mice (Fut2+/+ and Fut2+/− mice) (n = 10) and Fut2-deficient mice (n = 14) (C), WT mice (n = 15) and Fut1-deficient mice (n = 16) (D), and FMT mice that received WT feces (n = 5) or IL-22–deficient feces (n = 5) and WT feces (n = 5) or Fut2-deficient feces (n = 5) (E), as measured by ELISA. Data are presented as mean ± SD **P < 0.01, *P < 0.05, n.s., not significant, Student’s t test. (B) Sections of ileum from WT mice (n = 5) and Fut2-deficient mice (n = 5) were subjected to hematoxylin and eosin staining to detect Paneth cell granules. (Scale bars, 20 µm.) A total of 10 crypts per mouse were examined, and Paneth cell granule area was measured with the ImageJ software. Representative images are shown. Data are presented as mean ± SD **P < 0.01, Student’s t test. (F) α-defensin concentration in the intestinal organoid culture medium was measured by ELISA. Intestinal organoids derived from WT mice were treated with 1 ng/mL recombinant IL-22 for 2 d, and granule secretion was induced by 5 ng/mL recombinant IFN-γ. Four samples per group were analyzed. Data are presented as mean ± SD; ***P < 0.001, Student’s t test. (G) Intestinal organoids derived from WT mice were treated with 1 ng/mL recombinant IL-22 for 2 d. Paneth cell granule secretion was visualized and quantified as previously described (31). Percent granule secretion was calculated using Paneth cell granule area measured before and 10 min after treatment of the organoids with 0.1 µM carbamylcholine, a cholinergic agonist that stimulates secretion of α-defensin by Paneth cells. A total of 10 Paneth cells in five organoids from each subject were analyzed. Data were pooled from four independent experiments and are presented as mean ± SD; **P < 0.01, Student’s t test. (H) Ileal organoids derived from WT or Fut2-deficient mice were cultured in Matrigel. Percent granule secretion was calculated using Paneth cell granule area measured before and 10 min after treatment of the organoids with 0.1 µM carbamylcholine. Six Paneth cells in three organoids from each subject were analyzed. Data were pooled from three independent experiments and are presented as mean ± SD; *P < 0.05, Student’s t test.
To determine the role of Fut2-induced α(1,2)fucose in Fut2+ Paneth cells, we analyzed Fut2-deficient mice and found that their Paneth cells had larger granules compared with those of WT mice (P < 0.01; Fig. 4B). A previous report has shown that abnormally shaped or sized Paneth cell granules can disrupt degranulation (26–28). Therefore, we measured the fecal α-defensin concentration in Fut2-deficient mice and found that it was significantly decreased compared with that in controls (Fut2+/+ and Fut2+/− mice) (P < 0.05; Fig. 4C). However, our results also demonstrated that the Paneth cell granules of Fut2-deficient mice still contained α-defensin, as detected by immunofluorescent staining (SI Appendix, Fig. S4B). To examine the relationship between Fut1-induced α(1,2)fucose and Paneth cell degranulation, we also examined the amount of fecal α-defensin in Fut1-deficient mice and found that the concentrations were comparable between Fut1-deficient mice and WT mice (Fig. 4D). Together, these findings suggest that Fut2, but not Fut1, is involved in Paneth cell degranulation but not in the production of granules containing α-defensin.
Because the gut microbiota influences the development and function of epithelial cells, including Paneth cells (29), we performed a fecal microbiota transfer (FMT) experiment to examine whether changes of the gut microbiota in IL-22–deficient mice (30) and Fut2-deficient mice (2) resulted in changes in the fecal concentration of α-defensin. Donor feces were obtained from IL-22–deficient mice or Fut2-deficient mice or their littermate WT mice and then administered orally to C57BL/6J mice that had had their microbiota abolished by pretreatment with antibiotics. The amount of α-defensin in the feces of mice that received feces from IL-22–deficient mice was comparable with that in the feces of mice that received feces from WT mice (Fig. 4E and SI Appendix, Fig. S5). Similarly, the amount of α-defensin in the feces of mice that received feces from Fut2-deficient mice was comparable with that in the feces of mice that received feces from WT mice, except for at 3 wk after FMT (Fig. 4E and SI Appendix, Fig. S5). We also examined the effects of the microbiota on the size of Paneth cell granules and number of Paneth cells in the crypt regions of FMT mice. No significant differences were observed between the FMT mice that received feces from WT mice and those that received feces from Fut2-deficient mice (SI Appendix, Fig. S6), and the numbers of Paneth cells per crypt were comparable between FMT mice that received feces from IL-22–deficient mice or WT mice (SI Appendix, Fig. S7), suggesting that changes in the microbiota do not affect Paneth cell granule size or the number of Paneth cells. Together, these results suggest that the amount of fecal α-defensin is not influenced by the microbiota in these gene-deficient mice and that IL-22– and Fut2-induced α(1,2)fucosylation may directly regulate the function of Paneth cells.
Building on our finding that IL-22 plays a direct role in the development of Fut2+ Paneth cells (Fig. 3), we next examined whether IL-22 also influences granule secretion by Paneth cells. To do this, we treated intestinal organoids derived from WT mice with recombinant IL-22 for 2 d and found that IL-22 significantly increased the concentration of α-defensin in the culture medium (Fig. 4F). To further examine the direct effect of IL-22 on the regulation of α-defensin granule secretion by Fut2+ Paneth cells, granule secretion was visualized and quantified as described previously (31); these experiments were conducted ex vivo, which allowed us to exclude the effects of the gut microbiota. Organoids treated with IL-22 showed greater granule secretion from Paneth cells compared with untreated organoids (Fig. 4G and Video S1). Together with our other observations showing that IL-22 induces Fut2 expression (SI Appendix, Fig. S2) and that Fut2-deficient mice had less α-defensin in their feces compared with that in the feces of controls (Fig. 4C), these present findings suggest that IL-22–mediated Fut2 is associated with the Paneth cell secretion mechanism. To further examine this association, we isolated ileal crypts from WT mice and Fut2-deficient mice and then used those crypts to obtain organoids; we found significantly less granule secretion in organoids from Fut2-deficient mice compared with that in organoids from WT mice (Fig. 4H and Video S2). Together, these results show that a lack of Fut2-induced α(1,2)fucosylation, not changes in the microbiota, is responsible for the observed decrease of α-defensin in Fut2-deficient mice. These results also demonstrate that IL-22 affects Fut2+ Paneth cell α-defensin secretion system via control of Fut2 expression.
IL-17a Regulates the Amount of α-Defensin Produced by Paneth Cells.
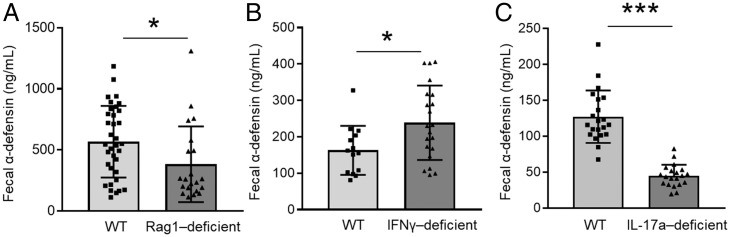
Fut2+ Paneth cells were detected by using UEA-1 lectin in Fut1-deficient mice (Figs. 1 and 2). To elucidate the involvement of the other cytokines in the governance of Fut2+ Paneth cells, Fut1/Rag1–double-deficient mice, which lack adaptive immune cells, including T and B cells (32), were generated and examined. These double-deficient mice were found to possess Fut2+ Paneth cells (SI Appendix, Fig. S8A). Rag1-deficient mice possessed ileal Paneth cells with mature granules containing α-defensin (SI Appendix, Fig. S8 B and C); however, the feces of these mice contained less α-defensin than did the feces of WT mice (P < 0.05; Fig. 5A). Transmission electron microscopy of crypt epithelial cells revealed greater expansion of rough endoplasmic reticulum cisternae stacks in the Paneth cells of Rag1-deficient mice compared with that in the Paneth cells of WT mice, although the Paneth cells of Rag1-deficient mice still possessed electron-dense secretory granules and ribosomes (SI Appendix, Fig. S9). We also noted significantly increased numbers of Paneth cells with irregularly expanded, rough endoplasmic reticulum cisternae stacks, a typical microarchitecture of abnormal Paneth cells (26–28), in the Rag1-deficient mice compared with in the WT mice (P < 0.01; SI Appendix, Fig. S9). These findings support our notion that the adaptive immune pathway is involved in the control of the α-defensin granule secretion machinery. Further supporting data were obtained from an analysis of the expression of Rab family genes, which are known to regulate the membrane trafficking of proteins (33). Among the Rab family genes involved in exocytic events, the expressions of Rab1b, Rab8a, and Rab37, but not of Rab3d, Rab26, or Rab27a, were significantly down-regulated in the ileal crypts of Rag1-deficient mice compared with that in WT mice (P < 0.001 to 0.01; SI Appendix, Fig. S10). Together, these results indicate that the adaptive immune pathway regulates the protein transport of α-defensin in Fut2+ Paneth cells.
Fig. 5.
IL-17a regulates fecal α-defensin produced by Paneth cells. α-defensin concentration in the feces of WT (n = 34) or Rag1-deficient (n = 22) mice (A), WT (n = 14) or IFN-γ–deficient (n = 21) mice (B), and WT (n = 21) or IL-17a–deficient (n = 20) mice (C) was measured with an ELISA. Data are presented as mean ± SD *P < 0.05, ***P < 0.001, Student’s t test.
To elucidate whether T cell cytokines are involved in α-defensin production by Fut2+ Paneth cells, we examined the role of IFN-γ, which is produced mainly by type-1 T helper (Th1) cells (34) and is known to trigger granule release by Paneth cells (35) (SI Appendix, Fig. S11). When enteroid cultures were exposed to IFN-γ, α-defensin production was elevated (SI Appendix, Fig. S11); however, fecal α-defensin was comparable or increased in IFN-γ–deficient mice compared with that in WT mice (P < 0.05; Fig. 5B). This discrepancy between our in vitro and in vivo data suggests that another T cell cytokine regulates α-defensin production by Paneth cells. We therefore examined the involvement of IL-17a in α-defensin production by Paneth cells, because IL-17a produced by Th17 cells has been shown to play a critical role in the regulation of the gut immune system (36). We found that the expression of IL-17a was significantly higher in ileal lamina propria cells than in duodenal lamina propria cells; thus, IL-17a expression was higher in the location of Fut2+ Paneth cells (SI Appendix, Fig. S12). We also found that IL-17a–deficient mice had a significantly lower amount of α-defensin in their feces compared with that in the feces of WT mice (P < 0.001; Fig. 5C). Together, our data suggest that IL-17a and potentially others are involved in the regulation of the amount of α-defensin produced in the gut.
Discussion
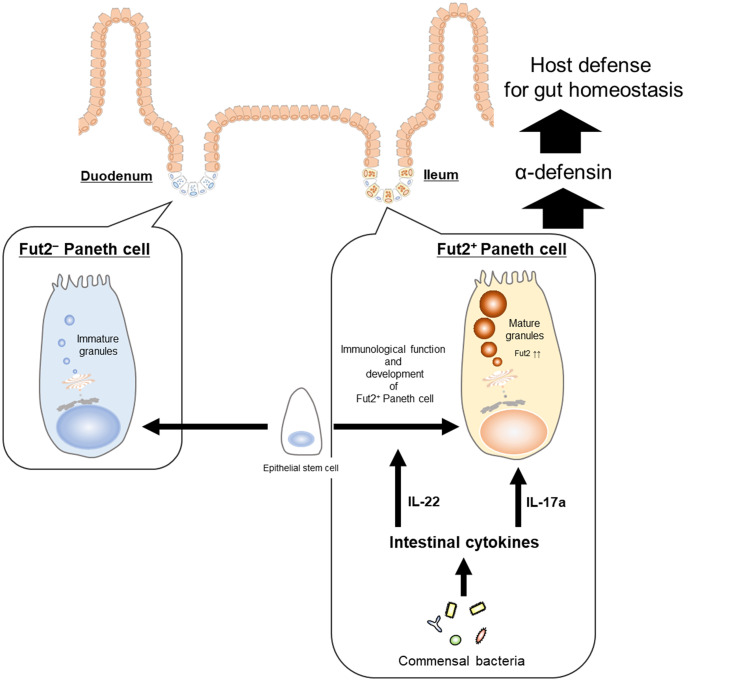
Here, we examined the murine Paneth cell population and found that it comprises two subsets: Fut2+ and Fut2− Paneth cells (Fig. 1). Although the cell and granule sizes were comparable between the two subtypes, Fut2+ Paneth cells were characterized by a higher SSC-H profile compared with Fut2− Paneth cells (Fig. 2 C and D), indicating that Fut2+ Paneth cells contain a higher density of granules compared with Fut2− Paneth cells (20). Given that Paneth cell granules contain antimicrobial molecules that are important for the host innate immune system (10), Fut2+ Paneth cells may be an important subtype for the production of antimicrobial molecules and maintenance of a healthy intestinal environment for symbiosis (Fig. 6). In this study, we therefore focused on the biological characterization of Fut2+ Paneth cells.
Fig. 6.
Intestinal commensal microbiota and cytokines regulate Fut2+ Paneth cell development and function. Paneth cells are separated into two subsets based on their production and utilization of Fut2. Fut2+ Paneth cells are preferentially located in ileum and are engaged in host defense by secreting α-defensin from granules. IL-22 plays important roles in the immunological function and development of Fut2+ Paneth cells, in response to stimuli from commensal bacteria. IL-17a induces the increase of α-defensin in the intestinal tract. Thus, intestinal cytokines (i.e., IL-22 and IL-17a) regulate the development and function of Fut2+ Paneth cells.
KEGG enrichment analysis revealed that in Fut2+ Paneth cells, genes related to the carbohydrate metabolism (e.g., Hkdc1, Lct, Mgam, Sis, and Enpp3) and secretion (e.g., Cftr and Clca1) systems were up-regulated (Fig. 2E and SI Appendix, Table S1). Our findings suggest that Fut2-induced α(1,2)fucose is related to granule secretion by Paneth cells (Fig. 4 B and C) and that other carbohydrate metabolism systems, including the glycosylation system, may also be involved in the control of granule secretion by Fut2+ Paneth cells. Consistent with this possibility, a recent study has reported a role of intestinal epithelial glycosylation in fostering the growth of beneficial commensal bacteria that compete with the pathogenic species Clostridioides difficile for the nutritional niche (37). Further studies to elucidate the dynamics of different subsets of epithelial cells and their glycosylation will further our understanding of the physiological and pathological roles of glycosylation and its associated molecules in the establishment and deterioration of intestinal homeostasis.
The Pigr gene, which encodes polymeric immunoglobulin receptor (pIgR), was found to be highly expressed in Fut2+ Paneth cells (Fig. 2F and SI Appendix, Table S1). pIgR delivers polymeric IgA and IgM in secretory form to the apical surface of the intestine through intestinal epithelial cells and is, therefore, an important protein for mucosal defense (38). Cleaved pIgR is released as a secretory component that associates with IgA to protect the function of secretory IgA (39). In addition, free secretory component is involved in the exclusion of enteric pathogens (39). Thus, it is possible that Fut2+ Paneth cells are involved in pIgR-mediated acquired immunity in addition to their role in innate immunity with antimicrobial peptides. In addition, human Paneth cells have been shown to express pIgR-specific messenger RNA and protein (40), suggesting that human Paneth cells may also comprise two subtypes.
α(1,2)fucosylation in the small intestine is regulated by both Fut1 and Fut2 (5). A previous study by another group has shown that the expression level of Fut1 is not changed between specific, pathogen-free, and germ-free mice (41), indicating that Fut1 is constitutively expressed without being influenced by bacterial stimulation. It has also been demonstrated that crypt regions are enriched with Paneth cells possessing α(1,2)fucose in antibiotic-treated or germ-free WT mice (2), suggesting that Fut1 is also constitutively expressed in Paneth cells without being influenced by bacterial stimulation. However, we previously reported that Fut2 gene expression and subsequent α(1,2)fucosylation in columnar epithelial cells in the small intestine was dramatically decreased in antibiotic-treated or germ-free mice compared with that in specific, pathogen-free mice, suggesting that Fut2 expression is influenced by bacterial stimulation (2). Further studies are needed to elucidate the differences between Fut1- and Fut2-induced α(1,2)fucosylation in Paneth cells and improve our understanding of Paneth cell biology related to host defense.
The findings of the present study, together with those of previous studies (42–44), provide a possible immunological overview of how commensal bacteria and IL-22–producing type 3 innate lymphoid cells (ILC3s) interact with Paneth cells for the establishment of gut homeostasis. For example, a previous study has shown that microbial colonization induces the production of antimicrobial peptides and host defense proteins by Paneth cells (11, 45). In addition, a recent study has demonstrated that rapid Paneth cell granule release is induced by direct recognition of bacterial stimuli (31). In the present study, our results suggest that commensal bacteria induce the differentiation of Fut2+ Paneth cells that possess a high density of granules containing α-defensin (Figs. 2 C and D and 3 A and B). The present results also suggest that the ILC3-associated cytokine IL-22 plays important roles in the immunological function and development of Fut2+ Paneth cells (Figs. 3 C–E and 4A and SI Appendix, Figs. S2 and S3). Because gut dendritic cells acquire bacterial antigens from the lumen and secrete IL-23, which in turn promotes IL-22 production by ILC3s (46), this could be an initial step in the triangular interaction among commensal bacteria, ILC3s, and Paneth cells. Chemokine receptor (CCR) 6+ ILC3s are located in cryptopatches, where the CCR6 ligand CCL20 is highly expressed (47, 48). Commensal bacteria may stimulate production of IL-22 by CCR6+ ILC3s, which then migrate to and accumulate deep within crypts enriched with Paneth cells, leading to differentiation of Fut2+ Paneth cells harboring large numbers of α-defensin–containing granules (Fig. 6).
In the present study, we found that the fecal α-defensin concentration in Fut2-deficient mice was lower than that in control mice, Paneth cells with abnormal granules (in vivo experiment), and Paneth cells with inhibited granule secretion (ex vivo experiment) (Fig. 4 B, C, and H and Video S2), suggesting that Fut2-induced α(1,2)fucose is involved in granule secretion by Paneth cells. Fut2-deficient mice are known to have an altered, intestinal microbiota (2), which may induce defective Paneth cell granule secretion because the gut microbiota is known to influence the function of epithelial cells, including Paneth cells (29). However, our FMT experiments using the feces of Fut2-deficient mice showed that although Fut2-induced α(1,2)fucose was important for the regulation of Paneth cell function, especially granule secretion, changes in the microbiota had no effect on Paneth cell function (Fig. 4E and SI Appendix, Fig. S6). The glycan system has been shown to play important roles in the control of epithelial cell secretion systems, especially that of goblet cells (49). Our present data show the critical role of glycosylation in the Paneth cell secretion system, although further studies are needed to elucidate the molecular and cellular mechanisms regulating Paneth cell secretion via the glycosylation pathway.
Paneth cell granules contain antimicrobial peptides, such as α-defensin (10, 12), that are part of the innate immune system protecting the gut epithelium. In mice, a lack of α-defensin has been shown to increase susceptibility to oral infection by intestinal pathogens such as Salmonella typhimurium (50). Considering that Fut2-deficient mice have increased susceptibility to S. typhimurium infection (2) and we found here that α-defensin secretion is decreased in Fut2-deficient mice (Fig. 4C), it is likely that malfunction of Fut2+ Paneth cell–mediated α-defensin release leads to increased susceptibility to infection by S. typhimurium. This suggests that Fut2+ Paneth cells play an essential role in the innate host defense against microbial invasion and the creation of an environment that promotes symbiosis.
The present data show that fecal α-defensin was increased in IFN-γ–deficient mice (Fig. 5B); however, this finding is inconsistent with a previous report showing that IFN-γ contributes to Paneth cell granule release (35) and an in vitro study we conducted that revealed an elevated concentration of α-defensin in intestinal organoid culture medium (SI Appendix, Fig. S11). Although we cannot explain these discrepancies, it has been reported that IFN-γ induces Paneth cell apoptosis (35, 51, 52). It is therefore possible that IFN-γ deficiency results in a lack of apoptosis in Paneth cells, leading to continuous production of α-defensin and to elevated fecal α-defensin in IFN-γ–deficient mice (Fig. 5B). Our results also show that IL-17a regulates the amount of fecal α-defensin produced by Paneth cells (Fig. 5C). Considering a previous report showing that disruption of IL-17–receptor signaling in the enteric epithelium results in a reduction of global α-defensin (pan-defensin) transcripts in the terminal ileum (53), IL-17a is regulating the amount of α-defensin in the gut homeostatic environment. It has been also reported that IL-17a induces Pigr expression in an enteroid culture (53). Because, in the present study, Fut2+ Paneth cells had higher-Pigr expression than did Fut2− Paneth cells (Fig. 2), IL-17a may also be responsible for the up-regulation of Pigr expression by Fut2+ Paneth cells. Taken together, our present results (Fig. 5) and those of previous studies (35, 51–53) suggest that not just a single cytokine but multiple intestinal cytokines regulate the function of Paneth cells.
In summary, the present data show that Paneth cells can be separated into Fut2+ and Fut2− subsets, which are preferentially located in anatomically distinct parts of the small intestine—the ileum and duodenum, respectively; furthermore, IL-22 and IL-17a are essential for commensal bacteria–dependent Fut2+ Paneth cell development and function for the establishment of gut defense (Fig. 6).
Materials and Methods
Mice.
C57BL/6 mice were purchased from CLEA Japan or Sankyo Laboratory service corporation. Mice with Fut1 or Fut2 loci replaced by the reporter gene LacZ (Fut1LacZ/LacZ and Fut2LacZ/LacZ mice), IL-22–deficient mice, IFN-γ–deficient mice, and IL-17a–deficient mice were generated as described previously (16, 54–56). Rag1-deficient were purchased from the Jackson Laboratory. Antibiotic-treated mice were administered a mixture of broad-spectrum antibiotics (i.e., ampicillin [1 g/L; Sigma], vancomycin [500 mg/L; Shionogi], neomycin [1 g/L; Sigma], and metronidazole [1 g/L; Sigma]) in their drinking water for 4 wk, as previously described (57). Mice were maintained under specific, pathogen-free conditions at the Institute of Medical Science, the University of Tokyo, and all experiments were conducted in accordance with the guidelines of the Animal Care and Use Committees of the University of Tokyo. In all experiments, mice were used at 6 to 24 wk of age.
Immunohistochemistry, Detection of Fut2 Expression by X-Gal Staining, Measurement of Paneth Cell Granule Size, and Transmission Electron Microscopy Analysis.
Observation samples were prepared as described in SI Appendix, Materials and Methods. To detect α-defensin, anti–α-defensin monoclonal antibody [clone 77-R63 (58)] was used. The detailed method and other information for observation were described in SI Appendix, Materials and Methods.
Cell Preparation for Flow Cytometry and Intestinal Organoid Culture.
Crypt region cells were isolated from small intestine, as previously described (8), and used for flow cytometry and preparation of intestinal organoids. The detailed method was described in SI Appendix, Materials and Methods.
Analysis of Purified Fut2+ Paneth Cells and Fut2− Paneth Cells.
The detailed method for the analysis of purified Fut2+ Paneth cells and Fut2− Paneth cells shown in Fig. 2 D–F and SI Appendix, Fig. S1 was described in SI Appendix, Materials and Methods.
Preparation of Fecal Samples for ELISA.
Preparation of fecal samples and sandwich ELISA were performed as previously described (25). The detailed method was described in SI Appendix, Materials and Methods.
Visualization and Quantification of Paneth Cell Granule Secretion.
Paneth cell granule secretion was visualized and quantified as previously described (31). The detailed method was described in SI Appendix, Materials and Methods.
FMT.
FMT was performed as reported previously (59). The detailed method was described in SI Appendix, Materials and Methods.
Isolation of RNA and Real-Time RT-PCR Analysis.
Total RNA from tissue and organoids was isolated with TRIzol reagent (Invitrogen), and complementary DNA (cDNA) was synthesized from the RNA by using a SuperScript VILO cDNA Synthesis Kit (Invitrogen). The specific primers (Hokkaido System Science, Co., Ltd) and universal probes (Roche) used for the real-time RT-PCR were described in SI Appendix, Materials and Methods.
Statistical Analysis.
Statistical analysis was performed using the unpaired, two-tailed Student’s t test. Statistical significance was established at P < 0.05. All statistical analyses were conducted with Prism 7 (GraphPad). No statistical methods were used to determine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during the experiments or outcome assessment. In the transcriptomics analysis, statistical analysis was performed using fold change per comparison pair; significant results were selected at |fc| ≥ 2.
Supplementary Material
Acknowledgments
We thank Dr. Hiroshi Sagara for helping with the electron microscope analyses; Dr. Nan Gao and Dr. Shiyan Yu for their technical advice related to immunohistochemistry; and Akemi Arakawa, Tomoyuki Yamanoue, Ruka Iijima, Sayuri Murasaki, Fujimi Arai, Kaoru Shimada, Amber Ablack, Yukie Tokura, Kayo Satonaga, and Keiko Warren for their technical assistance. We also thank University of California San Diego (UCSD) Microscopy Core and their grant NINDS NS047101. This work was supported by Grants-in-Aid for Scientific Research (S) (18H05280 to H.K.), Scientific Research (B) (18H02788 to T.A.), Scientific Research (C) (26462831 to K.N.), Young Scientists (A) (16H06229 to Y.G. and 16H06243 to Y.K.), Grant-in-Aid for Early-Career Scientists (19K17452 to M.K.), and Grant-in-Aid for Japan Society for the Promotion of Science (JSPS) Fellows (16J06100 to M.K.), Funds for the Promotion of Joint International Research (18KK0432 to Y.K.), the Institute of Medical Science, the University of Tokyo International Joint Research Project (K003 to P.B.E.) from JSPS; as well as by grants from the Core Research for Evolutional Science and Technology Program of the Japan Science and Technology Agency (JP15gm0410004h0006 to H.K.); Japan Agency for Medical Research and Development (AMED) (JP20gm1010006h004 to J.K.); AMED–the Japan Agency for Medical Research and Development-Science and Technology Research Partnership for Sustainable Development (JP20jm0110012h0006 to H.K.); AMED–Practical Research Project for Allergic Diseases and Immunology (JP18ek0410032h0003 to H.K. and J.K.); AMED–Precursory Research for Innovative Medical Care (PRIME) (JP19gm6010005h0004 to Y.G. and JP20gm6010012h0004 and JP20gm6210024h0001 to Y.K.); AMED–Science and Technology Platform Program for Advanced Biological Medicine (JP21am0401029h0001 to H.K.); AMED–Japan Program for Infectious Diseases Research and Infrastructure (JP20fk0108122h0001 to H.K.); AMED–Acceleration Transformative Research for Medical Innovation (JP19im0210623h0001 to H.K.); AMED–the Research Program on Emerging and Re-emerging Infectious Diseases (JP19fk0108112j0001 to H.K.); AMED–Project Focused on Developing Key Technology for Discovering and Manufacturing Drugs for Next-Generation Treatment and Diagnosis (to H.K.); the Center of Innovation Program from Japan Science and Technology Agency (to K.N.); the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (to J.K.); the Ministry of Health and Welfare of Japan and Public/Private R&D Investment Strategic Expansion Program (20AC5004 to J.K.); the Chiba University-UCSD Center for Mucosal Immunology, Allergy, and Vaccines and UCSD San Diego Digestive Disease Research Center (to Y.K., P.B.E., and H.K.); NIH RO1 (AI079145-08 to P.B.E); 3M Global Giving Donation Fund (to H.K.); the Grant for Joint Research Project of the Institute of Medical Science, the University of Tokyo (to H.K. and J.K.); and Japanese Society for Immunology-Outstanding Young Women Immunology Researcher Award (to M.K.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115230119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Barker N., Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 15, 19–33 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Goto Y., et al. , Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 345, 1254009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pickard J. M., et al. , Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 514, 638–641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pham T. A. N., et al. ; Sanger Mouse Genetics Project, Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe 16, 504–516 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terahara K., et al. , Distinct fucosylation of M cells and epithelial cells by Fut1 and Fut2, respectively, in response to intestinal environmental stress. Biochem. Biophys. Res. Commun. 404, 822–828 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Clevers H. C., Bevins C. L., Paneth cells: Maestros of the small intestinal crypts. Annu. Rev. Physiol. 75, 289–311 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Barker N., et al. , Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Sato T., et al. , Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farin H. F., Van Es J. H., Clevers H., Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143, 1518–1529.e7 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Porter E. M., Bevins C. L., Ghosh D., Ganz T., The multifaceted Paneth cell. Cell. Mol. Life Sci. 59, 156–170 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayabe T., et al. , Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 1, 113–118 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Selsted M. E., Ouellette A. J., Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6, 551–557 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Nakamura K., Sakuragi N., Takakuwa A., Ayabe T., Paneth cell α-defensins and enteric microbiota in health and disease. Biosci. Microbiota Food Health 35, 57–67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haber A. L., et al. , A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farr A. G., Anderson S. K., Epithelial heterogeneity in the murine thymus: Fucose-specific lectins bind medullary epithelial cells. J. Immunol. 134, 2971–2977 (1985). [PubMed] [Google Scholar]
- 16.Domino S. E., Zhang L., Gillespie P. J., Saunders T. L., Lowe J. B., Deficiency of reproductive tract alpha(1,2)fucosylated glycans and normal fertility in mice with targeted deletions of the FUT1 or FUT2 alpha(1,2)fucosyltransferase locus. Mol. Cell. Biol. 21, 8336–8345 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu S., et al. , Paneth cell multipotency induced by notch activation following injury. Cell Stem Cell 23, 46–59.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong V. W. Y., et al. , Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat. Cell Biol. 14, 401–408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayase E., et al. , R-Spondin1 expands Paneth cells and prevents dysbiosis induced by graft-versus-host disease. J. Exp. Med. 214, 3507–3518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shapiro H., Practical Flow Cytometry (John Wiley & Sons, 2003). [Google Scholar]
- 21.Trapnell C.et al., . Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M., New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47, D590–D595 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sartor R. B., Microbial influences in inflammatory bowel diseases. Gastroenterology 134, 577–594 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Lindemans C. A., et al. , Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 528, 560–564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura K., Sakuragi N., Ayabe T., A monoclonal antibody-based sandwich enzyme-linked immunosorbent assay for detection of secreted α-defensin. Anal. Biochem. 443, 124–131 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Kaser A., et al. , XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134, 743–756 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cadwell K., et al. , A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cadwell K., Patel K. K., Komatsu M., Virgin H. W. IV, Stappenbeck T. S., A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy 5, 250–252 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salzman N. H., Bevins C. L., Dysbiosis–A consequence of Paneth cell dysfunction. Semin. Immunol. 25, 334–341 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Sonnenberg G. F., et al. , Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science 336, 1321–1325 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yokoi Y., et al. , Paneth cell granule dynamics on secretory responses to bacterial stimuli in enteroids. Sci. Rep. 9, 2710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mombaerts P., et al. , RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 (1992). [DOI] [PubMed] [Google Scholar]
- 33.Stenmark H., Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Serre K., Silva-Santos B., Molecular mechanisms of differentiation of murine pro-inflammatory γδ T cell subsets. Front. Immunol. 4, 431 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farin H. F., et al. , Paneth cell extrusion and release of antimicrobial products is directly controlled by immune cell-derived IFN-γ. J. Exp. Med. 211, 1393–1405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miossec P., Kolls J. K., Targeting IL-17 and TH17 cells in chronic inflammation. Nat. Rev. Drug Discov. 11, 763–776 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Nagao-Kitamoto H., et al. , Interleukin-22-mediated host glycosylation prevents Clostridioides difficile infection by modulating the metabolic activity of the gut microbiota. Nat. Med. 26, 608–617 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mostov K., The polymeric immunoglobulin receptor. Semin. Cell Biol. 2, 411–418 (1991). [PubMed] [Google Scholar]
- 39.Phalipon A., Corthésy B., Novel functions of the polymeric Ig receptor: Well beyond transport of immunoglobulins. Trends Immunol. 24, 55–58 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Tang Q.-J., et al. , Expression of polymeric immunoglobulin receptor mRNA and protein in human Paneth cells: Paneth cells participate in acquired immunity. Am. J. Gastroenterol. 101, 1625–1632 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Lin B., et al. , GDP-fucose: Beta-galactoside alpha1,2-fucosyltransferase, MFUT-II, and not MFUT-I or -III, is induced in a restricted region of the digestive tract of germ-free mice by host-microbe interactions and cycloheximide. Biochim. Biophys. Acta 1487, 275–285 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Satoh-Takayama N., et al. , Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29, 958–970 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Sanos S. L., et al. , RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol. 10, 83–91 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonnenberg G. F., Fouser L. A., Artis D., Border patrol: Regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat. Immunol. 12, 383–390 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Kurashima Y., Kiyono H., Mucosal ecological network of epithelium and immune cells for gut homeostasis and tissue healing. Annu. Rev. Immunol. 35, 119–147 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Goto Y., et al. , Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 40, 594–607 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cella M., et al. , A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457, 722–725 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klose C. S. N., et al. , A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature 494, 261–265 (2013). [DOI] [PubMed] [Google Scholar]
- 49.McGuckin M. A., Lindén S. K., Sutton P., Florin T. H., Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 9, 265–278 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Wilson C. L., et al. , Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 286, 113–117 (1999). [DOI] [PubMed] [Google Scholar]
- 51.Eriguchi Y., et al. , Essential role of IFN-γ in T cell-associated intestinal inflammation. JCI Insight 3, e121886 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yokoi Y., et al. , Simultaneous real-time analysis of Paneth cell and intestinal stem cell response to interferon-γ by a novel stem cell niche tracking method. Biochem. Biophys. Res. Commun. 545, 14–19 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Kumar P., et al. , Intestinal interleukin-17 receptor signaling mediates reciprocal control of the gut microbiota and autoimmune inflammation. Immunity 44, 659–671 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kreymborg K., et al. , IL-22 is expressed by Th17 cells in an IL-23-dependent fashion, but not required for the development of autoimmune encephalomyelitis. J. Immunol. 179, 8098–8104 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Nakae S., et al. , Antigen-specific T cell sensitization is impaired in IL-17-deficient mice, causing suppression of allergic cellular and humoral responses. Immunity 17, 375–387 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Tagawa Y., Sekikawa K., Iwakura Y., Suppression of concanavalin A-induced hepatitis in IFN-gamma(-/-) mice, but not in TNF-alpha(-/-) mice: Role for IFN-gamma in activating apoptosis of hepatocytes. J. Immunol. 159, 1418–1428 (1997). [PubMed] [Google Scholar]
- 57.Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., Medzhitov R., Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Eriguchi Y., et al. , Decreased secretion of Paneth cell α-defensins in graft-versus-host disease. Transpl. Infect. Dis. 17, 702–706 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Matsuo K., et al. , Fecal microbiota transplantation prevents Candida albicans from colonizing the gastrointestinal tract. Microbiol. Immunol. 63, 155–163 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.