Abstract
The precise assembly and disassembly of actin filaments is required for several cellular processes, and their regulation has been scrutinized for decades. Twenty years ago, a handful of studies marked the advent of a new type of experiment to study actin dynamics: using optical microscopy to look at individual events, taking place on individual filaments in real time. Here, we summarize the main characteristics of this approach and how it has changed our ability to understand actin assembly dynamics. We also highlight some of its caveats and reflect on what we have learned over the past 20 years, leading us to propose a set of guidelines, which we hope will contribute to a better exploitation of this powerful tool.
Keywords: cytoskeleton, microscopy, TIRF, biochemistry, biophysics
Actin filaments and networks play such key and versatile roles in cells that we must understand every aspect of their assembly. We thus need to accurately describe the mechanisms that regulate actin assembly, and quantify their reaction rates.
A lot of our early knowledge of actin, following its identification in the 1940s as a substrate required to “activate” myosin motors (ref. 1 has a review), comes from electron microscopy (EM), which provided detailed images of fixed filaments (2). Filaments could not be imaged “live,” but in the early 1980s, by taking EM snapshots of individual filaments at different time points, Pollard and Mooseker (3, 4) managed to derive kinetic rates of actin assembly. At that time, bulk solution studies were emerging as the tool of choice to study actin biochemistry: in particular, cosedimentation assays to investigate the binding of proteins to filaments, and spectrofluorimetry exploiting the enhanced fluorescence of pyrenyl-labeled actin incorporated into filaments (5) to quantify assembly kinetics. These bulk solution assays are still extensively used today.
Also in the 1980s, fluorescent microscopy emerged as another method to study actin filaments in vitro, first by labeling the filament-stabilizing drug phalloidin (6) and later by directly labeling actin (7, 8) with a fluorophore such as rhodamine. In principle, the availability of fluorescently labeled actin allowed one to directly monitor dynamic filaments. Yet, until the turn of the century, only stabilized actin filaments were observed in vitro using light microscopy. These experiments nonetheless provided important measurements regarding, for example, the mechanical properties of actin filaments and of Arp2/3 branch junctions (8–11), the severing of phalloidin-stabilized filaments by actophorin, the cofilin ortholog in amoeba (12), and the properties of myosin motor proteins (6, 13, 14).
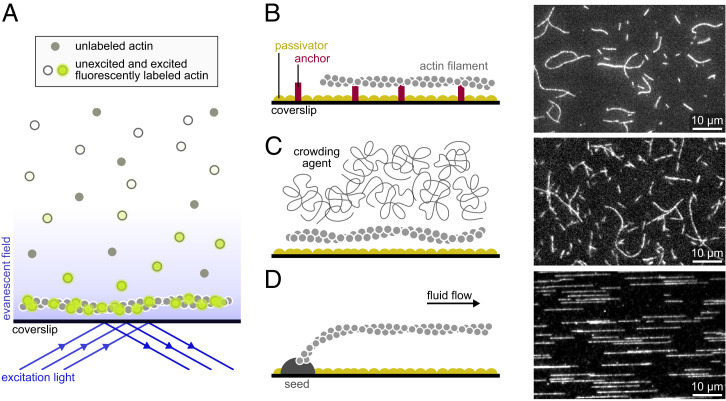
A key limitation to monitor dynamic actin filaments is the background signal from fluorescently labeled proteins in solution (in particular, actin monomers if one wishes to monitor elongating filaments). This problem can be solved by total internal reflection fluorescence (TIRF) microscopy, a technique where only a shallow region above the coverslip is illuminated (Fig. 1A), which has been used to image cells since the early 1980s (15). In the 1990s, TIRF was also used to monitor individual proteins in vitro, like myosins (16), interacting with stabilized actin filaments (17). In 2001 and 2002, for the first time, dynamic actin filaments were visualized in real time using TIRF (18–20) and confocal microscopy (21). These pioneer studies provided the first live observations of Arp2/3-mediated branching (18, 19, 21) and of filament treadmilling (20). Most importantly, they mark the birth of a new powerful assay to directly monitor actin dynamics.
Fig. 1.
Main single-filament techniques used to study actin dynamics. (A) In TIRF microscopy, a shallow region of the sample, a few hundred nanometers above the coverslip, is illuminated by the evanescent wave resulting from the total reflection of the incoming light. Note that if the concentration of labeled proteins is low, TIRF may not be necessary, and epifluorescence can be used. (B–D) Actin filaments can be maintained close to the surface by specific anchors (A), by a crowding agent present in solution (B), or by a microfluidic flow while they are anchored by one end only (here, via a seed; C). (B, Right; C, Right; and D, Right) Actin filaments (10% labeled with AlexaFluor-488 and 1% biotinylated actin subunits on surface lysines) anchored to a streptavidin functionalized coverslip (A), actin filaments (10% labeled with AlexaFluor-568) with 0.3% methylcellulose (4,000 cP at 2%; B), and actin filaments (10% labeled with AlexaFluor-488) polymerized from spectrin-actin seeds inside a microfluidic chamber (C). All filaments are made with rabbit alpha-skeletal actin, imaged in the same buffer (5 mM Tris⋅HCl, pH 7.4, 50 mM KCl, 1 mM MgCl2, 0.2 mM EGTA, 0.2 mM ATP, 10 mM DTT, 1 mM DABCO). EGTA, ethylene glycol tetraacetic acid; DTT, dithiothreitol; DABCO, diazabicyclooctane.
Live single-actin-filament assays can have technical differences (Fig. 1), but they all exhibit the following basic features. First, actin and actin-binding proteins (ABPs) are purified to be used outside of the cell. This provides control over the nature and concentrations of proteins regulating actin assembly. Purified proteins can be supplemented by cell extracts (22). Second, actin (and sometimes ABPs) is fluorescently labeled in order to visualize filaments directly. Techniques such as high-speed atomic force microscopy can be used to avoid this requirement, but their use on actin filaments remains scarce (23, 24). Third, filaments are constrained in a region close to the coverslip surface. Various techniques can be used to achieve this quasi-two-dimensional confinement: multiple anchoring points (Fig. 1B), crowding agents (Fig. 1C), and microfluidics (Fig. 1D). This feature is required to maintain the filaments in the focal plane of the microscope, so that they are entirely visible and can be monitored over time. While epifluorescence can be used in many cases, this confinement near the coverslip also enables the use of TIRF microscopy, which is today the most widely used technique to monitor single filaments live (25), as it provides high-quality images and is now easy to use thanks to the development of objective-based TIRF (16, 26). Other techniques, such as optical traps, allow one to manipulate filaments in a plane farther above the surface (14), but they make it difficult to observe elongation or depolymerization from either end.
Live Single-Filament Studies Have Changed Our Ability to Understand Actin Dynamics
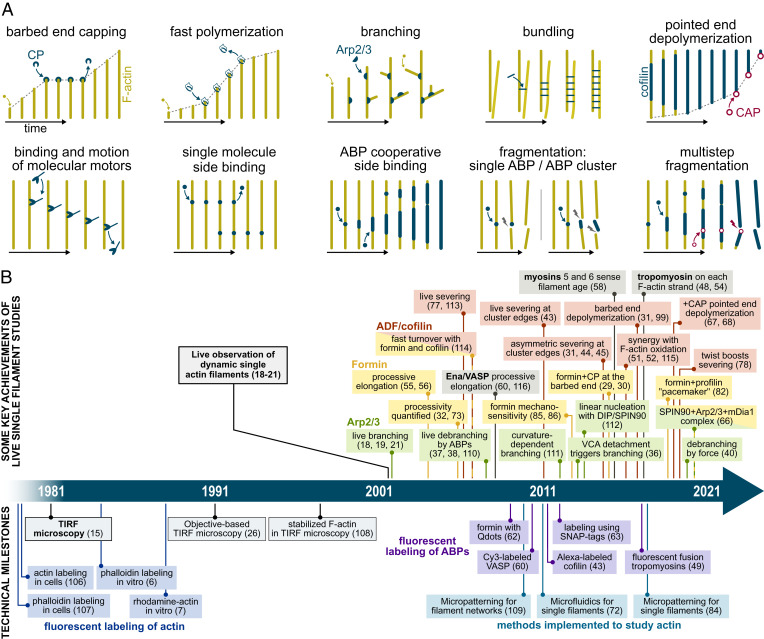
In vitro studies, using purified proteins, allow us to decipher the regulatory mechanisms that control actin assembly. On their own, they are not enough to explain how actin works in cells, but they are a necessary piece of the puzzle. In that respect, being able to observe individual reactions on individual filaments is a major step forward and opens new avenues to study actin dynamics (Fig. 2).
Fig. 2.
A selection of key live single-actin-filament assays and discoveries. (A) Sketches of typical live single-filament experiments. For clarity, filaments (F-actin) are depicted straight, with time passing from left to right. (B) A time line presenting some of the key scientific achievements of single-actin-filament studies and important technical milestones that led to these discoveries. ADF, actin depolymerizing factor; CAP, cyclase-associated protein; CP, capping protein; DIP, dia-interacting protein; SPIN90, SH3 protein interacting with Nck; VASP, vasodilator-stimulated phosphoprotein; VCA, verprolin homology, central and acidic domains. The timeline contains additional references (106–115).
Qualitatively, a great asset of single-filament observations, sometimes reinforced by the observation of single fluorescent ABPs, is that they provide direct information. One can directly assess where events take place on the filaments and what basic architectural elements (e.g., side branching) emerge from these events. One can disentangle the different reactions involving an ABP rather than simply observing their collective outcome. One can readily distinguish severing from depolymerization or separate the contributions of nucleation and elongation, which can be very difficult with solution assays.
Quantitatively, as single-filament studies allow us to monitor events over time, they are also a powerful means to determine reaction rates (27). When observing individual events, such as the barbed end binding and unbinding by a capping protein (28–31) or by a formin (29, 30, 32, 33), rates can be extracted from exponential fits of the survival functions, and reliable numbers can be derived from the observations of only a few tens of events. Since hundreds of individual filaments can be observed, these reasonable requirements make it possible to quantify several individual reactions in several conditions.
Thanks to these assets, live single-actin-filament observations have quickly confirmed rates previously measured using other techniques, such as the elongation and depolymerization rates of barbed and pointed ends (18, 34, 35) and the rate at which capping proteins bind and unbind barbed ends (28, 29, 31). The live observation of filament branching by the Arp2/3 complex (18, 19, 21) confirmed that branches grew off the sides of existing filaments, as already concluded from experiments using stabilized filaments (11). Subsequent experiments visualizing live branching events went further and were able, notably, to decipher the molecular steps leading to branch formation (36) and to demonstrate the acceleration of debranching by proteins (37–39) and more recently, by mechanical forces (40). The cooperative formation of cofilin clusters on the side of actin filaments and the severing of the filament at the boundaries of these clusters had been deduced from a number of studies using other techniques (41, 42) and were later observed directly on single filaments using fluorescently labeled cofilin (43). In addition, such direct observations further revealed information that seems inaccessible to other methods: that cofilin-induced severing occurred preferentially at the cluster boundary located toward the pointed end of the filament (31, 44, 45), how fast severing per cofilin cluster occurred (31), and how the action of cofilin is affected by other proteins (22, 44, 46–51) or by the oxidation of actin filaments (51, 52). The cooperative formation of tropomyosin clusters was also confirmed on single filaments (48, 49, 53), and single-filament studies further revealed that tropomyosins independently decorate the two filament strands (48, 54). Another example of key information provided by live single-filament observation is the assessment of formin processivity at the barbed end of a growing filament (32, 55, 56). Revisiting classical assays with nonstabilized filaments can yield unexpected results, such as myosin 1b’s ability to enhance barbed end depolymerization in a standard gliding assay (57) and myosins’ preference for either ADP- or ADP-Pi–rich actin filaments revealed by monitoring individual motors on dynamic filaments (58).
Advances in the observation of single fluorescent molecules have reinforced the potency of single-filament studies. Notably, they provide a direct means to detect the presence of individual ABPs at either end or on the side of the filaments, quantify their interactions, and shed light on their regulatory mechanisms. Key molecular details were thus discovered by monitoring individual fluorescently labeled Arp2/3 complexes (36, 59), capping protein (30), Ena/VASP (60), VopF/VopL (61), formins (30, 62, 63), DIP/SPIN90 (64–66), and Srv2/CAP (67, 68).
The results provided by live single-actin-filament studies are too numerous to be all listed here. The aforementioned examples (some of which are sketched in Fig. 2A) illustrate the enormous impact that this technique has had on our understanding of the molecular processes regulating actin assembly. A partial selection of findings, which would not have been possible without observing single filaments in real time, is put on a time line in Fig. 2B.
Requirements of Live Single-Filament Studies
As for any biochemical experiment carried out in vitro, great care must be taken to ensure the quality and stability of the purified proteins (69). In addition, when biological objects are put in an artificial context, one must make sure that this context is not changing their behavior. Fortunately, single-filament techniques are quantitative, and experimental conditions can be controlled well enough so that potential artifacts can be readily identified and quantified. Early live single-filament studies, including the first one by Amann and Pollard (18), already identified the two probably most important caveats: the potential impact of fluorescent labeling and of surface–filament interactions. They can be minimized and taken into account.
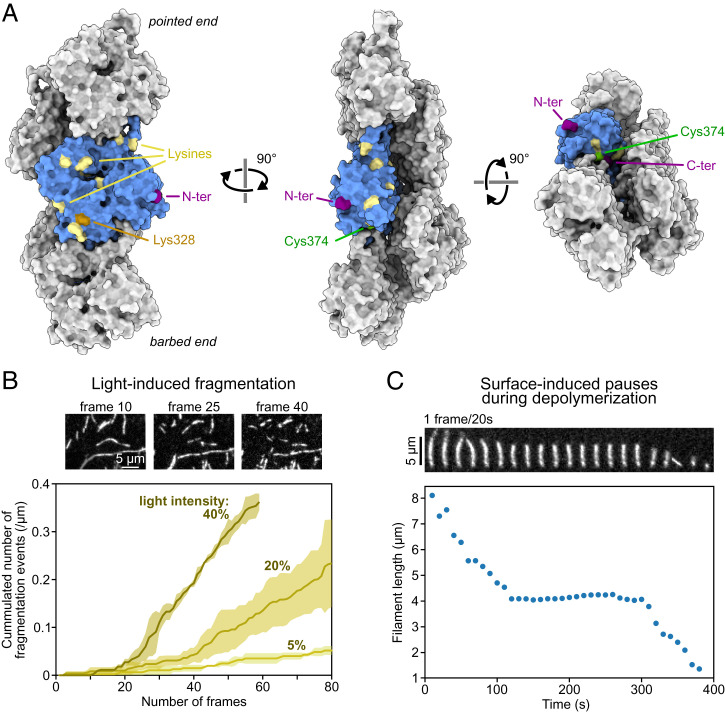
Perhaps the best-known and most dramatic artifact that can occur when observing an actin filament in fluorescent microscopy is photo-induced severing (Fig. 3B). Other less obvious artifacts include the formation of photo-induced covalent actin dimers within filaments (70) and the supercoiling of phalloidin-decorated filaments exposed to strong illumination (71). Even in the absence of illumination, the presence of the fluorescent label itself can have an impact on protein–protein interactions. The first live single-filament studies established that actin monomers labeled on Cys374 with Rhodamine or Oregon Green dyes hardly integrated the filaments (18, 34). The location of Cys374, on the barbed face of the actin monomer (Fig. 3A), also impedes the binding to profilin, and as a result, filaments elongated by formins in the presence of profilin were shown to incorporate even lower amounts of labeled actin (32), a feature that could actually be used as a means to identify formin-elongated filaments. In contrast, labeling actin on surface lysines with AlexaFluor dyes appeared to have no impact on free barbed end dynamics (33, 72) and a moderate impact on formin-assisted barbed end elongation (33, 73). Several lysines are available on the surface of polymerized actin and could potentially bind the AlexaFluor dye, but mass spectrometry revealed that Lys328 was the preferred labeling site (74) (Fig. 3A). Fluorescently labeling ABPs can also be an issue and has long hindered the visualization of functional cofilin and tropomyosins. The nature of the fluorescent label can have subtle consequences; for example, the binding of cofilin-1 to the filament sides is barely affected by the N-terminal fusion of enhanced green fluorescent protein, while mCherry-cofilin-1 binds 30% slower than unlabeled cofilin-1 (31).
Fig. 3.
Surface anchoring and fluorescent labeling are common sources of artifacts. (A) Five actin subunits in a filament based on the cryo-EM structure of ADP-actin filaments from ref. 103 (Protein Data Bank ID code 6DJO) shown from different angles using ChimeraX (104). One actin subunit is colored in blue, and the amino acids commonly targeted for labeling with synthetic dyes are highlighted. Actin can also be fused to a fluorescent protein at its C or N terminus (for cell studies mostly). (B) Fragmentation, induced by illumination, of actin filaments (10% labeled with AlexaFluor-568 on surface lysines). Images are acquired with the power of the lamp (Xcite, Lumen Dynamics) set at 40% of its maximum. The curves were obtained by observing the severing of ≥10 filaments per experiment. Lines and shaded surfaces represent the average and SD over three independent experiments. Note the lag, which indicates that photo-induced severing is not a simple first-order reaction. (C) Depolymerization of an actin filament (10% labeled with AlexaFluor-488 and 1% biotinylated actin subunits on surface lysines) anchored to a streptavidin-functionalized coverslip, at 25 °C. Filament length was measured using the JFilament ImageJ plugin (105). A pause occurs when the depolymerizing barbed end reaches an anchoring point, which can be identified as a static and brighter point along the filament. In B and C, filaments are made with rabbit alpha-skeletal actin and imaged in the same buffer (5 mM Tris⋅HCl, pH 7.4, 50 mM KCl, 1 mM MgCl2, 0.2 mM EGTA, 0.2 mM ATP, 10 mM DTT, 1 mM DABCO) supplemented with 0.3% (B) and 0.5% (C) methylcellulose (4,000 cP at 2%). Data from B and C can be found in Dataset S1. EGTA, ethylene glycol tetraacetic acid; DTT, dithiothreitol; DABCO, diazabicyclooctane.
Early single-filament studies observed pauses, randomly occurring during both elongation and depolymerization of surface-anchored filaments, and attributed them to interactions between the filament end and the surface either via anchoring points or via nonspecific interactions (34, 75). One can recover the correct depolymerization rate constant by excluding these pauses from the analysis (34, 76). Fig. 3C illustrates this artifact. It was also reported that the density of anchoring points enhanced cofilin-induced severing (77). It was recently shown that the rate of severing at the edge of a cofilin cluster was enhanced up to 100-fold between anchoring points because of the filament’s inability to rotate and relax torsional strain (78). Since actin filaments in cells are often interconnected, this is a relevant configuration. What was first identified as an artifact led to the discovery of a mechanism of physiological importance.
Single-filament studies, thanks to the direct observation and the control they provide, offer the possibility to identify artifacts and sometimes even exploit them. In a way, these assays contain their own antidote. They can also be used to detect and quantify artifacts that may take place in cells, with widely used probes like GFP-Lifeact (79), and that would otherwise remain unnoticed.
Where Do We Stand Now That This Field Is 20 Years Old?
Today, looking at single events live on actin filaments is one of the most powerful tools in our arsenal to study actin assembly dynamics. Putting key dates on a time line (Fig. 2B), it is striking to see how the advent of this technique, 20 years ago, has triggered an explosion of results that would have otherwise largely remained inaccessible. It is also notable that TIRF microscopy was first applied to study dynamic actin filaments several years after its invention. This highlights the importance of all the incremental improvements that constitute the backdrop of such a time line; fluorophores, cameras, microscope objectives, and computers all got better over the years and contributed to making the technique more sensitive, more reliable, and easier to use.
Importantly, these technical improvements are still going on today and directly benefit single-actin-filament studies. New tools for the production and characterization of proteins should provide a better control of which isoform is studied and what posttranslational modifications it harbors (80, 81). New ways to fluorescently label actin filaments have been proposed, offering interesting possibilities; the fluorescently labeled Calponin Homology domains of utrophin, which efficiently decorate actin filaments, have been used to successfully monitor the rapid elongation of filaments exposed to over 100 µM unlabeled monomeric actin (i.e., concentrations where the background signal from labeled monomers would preclude imaging filaments, even in TIRF) (82). Another study has shown that the fluorescent nucleotide analog ATP-ATTO-488 could be used as an indirect yet reliable way to label actin monomers, with little impact on their kinetic rates and interaction with ABPs (83). Additional developments are likely to emerge in the future, such as new fluorescent probes or fully automated image analysis, and will certainly continue to expand our ability to monitor individual reactions on dynamic actin filaments.
Improvements also come from the addition of independent technical features to basic single-filament assays. These assays, which can already be performed in different ways (Fig. 1), can be complemented by other approaches, such as surface micropatterning in order to restrict protein activity to a specific region (84). They can be used to apply mechanical stress to several filaments simultaneously, thereby adding a new dimension to the questions they can address, thanks to microfluidics (33, 40, 78, 85, 86), or to a combination of myosins with opposite polarities (87). In addition, the single-filament realm is expanding to include filament bundles (48, 88–91), and in vitro studies using purified proteins now cover multiple scales, from single-molecule assays to the construction of filament networks with diverse architectures (44, 92–94).
Thanks to these ongoing technical improvements, live single-filament techniques are more powerful than ever and will certainly provide essential results in the next decades.
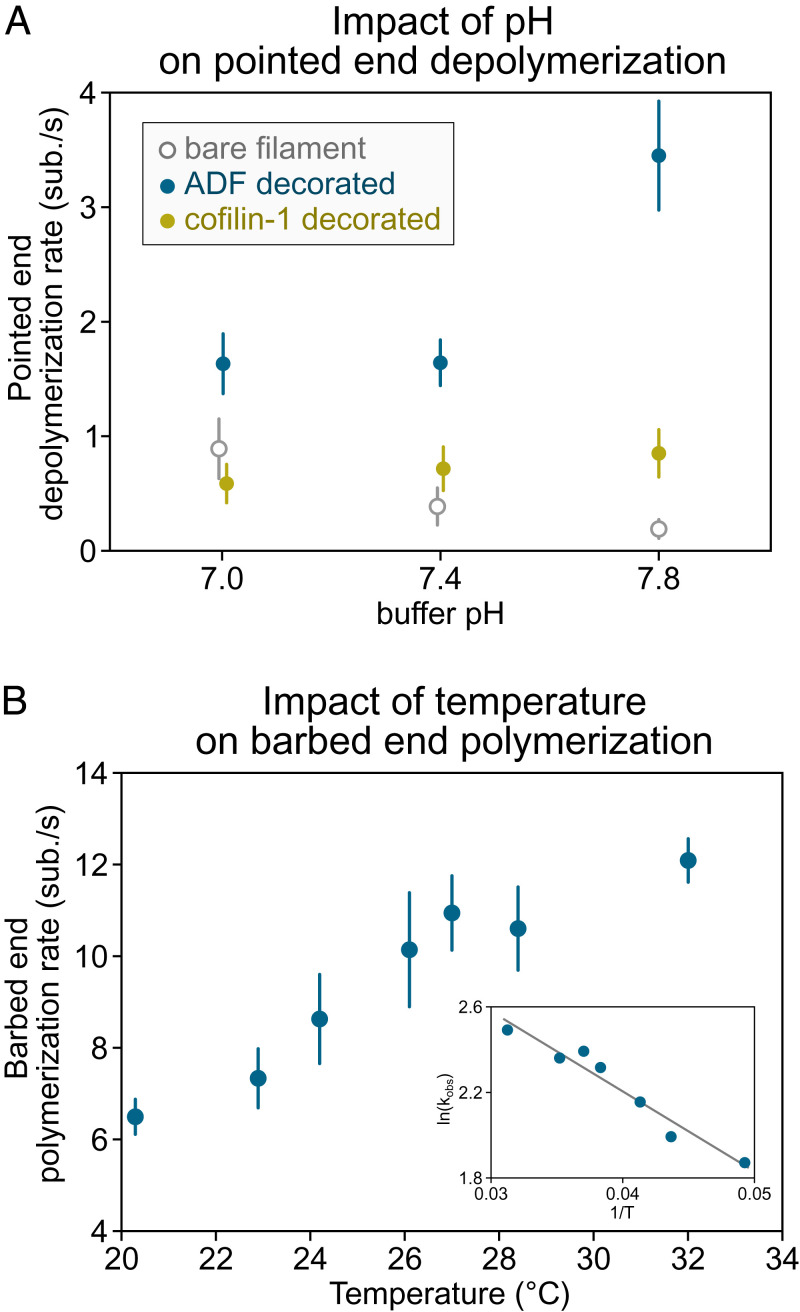
Another thing we have learned over the past 20 years is that apparent discrepancies in single-actin-filament studies usually come from differences in experimental conditions (such as pH, salt, and temperature) (Fig. 4) or from overlooked artifacts (Fig. 3). Knowing this, the results from different studies are remarkably consistent. Apparent discrepancies are eventually solved, yielding valuable insights, and it is worth addressing them by repeating existing experiments and comparing results. Recent work on the action of twinfilin at the barbed end is a good example of what can be gained by collectively repeating assays until a consensus is reached (76, 95). In time, by repeating experiments, we should be able to clarify whether aip1 causes filaments decorated by cofilin and coronin to disassemble in dramatic “bursts” (46, 96) or if it simply causes their severing (47). We should also find out why, depending on how tension is applied to a filament, it appears to provide protection against cofilin-induced severing (97) or not (78).
Fig. 4.
Impact of pH and temperature on reaction rates. (A) Depolymerization of actin filament pointed ends (15% labeled with AlexaFluor-568 on surface lysines) at different buffer pH (5 mM Tris⋅HCl, 50 mM KCl, 1 mM MgCl2, 0.4 mM CaCl2, 0.2 mM EGTA, 0.2 mM ATP, 10 mM DTT, 1 mM DABCO) supplemented with 1 to 2 µM ADF or cofilin-1 as indicated. Filaments are attached to the coverslip at their barbed end by gelsolin in a microfluidics chamber. Depolymerization of bare and ADF/cofilin-decorated filaments was recorded for up to 30 min at one frame every 30 to 60 s. Each data point represents the mean and SD over 14 to 36 filaments (from one experiment). Data adapted from ref. 99 with permission from the American Chemical Society (ACS), further permission should be directed to the ACS. (B) Elongation of the actin filament barbed end from anchored spectrin-actin seeds in a microfluidics flow, at different temperatures, exposed to 1 µM Mg-ATP-actin (10% AlexaFluor-488 labeled on surface lysines) in buffer (5 mM Tris⋅HCl, pH 7.4, 50 mM KCl, 1 mM MgCl2, 0.2 mM EGTA, 0.2 mM ATP, 10 mM DTT, 1 mM DABCO). Elongation was recorded for 5 min at one frame every 10 s, and n > 30 filaments were analyzed for each condition. (Inset) The slope of the linear relation of log(elongation rate) as a function of the inverse of the temperature (Arrhenius plot) allows us to estimate the free energy associated with actin monomer addition at the barbed end to be 37.8 kJ/mol (9.03 kcal/mol), similar to the value reported by Drenckhahn and Pollard (100) using EM. Data can be found in Dataset S1. ADF, actin depolymerizing factor; EGTA, ethylene glycol tetraacetic acid; DTT, dithiothreitol; DABCO, diazabicyclooctane.
Five Golden Rules for Single-Actin-Filament Studies
In order to further improve the reliability of single-filament results and to facilitate the fruitful comparison of results from different studies, we propose here a set of guidelines. The following five “golden rules” are often basic reminders of good scientific practice, applicable to other types of experiments. Yet, most reports of live single-filament studies (including our own, admittedly) follow only some of these rules, which is a pity because they are particularly meaningful for this type of experiment.
Be Explicit about Experimental Conditions.
Seemingly identical experiments may actually differ in a number of ways. Proteins may be different isoforms, with partial truncations and with different tags. Buffers may differ in pH and ionic strength. These details often matter. For instance, pH is known to impact several reaction rates (98, 99). Whether the pH is 7.0 or 7.8, one can reach opposite conclusions regarding the acceleration of pointed end depolymerization by cofilin, for example (Fig. 4A). Experiments are usually carried out “at room temperature,” which can effectively be anywhere between 18 and 28 °C, depending on the efficiency of air conditioning and on the number of electrical devices running nearby. Over this temperature range, reaction rates can vary by a factor of two (100) (Fig. 4B) and certainly more for higher-order reactions, like filament nucleation. If temperature cannot be accurately controlled, it can at least be measured near the sample and reported.
Readers should be able to find all these details rapidly. Summarizing them in a couple of sentences in the text and reminding of them in the figures can be very helpful.
Hunt Down Potential Artifacts.
Seeing an individual reaction take place live, before our own eyes, can give the compelling impression that we are looking at the naked truth. We are not. Proteins are observed in an artificial context, and as discussed earlier, several artifacts (in particular, caused by labeling, illumination, and surface interactions) (Fig. 3) can lead to the misinterpretation of single-filament data. Thankfully, these assays provide the means to vary conditions and track down potential artifacts. This should be systematically done, as much as possible.
Any Experiment Bears Repeating.
Cell biologists have long known the importance of repeating experiments, and are now improving ways to present these datasets [e.g., by using “superplots” (101, 102)]. In vitro single-filament studies have not yet adopted such standards. Results are often presented based on several observations (several individual events or several filaments) from a single experiment. However, many parameters escape our control (pipetting errors, variations in surface passivation), and the only way to assess reproducibility is to repeat the experiment.
Importantly, it is also worth repeating experiments from previous papers, whether from your laboratory or other laboratories. The trustworthiness of a result and its benefit to the community are greatly enhanced when it has been observed on independent occasions. If discrepancies arise, they are likely due to differences in the way the experiment was carried out and analyzed, and identifying them will drive the field forward.
Show Your Hand (Raw Data and Methodology).
In order to properly reproduce your results, readers need to know in detail how you performed your experiments (beyond the basic biochemical conditions listed in the first point). They should also be able to compare their data with your data. Others may obtain new results by analyzing your data in a different way. This is beneficial to all, and you can make it easier by sharing detailed protocols and by sharing your raw data (movies, kymographs), at least for key representative experiments. Nowadays, all this is easily done thanks to online supplementary data, online data repositories, video protocols, and even your personal website.
Studying Single Filaments Is a Collective Endeavor.
Following the previous rules should make it easier for others to reproduce your results and to compare their data with your data. The corollary is to put your new results in the perspective of previous work. If your results differ from published work, try to repeat that previous experiment in order to identify where the difference comes from. Earlier studies are not necessarily right, but if you find rates that disagree with well-established values obtained by independent laboratories (Table 1 shows some examples), it is probably worth taking a closer look at your experiment (rules 2 and 3).
Table 1.
Compilation of reaction rate constants confirmed by independent studies
| Substrate | On/off rate | Value range* | Single-filament refs. | Other techniques refs. | |
| Actin monomers (Mg-ATP-actin) | Barbed end | On rate | 7–12 µM−1⋅s−1 | 18, 73, 84, 116 | 4, 117 |
| Profilin-actin (Mg-ATP-actin) | Barbed end | On rate | 6–9 µM−1⋅s−1 | 33, 56, 73, 116 | |
| Actin monomers (Mg-ADP-actin) | Barbed end | On rate | 3–5 µM−1⋅s−1 | 35 | 4, 117 |
| Actin subunit (Mg-ADP-actin) | Barbed end | Off rate | 4–12 s−1 | 34, 35, 76, 95, 99 | 4, 117 |
| Profilin-actin (Mg-ADP-actin) | Barbed end | Off rate | 52 s−1 | 72 | 118 |
| Actin monomers (Mg-ATP-actin) | Pointed end | On rate | 0.5–1.5 µM−1⋅s−1 | 34, 35 | 4 |
| Actin subunit (Mg-ADP-actin) | Pointed end | Off rate | 0.1–0.4 s−1 | 34, 35, 67, 68, 99 | 4, 119 |
| Pi (release from ADP-Pi-actin) | Within filament | Off rate | 2–7⋅10−3 s−1 | 35, 72 | 120, 121 |
| Profilin-actin (Mg-ATP-actin) | mDia1-bound barbed end | On rate | 40–60 µM−1⋅s−1 | 63, 73, 85, 122, 123 | |
| Bni1p-bound barbed end | On rate | 15–20 µM−1⋅s−1 | 73, 124 | ||
| Capping protein | Barbed end | On rate | 3–19 µM−1⋅s−1 | 29, 31, 34 | 125, 126 |
| Off rate | 3–11⋅10−4 s−1 | 28, 29, 31 | 125, 126 |
*Reported values from single-filament studies vary within the indicated range due to different experimental conditions: pH 7.0 to 7.8, with 50 to 100 mM KCl, at an unspecified “room temperature.”
Conclusion
Being able to observe reactions taking place on individual actin filaments, live, has greatly expanded our understanding of the molecular processes that regulate actin dynamics. This experimental saga started decades ago, and we celebrate here the 20th anniversary of some of its landmark publications. In the coming years, we expect that this technique will provide many more results and will expand the list of quantified reaction rates, confirmed by independent measurements (we have compiled some in Table 1). These numbers should serve as references for experiments, provide rate constants for theoretical models, and guide the interpretation of cell data.
Supplementary Material
Acknowledgments
Live single-actin-filament studies are too numerous to be all cited here, and we apologize to the authors whose work was left out. We thank Ikram Fyfy for her help with some of the experimental data presented in this article and Laurent Blanchoin and Beata Bugyi for interesting discussions. We acknowledge funding from European Research Council Grant StG-679116 (to A.J.) and from Agence Nationale de la Recherche Grants Muscactin and Conformin (to G.R.-L.)
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109506119/-/DCSupplemental.
Data Availability
Previously published data were used for this work (99). All data are included in the manuscript and/or supporting information.
References
- 1.Bugyi B., Kellermayer M., The discovery of actin: “To see what everyone else has seen, and to think what nobody has thought.” J. Muscle Res. Cell Motil. 41, 3–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Depue R. H. Jr., Rice R. V., F-actin is a right-handed helix. J. Mol. Biol. 12, 302–303 (1965). [DOI] [PubMed] [Google Scholar]
- 3.Pollard T. D., Mooseker M. S., Direct measurement of actin polymerization rate constants by electron microscopy of actin filaments nucleated by isolated microvillus cores. J. Cell Biol. 88, 654–659 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pollard T. D., Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 103, 2747–2754 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kouyama T., Mihashi K., Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur. J. Biochem. 114, 33–38 (1981). [PubMed] [Google Scholar]
- 6.Yanagida T., Nakase M., Nishiyama K., Oosawa F., Direct observation of motion of single F-actin filaments in the presence of myosin. Nature 307, 58–60 (1984). [DOI] [PubMed] [Google Scholar]
- 7.Kellogg D. R., Mitchison T. J., Alberts B. M., Behaviour of microtubules and actin filaments in living Drosophila embryos. Development 103, 675–686 (1988). [DOI] [PubMed] [Google Scholar]
- 8.Isambert H., et al. , Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J. Biol. Chem. 270, 11437–11444 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Kishino A., Yanagida T., Force measurements by micromanipulation of a single actin filament by glass needles. Nature 334, 74–76 (1988). [DOI] [PubMed] [Google Scholar]
- 10.Gittes F., Mickey B., Nettleton J., Howard J., Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol. 120, 923–934 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanchoin L., et al. , Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404, 1007–1011 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Maciver S. K., Zot H. G., Pollard T. D., Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J. Cell Biol. 115, 1611–1620 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kron S. J., Spudich J. A., Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl. Acad. Sci. U.S.A. 83, 6272–6276 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finer J. T., Simmons R. M., Spudich J. A., Single myosin molecule mechanics: Piconewton forces and nanometre steps. Nature 368, 113–119 (1994). [DOI] [PubMed] [Google Scholar]
- 15.Axelrod D., Cell-substrate contacts illuminated by total internal reflection fluorescence. J. Cell Biol. 89, 141–145 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tokunaga M., Kitamura K., Saito K., Iwane A. H., Yanagida T., Single molecule imaging of fluorophores and enzymatic reactions achieved by objective-type total internal reflection fluorescence microscopy. Biochem. Biophys. Res. Commun. 235, 47–53 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Rock R. S., et al. , Myosin VI is a processive motor with a large step size. Proc. Natl. Acad. Sci. U.S.A. 98, 13655–13659 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amann K. J., Pollard T. D., Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc. Natl. Acad. Sci. U.S.A. 98, 15009–15013 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiwara I., Suetsugu S., Uemura S., Takenawa T., Ishiwata S., Visualization and force measurement of branching by Arp2/3 complex and N-WASP in actin filament. Biochem. Biophys. Res. Commun. 293, 1550–1555 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Fujiwara I., Takahashi S., Tadakuma H., Funatsu T., Ishiwata S., Microscopic analysis of polymerization dynamics with individual actin filaments. Nat. Cell Biol. 4, 666–673 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Ichetovkin I., Grant W., Condeelis J., Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 12, 79–84 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Nadkarni A. V., Brieher W. M., Aip1 destabilizes cofilin-saturated actin filaments by severing and accelerating monomer dissociation from ends. Curr. Biol. 24, 2749–2757 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ngo K. X., Kodera N., Katayama E., Ando T., Uyeda T. Q. P., Cofilin-induced unidirectional cooperative conformational changes in actin filaments revealed by high-speed atomic force microscopy. eLife 4, e04806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuda K., Sugawa M., Yamagishi M., Kodera N., Yajima J., Visualizing dynamic actin cross-linking processes driven by the actin-binding protein anillin. FEBS Lett. 594, 1237–1247 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Winterhoff M., Brühmann S., Franke C., Breitsprecher D., Faix J., Visualization of actin assembly and filament turnover by in vitro multicolor TIRF microscopy. Methods Mol. Biol. 1407, 287–306 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Hellen E. H., Axelrod D., Kinetics of epidermal growth factor/receptor binding on cells measured by total internal reflection/fluorescence recovery after photobleaching. J. Fluoresc. 1, 113–128 (1991). [DOI] [PubMed] [Google Scholar]
- 27.Pollard T. D., De La Cruz E. M., Take advantage of time in your experiments: A guide to simple, informative kinetics assays. Mol. Biol. Cell 24, 1103–1110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhn J. R., Pollard T. D., Single molecule kinetic analysis of actin filament capping. Polyphosphoinositides do not dissociate capping proteins. J. Biol. Chem. 282, 28014–28024 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Shekhar S., et al. , Formin and capping protein together embrace the actin filament in a ménage à trois. Nat. Commun. 6, 8730 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bombardier J. P., et al. , Single-molecule visualization of a formin-capping protein ‘decision complex’ at the actin filament barbed end. Nat. Commun. 6, 8707 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wioland H., et al. , ADF/cofilin accelerates actin dynamics by severing filaments and promoting their depolymerization at both ends. Curr. Biol. 27, 1956–1967.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paul A. S., Pollard T. D., The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 18, 9–19 (2008). Correction in: Curr. Biol. 18, 233 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao L., et al. , Modulation of formin processivity by profilin and mechanical tension. eLife 7, e34176 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhn J. R., Pollard T. D., Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88, 1387–1402 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujiwara I., Vavylonis D., Pollard T. D., Polymerization kinetics of ADP- and ADP-Pi-actin determined by fluorescence microscopy. Proc. Natl. Acad. Sci. U.S.A. 104, 8827–8832 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith B. A., et al. , Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. eLife 2, e01008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai L., Makhov A. M., Schafer D. A., Bear J. E., Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell 134, 828–842 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan C., Beltzner C. C., Pollard T. D., Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 19, 537–545 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gandhi M., et al. , GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr. Biol. 20, 861–867 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandit N. G., et al. , Force and phosphate release from Arp2/3 complex promote dissociation of actin filament branches. Proc. Natl. Acad. Sci. U.S.A. 117, 13519–13528 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De La Cruz E. M., Cofilin binding to muscle and non-muscle actin filaments: Isoform-dependent cooperative interactions. J. Mol. Biol. 346, 557–564 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Cao W., Goodarzi J. P., De La Cruz E. M., Energetics and kinetics of cooperative cofilin-actin filament interactions. J. Mol. Biol. 361, 257–267 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Suarez C., et al. , Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr. Biol. 21, 862–868 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gressin L., Guillotin A., Guérin C., Blanchoin L., Michelot A., Architecture dependence of actin filament network disassembly. Curr. Biol. 25, 1437–1447 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Chin S. M., Jansen S., Goode B. L., TIRF microscopy analysis of human Cof1, Cof2, and ADF effects on actin filament severing and turnover. J. Mol. Biol. 428, 1604–1616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kueh H. Y., Charras G. T., Mitchison T. J., Brieher W. M., Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J. Cell Biol. 182, 341–353 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jansen S., et al. , Single-molecule imaging of a three-component ordered actin disassembly mechanism. Nat. Commun. 6, 7202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christensen J. R., et al. , Competition between Tropomyosin, Fimbrin, and ADF/Cofilin drives their sorting to distinct actin filament networks. eLife 6, e23152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gateva G., et al. , Tropomyosin isoforms specify functionally distinct actin filament populations in vitro. Curr. Biol. 27, 705–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jansen S., Goode B. L., Tropomyosin isoforms differentially tune actin filament length and disassembly. Mol. Biol. Cell 30, 671–679 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wioland H., et al. , Actin filament oxidation by MICAL1 suppresses protections from cofilin-induced disassembly. EMBO Rep. 22, e50965 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grintsevich E. E., et al. , Catastrophic disassembly of actin filaments via Mical-mediated oxidation. Nat. Commun. 8, 2183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsiao J. Y., Goins L. M., Petek N. A., Mullins R. D., Arp2/3 complex and cofilin modulate binding of tropomyosin to branched actin networks. Curr. Biol. 25, 1573–1582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bareja I., et al. , Dynamics of Tpm1.8 domains on actin filaments with single molecule resolution. Mol. Biol. Cell 30, 2452–2462 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kovar D. R., Pollard T. D., Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. U.S.A. 101, 14725–14730 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romero S., et al. , Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell 119, 419–429 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Pernier J., et al. , Myosin 1b is an actin depolymerase. Nat. Commun. 10, 5200 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zimmermann D., Santos A., Kovar D. R., Rock R. S., Actin age orchestrates myosin-5 and myosin-6 run lengths. Curr. Biol. 25, 2057–2062 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith B. A., Daugherty-Clarke K., Goode B. L., Gelles J., Pathway of actin filament branch formation by Arp2/3 complex revealed by single-molecule imaging. Proc. Natl. Acad. Sci. U.S.A. 110, 1285–1290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansen S. D., Mullins R. D., VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J. Cell Biol. 191, 571–584 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burke T. A., Harker A. J., Dominguez R., Kovar D. R., The bacterial virulence factors VopL and VopF nucleate actin from the pointed end. J. Cell Biol. 216, 1267–1276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paul A. S., Pollard T. D., Energetic requirements for processive elongation of actin filaments by FH1FH2-formins. J. Biol. Chem. 284, 12533–12540 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Breitsprecher D., et al. , Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science 336, 1164–1168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Balzer C. J., Wagner A. R., Helgeson L. A., Nolen B. J., Dip1 co-opts features of branching nucleation to create linear actin filaments that activate WASP-bound Arp2/3 complex. Curr. Biol. 28, 3886–3891.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balzer C. J., Wagner A. R., Helgeson L. A., Nolen B. J., Single-turnover activation of Arp2/3 complex by Dip1 may balance nucleation of linear versus branched actin filaments. Curr. Biol. 29, 3331–3338.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cao L., et al. , SPIN90 associates with mDia1 and the Arp2/3 complex to regulate cortical actin organization. Nat. Cell Biol. 22, 803–814 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Kotila T., et al. , Mechanism of synergistic actin filament pointed end depolymerization by cyclase-associated protein and cofilin. Nat. Commun. 10, 5320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shekhar S., Chung J., Kondev J., Gelles J., Goode B. L., Synergy between Cyclase-associated protein and Cofilin accelerates actin filament depolymerization by two orders of magnitude. Nat. Commun. 10, 5319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oliveira C., Domingues L., Guidelines to reach high-quality purified recombinant proteins. Appl. Microbiol. Biotechnol. 102, 81–92 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Niedermayer T., et al. , Intermittent depolymerization of actin filaments is caused by photo-induced dimerization of actin protomers. Proc. Natl. Acad. Sci. U.S.A. 109, 10769–10774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanchez T., Kulic I. M., Dogic Z., Circularization, photomechanical switching, and a supercoiling transition of actin filaments. Phys. Rev. Lett. 104, 098103 (2010). [DOI] [PubMed] [Google Scholar]
- 72.Jégou A., et al. , Individual actin filaments in a microfluidic flow reveal the mechanism of ATP hydrolysis and give insight into the properties of profilin. PLoS Biol. 9, e1001161 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N., Pollard T. D., Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124, 423–435 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Tóth M. Á., et al. , Biochemical activities of the Wiskott-Aldrich syndrome homology region 2 domains of sarcomere length short (SALS) protein. J. Biol. Chem. 291, 667–680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vavylonis D., Yang Q., O’Shaughnessy B., Actin polymerization kinetics, cap structure, and fluctuations. Proc. Natl. Acad. Sci. U.S.A. 102, 8543–8548 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hakala M., et al. , Twinfilin uncaps filament barbed ends to promote turnover of lamellipodial actin networks. Nat. Cell Biol. 23, 147–159 (2021). [DOI] [PubMed] [Google Scholar]
- 77.Pavlov D., Muhlrad A., Cooper J., Wear M., Reisler E., Actin filament severing by cofilin. J. Mol. Biol. 365, 1350–1358 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wioland H., Jegou A., Romet-Lemonne G., Torsional stress generated by ADF/cofilin on cross-linked actin filaments boosts their severing. Proc. Natl. Acad. Sci. U.S.A. 116, 2595–2602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Courtemanche N., Pollard T. D., Chen Q., Avoiding artefacts when counting polymerized actin in live cells with LifeAct fused to fluorescent proteins. Nat. Cell Biol. 18, 676–683 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hatano T., Sivashanmugam L., Suchenko A., Hussain H., Balasubramanian M. K., Pick-ya actin: A method to purify actin isoforms with bespoke key post-translational modifications. J. Cell Sci. 133, jcs241406 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abella J. V. G., et al. , Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat. Cell Biol. 18, 76–86 (2016). [DOI] [PubMed] [Google Scholar]
- 82.Funk J., et al. , Profilin and formin constitute a pacemaker system for robust actin filament growth. eLife 8, e50963 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Colombo J., et al. , A functional family of fluorescent nucleotide analogues to investigate actin dynamics and energetics. Nat. Commun. 12, 548 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bieling P., et al. , WH2 and proline-rich domains of WASP-family proteins collaborate to accelerate actin filament elongation. EMBO J. 37, 102–121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jégou A., Carlier M.-F., Romet-Lemonne G., Formin mDia1 senses and generates mechanical forces on actin filaments. Nat. Commun. 4, 1883 (2013). [DOI] [PubMed] [Google Scholar]
- 86.Courtemanche N., Lee J. Y., Pollard T. D., Greene E. C., Tension modulates actin filament polymerization mediated by formin and profilin. Proc. Natl. Acad. Sci. U.S.A. 110, 9752–9757 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mei L., et al. , Molecular mechanism for direct actin force-sensing by α-catenin. eLife 9, e62514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Winkelman J. D., et al. , Fascin- and α-actinin-bundled networks contain intrinsic structural features that drive protein sorting. Curr. Biol. 26, 2697–2706 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Christensen J. R., et al. , Cooperation between tropomyosin and α-actinin inhibits fimbrin association with actin filament networks in fission yeast. eLife 8, e47279 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Suzuki E. L., et al. , Geometrical constraints greatly hinder formin mDia1 activity. Nano Lett. 20, 22–32 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Breitsprecher D., et al. , Cofilin cooperates with fascin to disassemble filopodial actin filaments. J. Cell Sci. 124, 3305–3318 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reymann A. C., et al. , Actin network architecture can determine myosin motor activity. Science 336, 1310–1314 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Murrell M. P., Gardel M. L., F-actin buckling coordinates contractility and severing in a biomimetic actomyosin cortex. Proc. Natl. Acad. Sci. U.S.A. 109, 20820–20825 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Köhler S., Bausch A. R., Contraction mechanisms in composite active actin networks. PLoS One 7, e39869 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shekhar S., Hoeprich G. J., Gelles J., Goode B. L., Twinfilin bypasses assembly conditions and actin filament aging to drive barbed end depolymerization. J. Cell Biol. 220, e202006022 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tang V. W., Nadkarni A. V., Brieher W. M., Catastrophic actin filament bursting by cofilin, Aip1, and coronin. J. Biol. Chem. 295, 13299–13313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hayakawa K., Tatsumi H., Sokabe M., Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. J. Cell Biol. 195, 721–727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Crevenna A. H., et al. , Electrostatics control actin filament nucleation and elongation kinetics. J. Biol. Chem. 288, 12102–12113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wioland H., Jegou A., Romet-Lemonne G., Quantitative variations with pH of actin depolymerizing factor/Cofilin’s multiple actions on actin filaments. Biochemistry 58, 40–47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Drenckhahn D., Pollard T. D., Elongation of actin filaments is a diffusion-limited reaction at the barbed end and is accelerated by inert macromolecules. J. Biol. Chem. 261, 12754–12758 (1986). [PubMed] [Google Scholar]
- 101.Lord S. J., Velle K. B., Mullins R. D., Fritz-Laylin L. K., SuperPlots: Communicating reproducibility and variability in cell biology. J. Cell Biol. 219, e202001064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kenny M., Schoen I., Violin SuperPlots: Visualizing replicate heterogeneity in large data sets. Mol. Biol. Cell 32, 1333–1334 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chou S. Z., Pollard T. D., Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides. Proc. Natl. Acad. Sci. U.S.A. 116, 4265–4274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pettersen E. F., et al. , UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smith M. B., et al. , Segmentation and tracking of cytoskeletal filaments using open active contours. Cytoskeleton (Hoboken) 67, 693–705 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Y. L., Taylor D. L., Distribution of fluorescently labeled actin in living sea urchin eggs during early development. J. Cell Biol. 81, 672–679 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wulf E., Deboben A., Bautz F. A., Faulstich H., Wieland T., Fluorescent phallotoxin, a tool for the visualization of cellular actin. Proc. Natl. Acad. Sci. U.S.A. 76, 4498–4502 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Saito K., Tokunaga M., Iwane A. H., Yanagida T., Dual-colour microscopy of single fluorophores bound to myosin interacting with fluorescently labelled actin using anti-Stokes fluorescence. J. Microsc. 188, 255–263 (1997). [DOI] [PubMed] [Google Scholar]
- 109.Reymann A.-C., et al. , Nucleation geometry governs ordered actin networks structures. Nat. Mater. 9, 827–832 (2010). [DOI] [PubMed] [Google Scholar]
- 110.Ydenberg C. A., et al. , GMF severs actin-Arp2/3 complex branch junctions by a cofilin-like mechanism. Curr. Biol. 23, 1037–1045 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Risca V. I., et al. , Actin filament curvature biases branching direction. Proc. Natl. Acad. Sci. U.S.A. 109, 2913–2918 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wagner A. R., Luan Q., Liu S.-L., Nolen B. J., Dip1 defines a class of Arp2/3 complex activators that function without preformed actin filaments. Curr. Biol. 23, 1990–1998 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Andrianantoandro E., Pollard T. D., Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24, 13–23 (2006). [DOI] [PubMed] [Google Scholar]
- 114.Michelot A., et al. , Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr. Biol. 17, 825–833 (2007). [DOI] [PubMed] [Google Scholar]
- 115.Grintsevich E. E., et al. , F-actin dismantling through a redox-driven synergy between Mical and cofilin. Nat. Cell Biol. 18, 876–885 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Breitsprecher D., et al. , Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 27, 2943–2954 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Carlier M. F., Pantaloni D., Korn E. D., Evidence for an ATP cap at the ends of actin filaments and its regulation of the F-actin steady state. J. Biol. Chem. 259, 9983–9986 (1984). [PubMed] [Google Scholar]
- 118.Bubb M. R., Yarmola E. G., Gibson B. G., Southwick F. S., Depolymerization of actin filaments by profilin. Effects of profilin on capping protein function. J. Biol. Chem. 278, 24629–24635 (2003). [DOI] [PubMed] [Google Scholar]
- 119.Korn E. D., Carlier M. F., Pantaloni D., Actin polymerization and ATP hydrolysis. Science 238, 638–644 (1987). [DOI] [PubMed] [Google Scholar]
- 120.Melki R., Fievez S., Carlier M.-F., Continuous monitoring of Pi release following nucleotide hydrolysis in actin or tubulin assembly using 2-amino-6-mercapto-7-methylpurine ribonucleoside and purine-nucleoside phosphorylase as an enzyme-linked assay. Biochemistry 35, 12038–12045 (1996). [DOI] [PubMed] [Google Scholar]
- 121.Blanchoin L., Pollard T. D., Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J. Biol. Chem. 274, 15538–15546 (1999). [DOI] [PubMed] [Google Scholar]
- 122.Maiti S., et al. , Structure and activity of full-length formin mDia1. Cytoskeleton (Hoboken) 69, 393–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kubota H., et al. , Biphasic effect of profilin impacts the formin mDia1 force-sensing mechanism in actin polymerization. Biophys. J. 113, 461–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Alioto S. L., Garabedian M. V., Bellavance D. R., Goode B. L., Tropomyosin and profilin cooperate to promote formin-mediated actin nucleation and drive yeast actin cable assembly. Curr. Biol. 26, 3230–3237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schafer D. A., Jennings P. B., Cooper J. A., Dynamics of capping protein and actin assembly in vitro: Uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 135, 169–179 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wear M. A., Yamashita A., Kim K., Maéda Y., Cooper J. A., How capping protein binds the barbed end of the actin filament. Curr. Biol. 13, 1531–1537 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Previously published data were used for this work (99). All data are included in the manuscript and/or supporting information.