Significance
As a master transcription factor, the tumor suppressor p53 regulates a variety of cellular processes by modulating target gene expression. Although well known as a transcriptional activator, p53 is also able to repress expression of a number of protein-coding genes. However, it remains largely unknown whether long noncoding RNA (lncRNA) gene repression is involved in the regulation of p53 function. Here, we show that p53 can transcriptionally repress expression of the lncRNA E2F1 messenger RNA (mRNA) stabilizing factor (EMS) and vice versa and that EMS can suppress p53 translation and negatively regulate p53 function. Our study reveals an important double-negative feedback loop that controls p53 activity and provides insights into the mechanisms whereby EMS promotes tumorigenesis.
Keywords: p53, lncRNA, CPEB2, translation, tumorigenesis
Abstract
p53 plays a central role in tumor suppression. Emerging evidence suggests long noncoding RNA (lncRNA) as an important class of regulatory molecules that control the p53 signaling. Here, we report that the oncogenic lncRNA E2F1 messenger RNA (mRNA) stabilizing factor (EMS) and p53 mutually repress each other’s expression. EMS is negatively regulated by p53. As a direct transcriptional repression target of p53, EMS is surprisingly shown to inhibit p53 expression. EMS associates with cytoplasmic polyadenylation element-binding protein 2 (CPEB2) and thus, disrupts the CPEB2–p53 mRNA interaction. This disassociation attenuates CPEB2-mediated p53 mRNA polyadenylation and suppresses p53 translation. Functionally, EMS is able to exert its oncogenic activities, at least partially, via the CPEB2–p53 axis. Together, these findings reveal a double-negative feedback loop between p53 and EMS, through which p53 is finely controlled. Our study also demonstrates a critical role for EMS in promoting tumorigenesis via the negative regulation of p53.
The tumor suppressor p53 plays an important role in tumor suppression (1, 2). p53 primarily functions as a transcription factor to regulate expression of downstream target genes that are involved in various cellular processes, including cell cycle progression, cellular senescence, apoptosis, ferroptosis, autophagy, and cell metabolism (3–8). In the majority of human tumors, p53 is often inactivated through either direct mutation or altered expression of regulatory proteins (9, 10). Moreover, p53-deficient mice are highly susceptible to spontaneous tumorigenesis (11). Therefore, inhibition of p53 activity is considered a crucial event during tumorigenesis.
It has been well recognized that p53 expression is primarily regulated at the level of protein stability. In unstressed cells, p53 is expressed at low levels due to the constant ubiquitination and proteasomal degradation mediated by several ubiquitin E3 ligases, including murine double minute 2 (Mdm2), Pirh2, and COP1 (12–15). Under stress conditions, such as DNA damage and oncogenic insults, p53 becomes rapidly stabilized and activated (16). Although it has been demonstrated that posttranslational modifications, including phosphorylation, acetylation, and ubiquitination, play an important role in controlling p53 activity, accumulating evidence indicates that p53 activity is also regulated by other mechanisms, particularly translational regulation (17, 18). For instance, p53 has been shown to be modulated by internal ribosome entry site (IRES)-mediated translation (19, 20). In addition, several RNA-binding proteins, including HuR, RPL26, nucleolin, RNPC1, cytoplasmic polyadenylation element-binding protein 1 (CPEB1), and cytoplasmic polyadenylation element-binding protein 2 (CPEB2), are able to regulate p53 translation (21–26).
Long noncoding RNAs (lncRNAs) represent a class of nonprotein-coding transcripts with a length of more than 200 nucleotides (27). Thus far, more than 17,000 human lncRNA genes have been documented in GENCODE v38 (28). Increasing evidence suggests that lncRNAs play a critical role in the regulation of gene expression at different levels (29). Of note, lncRNAs have been shown to translationally regulate gene expression by base pairing with target messenger RNA (mRNA), modulating the translational factors, or acting as decoys of RNA-binding proteins or microRNAs (30). Recently, it has been reported that lncRNAs participate in the control of p53 signaling (31). For example, lncRNAs, including PURPL, PARL, and MEG3, are able to posttranslationally regulate p53 expression and thus, exert their tumor-regulatory effects (32–34). In addition, p53 is capable of transcriptionally activating expression of a number of lncRNAs, such as lincRNA-p21, PANDA, damage induced noncoding (DINO), and GUARDIN, to regulate different cellular processes, including apoptosis, cell cycle progression, and genomic stability maintenance (35–38). Although well known as a transcriptional activator (39), p53 is also shown to suppress expression of multiple protein-coding genes (40). In addition to gene activation, gene repression is also considered as an essential part of the p53 cellular response (41). However, it remains poorly understood whether p53 could repress lncRNAs expression and whether these lncRNAs are functionally involved in the regulation of p53 tumor-suppressive activity.
Our recent study identified an oncogenic lncRNA E2F1 mRNA stabilizing factor (EMS) (42). As a transcriptional target of c-Myc, EMS was shown to connect c-Myc to cell cycle control and tumorigenesis via the stabilization of E2F1 mRNA. We noticed that induction of E2F1 only partially rescued the inhibitory effect of EMS knockdown on tumor cell growth, indicating that EMS may promote tumorigenesis via additional mechanism(s). In the present study, we show that EMS is transcriptionally repressed by p53. On the other hand, EMS functions as a negative regulator of p53. EMS associates with the RNA-binding protein CPEB2 to suppress p53 expression at the translational level. Furthermore, EMS is able to exert its oncogenic effects through the regulation of the CPEB2–p53 axis. These findings indicate the existence of a double-negative feedback loop between EMS and p53 and provide insights into the mechanisms of how EMS promotes tumorigenesis.
Results
EMS Is Negatively Regulated by p53.
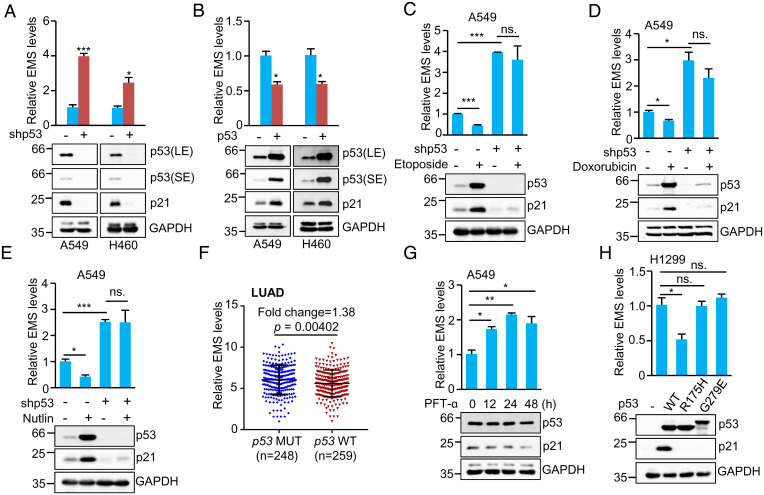
In our recent study, we noticed that p53-deficient H1299 lung adenocarcinoma (LUAD) cells exhibited much higher copy numbers of EMS than p53 wild-type A549 LUAD cells (42). This prompted us to investigate whether p53 could regulate EMS expression. The results showed that knockdown of p53 increased, whereas overexpression of p53 decreased EMS levels in both A549 and H460 cells harboring wild-type p53 (Fig. 1 A and B). Treatment of A549 cells with p53-inducing agent etoposide or doxorubicin resulted in decreased expression of EMS (Fig. 1 C and D). However, when p53 was knocked down in these cells, treatment with etoposide or doxorubicin no longer inhibited the expression of EMS (Fig. 1 C and D), indicating the involvement of p53 in the decreased expression of EMS caused by etoposide or doxorubicin treatment. Moreover, the p53 activator Nutlin-3 was shown to suppress EMS expression in control A549 cells but not in p53 knockdown A549 cells (Fig. 1E). Collectively, these findings suggest that EMS is negatively regulated by p53. In accordance, analysis of The Cancer Genome Atlas (TCGA) database showed that EMS was expressed at relatively higher levels in p53 mutated than in p53 wild-type LUAD (Fig. 1F), although the difference between these two groups was not dramatic.
Fig. 1.
EMS is negatively regulated by p53. (A) Real-time RT-PCR analysis of EMS levels in A549 and H460 cells infected with lentiviruses expressing control shRNA or p53 shRNA. Data shown are mean ± SD (n = 3). LE and SE indicate long-time exposure and short-time exposure, respectively. *P < 0.05; ***P < 0.001. (B) Real-time RT-PCR analysis of EMS levels in A549 and H460 cells infected with lentiviruses expressing control (pCDH) or p53 (pCDH–p53). pCDH is a lentiviral expression vector. Data shown are mean ± SD (n = 3). *P < 0.05. (C–E) A549 cells expressing control shRNA or p53 shRNA were treated with (C) 10 μM Etoposide, (D) 0.5 μg/mL Doxorubicin, or (E) 10 μM Nutlin-3 for 24 h. Total RNA and cell lysates were then analyzed by real-time RT-PCR and western blotting, respectively. Data shown are mean ± SD (n = 3). *P < 0.05; ***P < 0.001. (F) EMS was expressed at relatively lower levels in LUAD with wild-type (WT) p53 (n = 259) than in those carrying mutated (MUT) p53 (n = 248). (G) A549 cells were treated with pifithrin-α (PFT-α) (20 μM) for the indicated periods of time, followed by real-time RT-PCR and western blot analyses. Data shown are mean ± SD (n = 3). *P < 0.05; **P < 0.01. (H) H1299 cells were transfected with plasmids encoding wild-type p53 or the indicated mutant forms of p53. Twenty-four hours after transfection, total RNA and cell lysates were analyzed by real-time RT-PCR and western blotting, respectively. Data shown are mean ± SD (n = 3). ns., no significance. *P < 0.05.
To determine whether EMS is transcriptionally repressed by p53, we first utilized pifithrin-α (PFT-α), an inhibitor of p53 transcriptional activity. EMS expression was indeed induced upon PFT-α treatment in a time-dependent manner (Fig. 1G). Additionally, we examined whether the tumor-derived p53 mutants (R175H and G279E), which lack the transcriptional activity of wild-type p53, could regulate EMS expression. The results showed that these two p53 mutants failed to down-regulate EMS expression in H1299 cells (Fig. 1H). Taken together, these data suggest that EMS is negatively regulated by p53 at the transcriptional level.
EMS Is a Transcriptional Target of p53.
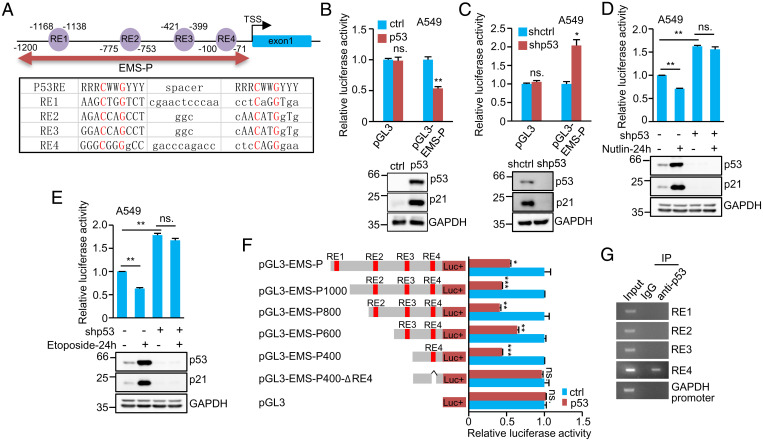
We next asked whether EMS is a transcriptional target of p53. By analyzing the promoter sequence of the EMS gene, we identified four putative repressive p53 response elements (RE) (43) (RE1, RE2, RE3, and RE4) within the promoter region spanning from −1,200 to −1 bp (EMS promoter [EMS-P]) (Fig. 2A). We, therefore, examined whether p53 could repress the activity of this EMS promoter region. The luciferase activity driven by EMS-P was indeed reduced by p53 overexpression but induced by p53 knockdown (Fig. 2 B and C). In addition, both Nutlin-3 and etoposide showed the inhibitory effects on the luciferase activity from pGL3-EMS-P in A549 cells, however, which was minimized by knockdown of p53 (Fig. 2 D and E). These data indicate that EMS-P is functional for the transcriptional repression of EMS by p53.
Fig. 2.
EMS is a direct target gene of p53. (A) Schematic illustration of four putative repressive p53 response elements (RE1, RE2, RE3, and RE4) in the EMS gene promoter. (B) A549 cells were cotransfected with control vector, Flag–p53, or together with the reporter constructs in the indicated combination. Twenty-four hours after transfection, reporter activity was measured. Data shown are mean ± SD (n = 3). **P < 0.01. (C) A549 cells expressing either control shRNA or p53 shRNA were transfected with the indicated reporter constructs. Twenty-four hours after transfection, reporter activity was measured. Data shown are mean ± SD (n = 3). *P < 0.05. (D and E) A549 cells expressing either control shRNA or p53 shRNA were cotransfected with pGL3-EMS-P and Renilla luciferase plasmid. Twenty-four hours after transfection, cells were treated with (D) 10 μM Nutlin or (E) 10 μM Etoposide for 24 h. Reporter activity was then measured. Data shown are mean ± SD (n = 3). **P < 0.01. (F) A549 cells were cotransfected with control vector, Flag–p53, or together with the indicated reporter constructs. Twenty-four hours after transfection, reporter activity was measured. Data shown are mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001. (G) Lysates from A549 cells were subjected to ChIP assay using anti-p53 antibody or an isotope-matched control IgG. ChIP products were amplified by PCR. ctrl, control; IgG, immunoglobulin G; IP, immunoprecipitation; ns., no significance.
To further determine the region of EMS-P that is responsible for p53-repressed EMS expression, a series of pGL3-based EMS-P deletion mutants was generated. The luciferase activities from pGL3-EMS-P, pGL3-EMS-P1000, pGL3-EMS-P800, pGL3-EMS-P600, and pGL3-EMS-P400 carrying the RE4 site were strongly decreased by ectopically expressed p53 (Fig. 2F). By contrast, the luciferase activity from pGL3-EMS-P400-ΔRE4 with deletion of the RE4 site showed no response to p53 induction (Fig. 2F). In accordance with these results, the chromatin immunoprecipitation (ChIP) assay revealed the association of p53 with the chromatin fragment containing the RE4 site but not the RE1, RE2, or RE3 site (Fig. 2G). Taken together, these data implicate EMS as a direct transcriptional repression target of p53.
EMS Suppresses p53 Expression at the Translational Level.
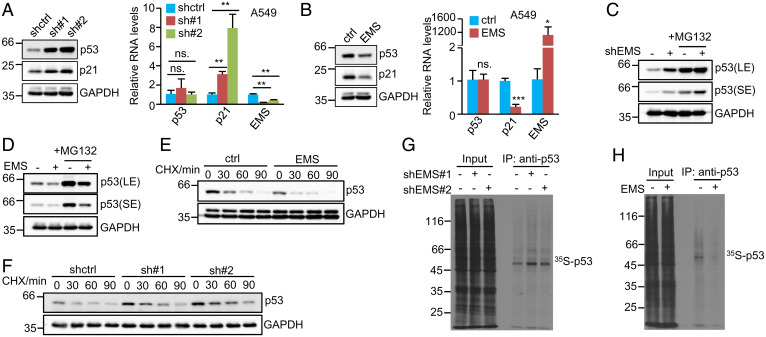
To better understand the relationship between EMS and p53, we extended our analysis to investigate the effect of EMS on p53 expression. Intriguingly, knockdown of EMS strongly increased, whereas ectopic expression of EMS markedly decreased the protein levels of p53 and its downstream target gene p21 in both A549 and H460 cells (Fig. 3 A and B and SI Appendix, Fig. S1 A and B). Of note, neither knockdown nor overexpression of EMS showed an obvious impact on p53 mRNA levels (Fig. 3 A and B and SI Appendix, Fig. S1 A and B). These data suggest that EMS may inhibit p53 expression at the posttranscriptional level.
Fig. 3.
EMS inhibits p53 expression at the translational level. (A) A549 cells were infected with lentiviruses expressing control shRNA, EMS shRNA#1, or EMS shRNA#2. (B) A549 cells were infected with lentiviruses expressing control (pSin) or EMS (pSin-EMS). pSin is a lentiviral expression vector. Forty-eight hours after infection, cell lysates and total RNA were analyzed by western blotting and real-time RT-PCR, respectively. Data shown are mean ± SD (n = 3). ns., no significance. *P < 0.05; **P < 0.01; ***P < 0.001. (C and D) (C) A549 cells were infected with lentiviruses expressing control shRNA or EMS shRNA. (D) A549 cells were infected with lentiviruses expressing control (pSin) or EMS (pSin-EMS). Forty-eight hours after infection, cells were treated with or without MG132 (20 μM) for 4 h. Cell lysates were then analyzed by western blotting. LE and SE indicate long-time exposure and short-time exposure, respectively. (E) A549 cells expressing control or EMS were treated with cycloheximide (CHX; 20 μg/mL) for the indicated periods of time, followed by western blot analysis. The band intensities were quantified using ImageJ software. The ratio of p53 to GAPDH is presented in SI Appendix, Fig. S1C. (F) A549 cells expressing control shRNA, EMS shRNA#1, or EMS shRNA#2 were treated with CHX (20 μg/mL) for the indicated periods of time, followed by western blot analysis. The band intensities were quantified using ImageJ software. The ratio of p53 to GAPDH is presented in SI Appendix, Fig. S1D. (G) A549 cells expressing control shRNA, EMS shRNA#1, or EMS shRNA#2 were pulse labeled with [35S]cysteine/methionine for 90 min. Cell lysates were then immunoprecipitated with anti-p53 antibody and analyzed by SDS-PAGE, followed by autoradiography. (H) A549 cells expressing control or EMS were pulse labeled with [35S]cysteine/methionine for 90 min. Cell lysates were then immunoprecipitated with anti-p53 antibody and analyzed by SDS-PAGE, followed by autoradiography. ctrl, control; IP, immunoprecipitation.
Considering that the ubiquitin proteasome pathway is crucial in controlling the cellular levels of p53, we first examined whether EMS affects p53 proteasomal degradation. A549 cells with EMS knockdown or overexpression were treated with the proteasome inhibitor MG132. It was evident that even in the presence of MG132, the protein levels of p53 were still induced by knockdown of EMS and reduced by ectopic expression of EMS (Fig. 3 C and D), implying that it is unlikely that EMS influences the proteasomal degradation of p53. In support of this notion, EMS knockdown or overexpression exhibited no effect on the half-life of p53 protein (Fig. 3 E and F and SI Appendix, Fig. S1 C and D). We next evaluated whether EMS could regulate p53 translation by tracing newly synthesized p53 (35S labeled). The results showed that knockdown of EMS clearly increased, whereas enforced expression of EMS dramatically decreased the levels of newly synthesized p53 (Fig. 3 G and H). Together, these data suggest that EMS represses p53 expression at the translational level.
EMS Exerts Its Oncogenic Function through the Negative Regulation of p53.
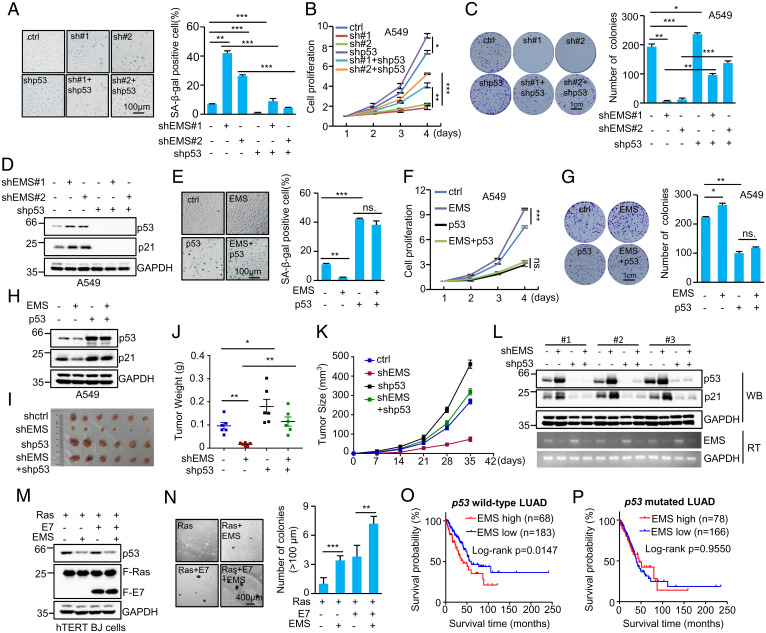
Given the ability of EMS to suppress p53 expression, we sought to determine whether EMS could exert its oncogenic function via the regulation of p53. EMS and p53 were knocked down individually or combined in A549 and H460 cells. Knockdown of EMS in these cells increased cellular senescence, reduced cell proliferation, and decreased the number of colonies, which could be greatly reversed by the simultaneous knockdown of p53 (Fig. 4 A–D and SI Appendix, Fig. S2 A–D). In addition, ectopic expression of EMS in A549 cells and H460 cells was shown to inhibit cellular senescence, accelerate cell proliferation, and increase the number of colonies (Fig. 4 E–H and SI Appendix, Fig. S2 E–H). However, EMS failed to show any of these effects when p53 was overexpressed (Fig. 4 E–H and SI Appendix, Fig. S2 E–H). By using a xenograft mouse model, we showed that EMS knockdown-inhibited in vivo xenograft tumor growth was able to be greatly recovered when p53 was concurrently knocked down (Fig. 4 I–L).
Fig. 4.
EMS exerts its oncogenic function through the negative regulation of p53. (A) Senescence-associated (SA) β-galactosidase staining in A549 cells expressing control shRNA, EMS shRNA#1, EMS shRNA#2, p53 shRNA, EMS shRNA#1 plus p53 shRNA, or EMS shRNA#2 plus p53 shRNA. The images are representative of three independent experiments. Data shown are mean ± SD (n = 3). **P < 0.01; ***P < 0.001. (B) The growth curves of A549 cells expressing control shRNA, EMS shRNA#1, EMS shRNA#2, p53 shRNA, EMS shRNA#1 plus p53 shRNA, or EMS shRNA#2 plus p53 shRNA. Data shown are mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001. (C) Colonies of A549 cells expressing control shRNA, EMS shRNA#1, EMS shRNA#2, p53 shRNA, EMS shRNA#1 plus p53 shRNA, or EMS shRNA#2 plus p53 shRNA were stained with crystal violet after 10 d of incubation. The shown images are representative of three independent experiments. Data shown are mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001. (D) Western blot analysis of lysates from A549 cells expressing control shRNA, EMS shRNA#1, EMS shRNA#2, p53 shRNA, EMS shRNA#1 plus p53 shRNA, or EMS shRNA#2 plus p53 shRNA. (E) SA β-galactosidase staining in A549 cells expressing control, EMS, p53, or both EMS and p53. The images are representative of three independent experiments. Data shown are mean ± SD (n = 3). **P < 0.01; ***P < 0.001. (F) The growth curves of A549 cells expressing control, EMS, p53, or both EMS and p53. Data shown are mean ± SD (n = 3). ***P < 0.001. (G) Colonies of A549 cells expressing control, EMS, p53, or both EMS and p53 were stained with crystal violet after 10 d of incubation. The shown images are representative of three independent experiments. Data shown are mean ± SD (n = 3). *P < 0.05; **P < 0.01. (H) Western blot analysis of lysates from A549 cells expressing control, EMS, p53, or both EMS and p53. (I–L) In total, 2 × 106 A549 cells transduced with lentiviruses expressing control, EMS shRNA, p53 shRNA, or both EMS shRNA and p53 shRNA were individually injected into nude mice (n = 6 for each group). (I) Xenograft tumors were taken 5 wk after injection. (J) Excised tumors were weighed. *P < 0.05; **P < 0.01. (K) Tumor sizes were measured at the indicated time points. (L) RNA and protein extracts from the excised xenografts were also analyzed by RT-PCR (RT) and western blotting (WB). (M and N) hTERT-immortalized BJ cells were sequentially infected with lentiviruses expressing EMS, E7, and KRAS G12V in the indicated combination. (M) Forty-eight hours later, cell lysates were analyzed by western blotting. (N) These cells were also subjected to soft agar colony formation assay; 20 d after seeding, colonies were stained with crystal violet, and images were acquired. Numbers of colonies in six randomly selected areas (40× magnification) were counted and averaged. Data shown are mean ± SD (n = 3). **P < 0.01; ***P < 0.001. (O and P) Kaplan–Meier analysis of the impact of EMS expression on overall survival of patients (O) with p53 wild-type LUAD or (P) with p53-mutated LUAD. The data were obtained from the TCGA database. ctrl, control; ns., no significance.
In general, inactivation of the p53 pathway, inactivation of the retinoblastoma protein (Rb) pathway, and induction of oncogenic RAS are required for the in vitro transformation of human cells (44). To further investigate whether EMS could confer tumorigenic potential on human cells, human telomerase reverse transcriptase (hTERT)-immortalized human foreskin fibroblasts (BJ cells) were utilized. Lentiviruses expressing EMS; the E7 protein of human papilloma virus type 16, which inactivates Rb; and the kirsten rat sarcoma viral oncogene homolog (KRAS) mutant G12V were sequentially introduced into hTERT-immortalized BJ cells, followed by the soft agar colony formation assay. Cells expressing EMS, E7, and KRAS G12V formed large colonies in soft agar, accompanied by the decreased levels of p53 (Fig. 4 M and N). The number and size of colonies were significantly higher and larger than those of cells expressing KRAS G12V plus either EMS or E7 (Fig. 4 M and N). Taken together, these data suggest that EMS exerts its oncogenic role through the negative regulation of p53. In support, analysis of TCGA database revealed that the high expression of EMS was associated with poor prognosis in patients with p53 wild-type LUAD but not those with p53-mutated LUAD (Fig. 4 O and P).
We have previously shown that EMS is able to promote tumorigenesis through increasing E2F1 expression (42). Consistent with our previous findings, induction of E2F1 in A549 cells could partially rescue the reduced cell proliferation and the decreased number of colonies caused by EMS knockdown (SI Appendix, Fig. S2 I–K). Importantly, the inhibitory effects of EMS knockdown on cell proliferation and colony formation were almost completely reversed by the simultaneous E2F1 overexpression and p53 knockdown (SI Appendix, Fig. S2 I–K). These data indicate that EMS promotes tumorigenesis through the regulation of both E2F1 and p53 expression.
EMS Coordinates with CPEB2 to Inhibit p53 Expression.
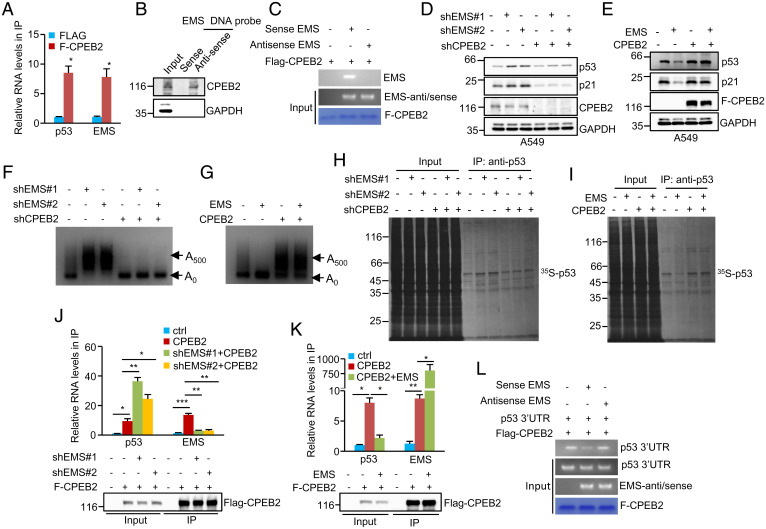
To explore the molecular mechanisms of EMS-mediated repression of p53, we sought to identify EMS-interacting proteins. Proteins pulled down by antisense DNA oligomers against EMS were analyzed by mass spectrometry. Of those potential EMS-interacting candidates (SI Appendix, Fig. S3A), CPEB2 attracted our particular attention because as an RNA-binding protein, CPEB2 has recently been implicated in the regulation of p53 translation (26). To validate the interaction between EMS and CPEB2, we first carried out an RNA immunoprecipitation experiment. EMS was indeed significantly enriched in the Flag–CPEB2 immunoprecipitates compared with control immunoprecipitates (Fig. 5A and SI Appendix, Fig. S3B). The CPEB2–EMS interaction was also verified by a biotin pull-down assay using a biotin-labeled antisense DNA oligomer against EMS, while EMS exhibited no binding to p53 mRNA (Fig. 5B and SI Appendix, Fig. S3C). Moreover, an in vitro binding assay revealed that CPEB2 directly interacted with EMS but not its antisense RNA (Fig. 5C). These combined data demonstrate CPEB2 as a binding partner for EMS.
Fig. 5.
EMS coordinates with CPEB2 to inhibit p53 expression. (A) Lysates from A549 cells expressing control or Flag–CPEB2 were immunoprecipitated with anti-Flag antibody. RNAs in immunoprecipitates were then analyzed by real-time RT-PCR to examine EMS and p53 RNA levels. Data shown are mean ± SD (n = 3). The input and immunoprecipitates were also analyzed by western blotting with anti-Flag antibody (SI Appendix, Fig. S3B). *P < 0.05. (B) Lysates from A549 cells were incubated with either sense or antisense biotin-labeled DNA oligomers corresponding to EMS, followed by the pull-down experiments using streptavidin-coated beads. The pulled-down complexes were analyzed by western blotting. The same complexes were also subjected to real-time RT-PCR analysis to examine EMS and p53 RNA levels (SI Appendix, Fig. S3C). (C) In vitro–synthesized EMS or its antisense RNA was incubated with purified recombinant Flag–CPEB2 bound with M2 beads. The inputs and beads-bound RNAs were analyzed by RT-PCR. The primers used for EMS antisense/sense detection do not discern between sense and antisense EMS. (D) A549 cells were infected with lentiviruses expressing EMS shRNA#1, EMS shRNA#2, or CPEB2 shRNA in the indicated combination. Forty-eight hours after infection, cell lysates were analyzed by western blotting. (E) A549 cells were infected with lentiviruses expressing control, EMS, Flag–CPEB2, or EMS plus Flag–CPEB2. Forty-eight hours later, cell lysates were analyzed by western blotting. (F) A549 cells were infected with lentiviruses expressing EMS shRNA#1, EMS shRNA#2, or CPEB2 shRNA in the indicated combination. (G) A549 cells were infected with lentiviruses expressing control, EMS, Flag–CPEB2, or EMS plus Flag–CPEB2. Forty-eight hours after infection, the poly(A) tail length of p53 mRNA was examined using the LM-PAT assay. (H) A549 cells were infected with lentiviruses expressing EMS shRNA#1, EMS shRNA#2, or CPEB2 shRNA in the indicated combination. (I) A549 cells were infected with lentiviruses expressing control, EMS, Flag–CPEB2, or EMS plus Flag–CPEB2. Forty-eight hours after infection, cells were pulse labeled with [35S]cysteine/methionine for 90 min. Cell lysates were then immunoprecipitated with anti-p53 antibody and analyzed by SDS-PAGE, followed by autoradiography. The band intensities of immunoprecipitated p53 in H and I were quantified using ImageJ software and are presented in SI Appendix, Fig. S3 F and I, respectively. (J) A549 cells were infected with lentiviruses expressing control, Flag–CPEB2, Flag–CPEB2 plus EMS shRNA#1, or Flag–CPEB2 plus EMS shRNA#2. Forty-eight hours later, cell lysates were immunoprecipitated with anti-Flag antibody. RNAs present in immunoprecipitates were analyzed by real-time RT-PCR. Data shown are mean ± SD (n = 3). The input and immunoprecipitates were also analyzed by western blotting. *P < 0.05; **P < 0.01; ***P < 0.001. (K) A549 cells were infected with lentiviruses expressing control, Flag–CPEB2, or Flag–CPEB2 plus EMS. Forty-eight hours later, cell lysates were immunoprecipitated with anti-Flag antibody. RNAs present in immunoprecipitates were analyzed by real-time RT-PCR. Data shown are mean ± SD (n = 3). The input and immunoprecipitates were also analyzed by western blotting. *P < 0.05; **P < 0.01. (L) Purified recombinant Flag–CPEB2 bound with M2 beads was incubated with in vitro–synthesized p53 3′-UTR, EMS, and its antisense RNA in the indicated combination. The inputs and beads-bound RNAs were analyzed by RT-PCR. The primers used for EMS antisense/sense detection do not discern between sense and antisense EMS. ctrl, control; IP, immunoprecipitation.
We next performed the rescue experiments to examine whether EMS inhibits p53 expression via CPEB2. Knockdown of EMS consistently led to an increase in p53 expression (Fig. 5D). However, this increased expression of p53 was greatly reversed by CPEB2 knockdown (Fig. 5D). Moreover, EMS-decreased p53 expression was shown to be restored by CPEB2 overexpression (Fig. 5E), supporting that CPEB2 indeed mediates the inhibitory effect of EMS on p53 expression. CPEB2 belongs to the CPEB family of proteins, which are able to mediate cytoplasmic polyadenylation of target mRNAs and modulate their translational efficiency (45, 46). We, therefore, hypothesized that EMS may affect the poly(A) tail length of p53 mRNA via CPEB2. To test this, we performed a ligation-mediated poly(A) test (LM-PAT) assay. Knockdown of EMS dramatically increased the poly(A) tail length of p53 mRNA (Fig. 5F and SI Appendix, Fig. S3D); however, this promoting effect on p53 mRNA poly(A) tail length was not observed in CPEB2 knockdown cells (Fig. 5F). In addition, ectopic expression of EMS clearly reduced the poly(A) tail length of p53 mRNA in control cells (Fig. 5G and SI Appendix, Fig. S3E) but not in CPEB2 overexpressing cells (Fig. 5G). These data suggest that EMS decreases p53 mRNA poly(A) tail length via CPEB2. Correlated with these findings, knockdown of EMS was able to increase the levels of newly synthesized p53 and induce luciferase expression from the p53 3′-untranslated region (UTR) reporter construct in control cells but not in CPEB2 knockdown cells (Fig. 5H and SI Appendix, Fig. S3 F–H). Moreover, the reduced levels of newly synthesized p53 and decreased luciferase expression from the p53 3′-UTR reporter construct caused by EMS overexpression could be recovered by ectopic expression of CPEB2 (Fig. 5I and SI Appendix, Fig. S3 I and J). These data suggest that EMS impairs CPEB2-mediated p53 translational control. To further support this, lysates from A549 cells expressing different EMS and CPEB2 levels were fractionated through sucrose gradients, followed by real-time RT-PCR analysis. Knockdown of either EMS or CPEB2 did not obviously change the polysome distribution profiles (SI Appendix, Fig. S3K), indicating that EMS and CPEB2 may not affect global translation. Interestingly, knockdown of EMS caused a shift in the distribution of p53 mRNA from low–molecular weight to high–molecular weight polysome fractions, which could be greatly reversed by the simultaneous knockdown of CPEB2 (SI Appendix, Fig. S3L). As a control, the distribution of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA remained unchanged under these conditions (SI Appendix, Fig. S3M). These data indicate that EMS specifically inhibits translational efficiency of p53 mRNA via CPEB2.
Our previous study has shown that EMS contains a poly-U stretch with 22 uridines that is required for the interaction with RNA-binding protein associated with lethal yellow mutation (RALY) (42). Reanalysis of the EMS sequence revealed that EMS also includes two UAUUUU sequences, which are inverted from the most common sequence of the cytoplasmic polyadenylation element (CPE; UUUUAU) (SI Appendix, Fig. S3N). An in vitro binding assay showed that compared with the wild-type EMS, mutant EMS depleted of either poly-U or CPE exhibited decreased binding to CPEB2 (SI Appendix, Fig. S3O). Strikingly, mutant EMS with deletion of both poly-U and CPE almost completely lost the CPEB2-binding ability (SI Appendix, Fig. S3O), implying that both poly-U and CPE of EMS are important for the CPEB2 interaction. Correlated with the decreased binding to CPEB2, mutant EMS depleted of either poly-U or CPE showed a weaker ability to reduce expression of p53 and its target gene p21 than wild-type EMS (SI Appendix, Fig. S3 P and Q). Moreover, mutant EMS with deletion of both poly-U and CPE, lacking the CPEB2-binding ability, exhibited no obvious effect on p53 and p21 expression (SI Appendix, Fig. S3 P and Q). These data indicate that the interaction with CPEB2 is required for EMS to inhibit p53 expression.
We finally extended our investigation to understand how EMS inhibits p53 expression via CPEB2. The RNA immunoprecipitation experiments showed that knockdown of EMS significantly increased the interaction of CPEB2 with p53 mRNA (Fig. 5J), while enforced expression of EMS strongly decreased the binding of CPEB2 to p53 mRNA (Fig. 5K), indicating that EMS competes with p53 mRNA for CPEB2 binding. In support of this, an in vitro binding assay revealed that EMS, but not its antisense RNA, was capable of inhibiting the interaction between CPEB2 and p53 3′-UTR (Fig. 5L). Taken together, these data suggest that EMS suppresses p53 translation by competing with p53 mRNA for CPEB2 binding.
EMS Regulates Cellular Senescence, Cell Proliferation, and Tumor Growth via CPEB2.
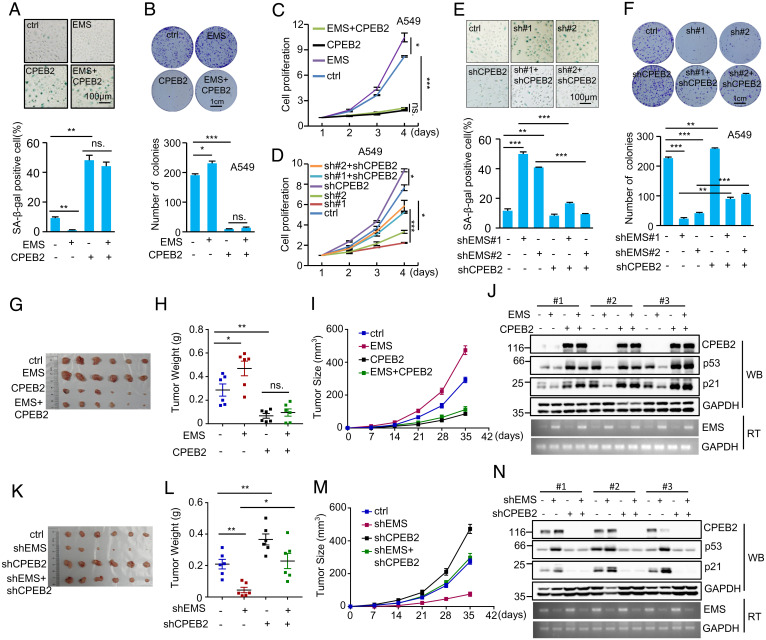
Given the above findings that the inhibitory effect of EMS on p53 expression is mediated by CPEB2, we sought to evaluate whether EMS regulates cellular senescence, cell proliferation, and tumor growth via CPEB2 by performing the rescue experiments. Ectopic expression of EMS consistently inhibited cellular senescence, accelerated cell proliferation, and increased the number of colonies (Fig. 6 A–C). However, EMS failed to show any of these effects when CPEB2 was overexpressed (Fig. 6 A–C). In addition, the increased cellular senescence, the reduced cell proliferation, and the decreased number of colonies caused by EMS knockdown could be markedly reversed by knockdown of CPEB2 (Fig. 6 D–F). These findings indicate the importance of CPEB2 in mediating EMS function. To further support this idea, the CPEB2 binding-defective mutant of EMS (ΔCPE-ΔpolyU) was used (SI Appendix, Fig. S3O). Correlating with the above finding that unlike wild-type EMS, EMS (ΔCPE-ΔpolyU) was not able to inhibit p53 expression (SI Appendix, Fig. S3P), this mutant EMS failed to show any promoting effects on cell proliferation, colony number, and cellular transformation in vitro (SI Appendix, Fig. S4 A–D). In addition, the enhancing effect of EMS on in vitro cellular transformation was greatly minimized by overexpression of CPEB2, correlating with the findings that ectopic expression of EMS decreased p53 levels in control hTERT-BJ cells but not in CPEB2 overexpressing hTERT-BJ cells (SI Appendix, Fig. S4 E and F). These data imply that EMS could exert its cellular function via the CPEB2–p53 axis. By using a xenograft mouse model, we showed that the ectopic expression of EMS promoted in vivo xenograft tumor growth of control A549 cells but not CPEB2 overexpressing A549 cells (Fig. 6 G–J). Moreover, the inhibitory effect of EMS knockdown on in vivo xenograft tumor growth was greatly rescued by CPEB2 knockdown (Fig. 6 K–N). Together, these data suggest that the effects of EMS on cellular senescence, cell proliferation, and tumor growth are mediated by CPEB2.
Fig. 6.
EMS regulates cellular senescence, cell proliferation, and tumor growth via CPEB2. (A) Senescence-associated (SA) β-galactosidase staining in A549 cells expressing control, EMS, CPEB2, or both EMS and CPEB2. The shown images are representative of three independent experiments. Data shown are mean ± SD (n = 3). **P < 0.01. (B) Colonies of A549 cells expressing control, EMS, CPEB2, or both EMS and CPEB2 were stained with crystal violet after 10 d of incubation. The images are representative of three independent experiments. Data shown are mean ± SD (n = 3). *P < 0.05; ***P < 0.001. (C) The growth curves of A549 cells expressing control, EMS, CPEB2, or both EMS and CPEB2. Data shown are mean ± SD (n = 3). *P < 0.05; ***P < 0.001. (D) The growth curves of A549 cells expressing control shRNA, EMS shRNA#1, EMS shRNA#2, CPEB2 shRNA, EMS shRNA#1 plus CPEB2 shRNA, or EMS shRNA#2 plus CPEB2 shRNA. Data shown are mean ± SD (n = 3). *P < 0.05; ***P < 0.001. (E) SA β-galactosidase staining in A549 cells expressing control shRNA, EMS shRNA#1, EMS shRNA#2, CPEB2 shRNA, EMS shRNA#1 plus CPEB2 shRNA, or EMS shRNA#2 plus CPEB2 shRNA. The images are representative of three independent experiments. Data shown are mean ± SD (n = 3). **P < 0.01; ***P < 0.001. (F) Colonies of A549 cells expressing control shRNA, EMS shRNA#1, EMS shRNA#2, CPEB2 shRNA, EMS shRNA#1 plus CPEB2 shRNA, or EMS shRNA#2 plus CPEB2 shRNA were stained with crystal violet after 10 d of incubation. The images are representative of three independent experiments. Data shown are mean ± SD (n = 3). **P < 0.01; ***P < 0.001. (G–J) In total, 2 × 106 A549 cells transduced with lentiviruses expressing control, EMS, CPEB2, or both EMS and CPEB2 were individually injected into nude mice (n = 6 for each group). (G) Xenograft tumors were taken 5 wk after injection. (H) Excised tumors were weighed. *P < 0.05; **P < 0.01. (I) Tumor sizes were measured at the indicated time points. (J) RNA and protein extracts from the excised xenografts were also analyzed by RT-PCR (RT) and western blotting (WB), respectively. (K–N) In total, 2 × 106 A549 cells transduced with lentiviruses expressing control, EMS shRNA, CPEB2 shRNA, or both EMS shRNA and CPEB2 shRNA were individually injected into nude mice (n = 6 for each group). (K) Xenograft tumors were taken 5 wk after injection. (L) Excised tumors were weighed. *P < 0.05; **P < 0.01. (M) Tumor sizes were measured at the indicated time points. (N) RNA and protein extracts from the excised xenografts were also analyzed by RT-PCR and western blotting, respectively. ctrl, control; ns., no significance.
Discussion
p53 is one of the most important tumor suppressor genes, and inactivation of p53 has been linked to a variety of human cancers (47, 48). Accumulating evidence suggests that lncRNAs are involved in the regulation of the p53 signaling pathway (49–57). In this study, we present evidence showing that as a transcriptional target of p53, EMS is able to repress p53 expression by suppressing CPEB2-mediated translational control of p53 mRNA. EMS is functionally shown to promote tumorigenesis via the CPEB2–p53 axis. Therefore, EMS is an important player in the regulation of p53 function.
We previously reported EMS as an oncogenic lncRNA that is transactivated by the oncogene c-Myc (42). Here, we further demonstrate that EMS is negatively regulated by the tumor suppressor gene p53. The importance of p53-repressed EMS expression is also supported by the observation that LUAD harboring mutant p53 exhibits relatively high expression of EMS compared with those harboring wild-type p53, although the difference between these two groups is not great. We think that the reason behind this is that the expression of EMS is also influenced by other cellular factors, such as c-Myc. By performing luciferase reporter and ChIP assays, we show that the promoter of EMS contains a functional repressive p53 response element. It has been reported that by binding to the repressive p53 response element(s), p53 is able to repress expression of multiple protein-coding genes (43). The proposed mechanisms underlying this direct transcriptional repression of protein-coding genes by p53 include the competition with other transcriptional activators and the recruitment of chromatin repressor factors (41, 58). Therefore, it would be interesting to investigate whether p53 suppresses the lncRNA EMS expression via these similar mechanisms in the future. Together with the recent finding of pluripotency-specific lncRNA (lncPRESS)1 as a p53-repressed lncRNA that safeguards pluripotency (59), our data imply that lncRNA gene repression may be an important part of the p53 cellular response.
Due to the strong antiproliferative effect of p53, the cellular levels of p53 are under strict control. In unstressed cells, the basal protein levels of p53 are low, which is mainly achieved by the E3 ubiquitin ligase Mdm2. Interestingly, Mdm2 itself is under transcriptional control by p53, establishing a p53–mdm2 negative feedback loop (60). In addition to protein factors, several p53-activated lncRNAs also play a role in the p53 autoregulatory network. For instance, PURPL associates with MYBBP1A and suppresses basal p53 levels (32). In addition, PiHL decreases p53 stability via the GRWD1–RPL11–Mdm2 axis (61). Unlike PURPL and PiHL, another p53-responsive lncRNA DINO is able to interact with p53 and promote its stabilization (37). In this study, we show that p53 and EMS can form a double-negative feedback loop. These findings suggest the importance and complexity of lncRNA in fine-tuning p53 function.
Although the posttranslational regulation is well recognized as a critical mechanism that controls p53 activity, increasing evidence suggests that p53 is also subjected to the translational modulation, particularly via RNA-binding proteins including HuR, RPL26, nucleolin, RNPC1, CPEB1, and CPEB2 (21–26). Of note, CPEB1 and CPEB2 belong to the CPEB family of proteins, which recognizes the CPE in the 3′-UTR of target mRNA (62). By binding to the CPE, CPEB nucleates a complex of factors that regulate target mRNA polyadenylation, thereby leading to translational repression or activation (46, 63). Among the CPEB family proteins, CPEB1 has been shown to promote p53 translation by increasing its mRNA polyadenylation (25). CPEB2 also exhibits the promoting effect on p53 translation (26), yet the underlying mechanism remains unclear. Here, we show that CPEB2 induction increases, whereas CPEB2 knockdown reduces the poly(A) length of p53 mRNA, implying that similar to CPEB1, CPEB2 also controls p53 mRNA polyadenylation-induced translation. In addition, we identify EMS as a CPEB2-binding partner. By binding to CPEB2, EMS decreases the interaction between CPEB2 and p53 3′-UTR, indicating that EMS competes with p53 mRNA for CPEB2 binding. Functionally, EMS indeed suppresses p53 translation and promotes tumorigenesis via CPEB2. As a translation regulatory factor, CPEB2 has been shown to modulate the translation of multiple target mRNAs, including HIF1α, Twist1, and UCP1 (64–66). Therefore, in the future, it would be interesting to investigate whether EMS also exerts its oncogenic function by regulating CPEB2-mediated translational control of other target mRNAs in addition to p53.
We have previously reported that EMS is able to promote tumorigenesis partially through increasing E2F1 expression (42). Here, we show that EMS also exerts its oncogenic effects through the negative regulation of p53. Importantly, the simultaneous E2F1 overexpression and p53 knockdown almost completely reverse the inhibitory effect of EMS knockdown on cell proliferation in p53 wild-type A549 cells, indicating that EMS promotes tumorigenesis through the regulation of both E2F1 and p53 expression. Because EMS is shown to regulate expression of E2F1 and p53 via the different mechanisms, hypothetically EMS may promote tumorigenesis in both p53 wild-type and p53-mutated LAUD. Interestingly, high expression of EMS is associated with poor prognosis in patients with p53 wild-type LUAD but not those with p53-mutated LUAD, although the reason behind this is not clear. By analyzing TCGA database, we show that p53 wild-type LUAD expresses relatively higher levels of CPEB2 than p53-mutated LUAD (SI Appendix, Fig. S4G). Correlated with this, it has been recently reported that as a transcriptional target of p53, CPEB2 is up-regulated by p53 (67). Together, these findings imply that the posttranscriptional regulation of p53 by the EMS–CPEB2 axis could be a common feature in p53 wild-type LUAD. Several p53-activating compounds are currently in clinical trials for the treatment of human cancer with wild-type p53 (68). Therefore, our finding of the association of EMS high expression with poor prognosis in patients with p53 wild-type LUAD suggests that therapeutic targeting of EMS could be beneficial to LUAD patients with wild-type p53.
Materials and Methods
Reagents and Antibodies.
The following reagents and antibodies used in this study were purchased from the indicated sources: lipofectamine 2000 (Invitrogen), streptavidin-coated agarose beads (Thermo Fisher Scientific), MG132 (Calbiochem; 20 μM), cycloheximide (Sigma; 20 μg/mL), doxorubicin (Sigma; 0.5 μg/mL), Nutlin-3 (Sigma; 10 μM), Etoposide (Sigma; 10 μM), complete ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor mixture (Roche Applied Science), antibodies against GAPDH (Santa Cruz; sc-166545; 1:5,000), Flag (Sigma; #F3165; 1:4,000), CPEB2 (Invitrogen; #PA5-30938; 1:1,000), p53 (Santa Cruz; sc-126; 1:1,000), p21 (Sigma; #P1484; 1:5,000), and horseradish peroxidase (HRP)-conjugated secondary antibodies against mouse (115-035-062) and rabbit (111-035-144; Jackson ImmunoResearch; 1:10,000).
Cell Culture.
The A549 cells were cultured in Nutrient Mixture F-12 Ham medium (Sigma) supplemented with 10% fetal bovine serum (FBS) and antibiotics. H460, hTERT-BJ, and HEK293T cell lines were cultured in Dulbecco's modified Eagle's medium (GIBCO) supplemented with 10% FBS and antibiotics. All cell lines were routinely tested for mycoplasma contamination before they were used for experiments.
Real-Time RT-PCR.
Real-time RT-PCR was performed as we previously described (42). The primer sequences are listed in SI Appendix, Table S1.
ChIP Assay.
The ChIP assay was performed as we previously described (69). The primer sequences are shown in SI Appendix, Table S1.
Generation of the Lentiviral Expression System.
To generate lentiviruses overexpressing EMS or the indicated proteins, HEK293T cells were transfected with pSin-EF1ɑ–based construct, pmd2.g, and pspax2. To generate lentiviruses expressing the indicated short hairpin RNAs (shRNAs), HEK293T cells were transfected with shRNA (cloned in PLKO.1), pREV, pGag/Pol/PRE, and pVSVG. For generation of the control virus, pSin empty vector or PLKO.1 containing control shRNA was used. Six hours after transfection, cells were cultured in fresh medium for an additional 24 h. After filtering through a 0.45-μm polyvinylidene fluoride (PVDF) filter (Millipore), the culture medium containing lentivirus particles was used for infection. The successful knockdown and overexpression were confirmed by western blot or real-time RT-PCR analysis in each experiment. In this study, two EMS-targeting shRNAs (sh-EMS#1 and sh-EMS#2) were used. Unless specified otherwise, sh-EMS#1 was used because sh-EMS#1 consistently achieved better knockdown efficiency. The shRNA target sequences were listed in SI Appendix, Table S1.
RNA Immunoprecipitation.
The A549 cells expressing control or Flag–CPEB2 were lysed in lysis buffer (50 mM trisaminomethane (Tris)⋅HCl, pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 0.5% Nonidet P-40) supplemented with 1× protease inhibitor mixture, ribonuclease (RNase) A inhibitor, and DNase I. Cell lysates were incubated with M2 beads at 4 °C for 6 h. The beads-bound immunocomplexes were eluted using elution buffer (50 mM Tris⋅HCl, pH 8.0, 1% sodium dodecyl sulfate (SDS), 10 mM EDTA) at 65 °C for 10 min. RNAs and proteins in the eluents were then analyzed by real-time RT-PCR and western blotting, respectively.
In Vitro Transcription of EMS and Its Antisense RNA.
The DNA template used in the transcription system was generated by RT-PCR using forward and reverse primers containing the T7 RNA polymerase promoter sequence (SI Appendix, Table S1). In vitro transcription was performed using the MaxiScript T7 kit (Ambion) according to the instructions provided by the manufacturer. To verify the interaction of EMS with CPEB2, in vitro–synthesized EMS or its antisense RNA was incubated with purified recombinant Flag–CPEB2 bound with M2 beads. After incubation and extensive washing, the beads-bound RNAs were eluted as templates for RT-PCR analysis.
Biotin Pull-Down Assay.
All of the process was performed in the RNase-free condition. Sense or antisense biotin-labeled DNA oligomers corresponding to EMS (1 μg) were incubated with lysates from 5 × 106 A549 cells. One hour after incubation, streptavidin-coated beads (Thermo Fisher Scientific) was added to isolate the RNA–protein complex, followed by western blot and real-time RT-PCR analyses.
Luciferase Reporter Assay.
To investigate whether EMS is transcriptionally repressed by p53, A549 cells expressing control shRNA or p53 shRNA or A549 cells expressing control or p53 proteins were transfected with the indicated pGL3-based constructs plus Renilla luciferase reporter plasmid. To examine the effect of EMS on p53 3′-UTR, A549 cells transduced with the indicated lentiviruses were transfected with psiCHECK2–p53 3′-UTR. Twenty-four hours after transfection, firefly and Renilla luciferase activities were measured by the Dual-Luciferase Reporter Assay System (Promega), and Renilla activity was used to normalize firefly activity.
p53 Labeling and Immunoprecipitation.
In total, 1 × 107 A549 cells were cultured for 60 min in cysteine/methionine-free media (Gibco) before 50 µCi [35S]cysteine/methionine (EasyTag; NEG772) was added. After 90 min of incubation, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) supplemented with 1× protease inhibitor mixture. Cell lysates were incubated with protein A/G beads (Pierce) coated with anti-p53 antibody at 4 °C for 6 h. The immunoprecipitates were subjected to sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE), followed by autoradiography.
LM-PAT Assay.
The LM-PAT assay was performed as previously described (70). Briefly, 200 ng of total RNA was annealed to phosphorylated oligo (dT)12–18 (40 ng), and the oligo (dT) anchor (200 ng) was targeted to the 3′ end of the poly(A) tail in the presence of T4 DNA Ligase. Then, reverse transcription was performed with SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer’s instructions, followed by PCR amplification using p53-specific oligomers and oligo (dT) anchor (SI Appendix, Table S1). The PCR products were analyzed on agarose gels.
Polysome Profiling Assay.
Forty-eight hours after infection with lentiviruses expressing the indicated shRNAs, 1 × 107 A549 cells were incubated with 100 µg/mL cycloheximide for 15 min and lysed in polysome lysis buffer (50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (Hepes), 0.1%Triton X-100, 100 mM KCl, 2 mM MgCl2, 10% glycerol, 100 mg/mL cycloheximide, 1 mM dithiothreitol (DTT), 20 units/mL RNase inhibitor, 1× EDTA-free protease inhibitor mixture). Cell lysates were then loaded onto 20 to 50% (wt/vol) sucrose density gradients (10 mM Tris⋅HCl, pH 7.5, 5 mM MgCl2, 100 mM NaCl, 1mM DTT, 20 to 50% RNase-free sucrose) and centrifuged at 38,000 rpm for 3 h at 4 °C in a Beckman SW41 Ti rotor. Fractions were acquired using the Gradient Fractionator (BioComp) with continuous recording of the absorbance at 254 nm. One-milliliter fractions were collected throughout, and total RNA extracted from each fraction was subjected to real-time RT-PCR analysis.
Cell Senescence Assay.
Senescence assay was performed using the senescence detection kit (Beyotime). Briefly, A549 or H460 cells were infected with the indicated lentiviruses. Ninety-six hours after infection, cells were fixed by fixative solution for 20 min at room temperature. After washing twice with PBS, cells were stained with 0.1% 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) solution for 48 h at 37 °C. The X-gal–stained cells were counted under microscope.
Xenograft Mouse Model.
Studies on mice were approved by the Animal Research Ethics Committee of the University of Science and Technology of China and were performed in accordance with relevant guidelines and regulations. For xenograft experiments, 2 × 106 A549 cells were injected into the left flank or right flank of 4-wk-old male athymic nude mice (Shanghai SLAC Laboratory Animal Co. Ltd.; n = 6 for each group). Mice were used in the experiment at random. Severn days after injection, tumor volumes were measured every 7 d with calipers and calculated using the equation: volume = length × width2 × 0.52. Five weeks after injection, mice were euthanized, and tumors were excised. During testing of the tumors' weights, the experimentalists were blinded to the information of tumor tissues. The excised tumors were homogenized for RNA and protein extraction. The extracted RNAs and proteins were subjected to RT-PCR and western blot analyses, respectively.
Reproducibility.
All the data were repeated at least three times with similar results. The western blot images are representative of three independent experiments.
Statistical Analysis.
Statistical analysis was carried out using Microsoft Excel software and GraphPad Prism to assess differences between experimental groups. Statistical significance was analyzed by the Student's t test and expressed as a P value. P values lower than 0.05 were considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Daxing Gao for providing us hTERT-immortalized BJ cells. This work was supported by Ministry of Science and Technology of China Grant 2019YFA0802600; National Natural Science Foundation of China Grants 91957108 and 31871440; Collaborative Innovation Programs of Hefei Science Center, Chinese Academy of Sciences Grant 2019HSC-CIP010; and Fundamental Research Funds for Central Universities Grants YD9100002012, WK9100000024, and YD2070002007.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111409119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Boutelle A. M., Attardi L. D., p53 and tumor suppression: It takes a network. Trends Cell Biol. 31, 298–310 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aylon Y., Oren M., The paradox of p53: What, how, and why? Cold Spring Harb. Perspect. Med. 6, a026328 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Labuschagne C. F., Zani F., Vousden K. H., Control of metabolism by p53 - Cancer and beyond. Biochim. Biophys. Acta Rev. Cancer 1870, 32–42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J., Zhang C., Hu W., Feng Z., Tumor suppressor p53 and metabolism. J. Mol. Cell Biol. 11, 284–292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tasdemir E., et al. , Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 10, 676–687 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hafner A., Bulyk M. L., Jambhekar A., Lahav G., The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 20, 199–210 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Jiang L., et al. , Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520, 57–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfister N. T., Prives C., Transcriptional regulation by wild-type and cancer-related mutant forms of p53. Cold Spring Harb. Perspect. Med. 7, a026054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freed-Pastor W. A., Prives C., Mutant p53: One name, many proteins. Genes Dev. 26, 1268–1286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogelstein B., Lane D., Levine A. J., Surfing the p53 network. Nature 408, 307–310 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Donehower L. A., et al. , Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356, 215–221 (1992). [DOI] [PubMed] [Google Scholar]
- 12.Haupt Y., Maya R., Kazaz A., Oren M., Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Kubbutat M. H., Jones S. N., Vousden K. H., Regulation of p53 stability by Mdm2. Nature 387, 299–303 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Leng R. P., et al. , Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112, 779–791 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Dornan D., et al. , The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429, 86–92 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Horn H. F., Vousden K. H., Coping with stress: Multiple ways to activate p53. Oncogene 26, 1306–1316 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Liu Y., Tavana O., Gu W., p53 modifications: Exquisite decorations of the powerful guardian. J. Mol. Cell Biol. 11, 564–577 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucchesi C., Zhang J., Chen X., Modulation of the p53 family network by RNA-binding proteins. Transl. Cancer Res. 5, 676–684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ray P. S., Grover R., Das S., Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 7, 404–410 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang D. Q., Halaby M. J., Zhang Y., The identification of an internal ribosomal entry site in the 5′-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene 25, 4613–4619 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Ofir-Rosenfeld Y., Boggs K., Michael D., Kastan M. B., Oren M., Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol. Cell 32, 180–189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazan-Mamczarz K., et al. , RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. U.S.A. 100, 8354–8359 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takagi M., Absalon M. J., McLure K. G., Kastan M. B., Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123, 49–63 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Zhang J., et al. , Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev. 25, 1528–1543 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burns D. M., Richter J. D., CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 22, 3449–3460 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tordjman J., et al. , Tumor suppressor role of cytoplasmic polyadenylation element binding protein 2 (CPEB2) in human mammary epithelial cells. BMC Cancer 19, 561 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quinn J. J., Chang H. Y., Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Frankish A., et al. , Gencode 2021. Nucleic Acids Res. 49 (D1), D916–D923 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchese F. P., Raimondi I., Huarte M., The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 18, 206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Statello L., Guo C. J., Chen L. L., Huarte M., Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhary R., Lal A., Long noncoding RNAs in the p53 network. Wiley Interdiscip. Rev. RNA 8, e1410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X. L., et al. , Long noncoding RNA PURPL suppresses basal p53 levels and promotes tumorigenicity in colorectal cancer. Cell Rep. 20, 2408–2423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou C. C., et al. , Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology 63, 850–863 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Zhou Y., et al. , Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 282, 24731–24742 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Hung T., et al. , Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 43, 621–629 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huarte M., et al. , A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt A. M., et al. , An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet. 48, 1370–1376 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu W. L., et al. , GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat. Cell Biol. 20, 492–502 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Sullivan K. D., Galbraith M. D., Andrysik Z., Espinosa J. M., Mechanisms of transcriptional regulation by p53. Cell Death Differ. 25, 133–143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laptenko O., Prives C., Transcriptional regulation by p53: One protein, many possibilities. Cell Death Differ. 13, 951–961 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Rinn J. L., Huarte M., To repress or not to repress: This is the guardian’s question. Trends Cell Biol. 21, 344–353 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Wang C., et al. , Long noncoding RNA EMS connects c-Myc to cell cycle control and tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 116, 14620–14629 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B., Xiao Z., Ren E. C., Redefining the p53 response element. Proc. Natl. Acad. Sci. U.S.A. 106, 14373–14378 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narisawa-Saito M., et al. , An in vitro multistep carcinogenesis model for human cervical cancer. Cancer Res. 68, 5699–5705 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Fernández-Miranda G., Méndez R., The CPEB-family of proteins, translational control in senescence and cancer. Ageing Res. Rev. 11, 460–472 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Richter J. D., CPEB: A life in translation. Trends Biochem. Sci. 32, 279–285 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Kastenhuber E. R., Lowe S. W., Putting p53 in context. Cell 170, 1062–1078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vousden K. H., Prives C., Blinded by the light: The growing complexity of p53. Cell 137, 413–431 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Zhang A., Xu M., Mo Y. Y., Role of the lncRNA-p53 regulatory network in cancer. J. Mol. Cell Biol. 6, 181–191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sánchez Y., et al. , Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat. Commun. 5, 5812 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olivero C. E., et al. , p53 activates the long noncoding RNA Pvt1b to inhibit Myc and suppress tumorigenesis. Mol. Cell 77, 761–774.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diaz-Lagares A., et al. , Epigenetic inactivation of the p53-induced long noncoding RNA TP53 target 1 in human cancer. Proc. Natl. Acad. Sci. U.S.A. 113, E7535–E7544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Léveillé N., et al. , Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat. Commun. 6, 6520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Q., et al. , LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 41, 4976–4987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marín-Béjar O., et al. , Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 14, R104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mello S. S., et al. , Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 31, 1095–1108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Y., et al. , TRMP, a p53-inducible long noncoding RNA, regulates G1/S cell cycle progression by modulating IRES-dependent p27 translation. Cell Death Dis. 9, 886 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beckerman R., Prives C., Transcriptional regulation by p53. Cold Spring Harb. Perspect. Biol. 2, a000935 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jain A. K., et al. , LncPRESS1 is a p53-regulated LncRNA that safeguards pluripotency by disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol. Cell 64, 967–981 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harris S. L., Levine A. J., The p53 pathway: Positive and negative feedback loops. Oncogene 24, 2899–2908 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Deng X., et al. , Long noncoding RNA PiHL regulates p53 protein stability through GRWD1/RPL11/MDM2 axis in colorectal cancer. Theranostics 10, 265–280 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivshina M., Lasko P., Richter J. D., Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu. Rev. Cell Dev. Biol. 30, 393–415 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Villalba A., Coll O., Gebauer F., Cytoplasmic polyadenylation and translational control. Curr. Opin. Genet. Dev. 21, 452–457 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Hägele S., Kühn U., Böning M., Katschinski D. M., Cytoplasmic polyadenylation-element-binding protein (CPEB)1 and 2 bind to the HIF-1alpha mRNA 3′-UTR and modulate HIF-1alpha protein expression. Biochem. J. 417, 235–246 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Nairismägi M. L., et al. , Translational control of TWIST1 expression in MCF-10A cell lines recapitulating breast cancer progression. Oncogene 31, 4960–4966 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Chen H. F., Hsu C. M., Huang Y. S., CPEB2-dependent translation of long 3′-UTR Ucp1 mRNA promotes thermogenesis in brown adipose tissue. EMBO J. 37, e99071 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di J., et al. , A p53/CPEB2 negative feedback loop regulates renal cancer cell proliferation and migration. J. Genet. Genomics 48, 606–617 (2021). [DOI] [PubMed] [Google Scholar]
- 68.Sanz G., Singh M., Peuget S., Selivanova G., Inhibition of p53 inhibitors: Progress, challenges and perspectives. J. Mol. Cell Biol. 11, 586–599 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao K., et al. , WDR63 inhibits Arp2/3-dependent actin polymerization and mediates the function of p53 in suppressing metastasis. EMBO Rep. 21, e49269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sallés F. J., Richards W. G., Strickland S., Assaying the polyadenylation state of mRNAs. Methods 17, 38–45 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.