Significance
Cystic fibrosis (CF) is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene, which can lead to respiratory failure. To date, there is no treatment for CF caused by the CFTR-W1282X mutation located on CFTR exon 23. Nonsense-mediated messenger RNA (mRNA) decay (NMD) degrades the CFTR-W1282X mRNA, leading to low levels of functional CFTR protein. We developed a cocktail of two antisense oligonucleotides (ASOs) that promotes the skipping of exon 23 of the CFTR-W1282X mRNA. The resulting mRNA is NMD resistant and preserves the reading frame. Its translation produces CFTR-Δex23 protein that improves CFTR activity in human bronchial epithelial cells. Our results set the stage for developing an ASO therapy for CF caused by the W1282X mutation.
Keywords: cystic fibrosis, antisense oligonucleotide, nonsense mutation, mRNA splicing, exon skipping
Abstract
Mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene cause cystic fibrosis (CF), and the CFTR-W1282X nonsense mutation causes a severe form of CF. Although Trikafta and other CFTR-modulation therapies benefit most CF patients, targeted therapy for patients with the W1282X mutation is lacking. The CFTR-W1282X protein has residual activity but is expressed at a very low level due to nonsense-mediated messenger RNA (mRNA) decay (NMD). NMD-suppression therapy and read-through therapy are actively being researched for CFTR nonsense mutants. NMD suppression could increase the mutant CFTR mRNA, and read-through therapies may increase the levels of full-length CFTR protein. However, these approaches have limitations and potential side effects: because the NMD machinery also regulates the expression of many normal mRNAs, broad inhibition of the pathway is not desirable, and read-through drugs are inefficient partly because the mutant mRNA template is subject to NMD. To bypass these issues, we pursued an exon-skipping antisense oligonucleotide (ASO) strategy to achieve gene-specific NMD evasion. A cocktail of two splice-site–targeting ASOs induced the expression of CFTR mRNA without the premature-termination-codon–containing exon 23 (CFTR-Δex23), which is an in-frame exon. Treatment of human bronchial epithelial cells with this cocktail of ASOs that target the splice sites flanking exon 23 results in efficient skipping of exon 23 and an increase in CFTR-Δex23 protein. The splice-switching ASO cocktail increases the CFTR-mediated chloride current in human bronchial epithelial cells. Our results set the stage for developing an allele-specific therapy for CF caused by the W1282X mutation.
Nonsense mutations account for about 20% of all known pathogenic genetic lesions within the coding region (1). Nonsense mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene cause cystic fibrosis (CF) by impairing chloride conductance in the epithelial tissues of multiple organs including the lungs, the gastrointestinal tract, and the reproductive organs (2). The CFTR-W1282X mutation is the sixth most common CF-causing mutation and the second most common CF-causing nonsense mutation; it is found in 1.2% of CF patients worldwide and causes a severe form of CF if homozygous or combined with another CF-causing allele (3, 4). Nonsense mutations, including the CFTR-W1282X mutation, lead to reduction of functional proteins due to premature termination of translation and degradation of the messenger RNA (mRNA) by nonsense-mediated mRNA decay (NMD) (5).
There have been significant strides in CF therapeutics development, with the most recent Food and Drug Administration (FDA)–approved treatment, Trikafta—a combination of CFTR correctors (elexacaftor and tezacaftor) and a potentiator (ivacaftor)—showing unprecedented improvements in the pulmonary exacerbations, survival, and quality of life for CF patients with at least one F508del mutation (6). Although almost 90% of CF patients may benefit from Trikafta (7), there are no approved targeted therapies for CF caused by nonsense mutations. Therefore, there is still a significant unmet need for CF treatments that can simultaneously address different classes of CFTR protein deficiencies, particularly the CFTR-W1282X mutation (8).
One potential therapeutic modality for CFTR nonsense mutations is read-through compounds (RTCs), which increase the level of full-length protein by reducing the fidelity of the ribosome at the premature termination codon (PTC) (9). Ataluren is a nonaminoglycoside RTC with a favorable safety profile that was investigated in the context of several genetic diseases. Although it showed a therapeutic effect in Duchenne muscular dystrophy caused by nonsense mutations (9), it was not able to improve forced expiratory volume in clinical trials in CF patients with various CFTR nonsense mutations (10). Gentamicin is an aminoglycoside RTC that can increase full-length CFTR protein in vitro, whose clinical efficacy in CF is limited by NMD (11). Recently, a new class of RTC that induces read-through by depleting the translation release factor eRF1 showed efficacy in vitro (12). Despite active research, a clinically viable read-through approach for CF caused by nonsense mutation does not yet exist.
Because the activity of CFTR-W1282X protein can be enhanced by CFTR modulators (4, 13, 14), combining NMD-inhibition strategies with CFTR modulators could potentially benefit patients. One such strategy involves antisense oligonucleotides (ASOs), which are short, synthetic, single-stranded DNA, RNA, or structural mimics of DNA/RNA that bind to complementary RNA to alter its functions (15). ASOs with uniformly modified nucleotides, such as 2′-O-methoxyethylribose-phosphorothioate (PS-MOE-ASOs), phosphorodiamidate morpholino oligomer (PMO), or 2′-O-methyl (2’-OMe), can sterically block the binding of proteins, nucleic acids, or other factors that are important for RNA metabolism and processing (15). On the other hand, chemically modified nucleotides in “gapmer” ASOs are spaced out by natural DNA nucleotides in order to induce RNase-H–mediated degradation of their target RNA (15). Antisense-mediated knockdown or small-molecule inhibition of SMG1 reduces NMD in human bronchial epithelial cells and improves expression and function of CFTR in cells harboring the W1282X mutation (16). However, general inhibition of NMD may not be the optimal strategy because the NMD machinery is essential for vertebrate development and regulates the expression of a subset of normal and physiologically functional mRNAs (17). Therefore, a targeted approach for restoring functional CFTR mRNA seems a more desirable therapeutic strategy.
According to the “55-nucleotide rule,” mRNA with an exon–exon junction >55 nucleotides (nt) downstream of a stop codon is degraded by NMD, due to the binding of exon-junction complexes (EJCs) near each of the downstream exon–exon junctions (18). Attempts have been made to target the multiple exon–exon junctions downstream of the PTC on the CFTR-W1282X mRNA to inhibit NMD. Deleting the genic region downstream of the W1282X mutation using CRISPR-Cas9 genome editing prevents NMD of CFTR-W1282X mRNA and increases the truncated protein levels in human airway cells (19). PS-MOE ASOs designed to prevent EJC binding downstream of a PTC can attenuate NMD in a gene-specific manner (20). Recently, we showed that a cocktail of three ASOs designed to prevent EJC binding downstream of the W1282X mutation specifically increases the expression of CFTR-W1282X mRNA and CFTR protein function in human bronchial epithelial cells (21).
An alternative strategy we describe here is to up-regulate a partially functional isoform of CFTR mRNA using ASOs that can modulate mRNA splicing. In recent years, ASOs drugs have been shown to be well suited for the treatment of various diseases (22). Recent FDA and European Medicines Agency regulatory approvals have demonstrated the clinical potential of ASOs for hereditary diseases. Several PMO ASOs approved to treat different causal mutations in Duchenne muscular dystrophy induce skipping of target exons in the mutant DMD pre-mRNA and promote the production of partially functional dystrophin protein (23). The FDA approval of nusinersen (Spinraza) in 2016 to treat spinal muscular atrophy and the rapid development of milasen, a personalized therapy for a single patient with neuronal ceroid lipofuscinosis 7 (CLN7, a form of Batten’s disease), further highlights the clinical potential of splice-switching ASOs (24, 25). Although there is no FDA-approved ASO therapy for CF at present, splice-switching ASOs designed to correct defective CFTR splicing have shown efficacy in patient-derived airway cells and nasal epithelial cells (26, 27).
If CFTR exon 23 with the W1282X mutation is skipped using splicing-switching ASOs, the resulting CFTR-Δex23 mRNA should not be targeted by NMD, as exon 23 is in frame. The intrinsic activity of CFTR-Δex23 has not been reported, but it is expected to be less active than wild-type (WT) CFTR, based on the deletion’s location in the second nucleotide-binding domain (NBD2). On the other hand, this protein is expected to retain partial activity by analogy to the truncated CFTR-W1282X protein. CF-causing exon 23 splice-site mutations (4005 + 1G > A and +2T > C) have been identified (3); however, the intrinsic activity of CFTR-Δex23 cannot be inferred from the phenotypes of these mutations because whether they cause exon skipping, cryptic splice-site activation, and/or intron retention is unknown (28).
As previously mentioned, CFTR modulators can increase the activity of a wide variety of mutant CFTR proteins (29), so CFTR-Δex23 protein’s chloride conductance may also be enhanced by CFTR modulators. Moreover, as low as 10% of normal CFTR function provides a significant therapeutic benefit for CF patients (30). Thus, increasing the expression of mutant CFTR protein with residual activity could be beneficial for CF patients. In this report, we show that CFTR-Δex23 protein retains partial activity. We also describe uniformly modified PS-MOE ASOs that target the splice sites flanking exon 23 and induce efficient exon 23 skipping, with the resulting CFTR-Δex23 protein increasing CFTR-mediated chloride conductance in human bronchial epithelial cells. This approach provides the basis for clinical development of antisense-directed exon skipping as a therapeutic strategy for CF caused by the W1282X mutation.
Results
Intrinsic Activity of CFTR-Δex23.
To assess the functionality of CFTR protein lacking the internal peptide (amino acids 1,240 through 1,291) encoded by exon 23 (CFTR-Δ ex23), we generated 16HBE-W1282X cells with doxycycline (dox)-inducible overexpression of CFTR-WT, CFTR-W1282X, or CFTR-Δex23 (dubbed 16HBEge-GFP-P2A-WT, 16HBEge-GFP-P2A-W1282X, or 16HBEge-GFP-P2A-Δex23) using a lentiviral vector that allows coexpression of Turbo GFP using a 2A self-cleaving peptide (Fig. 1A and SI Appendix, Fig. S1A). Dose-dependent induction of the recombinant gene expression by dox was confirmed by fluorescence microscopy (Fig. 1 B–D and SI Appendix, Fig. S1B). Since the Turbo GFP and CFTR proteins are translated initially as the same polypeptide chain, GFP levels can be used to control for expression from the integrated plasmid (31). Treating the 16HBEge-GFP-P2A-WT/W1282X/Δex23 cells with 2 μg/mL dox resulted in similar levels of GFP signal, indicating that the expression of recombinant CFTR proteins is induced at similar levels (SI Appendix, Fig. S1C).
Fig. 1.
Intrinsic activity of CFTR-Δex23 protein. (A) Schematic of recombinant CFTR expression system. (B–D) Fluorescence microscopy images of (B) 16HBE-W1282X cells, (C) 16HBEge-GFP-P2A-WT, and (D) 16HBEge-GFP-P2A-Δex23 cells treated with dox for 48 h. (Scale bars, 120 μm.) (E and F) Average traces from Ussing chamber assays of (E) 16HBEge-GFP-P2A-WT and (F) 16HBEge-GFP-P2A-Δex23 cells treated with dox and Trikafta. The traces shown are the averages of three replicates. Black error bars on each point show SD. (G) The total AUC of E and F between minutes 20 and 40. (H) Quantification of response to acute treatment with CFTR-inh172 based on E and F. For all treatments in E–H, n = 3 independent treatments. ***P < 0.001, Student’s t test. Error bars show SDs. TRE = tetracycline-response-element. CMV = CMV promoter. P2A = self-cleaving 2A peptide. PuroR = puromycin-resistance gene. rtTA = reverse tetracycline transactivator. fsk = forskolin.
We verified the expression of CFTR-WT, W1282X, and Δex23 proteins tagged with T7-epitope peptide (1 kDa) in 16HBEge-GFP-P2A-WT/W1282X/Δex23 cells treated with or without dox. Different glycosylation states of CFTR-WT are visible on a Western blot: nonglycosylated A-band (127 kDa), core-glycosylated B-band (131 kDa), and fully mature glycosylated C-band (160 to 170 kDa) (32). The predicted size of the fully glycosylated CFTR-Δex23 C-band is about 160 kDa; the predicted size of the CFTR-W1282X C-band is about 145 kDa. CFTR-WT, W1282X, and Δex23 proteins were detected by anti-T7 and anti-CFTR antibodies in dox-treated cell extracts; the recombinant CFTR proteins were not detected in parental cells or non–dox-treated cells (SI Appendix, Fig. S1 D and E). T7-CFTR-W1282X protein migrated slightly faster than T7-CFTR-Δex23 protein, as expected.
Confocal immunofluorescence microscopy showed that the T7-tagged recombinant CFTR-WT and CFTR-Δex23 proteins are localized to the cytoplasm and plasma membrane (SI Appendix, Fig. S2 A–C). We compared the chloride currents with CFTR-WT or CFTR-Δex23 overexpression by Ussing chamber assays. 16HBEge-GFP-P2A-WT or 16HBEge-GFP-P2A-Δex23 cells treated with dox and Trikafta (VX-445, VX-661, and VX-770) led to significant increases in chloride current compared to Trikafta-only treatment, as quantified by the total area under the curve (AUC) and by the response to acute CFTR-inh172 treatment (Fig. 1 E–H). The total AUC of 16HBEge-GFP-P2A-Δex23 after VX-770 treatment was about 53% and 38% of the total AUC before and after VX-770 treatment in 16HBEge-GFP-P2A-WT cells, respectively (SI Appendix, Fig. S3A). The acute response to VX-770 was slightly higher in 16HBEge-GFP-P2A-Δex23 cells (SI Appendix, Fig. S3B). The background CFTR activity in 16HBE-W1282X cells due to endogenous CFTR-W1282X expression is negligible, as the chloride currents of the no-dox-treatment 16HBEge-GFP-P2A-WT (red traces in Fig. 1E) and 16HBEge-GFP-P2A-Δex23 (red traces in Fig. 1F) cells are substantially lower than those of the dox-treated cells (blue traces in Fig. 1 E and F). We conclude that the CFTR-Δex23 protein is partially active.
Design and Testing of Candidate ASOs.
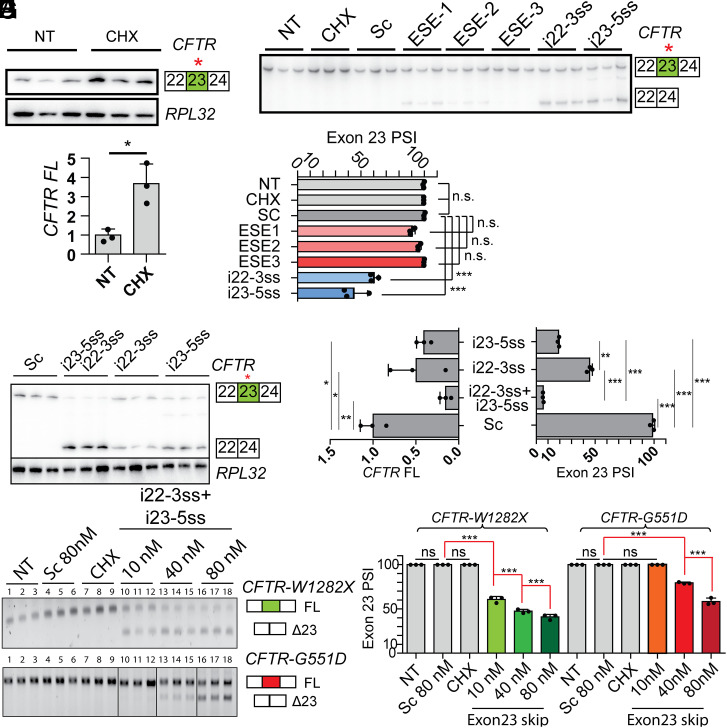
ASOs targeting exonic splicing enhancers (ESEs) or splice sites can induce skipping of alternative or constitutive exons (33, 34). We designed two PS-MOE ASOs targeting the splice sites flanking exon 23 (i22-3ss and i23-5ss). We used splicing-factor binding-site predictions by SFmap (SI Appendix, Fig. S4A) and ESEfinder (SI Appendix, Fig. S4B) to design three additional ASOs targeting potential ESEs (35, 36). We tested these ASOs in 16HBE-W1282X cells in which the endogenous CFTR-W1282X mRNA is degraded by NMD (Fig. 2 A and B). The 16HBE-W1282X cells transfected with the splice-site–targeting ASOs increased exon 23 skipping significantly compared to the scramble negative control ASO (Sc). The identity of the exon-23-skipped bands detected by RT-PCR was confirmed by Sanger sequencing (SI Appendix, Fig. S4C). Though we could detect exon 23 skipping in the presence of ESE-1 and ESE-2 ASOs, the decrease in exon-23-included mRNA was not statistically significant for any of the three ASOs designed to target potential ESEs (Fig. 2 C and D). These results suggest that the target sites for ASOs ESE-1/2/3 are not strong binding sites for splicing activators or are not individually essential for exon recognition. Thus, we selected the ASOs that target the 3′ splice site (3′ss) and 5′ splice site (5′ss) as the lead ASOs.
Fig. 2.
Designing ASOs that induce exon 23 skipping. (A) RT-PCR of CFTR-W1282X mRNA in 16HBE-W1282X cells with no treatment and cycloheximide treatment using RPL32 as an internal reference. (B) Quantification of A. (C) RT-PCR of CFTR mRNA in 16HBE-W1282X transfected with the ASOs complementary to the exon 23 ESEs or the splice sites flanking exon 23 at a nominal concentration of 80 nM. (D) Quantification of exon 23 inclusion based on C. (E) RT-PCR comparing the exon 23 PSI between the transfection of individual splice site–targeting ASOs (80 nM) and the ASO cocktail (40 nM each ASO). (F) Exon 23 PSI and full-length CFTR-W1282X mRNA based on E. (G) Nonradioactive RT-PCR showing dose-dependent exon 23 skipping in 16HBE-W1282X and 16HBE-G551D cells treated with splice-site ASO cocktail. For the image at the bottom, all samples were run on the same gel, but lanes 10–12 and 13–15 were subsequently reordered for clarity. (H) Exon 23 PSI based on G. RPL32 served as a loading control in A and E and was used for the calculations in B and F. For all treatments, n = 3 independent transfections. For all statistical tests, n.s. P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001. For B: Student’s t test. For F, Right: one-way ANOVA with Tukey’s post hoc test. For F, Left: one-way ANOVA with Dunnett’s post hoc test. For H: one-way ANOVA with Tukey’s post hoc test. Error bars are SDs. Sc = scramble ASO. NT = No-treatment control. CHX = cycloheximide treatment.
Combination of Splice-Site ASOs Induces Efficient Exon 23 Skipping.
To test whether simultaneously targeting the 3′ss and 5′ss promotes efficient exon 23 skipping, we transfected 16HBE-W1282X cells with ASOs i22-3ss and i23-5ss individually or as a cocktail. The splice-site ASO cocktail resulted in almost complete skipping of exon 23 compared to the partial skipping elicited by the individual ASOs and significantly reduced the level of full-length CFTR mRNA compared to the scramble ASO (Fig. 2 E and F). The splice-site ASO cocktail increased exon 23 skipping of 16HBE-W1282X cells in a dose-dependent manner (Fig. 2 G and H). As the W1282X (TGG → TGA) mutation disrupts potential binding sites for SRSF1 and SRSF2, according to ESEfinder (SI Appendix, Fig. S4 A and B), we tested if the ASO cocktail promotes exon 23 skipping in CFTR-G551D mRNA, which does not have a mutation on exon 23. The lead ASO cocktail also promoted dose-dependent exon 23 skipping in 16HBE-G551D cells, but the extent of exon 23 skipping was lower in these cells (Fig. 2H). Treating the 16HBE-W1282X cells with 10 μM ASO cocktail by free uptake also induced efficient exon skipping (SI Appendix, Fig. S5).
However, calculating the proportion of exon 23 inclusion without taking mRNA degradation into account can overestimate the splice-site ASO cocktail’s efficacy. Because the full-length CFTR-W1282X mRNA is an NMD target, we compared the extent of exon 23 inclusion between 16HBE-W1282X cells treated with the ASO cocktail only versus the ASO cocktail plus cycloheximide (to inhibit NMD). The combined treatment significantly increased the exon 23 percent-spliced-in (PSI) compared to the ASO cocktail treatment alone (Fig. 3 A and B). Compared to the controls, the full-length CFTR-W1282X mRNA levels were significantly lower for the ASO cocktail alone or in combination with cycloheximide (Fig. 3C). RT-qPCR using a primer pair complementary to CFTR exons 26 and 27 to amplify both full-length and exon-23–skipped isoforms shows that the ASO cocktail increased the total CFTR mRNA in 16HBE-W1282X cells (Fig. 3D). Combining the ASO cocktail with cycloheximide also increased the total CFTR mRNA (Fig. 3D). These results show that the splice-site ASO cocktail efficiently induces CFTR exon 23 skipping, bypassing NMD and increasing the total CFTR mRNA levels (Fig. 3E).
Fig. 3.
RT-PCR and RT-qPCR analysis of total CFTR mRNA and exon 23 splicing after treatment with the splice-site ASO cocktail alone or in combination with NMD inhibition. (A) NMD inhibition was combined with ASO treatments. (B and C) Quantification of (B) exon 23 PSI and (C) full-length CFTR-W1282X mRNA based on A. (D) Total CFTR mRNA levels (full-length CFTR-W1282X and CFTR-Δex23 mRNA) were measured by RT-qPCR. (E) The full-length CFTR-W1282X mRNA levels (from C) were plotted against the total CFTR mRNA levels (from D). Circle dots show no cycloheximide (CHX) treatment, and triangle dots show CHX treatment. RPL32 mRNA levels were used as an internal reference for C and D. For all treatments and statistical tests, n = 3 independent transfections, n.s. P > 0.05, **P < 0.01, and ***P < 0.001, one-way ANOVA with Tukey’s post hoc test. Error bars are SDs. Abbreviations are as in Fig. 2.
The Splice-Site ASO Cocktail Increases CFTR-Δex23 Protein and CFTR Activity.
As the splice-site ASO cocktail effectively promoted CFTR exon 23 skipping, we next analyzed the expression and function of the resulting CFTR-Δex23 protein. Transfection or free-uptake delivery of the splice-site ASO cocktail into 16HBE-W1282X cells increased the level of CFTR protein (Fig. 4 A–D). Whereas the DLD1 cell-lysate sample shows two CFTR-WT bands (C-band and B-band), the protein samples from the 16HBE-W1282X cells treated with the splice-site ASO cocktail show one predominant band that is presumably a CFTR-Δex23 B-band based on its size (Fig. 4B). An antibody that can specifically detect the part of CFTR encoded by exon 23 does not exist, to our knowledge. However, as shown in Fig. 3C, the splice-site ASO cocktail decreased the full-length CFTR-W1282X mRNA, so the increased CFTR protein is most likely the CFTR-Δex23 protein.
Fig. 4.
Combining the splice-site ASO cocktail and CFTR modulators increases CFTR-Δex23 protein levels and CFTR activity. (A and B) Western blot of 16HBE-W1282X cells treated with the splice-site ASO cocktail by (A) transfection or (B) free uptake. Na/K-ATPase was used as an internal reference. The open and filled arrowheads represent core-glycosylated and complex-glycosylated CFTR, respectively. (C and D) Quantification of A and B. (E) The data points represent the mean Isc tracings of 16HBE-W1282X cells treated with scramble ASO (red) or the splice-site ASO cocktail (blue). Traces shown are the average of the three replicates of each treatment. Both ASO treatments were combined with 10 μM VX-770 and 5 μM VX-809 treatments. (F) The total AUC of E. (G) Quantification of response to acute treatment with CFTR-inh172, based on E. For all treatments, n = 3 independent treatments. For all statistical tests, n.s. P > 0.05, **P < 0.01, and ***P < 0.001. For panel C: one-way ANOVA with Tukey’s post hoc test. For D, F, and G: Student’s unpaired t test versus Scramble ASO. Error bars are SDs. Abbreviations are as in Fig. 2.
We measured the CFTR function in 16HBE-W1282X cells treated with the splice-site ASO cocktail by Ussing chamber assay. The 16HBE-W1282X cells treated with the splice-site ASO cocktail by free uptake in combination with VX-809 and VX-770 showed increased CFTR-mediated chloride current compared to the cells treated with scramble 18mer ASO (Fig. 4 E–G). The increase in CFTR activity was similar to what we previously observed by treatment with an EJC-targeting triple-ASO cocktail, plus VX-809 and VX-770 (21). Based on the previously published CFTR activity in 16HBE14o- cells in Valley et al. (38), CFTR function in 16HBE-W1282X treated with the ASO cocktail plus VX-809 and VX-770 is probably <10% of the WT CFTR activity. However, we anticipate that combining the ASO cocktail with Trikafta may further increase the CFTR activity.
Discussion
Although major advances have been made for small-molecule therapies for CF that predominantly benefit patients with at least one CFTR-F508del allele, therapeutic options are lacking for patients harboring nonsense mutations in CFTR (6, 39). CFTR nonsense mutations, including CFTR-W1282X, lead to low levels of functional protein due to NMD and premature termination of translation. Overcoming NMD without disrupting the physiological gene-regulatory role of this pathway is a key objective (17). Although allele-specific NMD-inhibition strategies were recently developed using genome editing with CRISPR-Cas9 or ASOs that block EJC deposition (19, 21), these preclinical studies have yet to be translated to the bedside. Thus, there is an unmet need for therapies for CF caused by nonsense mutations. As low as 10% of normal CFTR function would provide a significant therapeutic benefit for CF patients who have a near-complete loss of CFTR function, as is the case for the W1282X mutation (30). Thus, increasing the expression of mutant CFTR protein with residual activity using splice-switching ASOs could benefit CF patients, as long as CFTR-Δex23 protein retains some function.
Compared to other gene-specific NMD-inhibition approaches using CRISPR-Cas9 genome editing or ASOs that inhibit EJC binding (GAIN) (19, 21), inducing expression of CFTR-Δex23 protein may have several advantages. CFTR-Δex23 protein retains carboxyl-terminal domain residues (1478TRL1480) important for posttranslational processing and CFTR gating, which are missing in the carboxyl terminal–truncated CFTR-W1282X protein (39, 40, 41). Adenosine triphosphate (ATP) binding and dimerization of NBD1 and NBD2 are critical for CFTR gating (42). Regions important for ATP binding, namely the Walker B motif (residues 1,366 through 1,372) and the NBD2 signature sequence (residues 1,346 through 1,350), are present in CFTR-Δex23 protein (which lacks residues 1,240 through 1,291) but are missing in the truncated W1282X mutant protein.
The two-ASO cocktail we describe here and our recently described EJC-targeting approach employing a three-ASO cocktail (21) can increase CFTR function. However, lead optimization of ASO cocktails is more difficult than optimizing a single lead ASO because cocktails require optimization of length, chemical modifications, and dose as well as analysis of toxicity for each of the ASOs in the cocktail. Going forward, an exon-skipping ASO strategy using a single ASO that targets an ESE or intronic splicing enhancer element may be more desirable than a two-ASO cocktail targeting two splice sites, from the clinical drug-development and regulatory perspectives, provided that such an ASO elicits efficient exon skipping. Our limited attempt to identify exon-skipping ASOs targeting putative ESEs in exon 23 was not successful, but the two-ASO cocktail may still be advantageous over the three-ASO cocktail we used in another study to block EJC deposition (21). The possibility still remains of finding a single ASO that blocks a splicing enhancer to efficiently promote exon skipping through a systematic ASO screen of exonic and intronic regions, as described previously for other pre-mRNAs (33, 34).
Based on the measurement of CFTR-Δex23 activity, while controlling for the plasmid expression levels, we estimate that CFTR-Δex23 protein has residual activity as high as 54% of WT when combined with Trikafta. We show that uniform PS-MOE ASOs targeting the pre-mRNA 3′ss or 5′ss flanking CFTR exon 23 induce exon 23 skipping. Targeting both splice sites simultaneously with an ASO cocktail induced more efficient exon 23 skipping than targeting each splice site with a single ASO. Measuring the exon 23 PSI after cycloheximide treatment to inhibit NMD showed that the lead ASO cocktail caused almost complete exon 23 skipping. The CFTR-Δex23 mRNA was not sensitive to NMD inhibition by cycloheximide, and it was translated into CFTR-Δex23 protein that retained partial function in human bronchial epithelial cells. Compared to the published activity of WT CFTR in 16HBE17o- cells (38), the 16HBE-W1282X cells treated with the splice-switching lead ASO cocktail combined with a CFTR corrector showed <10% of the WT activity. We anticipated higher CFTR activity in these cells, given our results with the recombinant CFTR-Δex23 expression experiments. Further optimizing various parameters, such as ASO delivery, chemical modifications, and combination with CFTR modulators, may improve the functional impact of the ASO cocktail.
ESEfinder and SFmap predictions are not sufficiently robust to obviate systematic ASO screening, but they can help to establish the mechanism of action of ASOs. We observed that transfecting the lead ASO cocktail at 10 nM caused more exon skipping in 16HBE-W1282X cells than in 16HBE-G551D cells (Fig. 2H). Although the extent of exon skipping in 16HBE-W1282X may have been overestimated, as we did not use any NMD inhibitors for this experiment, this alone does not explain the lack of exon-skipped isoform in 16HBE-G551D cells at the low ASO concentration. ESEfinder analysis predicts that the W1282X mutation may lead to the loss of SRSF1 and SRSF2 binding sites (SI Appendix, Fig. S4B), which, in turn, suggests a sensitized background with a reduced threshold to undergo exon skipping in response to ASOs targeting the splice sites.
CFTR nonsense mutations account for about 7% of all CF-causing mutations listed in the CFTR2 database (3). More common nonsense mutations, including G542X and W1282X, are known to trigger NMD (21), and less common nonsense mutations—except for S1455X and Q1476X, which are located on the terminal exon—likely trigger NMD, according to the 55-nt rule (18). Among these, the nonsense mutations that occur on in-frame exons may benefit from a similar ASO-mediated exon-skipping strategy to avoid NMD and increase partially active CFTR. However, this would depend on whether the skipped exon encodes a peptide that is dispensable for CFTR folding, localization, and activity. Inducing exon 23 skipping for other CFTR mutations without a nonsense mutation in exon 23 may not be desirable, as CFTR-Δex23 lacks amino acid residues in NBD2. The overall functional impact of the ASO cocktail in human bronchial cells that are compound heterozygous for CFTR-W1282X and another CF-causing CFTR mutation, such as the F508del mutation, remains to be determined.
The next stage in the development of the splice-switching ASOs as a potential therapy will require optimization of the delivery to the target tissues in mice. CF affects multiple organ systems, including the respiratory, gastrointestinal, and reproductive organs. ASO delivery to these tissues by parenteral administration is not effective, with the exception of intestinal tissues (43). Systemic delivery is not optimal for delivery of ASOs to the main target tissue for our approach, which is the airway, specifically, ciliated airway epithelial cells in the upper airway. In mice, intravenous (IV) bolus injection of 5 mg/kg ASO yields a concentration in the lung of only 2 µg/g tissue (43). To test whether our lead ASO cocktail increases CFTR expression and function in the airway epithelial cells of mice, two modes of ASO delivery can be used: intratracheal instillation and aerosolized ASO inhalation. In intratracheal instillation, a tested compound is instilled directly in the airway using a bulb-tipped gavage needle or aerosolizing microsprayer, with the assistance of an operating otoscope (44). Intratracheal instillation is easier to dose than aerosolized drug delivery.
Aerosolized ASO generated with a commercial nebulizer can be effectively delivered to the lung tissue and is a more clinically relevant mode of delivery (45). The bioavailability of an aerosolized MOE ASO was 1,260% compared to that of IV administration in mice, and reported ASO half-lives in the lung are 4 and >7 d in mice and monkeys, respectively (43). Thus, infrequent aerosolized dosing may maintain a sufficient tissue concentration of ASOs. Aerosolized gapmer ASO efficiently knocks down target genes in the mouse airway without significant systemic exposure or overt toxicity (45, 46). In a phase-1 clinical trial, the PS-MOE gapmer AIR645 (Altair/Ionis), which targets IL-4/IL-13, was safely administered aerosolized at 30 mg/kg, with some adverse events (47). As the proinflammatory responses are sequence- and chemistry-specific, careful selection of ASOs will be necessary to minimize side effects.
There are currently three inhaled ASOs developed by three companies in active clinical trials. These developments demonstrate that aerosolization is potentially a viable delivery method to target the CF airway with our lead ASO cocktail. IONIS-ENaCRx is a constrained ethyl phosphorothioate (PS) gapmer ASO targeting SCNNA1, which completed a phase-1/2a clinical trial. TPI ASM8, from Topigen, is a combination of two PS ASOs, TOP004 and TOP005, which simultaneously target CCR-3, IL-3, IL-5, and GMCSF, which are relevant targets for asthma (48). Phase-1 and phase-2 clinical trials of TPI ASM8 were completed in 2013. These ASOs all showed in vivo activity without severe side effects (43). QR-010 is a uniformly 2′-OMe PS-modified ASO developed by ProQR. QR-010 converts CFTR-F508del mRNA into WT CFTR mRNA by inserting the three missing bases in the mutant mRNA through an unknown mechanism (49). This drug is currently in a phase-2 clinical trial. The results of these studies will provide useful information about nucleic-acid therapeutics for CF and other lung diseases.
In summary, we have shown that ASO treatment directed to the 3′ss and 5′ss flanking the constitutive exon 23 of CFTR-W1282X pre-mRNA increases CFTR protein missing residues 1,240 through 1,291 and, consequently, increases CFTR activity. Further optimization of the ASOs for delivery, dosage, chemical modifications, and sequence may increase CFTR activity to a level predicted to be clinically beneficial in CF patients. This conclusion is supported by our results comparing the relative activities of CFTR-WT and CFTR-Δex23 proteins in 16HBE-W1282X cells, which showed that the CFTR-Δex23 protein in combination with Trikafta has substantial activity. Our results thus provide an avenue for developing a therapeutic strategy based on splice-switching ASO, in the era of CFTR-modulator therapy.
Materials and Methods
CFTR Expression Plasmid.
The plasmid pcDNA5.FRT-wtCFTR containing the coding sequence for the WT CFTR gene was kindly provided by the Cystic Fibrosis Foundation Therapeutics (CFFT) Lab. The plasmid contains the CFTR gene coding sequence in the pCDNA5 FRT/TO plasmid backbone (Life Technologies). The CFTR coding sequence with a carboxyl-terminal T7 tag from pcDNA5.FRT-wtCFTR was cloned into the multiple cloning site of the pCW57-GFP-2A-MCS plasmid, resulting in the plasmid pGFP-P2A-CFTR-WT-T7. The pCW57-GFP-2A-MCS plasmid was a gift from Adam Karpf, Eppley Institute for Cancer Research, University of Nebraska Medical Center, Omaha, NE (Addgene plasmid No. 71783; n2t.net/addgene:71783; RRID: Addgene_71783). pGFP-P2A-CFTR-WT-T7 was mutagenized to delete the coding sequence in CFTR exon 23, resulting in pGFP-P2A-CFTR-Δex23-T7. pGFP-P2A-CFTR-WT-T7 was mutagenized to introduce a PTC at amino acid position 1282 (W1282X) to generate the pGFP-P2A-CFTR-W1282X-T7 plasmid. Plasmid constructions were conducted by GenScript. The plasmids pGFP-P2A-CFTR-WT-T7, pGFP-P2A-CFTR-W1282X-T7, and pGFP-P2A-CFTR-Δex23-T7 encode recombinant GFP-P2A-CFTR proteins, which are Turbo GFP and T7-tagged CFTR polypeptides linked by a self-cleaving 2A peptide (31).
Tissue Culture.
Gene editing of 16HBE14o- parental cells at the endogenous CFTR locus yielded 16HBEge cell lines CFF-16HBEge CFTR W1282X and G551D, homozygous for the CFTR-W1282X or G551D mutation, respectively. These cells were kindly provided by the CFFT Lab (38). Throughout the text, these cells are referred to as 16HBE-W1282X and 16HBE-G551D, respectively. 16HBE-W1282X cells stably expressing pGFP-P2A-CFTR-WT-T7, pGFP-P2A-CFTR-W1282X-T7, or pGFP-P2A-CFTR-Δex23-T7 are dubbed 16HBEge-GFP-P2A-WT, 16HBEge-GFP-P2A-W1282X, or 16HBEge-GFP-P2A-Δex23, respectively. The 16HBEge cells were cultured in minimal essential medium (Thermo Fisher, 11095072) with 10% fetal bovine serum (FBS) (Thermo Fisher, 26400044). DLD1 cells were cultured in Dulbecco’s modified Eagle medium with 10% FBS. The human embryonic kidney 293T (HEK293T) cell line was obtained from the American Type Culture Collection and cultured according to the provider’s instructions. All cells were incubated at 37 °C and 5% CO2. NMD inhibition by cycloheximide was performed by treating the cells for 1 h with cycloheximide (Sigma, 100 μg/mL).
Generation of 16HBEge-GFP-P2A-WT/W1282X/Δex23 Cells.
HEK293T cells were transfected with CFTR-containing lentiviral plasmids and packaging vector to produce retroviral particles carrying the plasmids pGFP-P2A-CFTR-WT-T7, pGFP-P2A-CFTR-W1282X-T7, or pGFP-P2A-CFTR-Δex23-T7. Cells were incubated for 18 h after transfection; then fresh medium was added to the cells. After 24 h, the medium containing the viral particles was collected and replaced with fresh medium. After another 24 h, the medium containing the viral particles was collected and pooled with the previously collected medium. The pooled medium was filtered through a 0.45-μm filter. PEG-it solution was added to the filtered medium with 1/5 final dilution (System Biosciences, LV810A-1), and the mixture was incubated at 4 °C for 6 h. The medium was centrifuged for 30 min at 1,500 × g, and the supernatant was added to the 16HBE-W1282X cells. After 18 h from the addition of viral particles, the transduced cells were selected with medium containing puromycin (0.2 μg/mL) for 1 wk (EMD Millipore, 540411). Successful transduction was confirmed by the dox-induced GFP signal in 16HBEge-GFP-P2A-WT/W1282X/Δex23 cells (see Microscopy).
Plasmid Transfection.
All plasmids were transfected using Lipofectamine 2000 (Life Technologies, 11668019) according to the manufacturer’s protocol.
Microscopy.
Fluorescence microscopy.
The expression of recombinant GFP-P2A-CFTR proteins in 16HBEge-GFP-P2A-WT/W1282X/Δex23 cells was induced with growth medium containing 0 to 2 μg/mL dox for 48 h (Research Products International Corp, D43020-100). The images were taken with a Revolve microscope and acquired by ECHO Pro version 6.0.1 software (Revolve, ECHO). GFP-intensity analysis from the images was performed with ImageJ2 software (version 2.2.0).
Immunohistochemical microscopy.
The 16HBE-W1282X cells were seeded on chamber slides (Corning 354118) and transfected with pGFP-P2A-CFTR-WT-T7 or pGFP-P2A-CFTR-Δex23-T7 plasmids using Lipofectamine 2000, according to the manufacturer’s protocol. At 18 h posttransfection, the culture medium was replaced with medium containing 2 μg/mL dox. After another 48 h, the cells were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde (Thermo Fisher, 28906). The fixed cells were washed with PBS and blocked with 5% goat serum for 1 h (Thermo Fisher, 31872). Mouse anti-T7 primary antibody (Millipore Sigma, 69522) was applied at a dilution of 1:1,000 for 2 h at room temperature. The slides were washed three times and incubated with goat anti-mouse IgG2b secondary antibody conjugated to Alexa Fluor 594 for 1 h (Thermo Fisher, A-21145). Nuclei were stained with DAPI. Images were captured with a confocal laser-scanning microscope (Zeiss LSM710).
ASO Transfection and Treatment.
All ASOs were 18mers uniformly modified with 2'-O-MOE ribose, PS linkages, and 5′-methylcytosine. They were obtained from Integrated DNA Technologies or Bio-Synthesis. The scramble control ASO has a scrambled sequence based on i23-5ss. All ASOs were dissolved in water and stored at −20 °C. Stock ASO concentrations were calculated based on the A260, and each ASO’s extinction coefficient e (mM−1 × cm−1 @ 260 nm). The sequences of all ASOs used in this study are listed in Table 1.
Table 1.
ASO sequences
| ASO type | ASO name | Sequence |
| Exon skip | i22-3ss | CTATAAGGTAAAAGTGAT |
| Exon skip | ESE-1 | CAAAGTACTCTTCCCTGA |
| Exon skip | ESE-2 | GGCTTTCCTCCACTGTTG |
| Exon skip | ESE-3 | TCACTCCAAAGGCTTTCC |
| Exon skip | i23-5ss | CTAAGTCCTTTTGCTCAC |
| Scramble control | Sc | ACTTCGTTCGCCTTAACT |
Cells were transfected with ASOs and plasmids using Lipofectamine 3000 (Life Technologies, L3000015) according to the manufacturer’s protocol and harvested 48 h posttransfection. For ASO treatment by free uptake, 1-mM stock ASO solutions were diluted into minimum essential medium with 10% FBS to the desired final concentrations, and the cells were cultured for 4 d before harvesting or Ussing chamber assays. NMD-reporter expression was induced with 2 μg/mL dox with culture-medium change 6 h after transfection.
RNA Extraction and RT-PCR.
Total RNA was extracted with TRIzol (Life Technologies) according to the manufacturer’s protocol. Oligo deoxythymidine (dT18) primed reverse transcription was carried out with ImProm-II Reverse Transcriptase (Roche). Semiquantitative radioactive PCR was carried out in the presence of 32P-dCTP with AmpliTaq DNA polymerase (Thermo Fisher), and real-time RT-qPCR was performed with Power Sybr Green Master Mix (Thermo Fisher). Primers used for RT-PCR and RT-qPCR are listed in Table 2. RT-PCR products were separated by 6% native polyacrylamide gel electrophoresis (PAGE), detected with a Typhoon FLA7000 phosphorimager, and quantitated using MultiGauge version 2.3 software (Fujifilm); RT-qPCR data were quantitated using the QuantStudio 6 Flex system.
Table 2.
RT-PCR and qRT-PCR primer sequences
| Target gene | Primer | Sequence |
| CFTR-exon26 | CFTR26F | GCACAGTAATTCTCTGTGAACACAGG |
| CFTR-exon27 | CFTR27R | TCCTCTCGTTCAGCAGTTTCTGG |
| RPL32 | RPL32_F | AGAGGCATTGACAACAGGGTT |
| RPL32 | RPL32_R | GTGAGCGATCTCGGCACAG |
| CFTR-exon22 | CFTR-22F-splice | GCGATCTGTGAGCCGAGTC |
| CFTR-exon24 | CFTR-24R-splice | CTTGATCACTCCACTGTTCATAGGGATC |
Protein Extraction and Western Blotting.
Cells were harvested with radioimmunoprecipitation assay buffer (150 mM NaCl, 50 mM Tris⋅HCl pH 8.0, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) and 2 mM ethylenediaminetetraacetic acid + protease inhibitor mixture (Roche) by sonicating for 5 min at medium power using a Bioruptor (Diagenode), followed by 15-min incubation on ice. Protein concentration was measured using the Bradford assay (Bio-Rad) with bovine serum albumin as a standard. Cell lysates were mixed with Laemmli buffer and incubated at 37 °C for 30 min. The protein extracts were separated by SDS-PAGE (6% or 6 to 20% gradient Tris-chloride gels) and then transferred onto a nitrocellulose membrane. CFTR bands C, B, and A were detected with antibody UNC-596 (J. Riordan laboratory, University of North Carolina). The C- and B-band intensities were measured together for the quantification of CFTR protein levels. T7-tagged CFTR proteins were detected with anti-T7 antibody (Millipore Sigma, 69522). Na/K-ATPase, detected with a specific antibody (Santa Cruz, sc-48345), was used as a loading control. IRDye 800CW or 700CW secondary antibody (LI-COR) was used for Western blotting, and the blots were imaged and quantified using an Odyssey Infrared Imaging System (LI-COR).
Ussing Chamber Assay.
The 16HBEge cells were grown as an electrically tight monolayer on Snapwell filter supports (Corning, 3801), as described (38), and both serosal and mucosal membranes were exposed to the ASOs for 4 d and to CFTR correctors VX-809, VX-661, or VX-445 for 24 h before the assays (Selleckchem, S1565, S7059, and S8851, respectively). The Snapwell inserts were transferred to an Ussing chamber (P2302, Physiologic Instruments, Inc.). The serosal side only was superfused with 5 mL HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]–buffered physiological saline buffer; on the mucosal side, 5 mL CF-HEPES–buffered physiological saline was used (137 mM Na-gluconate, 4 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, and 10 mM glucose; pH adjusted to 7.4 with N-methyl-d-glucamine) to create a transepithelial chloride-ion gradient. After clamping the transepithelial voltage to 0 mV, the short-circuit current (ISC) was measured with a Physiologic Instruments VCC MC6 epithelial voltage clamp while maintaining the buffer temperature at 37 °C. Baseline activity was recorded for 20 min before agonists (final concentrations: 10 µM forskolin [Sigma, F6886] and 10 µM VX-770 [Selleckchem, S1144]) and inhibitor (final concentration: 20 µM CFTRinh-172 [Sigma, C2992]) were applied sequentially at 10-min intervals to both serosal and mucosal surfaces. Agonists/inhibitors were added from 200× to 1,000× stock solutions. Data acquisition was performed with ACQUIRE & ANALYZE Revision II (Physiologic Instruments).
ESE Motif Analysis.
Potential serine- and arginine-rich protein binding sites were analyzed by ESEfinder and SFmap (35, 36).
Statistical Analyses.
Statistical analyses were performed with GraphPad Prism 5. Statistical parameters are indicated in the figures and legends. For the two-tailed t test or one-way ANOVA with Tukey’s or Dunnett’s post hoc test, P < 0.05 was considered significant. The asterisks mark statistical significance as follows: n.s. P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Martin Mense and Hermann Bihler (CFFT, Lexington, MA) for generously sharing experimental protocols and advice. This work was supported by NIH Grant R37GM42699 and a grant from Emily’s Entourage to A.R.K. Y.J.K. was supported by NIH Grants F30HL137326-04 and T32GM008444. We acknowledge assistance from Cold Spring Harbor Laboratory Shared Resources, funded in part by National Cancer Institute Cancer Center Support Grant 5P30CA045508.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114858118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Mort M., Ivanov D., Cooper D. N., Chuzhanova N. A., A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 29, 1037–1047 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Cutting G. R., Cystic fibrosis genetics: From molecular understanding to clinical application. Nat. Rev. Genet. 16, 45–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CFTR2, Clinical and functional translation of CFTR (CFTR2. http://cftr2.org (2020) (Accessed, January 20, 2020). Accessed 20 January 2020.
- 4.Rowe S. M., et al. , Restoration of W1282X CFTR activity by enhanced expression. Am. J. Respir. Cell Mol. Biol. 37, 347–356 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurosaki T., Popp M. W., Maquat L. E., Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 20, 406–420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Middleton P. G., et al. , VX17-445-102 Study Group, Elexacaftor-tezacaftor-ivacaftor for cystic fibrosis with a single Phe508del allele. N. Engl. J. Med. 381, 1809–1819 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.US Food and Drug Administration, Highlights of prescribing information for Trikafta. https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/212273s000lbl.pdf. Accessed 8 October 2021.
- 8.Maiuri L., Raia V., Kroemer G., Strategies for the etiological therapy of cystic fibrosis. Cell Death Differ. 24, 1825–1844 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pascual-Morena C., et al. , Restorative treatments of dystrophin expression in Duchenne muscular dystrophy: A systematic review. Ann. Clin. Transl. Neurol. 7, 1738–1752 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zainal Abidin N., Haq I. J., Gardner A. I., Brodlie M., Ataluren in cystic fibrosis: Development, clinical studies and where are we now? Expert Opin. Pharmacother. 18, 1363–1371 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Linde L., et al. , Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J. Clin. Invest. 117, 683–692 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma J., et al. , A small molecule that induces translational readthrough of CFTR nonsense mutations by eRF1 depletion. Nat. Commun. 12, 4358 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W., Hong J. S., Rab A., Sorscher E. J., Kirk K. L., Robust stimulation of W1282X-CFTR channel activity by a combination of allosteric modulators. PLoS One 11, e0152232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haggie P. M., et al. , Correctors and potentiators rescue function of the truncated W1282X-CFTR translation product. J. Biol. Chem. 292, 771–785 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khorkova O., Wahlestedt C., Oligonucleotide therapies for disorders of the nervous system. Nat. Biotechnol. 35, 249–263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keenan M. M., et al. , Nonsense-mediated RNA decay pathway inhibition restores expression and function of W1282X CFTR. Am. J. Respir. Cell Mol. Biol. 61, 290–300 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Nasif S., Contu L., Mühlemann O., Beyond quality control: The role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin. Cell Dev. Biol. 75, 78–87 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Nagy E., Maquat L. E., A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem. Sci. 23, 198–199 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Erwood S., et al. , Allele-specific prevention of nonsense-mediated decay in cystic fibrosis using homology-independent genome editing. Mol. Ther. Methods Clin. Dev. 17, 1118–1128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nomakuchi T. T., Rigo F., Aznarez I., Krainer A. R., Antisense oligonucleotide-directed inhibition of nonsense-mediated mRNA decay. Nat. Biotechnol. 34, 164–166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y. J., Nomakuchi T., Papaleonidopoulou F., Krainer A. R., Gene-specific nonsense-mediated mRNA decay targeting for cystic fibrosis therapy. bioRxiv [Preprint] (2021). 10.1101/2021.07.13.452144(Accessed 18 July 2021). [DOI] [PMC free article] [PubMed]
- 22.Rüger J., Ioannou S., Castanotto D., Stein C. A., Oligonucleotides to the (Gene) rescue: FDA approvals 2017-2019. Trends Pharmacol. Sci. 41, 27–41 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Aartsma-Rus A., Corey D. R., The 10th oligonucleotide therapy approved: Golodirsen for Duchenne muscular dystrophy. Nucleic Acid Ther. 30, 67–70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.US Food and Drug Administration, FDA approves first drug for spinal muscular atrophy. https://www.fda.gov/news-events/press-announcements/fda-approves-first-drug-spinal-muscular-atrophy. Accessed 10 December 2021.
- 25.Kim J., et al. , Patient-customized oligonucleotide therapy for a rare genetic disease. N. Engl. J. Med. 381, 1644–1652 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michaels W. E., Bridges R. J., Hastings M. L., Antisense oligonucleotide-mediated correction of CFTR splicing improves chloride secretion in cystic fibrosis patient-derived bronchial epithelial cells. Nucleic Acids Res. 48, 7454–7467 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oren Y. S., et al. , Antisense oligonucleotide-based drug development for cystic fibrosis patients carrying the 3849 + 10 kb C-to-T splicing mutation. J. Cyst. Fibros. 20, 865–875 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anna A., Monika G., Splicing mutations in human genetic disorders: Examples, detection, and confirmation. J. Appl. Genet. 59, 253–268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han S. T., et al. , Residual function of cystic fibrosis mutants predicts response to small molecule CFTR modulators. JCI Insight 3, 1021–1022 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferec C., Cutting G. R., Assessing the disease-liability of mutations in CFTR. Cold Spring Harb. Perspect. Med. 2, a009480 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu D., Yates P. A., Zhang H., Cao K., Comparing lamin proteins post-translational relative stability using a 2A peptide-based system reveals elevated resistance of progerin to cellular degradation. Nucleus 7, 585–596 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trouvé P., et al. , Rapid detection of the mature form of cystic fibrosis transmembrane regulator by surface plasmon resonance. Anal. Methods 7, 226–236 (2015). [Google Scholar]
- 33.Aartsma-Rus A., van Ommen G.-J. B., Antisense-mediated exon skipping: A versatile tool with therapeutic and research applications. RNA 13, 1609–1624 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahashi K., et al. , TSUNAMI: An antisense method to phenocopy splicing-associated diseases in animals. Genes Dev. 26, 1874–1884 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paz I., Akerman M., Dror I., Kosti I., Mandel-Gutfreund Y., SFmap: A web server for motif analysis and prediction of splicing factor binding sites. Nucleic Acids Res. 38, W281-5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cartegni L., Wang J., Zhu Z., Zhang M. Q., Krainer A. R., ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31, 3568–3571 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valley H. C., et al. , Isogenic cell models of cystic fibrosis-causing variants in natively expressing pulmonary epithelial cells. J. Cyst. Fibros. 18, 476–483 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Keating D., et al. , VX16-445-001 Study Group, VX-445-Tezacaftor-Ivacaftor in patients with cystic fibrosis and one or two Phe508del alleles. N. Engl. J. Med. 379, 1612–1620 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aksit M. A., et al. , Decreased mRNA and protein stability of W1282X limits response to modulator therapy. J. Cyst. Fibros. 18, 606–613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quon B. S., Rowe S. M., New and emerging targeted therapies for cystic fibrosis. BMJ 352, i859 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guggino W. B., Stanton B. A., New insights into cystic fibrosis: Molecular switches that regulate CFTR. Nat. Rev. Mol. Cell Biol. 7, 426–436 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Hwang T.-C., Sheppard D. N., Gating of the CFTR Cl- channel by ATP-driven nucleotide-binding domain dimerisation. J. Physiol. 587, 2151–2161 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crooke S. T., Antisense Drug Technology: Principles, Strategies, and Applications (CRC Press, 2007). [Google Scholar]
- 44.Hasegawa-Baba Y., Kubota H., Takata A., Miyagawa M., Intratracheal instillation methods and the distribution of administered material in the lung of the rat. J. Toxicol. Pathol. 27, 197–204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Templin M. V., et al. , Pharmacokinetic and toxicity profile of a phosphorothioate oligonucleotide following inhalation delivery to lung in mice. Antisense Nucleic Acid Drug Dev. 10, 359–368 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Crosby J. R., et al. , Inhaled ENaC antisense oligonucleotide ameliorates cystic fibrosis-like lung disease in mice. J. Cyst. Fibros. 16, 671–680 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Hodges M. R., et al. , “Randomized, double-blind, placebo controlled first in human study of inhaled AIR645, an IL-4Rα oligonucleotide, in healthy volunteers” in American Thoracic Society International Conference Abstracts: B92. New Treatment Approaches for Asthma and Allergy (American Thoracic Society, 2009), p. A3640.
- 48.Allakhverdi Z., Allam M., Renzi P. M., Inhibition of antigen-induced eosinophilia and airway hyperresponsiveness by antisense oligonucleotides directed against the common β chain of IL-3, IL-5, GM-CSF receptors in a rat model of allergic asthma. Am. J. Respir. Crit. Care Med. 165, 1015–1021 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Zamecnik P. C., Raychowdhury M. K., Tabatadze D. R., Cantiello H. F., Reversal of cystic fibrosis phenotype in a cultured Delta508 cystic fibrosis transmembrane conductance regulator cell line by oligonucleotide insertion. Proc. Natl. Acad. Sci. U.S.A. 101, 8150–8155 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.