Significance
Schizophrenia (SCZ) is a complex, highly heritable, neurodevelopmental disorder with marked clinical heterogeneity and no clear pathological mechanism or cellular pathology. The polygenic nature of the disorder has hindered our ability to model the disorder in the laboratory. Prior studies of cortical neurons differentiated from SCZ patient and control hiPSCs have identified interesting differences but their relevance to clinical illness in adults remains unclear. We now identify electrophysiological measures that associate with diagnosis and/or predict the severity of clinical and cognitive features of individual adult donors. These results demonstrate neurophysiological measures that are related to the patient’s personal clinical characteristics, which may help with patient stratification and the development of novel biomarkers and therapeutic targets.
Keywords: schizophrenia, stem cell, sodium channel, electrophysiology
Abstract
Neurons derived from human induced pluripotent stem cells (hiPSCs) have been used to model basic cellular aspects of neuropsychiatric disorders, but the relationship between the emergent phenotypes and the clinical characteristics of donor individuals has been unclear. We analyzed RNA expression and indices of cellular function in hiPSC-derived neural progenitors and cortical neurons generated from 13 individuals with high polygenic risk scores (PRSs) for schizophrenia (SCZ) and a clinical diagnosis of SCZ, along with 15 neurotypical individuals with low PRS. We identified electrophysiological measures in the patient-derived neurons that implicated altered Na+ channel function, action potential interspike interval, and gamma-aminobutyric acid–ergic neurotransmission. Importantly, electrophysiological measures predicted cardinal clinical and cognitive features found in these SCZ patients. The identification of basic neuronal physiological properties related to core clinical characteristics of illness is a potentially critical step in generating leads for novel therapeutics.
Schizophrenia (SCZ) is a complex, highly heritable, “polygenic” disorder with considerable cost to affected individuals, their families, and society (1). Marked clinical heterogeneity supports the idea that SCZ is a spectrum of disorders with overlapping symptomatology, rather than a single disease. SCZ is characterized by positive symptoms including hallucinations and delusions, negative symptoms such as social and emotional withdrawal, and cognitive symptoms including deficits in executive function and decision making. Negative symptoms (2) and cognitive deficits (3) in SCZ are still only poorly addressed by current treatments, all of which have side effects that often result in poor medication adherence. Although SCZ is not typically diagnosed until early adulthood, versions of a “neurodevelopmental hypothesis” (4) propose that altered signaling during early brain development leads to altered connectivity/function of neural circuits in later critical maturational periods, and eventually leads to onset of clinical symptoms and cognitive deficits.
Substantial genetic and environmental complexity and heterogeneity are hallmarks of common, polygenic disorders, posing serious challenges to the mechanistic understanding necessary for advances in drug design. Genome-wide association studies (GWASs) have implicated at least 270 risk loci for SCZ, with each risk-associated variant tagging multiple haplotypes containing functional variants that confer only a small degree of risk (5), and it is predicted, at least at the population level, that there exist thousands of such risk-related genes. Derived from GWASs, the polygenic risk score (PRS) (6, 7) is a cumulative index of relative genomic risk for disease, calculated for each individual based on independent genetic risk markers. PRS is based on common genetic variation, and has become an important tool in SCZ research that has the potential to identify undiagnosed individuals for whom the risk of developing SCZ is relatively high, as well as in choosing subjects for molecular and clinical studies. However, very little is known about how these common variants and risk genes interact with each other and with the environment during neurodevelopment to initiate pathophysiology. Identification of simpler and more tractable, genome type–cellular phenotype relationships may facilitate progress in discovering biological mechanisms by which genomic risk for SCZ translates into more complex phenotypic states, potentially including diagnostic symptoms and cognitive deficits.
The advent of human induced pluripotent stem cell (hiPSC) technology to model biological aspects of neuropsychiatric disease (8) has enabled specific and detailed characterization and manipulation of human neural cells in vitro, without some of the limitations inherent in animal models and postmortem human brain studies. Several previous studies have used hiPSC technologies to model biological aspects of SCZ in a variety of downstream cell types including neural progenitors, glial cells, and both excitatory and inhibitory neurons (9–18). Although PRS was not used as a criterion for sample selection in those studies, associations between cellular phenotypes and SCZ diagnosis were observed, suggesting that the hiPSC model has considerable potential to identify biological dysfunction that might contribute to disease risk. However, a major uncertainty that remains in all published studies is whether any of the observed cellular phenotypes have any meaningful relationship to specific clinical characteristics of individual donors. This is an especially critical consideration in efforts to validate potential cell models of neuropsychiatric disorders because such illnesses do not have pathognomonic cellular pathology.
In this study, we investigated the cellular impact of common genetic risk for SCZ to better understand its role in early neural development, and to possibly identify cellular phenotypes that might relate to cardinal clinical and cognitive features of adult donors. Because the vast majority of cases of SCZ are caused by common variant risk, we focused on PRS to contrast high–genetic risk patients with neurotypicals of low genetic risk. Based on PRS, we selected patient fibroblast lines from participants in the National Institute of Mental Health (NIMH) Clinical Brain Disorders Branch Sibling Study of Schizophrenia (D.R.W., principal investigator [PI]), which captured extensive clinical and cognitive data on each subject. Here, we describe the design, execution, and results of our investigation into the relationships between the molecular, cellular, and physiological properties of hiPSC-derived neural progenitor cells (NPCs) and cortical neurons and clinical status, clinical symptoms, and cognitive performance. Importantly, we look beyond case–control differences, and identify within-case patterns of association that could serve to illuminate dimensions of illness and subgroups of patients who may be suitable for targeted treatments. The results demonstrate that common genetic variants associated with SCZ converge on select electrophysiological mechanisms that relate to clinically relevant features of SCZ, underscoring the potential of the findings for biomarker identification and perhaps downstream drug development.
Results
Reprogramming and Characterization of hiPSC Lines.
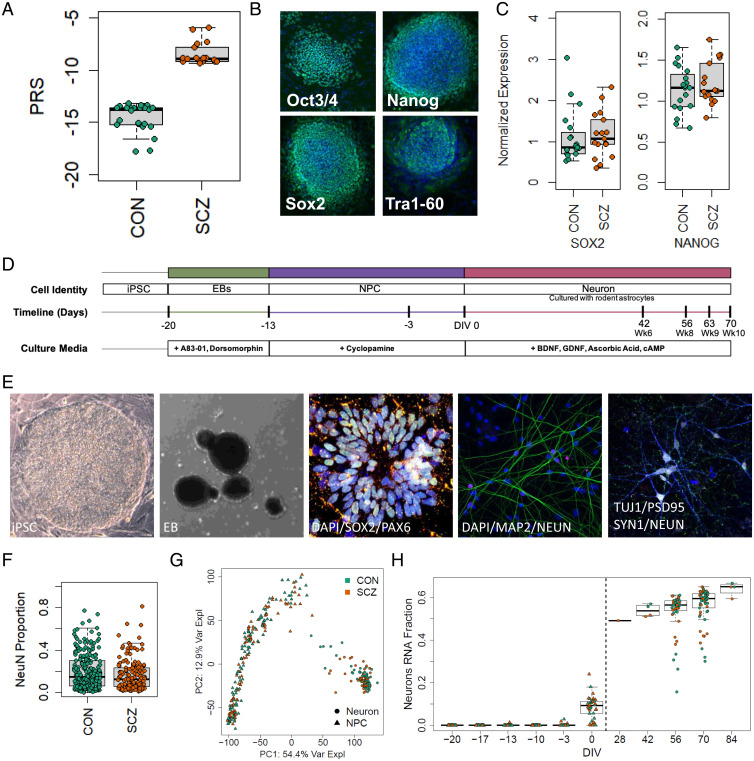
The study included 13 high-PRS individuals with SCZ (16 total cell lines: two lines each for 3 donors and one line each for 10 donors) and 15 low-PRS neurotypical (CON) individuals (20 total cell lines: two lines each for 5 donors and one each for 10 donors) (Fig. 1A and Dataset S1). These hiPSC lines were generated as previously described (19), and no differences in pluripotency were observed between SCZ and CON lines (Fig. 1 B and C and SI Appendix, Fig. S1). The hiPSCs were first differentiated into SOX2- and PAX6-expressing NPCs, and then further differentiated into forebrain-specific cortical neurons, which were cocultured on rat astrocytes as previously described (Fig. 1 D and E) (9, 12).
Fig. 1.
Description of experimental lines and experiment overview. (A) PRS of neurotypical controls and individuals with SCZ included in this study (CON, −14.829 ± 1.535, n = 15; SCZ, −8.375 ± 1.1736, n = 13; P = 1.78E-12). (B) Pluripotency of reprogrammed lines was verified by identification of NANOG+, SOX2+, OCT3/4+, and TRA-1-60+ colonies colocalized with DAPI using immunocytochemistry. (C) qPCR quantification of stem cell markers SOX2 (CON, 1.152 ± 0.151; SCZ, 1.162 ± 0.133; P = 0.96, n = 35 from 28 genomes) and NANOG (CON, 1.133 ± 0.062; SCZ, 1.222 ± 0.063; P = 0.32, n = 35 from 28 genomes), normalized to embryonic stem cell line H1. (D) hiPSCs were differentiated into neurons in multiple batches. (E) Representative images of induced pluripotent stem cell colony (bright field), EB (bright field), immunofluorescence of neural rosettes at 2 d, DAPI (blue), SOX2 (green), PAX6 (red); immunofluorescence of neurons at DIV56, DAPI (blue), MAP2 (green), NeuN (red); immunofluorescence of neurons showing synaptic puncta at DIV70, Tuj1 (blue), PSD95 (green), SYNAPSIN1 (red), NeuN (white). (F) Quantification of the proportion of NeuN+ cells to the number of DAPI-stained nuclei shows no difference in neuronal differentiation or survival by diagnosis (CON, 0.186 ± 0.01, n = 229 cultures from 18 lines; SCZ, 0.167 ± 0.012, n = 156 cultures from 14 lines; P = 0.43). (G) PCA of RNA-seq at multiple time points during culture differentiation (hiPSC, NPC, DIV56, and DIV70 neurons) demonstrates that the largest source of variation (top PC, PC1, representing 54.4% of the variance explained) corresponds to cell state. (H) The fraction of RNA attributed to neurons increases across differentiation stages in CON and SCZ lines.
Generation of Cortical Neurons and Molecular Profiling.
For all experiments, differentiation and data collection were performed blinded to diagnosis and to clinical and cognitive measures. We quantified the cellular composition of cultures across four rounds of differentiation and observed no differences in number of neurons (Fig. 1F) or neuronal types by diagnosis (SI Appendix, Fig. S2) or by line (SI Appendix, Fig. S3). Additionally, gene expression corresponding to neuron identity and maturity was assessed by RNA sequencing (RNA-seq) (9), where the largest source of variation (54.4%) corresponded to cell state (Fig. 1G and SI Appendix, Fig. S4A). The fraction of RNA from neurons increased over time (Fig. 1H), while the hiPSC fraction declined over time (SI Appendix, Fig. S4C) and NPC fraction peaked at days in vitro 0 (DIV0; SI Appendix, Fig. S4D). Other cell states of prenatal neuronal maturation showed similar developmental dynamics (SI Appendix, Fig. S4 E and F). We lastly compared our time-course data with three other public datasets (12, 20, 21) by performing principal-component analysis (PCA) on our data and then projecting other datasets into this latent space to assess comparability and relative maturity (SI Appendix, Fig. S4G). Our samples showed relatively higher neuronal maturation than these previous studies, via PCA (SI Appendix, Fig. S4H) as well as RNA deconvolution (SI Appendix, Fig. S4I).
We performed linear mixed-effects modeling on 23,119 expressed genes assessing for a main effect of SCZ (22), adjusting for fixed effects corresponding to cellular maturity, human alignment fraction, human gene assignment rate, and DIV of the neurons (binary, corresponding to 8 or 10 wk), and treating each donor as a random intercept to account for repeated measures. The full set of differential expression results for NPCs is in Dataset S6 and for neurons is in Dataset S7. We identified 68 differentially expressed genes (DEGs) at a false discovery rate (FDR) <0.1, with median absolute fold changes (FCs) of 1.8 (interquartile range 1.34 to 2.45; SI Appendix, Fig. S5A). We verified that no DEGs for SCZ were found in the rat genome—corresponding to effects in astrocytes—at an FDR <0.99 using an analogous statistical model. We then performed gene set enrichment analysis using a larger set of DEGs (at P < 0.005) and predefined Gene Ontology sets. We identified enrichment in pathways related to WNT signaling and forebrain development (SI Appendix, Fig. S5B). We further found that SCZ DEGs correlated across cell states, such that DEGs in NPCs were preserved as DEGs in neurons. This developmental overlap in SCZ DEGs suggests that expression differences found in our neurons started earlier in development/differentiation and, that for some genes, differences in NPCs may predict differences in neurons (SI Appendix, Fig. S5 C–E) (10). Finally, we compared DEGs in our hiPSC-derived neurons with SCZ state-related DEGs and risk-related transcriptome-wide association studies (TWASs) in the postmortem dorsolateral prefrontal cortex (DLPFC; SI Appendix, Fig. S5 F and G) and, similar to a previous study (23), we found no association between SCZ illness state (DEGs) and SCZ genetic risk (TWASs). Together, these results indicate that diagnosis or DEGs associated with diagnosis do not significantly impact the generation of subtypes of cortical forebrain neurons, thus enabling downstream cellular and physiological comparisons between genomes and diagnosis.
Initial Physiological Characterization.
We assessed cortical neuron function using Ca2+ imaging and whole-cell electrophysiology. Our differentiation protocol consistently generated electrically active neurons that displayed spontaneous and synchronous network activity (SI Appendix, Fig. S6). We quantified Ca2+ transients at DIV42 and DIV63 from 13,602 individual neurons across 871 videos, and observed no effect of diagnosis on the proportion of active cells (SI Appendix, Figs. S6 A–C and S7A), frequency of events (SI Appendix, Fig. S7 B and C), or synchronicity of events (SI Appendix, Fig. S7 D–F). Whole-cell recordings from over 1,000 neurons at DIV56 and DIV70 from multiple differentiation runs confirmed appropriate developmental maturation of intrinsic membrane properties (SI Appendix, Fig. S8), the ability to fire trains of action potentials (APs; see Fig. 4D), and spontaneous synaptic transmission (SI Appendix, Fig. S6 E–G), collectively indicating that our directed differentiation protocol generates cortical neurons that display intrinsic electrical activity and synaptic connectivity that result in functional neuronal networks.
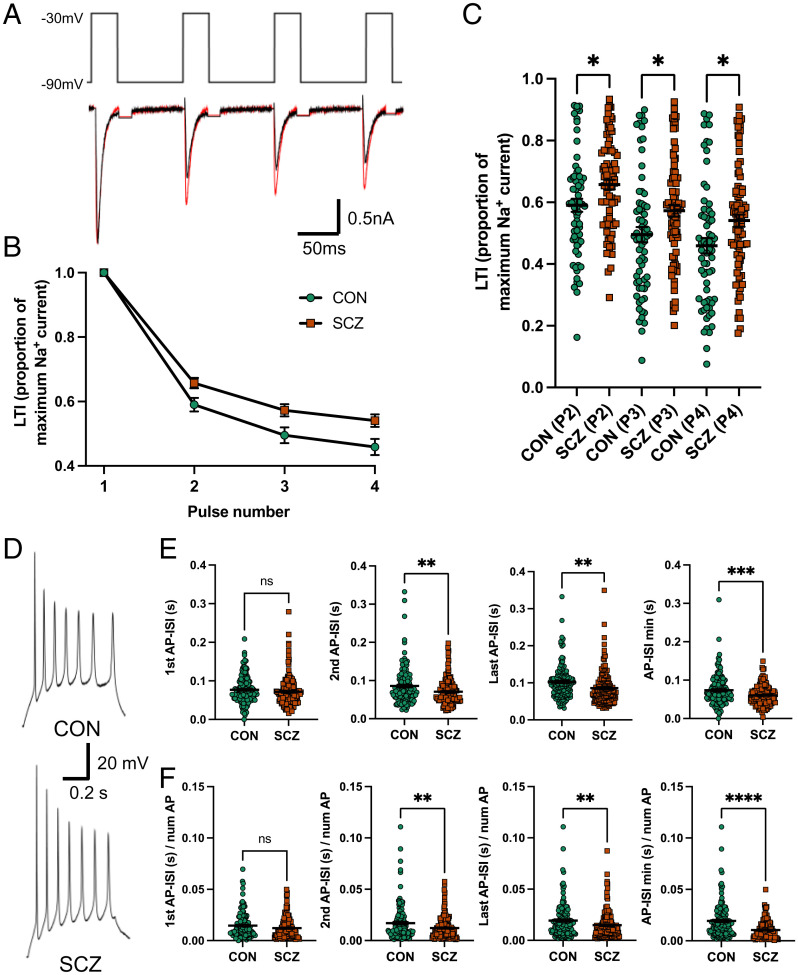
Fig. 4.
Reduced inactivation in SCZ is reflected in the AP-ISI in 13 SCZ compared with 15 CON. (A) Representative Na+ currents in response to four consecutive voltage steps from a holding potential of −90 to −30 mV in an SCZ (red) and CON (black) neuron. Capacitance transients are blanked out. (B) Group data of LTI. (C) Summary LTI of Na+ currents for each voltage step normalized by the currents generated by the first voltage step showing reduced LTI in SCZ compared with CON neurons (CON P2/P1 = 0.59 ± 0.02; SCZ P2/P1 = 0.66 ± 0.024, post hoc P = 0.024; CON P3/P1 = 0.50 ± 0.02; SCZ P3/P1 = 0.57 ± 0.02, *P = 0.023, post hoc P = 0.023; CON P4/P1 = 0.46 ± 0.03; SCZ P4/P1 = 0.54 ± 0.021, post hoc P = 0.021; mixed-effects ANOVA P < 0.0001, n = 148 from 11 genomes). (D) Example AP trains generated in SCZ and CON neurons. (E) Group data showing the AP-ISI for the second (CON 0.086 ± 0.004 s, SCZ 0.071 ± 0.003 s, **P = 0.0018, n = 297 from 28 genomes), last (CON 0.102 ± 0.004 s, SCZ 0.086 ± 0.004 s, **P = 0.002, n = 297 from 28 genomes), and minimum AP-ISI (CON 0.074 ± 0.003 s, SCZ = 0.060 ± 0.002 s, ***P = 0.0006, n = 297 from 28 genomes), but not the first AP-ISI (CON 0.077 ± 0.003 s, SCZ = 0.07 ± 0.003 s, P = 0.245, n = 297), are significantly shorter in SCZ versus CON neurons. (F) Group data showing the second (CON 0.017 ± 0.001 s, SCZ = 0.012 ± 0.0008 s, **P = 0.0042, n = 297 from 28 genomes), last (CON 0.0193 ± 0.001 s, SCZ 0.015 ± 0.001 s, **P = 0.0098, n = 297 from 28 genomes), and minimum AP-ISI (CON 0.019 ± 0.001 s, SCZ 0.010 ± 0.0007 s, ****P < 0.0001, n = 297 from 28 genomes), but not the first AP-ISI (CON 0.015 ± 0.001 s, SCZ 0.012 ± 0.0008 s, P = 0.062, n = 297 from 28 genomes), remain significantly reduced in SCZ versus CON neurons when normalized by the number of APs. All data represented as mean ± standard error of the mean. ns, not significant.
Analysis of Association between Electrophysiological Measures and Diagnosis.
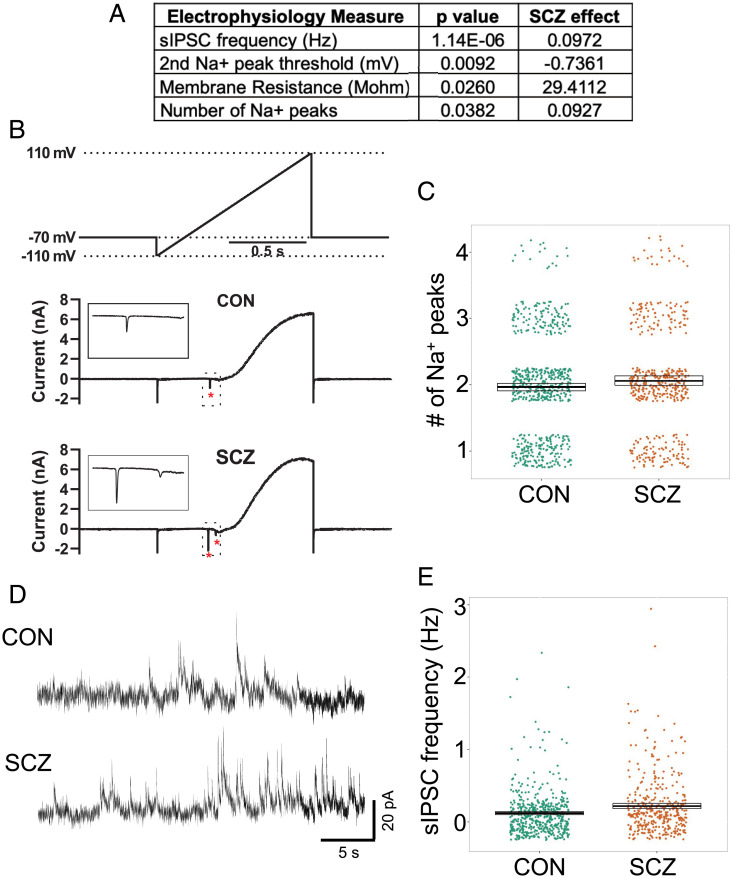
Three electrophysiological measures related to Na+ channel function were associated (P < 0.05) with diagnosis (Fig. 2A and Dataset S2). Lines derived from SCZ donors (SCZ lines) showed an increase in membrane resistance (Fig. 2A; P = 0.025), an increased number of Na+ current peaks in response to a voltage-ramp protocol (P = 0.038; Fig. 2 A–C and SI Appendix, Fig. S9 A and B), and a decrease in the activation threshold of the second Na+ peak (P = 0.009; Fig. 2A). Further examination of Na+ currents generated during the ramp protocol showed they are tetrodotoxin (TTX)-sensitive (SI Appendix, Fig. S10 A and B), and that their frequency correlates with physiologically relevant indices indicative of neuronal maturation (SI Appendix, Fig. S10 C–E). Interestingly, increased Na+ peaks are related to more mature AP kinetics, even though SCZ lines show increased membrane resistance, which is typical of a less mature neuron that is expressing fewer ion channels on its plasma membrane. In addition, we observed that the frequency of spontaneous inhibitory postsynaptic currents (sIPSCs) was increased in SCZ lines (P = 1.12E-06; Fig. 2 A, D, and E and SI Appendix, Fig. S9 E and F), while spontaneous excitatory postsynaptic transmission (sEPSC) was unchanged (SI Appendix, Fig. S6 F and G and Dataset S2), suggesting altered excitatory–inhibitory balance. The enhanced sIPSC frequency appears to be related to an increase in the number of gamma-aminobutyric acid–ergic (GABAergic) synapses and/or altered presynaptic probability of release, since the number of GABAergic cells (SI Appendix, Fig. S2), relative expression of SLC12A2 (protein NKCC1, P = 0.66) and SLC12A5 (protein CC2, P = 0.80), and frequency of network activity (SI Appendix, Fig. S7 B and C) did not differ by diagnosis. Together, these results identified associations between diagnosis and specific physiological phenotypes that are related to Na+ channel function and GABAergic transmission.
Fig. 2.
Electrophysiological differences between CON and SCZ lines. (A) Table of electrophysiological phenotypes observed between CON and SCZ lines. (B) Representative traces showing Na+ currents (asterisks and Insets) in response to a voltage-ramp protocol (above). (C) Summary data showing significantly more Na+ currents in SCZ neurons versus CON neurons across all genomes (SCZ effect, 0.093, P = 0.038, n = 1,074 from 28 genomes). (D) Representative traces of sIPSCs recorded from CON and SCZ neurons at 0 mV. (E) Summary data showing significantly increased sIPSC frequency in SCZ neurons versus CON neurons across all genomes (SCZ effect, 0.097, P = 1.14E-6, n = 903 from 28 genomes).
Sodium Channel Kinetics Associated with SCZ.
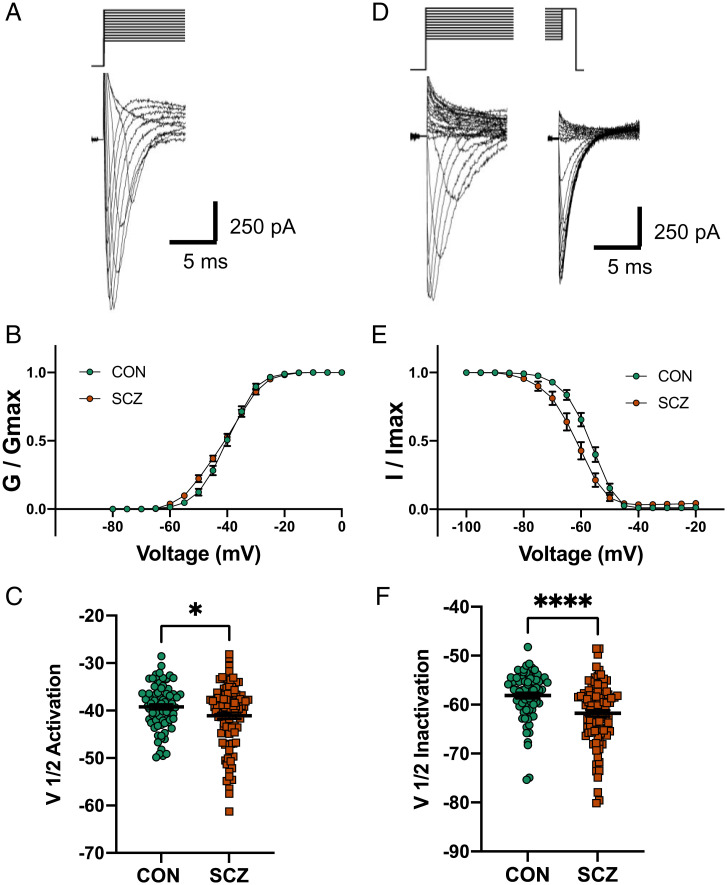
Given that three electrophysiological measures are directly related to Na+ channel function (Fig. 2A) and are associated with diagnosis, we performed a more detailed electrophysiological analysis of Na+ channel kinetics. To improve our voltage clamp of Na+ currents, we used a modified internal recording solution and measured the voltage dependence of Na+ channel activation and inactivation on a subset (Dataset S1) of SCZ (seven lines from six genomes) and CON (seven lines from seven genomes) genomes (Fig. 3 A and D). Using standard voltage-clamp protocols (24, 25), we observed that SCZ lines showed a significantly hyperpolarized shift in the voltage dependence of Na+ channel activation (Fig. 3B and SI Appendix, Fig. S11 A and B) and V1/2 activation voltage (P = 0.039; Fig. 3C). In addition, SCZ lines showed a significant hyperpolarized shift in the voltage dependence of inactivation (Fig. 3E and SI Appendix, Fig. S11 C and D) and V1/2 inactivation voltage (P < 0.0001, adjusted P < 0.0035; Fig. 3F). The hyperpolarized shift in activation supports the diagnosis association with the threshold for activation of the second Na+ peak (Fig. 2A) and the hyperpolarized shift in inactivation supports the higher membrane resistance observed in SCZ lines (Fig. 2A), because more channels are in a nonconducting state at resting membrane potentials. We also quantified Na+ channel long-term inactivation (LTI) by activating four consecutive Na+ currents with 16-ms (40-ms interval) voltage steps from −90 to −30 mV (Fig. 4 A and B) on a subset (Dataset S1) of SCZ (six lines from five genomes) and CON (six lines from six genomes) lines. SCZ neurons showed a significant reduction in the amount of LTI compared with CON neurons (Fig. 4C and SI Appendix, Fig. S11E; mixed-effects ANOVA P < 0.0001; post hoc t test: P2/P1, P = 0.024; P3/P1, P = 0.023; P4/P1, P = 0.021), which suggests more Na+ channels are available for repetitive rounds of activation in SCZ lines.
Fig. 3.
Altered Na+ channel activation and inactivation in six SCZ compared with seven CON. (A) Representative Na+ current traces in response to voltage steps (−100 to +20 mV). (B) Voltage dependence of steady-state activation of Na+ channels in SCZ and CON neurons. (C) Summary data showing a significantly hyperpolarized shift in the activation of Na+ channels in SCZ neurons versus CON neurons (CON, V1/2 = −39.2 ± 0.6 mV; SCZ, V1/2 = −41.1 ± 0.7 mV; Delta = 1.9 mV, *P = 0.039, n = 173 from 13 genomes). (D) Representative Na+ current traces in response to a steady-state inactivation protocol (initial voltage steps from −130 to 20 mV; test voltage steps, −20 mV) to measure channel inactivation. (E) Voltage dependence of steady-state inactivation of Na+ channels in SCZ and CON neurons. (F) Summary data showing a significantly hyperpolarized shift in the inactivation of Na+ channels in SCZ neurons versus CON neurons (CON, V1/2 = −58.1 ± 0.6 mV; SCZ, V1/2 = −61.8 ± 0.7 mV; Delta = 3.7 mV, ****P < 0.0001, adjusted P < 0.0035, n = 162 from 13 genomes). All data represented as mean ± standard error of the mean.
Together, these results indicate that Na+ channel gating characteristics are altered in cortical neurons derived from SCZ donors in comparison with CON and will likely have an impact on cellular excitability. In particular, LTI is known to regulate the AP interspike interval (AP-ISI) by decreasing the amount of Na+ current and making it harder for a neuron to spike at high frequency (26). Therefore, we analyzed AP-ISI in neurons from the 13 SCZ and 15 CON genomes (Dataset S1) that generated three or more APs in response to current injection (Fig. 4D). As predicted from LTI data, SCZ neurons showed a significant reduction in the second (P = 0.0018), last (P = 0.002), and minimum (P = 0.0006) AP-ISI compared with CON neurons (Fig. 4E and SI Appendix, Fig. S12). Moreover, the first AP-ISI (P = 0.245) was not different between SCZ and CON neurons because the effect of LTI on AP-ISI is more impactful after each consecutive round of Na+ channel activation. For this analysis, we only analyzed AP-ISI from the trace showing the maximum number of consecutive APs that reached a 4 mV/ms threshold. Importantly, the reduced AP-ISI in SCZ lines was not related to the maximum number of APs analyzed because AP-ISI remained significantly different when normalized by AP number (Fig. 4F). Overall, these results indicate the kinetics of Na+ channels and AP firing patterns are different in SCZ neurons compared with CON neurons. We show the altered Na+ channel function can impact the fidelity of AP generation and spike timing, which is a critical process for proper brain development and function, and therefore potentially represents an underlying etiological mechanism in SCZ.
Association of Electrophysiological Measures with Clinical and Cognitive Data.
While differences in electrophysiological characteristics between patient and control neurons may reflect a pathophysiological feature of SCZ neurons, they may also be due in part to context, that is, experiment-specific, and unrelated to the actual clinical syndrome in affected individuals. In an attempt to differentiate between these possibilities, we correlated electrophysiological measures with clinical symptomatology and cognitive performance of the cell-line donors. We focused on seven electrophysiological measures that either showed association with SCZ diagnosis (Fig. 2A) or were related to spontaneous synaptic transmission. Ratings for positive and negative syndrome scale items (PANSSs; 7 positive, 7 negative, 16 general) captured classic diagnostic symptomatology. Regarding cognition, we tested composite factor scores for six cognitive domains (verbal memory, N-back, visual memory, processing speed, card sorting, and digit span), as well as estimated IQ and a general cognitive ability composite (“g”) (27–32). Correlations within CON, who show minimal variation on symptom ratings but normal cognitive test score variation, were either very weak or absent (Dataset S4).
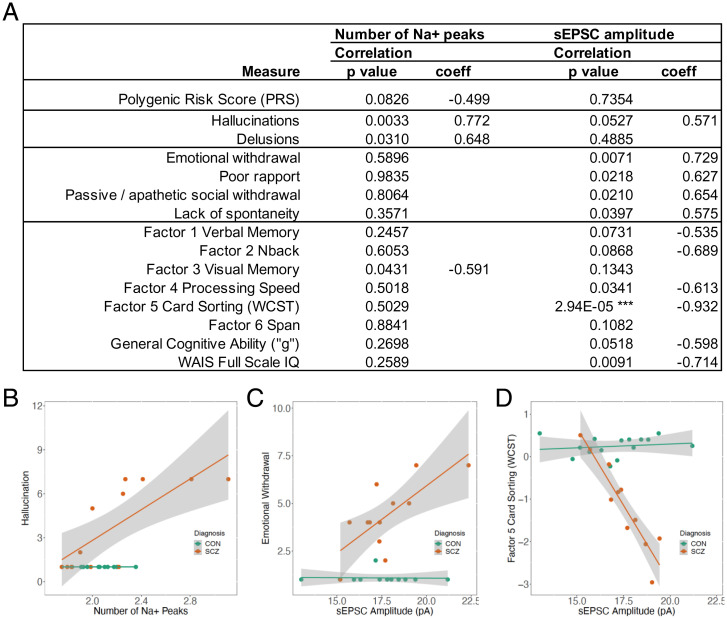
In SCZ, however, two electrophysiological variables showed a striking and divergent pattern of associations with the clinical and cognitive variables, echoing a frequent divergence of these characteristics in patients (Fig. 5A). The number of Na+ peaks showed directionally consistent, positive associations with canonical positive symptom ratings (for hallucinations and delusions), whereas there were no correlations with negative symptom ratings or with seven of nine cognitive measures. The relationship between the number of Na+ peaks and PANSS hallucinations is shown in Fig. 5B (unadjusted P = 0.0033, coefficient 0.77). In striking contrast, sEPSC amplitude showed no correlation with positive symptoms but was positively correlated with ratings for multiple negative symptoms, and negatively correlated with factor 4 processing speed, factor 5 card sorting (WCST), and IQ. Fig. 5C shows the positive correlation between sEPSC amplitude and PANSS emotional withdrawal (unadjusted P = 0.0071, coefficient 0.73), and Fig. 5D shows the negative correlation (unadjusted P = 2.94E-05, coefficient −0.93) between sEPSC amplitude and performance on the WCST. Note that these results appear independent of PRS, which is uncorrelated with sEPSC amplitude and weakly negatively correlated with the number of Na+ peaks (Fig. 5A). This lack of correlation is not surprising given the compressed range of variation of PRS in patients. Results for SCZ and CON, including the adjusted P values, for seven electrophysiological measures and all clinical and cognitive phenotypes tested are shown in Datasets S3 and S4. Linear regression results for CON and SCZ together are shown in Dataset S5. Most notable is the association between sEPSC amplitude and WCST in the combined SCZ+CON sample, with a highly significant main effect across groups (P = 7.75E-08, adjusted R2 = 0.8947) and interaction of sEPSC amplitude with diagnostic group (P = 1.23E-08). Given the small sample sizes, and the fact that subject selection and generation of cellular measures were blind to patient phenotypes, the strength and consistent linearity of some of the observed associations, and their specificity to SCZ, is noteworthy.
Fig. 5.
Association between two electrophysiological phenotypes and selected clinical and cognitive phenotypes. (A) Correlation results in 13 SCZ. Shown are all eight cognitive phenotypes tested, along with the clinical phenotypes with unadjusted P < 0.05 with either electrophysiological measure. ***Adjusted P = 0.00014. Results for SCZ and CON, including the adjusted P values, for seven electrophysiological measures and all clinical and cognitive phenotypes tested are shown in Datasets S3 and S4. (WAIS denotes Weschler Adult Intelligence Scale). (B) Relationships between the number of Na+ peaks and a positive symptom, hallucination (CON, n = 12, P, not calculated; SCZ, n = 12, R = 0.77, P = 0.0033). (C) Relationships between sEPSC amplitude and a negative symptom, emotional withdrawal (CON, n = 12, R = −0.034, P = 0.92; SCZ, n = 12, R = 0.73, P = 0.0071). (D) Relationships between sEPSC amplitude and WCST performance (CON, n = 15, R = 0.16, P = 0.58; SCZ, n = 11, R = −0.93, P = 2.94E-05).
Discussion
hiPSC-derived cellular models have revolutionized the experimental biology of human disease. In recent years, an increasing number of studies of a variety of disorders with pathognomonic cellular pathology have used hiPSCs to elucidate etiologic and pathogenic mechanisms (33–35). In the case of psychiatric disorders, however, the challenge to model illness at the cellular and circuit level is particularly difficult because there are no well-established pathological cellular characteristics, no evidence of autophagy or apoptosis, and no inclusion bodies or even consistent molecular associations. Moreover, no prior study has demonstrated a robust association between an hiPSC-derived neuronal measure and clinically relevant features of the psychiatric disorder being modeled.
In the present report, in an effort to bridge the gap between experimental measures and clinically meaningful characteristics of the individual donors, we have taken a classic translational approach, exploring a diverse landscape of physiological measures in hiPSC-derived neurons from individuals with SCZ who have been deeply characterized clinically. To maximize the biological distance between groups, we used genomic information to select individuals in the SCZ and neurotypical groups such that their PRS distributions did not overlap. We report several illness-associated physiological phenotypes in hiPSC-derived neurons that predict cardinal clinical manifestations of illness. Further, these associations aggregate in patterns that are consistent with clinical experience and well-supported in clinical research.
We observed no significant, reproducible differences between SCZ and CON lines in many important cellular phenotypes including the number and type of neurons produced and network activity as measured by Ca2+ imaging. However, multiple electrophysiological measures did show differences by diagnosis, which when taken together suggest that SCZ neurons display altered Na+ channel function. These phenotypes included increased membrane resistance, decreased Na+ current threshold, increased frequency of Na+ currents generated in response to a voltage ramp, hyperpolarized shifts in the activation and inactivation kinetics of Na+ currents, and reduced LTI, collectively indicating Na+ channel function is altered in hiPSC-derived cortical neurons from individuals with SCZ compared with CON. These physiological phenotypes are consistent with several previous transcriptomic studies from postmortem human brains, where DEGs were enriched in pathways associated with ion channel function (5, 36–38); however, none of our 68 differentially expressed genes were ion channel subunits.
Na+ channel LTI is known to regulate AP spike timing (26) and this relationship was reflected in our data, as SCZ neurons showed less LTI and progressively reduced AP-ISIs (Fig. 4). An analogous AP phenotype is reported in two studies characterizing hiPSC-derived neurons from monozygotic twins discordant for SCZ and from patients with 22q11.2 deletion syndrome (39, 40); however, in the case of 22q11.2, the increased excitability is associated with abnormal Ca2+ channel inactivation. Our observed differences in Na+ channel function and AP-ISI match well with a prevalent hypothesis for the etiology of SCZ which suggests SCZ is due to altered connectivity and plasticity during the early assembly of relevant neural circuits (41). Moreover, these phenotypes may be related to the clinical phenomenology of SCZ which is possibly associated with altered neuronal excitability (42). Although these phenotypes implicate Na+ channel dysfunction, whether the observed effects reflect differential α- and β-subunit expression and/or differential channel modulation via intracellular signaling pathways remains to be determined.
In addition to changes in Na+ channel function and AP spiking, we observed the frequency of GABA transmission (sIPSC) was increased in neurons from SCZ individuals while glutamatergic transmission remained unchanged (sEPSC), suggesting alteration in synaptic excitatory–inhibitory balance. Given that the fraction of GABAergic cells, expression of chloride transporters, and frequency of network activity did not differ by diagnosis, we hypothesize that this phenotype may reflect an increase in the number of inhibitory GABAergic synapses or an alteration in the probability of transmitter release. These findings are consistent with a previous SCZ hiPSC study showing that neuronal DEGs were associated with an up-regulation of GABAergic synapses (18). In particular, Tiihonen and colleagues highlighted CHL1 and CNTN4 as two prominent DEGs related to both SCZ risk and GABA transmission (18, 43). In our study, we similarly observed up-regulation of CHL1 (logFC = 2.93, P = 0.001, adjusted P = 0.239) and CNTN4 (logFC = 1.70, P = 0.013, adjusted P = 0.548). Although these two genes did not survive statistical adjustment, their directionality was consistent with the prior study, and their effect sizes ranked in the top 20 of all genes we measured in neurons.
In addition to the physiological phenotypes, we identified 68 DEGs, many of which were previously identified in other studies using hiPSCs. We observed dysregulation of two protocadherins (PCDHA5 up, PCDHA6 down), members of a family of genes previously associated with SCZ (43) and shown to be disrupted in cortical interneurons derived from SCZ patients (16). Knockout of the entire Pcdha locus in mice reduced the number of inhibitory synapses (16), and hence up-regulation of PCDHA5 in SCZ may contribute to the observed increase in sIPSC frequency. Gene set enrichment analyses suggested involvement of pathways related to WNT signaling and forebrain development. Dysregulation of both of these pathways has been consistently observed in previous studies of SCZ using hiPSCs (16, 44–46). In addition, we found that NPC DEGs correlated with neuronal DEGs, which is also consistent with previous findings (10). Together, these results provide further support for an early neurodevelopmental component to SCZ (41), and for WNT signaling and protocadherin biology as candidate pathways contributing to SCZ etiology.
A central advantage of this study was the unique use of legacy data where we conducted extensive and rigorous clinical characterization and tests of cognitive performance (27, 29). The exploratory study design reported here allowed us to test associations of cellular phenotypes in patient cell lines with the behavioral and cognitive phenotypes of the actual donors. Due to practical limitations, the sample sizes were small and, because there were no similar prior studies, we could not reasonably estimate expected effect sizes or error rates. Based on the correlation structure of the variables, we adjusted the P values for the effective number of tests performed. After this adjustment (and similarly, after Bonferroni correction), only three associations remained with P < 0.05: sIPSC frequency with diagnosis (adjusted P = 3.86E-05; Dataset S2), Na+ inactivation with diagnosis (adjusted P < 0.0035; Fig. 3F), and sEPSC amplitude with Wisconsin Card Sort Test (WCST) performance in SCZ (adjusted P = 0.0001; Dataset S3), with the latter showing a remarkably clear, almost monotonic, relationship. Nevertheless, it is the broader pattern of association among cognitive, positive, and negative symptom variables (including those that did not “survive correction”) that is noteworthy and merits further investigation. Importantly, the secondary analyses and the clustering of phenotypes pointing to Na+ channel kinetics strengthen our interpretation of the findings.
The number of Na+ peaks showed associations with two (correlated) canonical positive symptoms of SCZ, hallucinations and delusions, but no association with negative symptomatology and little with cognitive performance. In contrast, sEPSC amplitude showed a broad association with negative symptoms and cognitive dysfunction, but no connection to positive symptoms. This divergence recalls a substantial body of work implicating relatively consistent clinical subgroupings of individuals with SCZ (including positive vs. negative/cognitive) and distinct biological underpinnings for these subgroups (47).
The study design also included elements designed to monitor and reduce unwanted variability and artifacts. By comparing high-PRS SCZ and low-PRS CON, we aimed to magnify cellular and network phenotypes that are driven by genomic risk for SCZ, since these phenotypes are specifically what we seek to disentangle at the mechanistic level. Subjects were males of European ancestry, chosen on PRS and general availability of genomic and phenotypic data, but there were no selection criteria applied to stratify by specific clinical or cognitive features. All experiments were conducted blind to diagnosis and cell lines were handled in random SCZ–CON pairs. To minimize batch effects, we conducted at least four rounds of differentiation, with two experimenters in rotation. These combined elements were designed to reduce experimental bias and thus increase confidence in the measurements obtained. Nevertheless, due to the small sample sizes and the number of tests performed, some of the associations observed may be spurious, so replication studies with larger samples are necessary to confirm and extend these findings.
In summary, these data provide putative associations in SCZ between specific electrophysiological measures in human neurons and cardinal clinical and cognitive phenotypes of the adult donors from whom the cells originated. Integration of insights from a wider range of data types may provide the synergy necessary for elucidation of biological risk mechanisms, as well as the identification of pathological molecular and cellular pathways and biomarkers as leads toward personalized medicine.
Methods
Subjects and Fibroblast Sampling.
Subjects were males of European ancestry from the Sibling Study of Schizophrenia at the National Institute of Mental Health in the Clinical Brain Disorders Branch (NIMH, Protocol 95-M-0150, NCT00001486, Annual Report no. ZIA MH002942053, D.R.W., PI) with additional support from the Clinical Translational Neuroscience Branch, NIMH (K.F.B., PI) (27), chosen solely on their PRS and the availability of clinical and cognitive data (29). All subjects were extensively screened by obtaining medical, psychiatric, and neurological histories, physical examinations, MRI scans, and genome-wide genotyping to rule out diagnosable clinical disorders. Following informed consent, skin fibroblasts were cultured from biopsies taken from the mesial aspect of the upper arm using a 3-mm-diameter punch under lidocaine intradermal anesthesia. The protocol for this study was reviewed and approved by the NIMH Institutional Review Board.
Genotyping and PRS Calculation.
Subjects were genotyped and genotypes were imputed as described (48). PRSs are a sample-dependent estimate of an individual’s genomic risk for SCZ. PRSs were calculated based on the most commonly used “PGC2” Psychiatric Genomics Consortium GWAS for SCZ (49). We obtained odds ratios of 102,217 index single-nucleotide polymorphisms (SNPs) from a meta-analysis of PGC2 GWASs using datasets excluding our contributed sample. These 102,217 SNPs are linkage disequilibrium (LD)-independent (R2 < 0.1) and span the genome. We then calculated the PRS, a weighted sum of risk alleles, by summing the imputation probability for the reference allele of the index SNP, weighted by the natural log of the odds ratio of association, as described (49). Ten PRSs (PRS1 to PRS10) were calculated using SNP subsets under different thresholds of the PGC2 GWAS P values of association with SCZ: 5E-08, 1E-06, 1E-04, 0.001, 0.01, 0.05, 0.1, 0.2, 0.5, and 1. SNPs in sets with lesser P values are contained in sets with greater P values (SNPs in PRS1 are included in PRS2, SNPs in PRS1 and PRS2 are included in PRS3, etc.). We chose fibroblast lines to reprogram based on PRS6, which was constructed using 24,670 SNPs with GWAS association P values less than 0.05, since this threshold had the greatest prediction accuracy for diagnostic status in multiple independent samples (49).
Reprogramming of Fibroblasts into hiPSCs.
Early-passage fibroblasts (less than passage 5) were thawed and cultured in cDMEM media (Dulbecco’s modified Eagle’s medium [DMEM], 10% fetal bovine serum [FBS], 1% nonessential amino acids [NEAA], beta-mercaptoethanol (b-ME), and 1% penicillin/streptomycin). According to the manufacturer’s protocol (50), all cell lines were reprogrammed using a CytoTune-iPS 2.0 Sendai Reprogramming Kit (A16517; Thermo Fisher Scientific). Clones obtained were cultured onto irradiated mouse embryonic fibroblast (ir-MEF) feeder layers in hESC media (DMEM/F-12, 20% knockout serum replacement, 1% NEAA, and b-ME) supplemented with FGF2 (10 ng/mL; 100-18B; PeproTech) with daily medium changes. iPS cells were passaged once a week (∼85 to 90% confluent wells) using collagenase, type IV (1 mg/mL; 17104019; Gibco). All iPS clones were banked between passage 9 and passage 11 following reprogramming. iPS clones were expanded and banked between passage 13 and passage 18 for all differentiation experiments.
Validation of hiPSC Pluripotency.
For stem cell quality control, immunocytochemistry of pluripotency markers (NANOG, SOX2, OCT3/4, and TRA-1-60), qPCR for pluripotency markers (NANOG, OCT3/4, SOX2, KLF4, and LIN28), karyotyping (G banding), Sendai virus staining, and PCR and alkaline phosphatase (APh) staining were performed on all iPS cell lines used in the study. For pluripotency marker staining, iPS cells were fixed with 4% paraformaldehyde for 15 min. Permeabilization and blocking were performed simultaneously with 0.3% Triton X-100 and 10% donkey serum (blocking buffer) by incubating for at least 45 min at room temperature. Primary antibodies against NANOG, SOX2, OCT3/4, and TRA-1-60 were diluted in blocking buffer and incubated overnight at 4 °C. After three 5-min Dulbecco’s phosphate-buffered saline (DPBS) washes, cells were incubated with corresponding fluorescence-conjugated secondary antibodies for 2 h at room temperature. Three 5-min DPBS washes were performed and cells were counterstained with DAPI. All imaging was performed at 10× on an Operetta (see below). For qPCR, total RNA was extracted from iPS cells using phenol:chloroform isolation with TRIzol followed by purification using the RNeasy Micro Prep Kit (Qiagen). Subsequently, complementary DNA (cDNA) was prepared using the High-Capacity cDNA Reverse Transcription Kit (4387406; Thermo Fisher Scientific). For karyotyping analysis, cells were grown on 25-cm2 tissue-culture flasks and karyotyping was performed at WiCell (a chromosome analysis service). Only iPSC lines with normal karyotype were selected for the study. For APh staining, cells were plated on 12-well plates. Live APh staining (A14353; Thermo Fisher Scientific) was performed and APh activity was imaged using a standard fluorescein isothiocyanate filter. Sendai virus staining (PD029; MBL) was performed to confirm clearance of Sendai virus in the iPS clones.
Differentiation into Cortical Neurons.
iPS cells were differentiated into cortical neurons as previously described (12) with modifications. Briefly, iPS colonies were detached from the ir-MEF feeder layer by incubating the cells with collagenase, type IV (1 mg/mL; Gibco) for 1 h at 37 °C. The detached colonies were cultured in suspension in ultra-low-attachment 10-cm culture dishes (Corning) in FGF2-free hESC media supplemented with dorsomorphin (2 μM) and A-83 (2 μM) (here on referred to as embryoid body [EB] media) for 4 d with daily medium changes. On day 5, EB media were replaced with neural induction (hNPC) media consisting of DMEM/F-12, N2 supplement, and NEAA supplemented with heparin (2 mg/mL) and cyclopamine (2 μM). On day 7, EBs growing in suspension were then transferred onto Matrigel-coated 10-cm dishes to form neural tube–like rosettes. The attached rosettes were cultured for 10 d with medium changes every other day. After 10 d, rosettes were mechanically picked using hand-pulled glass Pasteur pipettes and transferred to ultra-low-attachment 6-well plates. Picked rosettes were cultured for an additional 3 d in hNPC medium containing B27 and penicillin/streptomycin, forming neural spheres. For neuronal differentiation, neural spheres were dissociated with Accutase (Gibco) at 37 °C for 10 min. Cells were passed through a 40-μm-mesh filter to obtain a single-cell suspension of NPCs. NPCs were then plated onto plates containing a confluent layer of rat primary astrocytes (see below). Neuronal cultures were treated with cytosine arabinoside treatment (AraC; 4 μM) after 1 wk in culture (DIV7) for 7 d to DIV14. Neuronal cultures were maintained for up to 10 wk (DIV70) with partial (1/2) medium changes twice a week with neuronal medium (Neurobasal media, 1% GlutaMAX, B27 supplemented with ascorbic acid [200 nM], cyclic adenosine monophosphate (cAMP) (1 μM), brain-derived neurotrophic factor (BDNF) (10 mg/mL), and glial cell-derived neurotrophic factor (GDNF) [10 mg/mL]). Neurons were plated on 24- or 96-well ibidi plates for subsequent live imaging or high-content imaging, on glass coverslips for electrophysiology or live imaging, or standard 24-well plates for biochemical assessments.
Primary Rat Astrocytes.
Primary astrocytes were prepared as previously described with modifications (51, 52). Cortices from a litter of 2- to 3-d-old pups were isolated. After dissection of the tissue and removal of the meninges, the cortices were triturated with a 1-mL pipette 10 times to break down the tissue into smaller pieces. The tissue was then incubated with 300 µL 2.5% trypsin-ethylenediaminetetraacetate (trypsin-EDTA) (Gibco) and 100 µL DNase (100 μg/mL; Sigma) on a shaker at 150 rpm at 37 °C for 30 to 45 min. Subsequently, the cells were passed through a 40-μm-mesh filter, and transferred in glial media (MEM, 5% FBS, glucose [20 mM], sodium bicarbonate [0.2 mg/mL], l-glutamine [2.5 mM], and 1% penicillin/streptomycin) to 75-cm2 tissue-culture flasks (five or six cortices per flask). After 24 h, the flasks were vigorously tapped and shaken to remove any loosely attached cells. The cells were cultured for 6 to 7 d with medium changes every other day. After 7 d, flasks were 80 to 85% confluent and were passaged onto poly-d-lysine/laminin–coated 24-well (seeding density 70,000 cells per well) or 96-well plates (seeding density 20,000 cells per well). Plated astrocytes were mitotically arrested via AraC (20 μM) 24 h after plating, prior to adding human NPCs.
Immunocytochemistry.
For neuronal staining, samples were fixed with ice-cold 4% paraformaldehyde for 10 min. Permeabilization and blocking were performed simultaneously with 3 to 5% normal goat serum. For neuronal identity markers (i.e., GAD67 and TH), 0.3% dimethyl sulfoxide was added. For cortical layer marker labeling (i.e., CTIP2, TBR1, FOXP2, SATB2, CUX1, and BRN2), an antigen-retrieval step was added by including a 60-min incubation in sodium citrate buffer at 80 °C, and the permeabilization buffer included 0.1% Triton X-100. Primary antibodies directed against NeuN, CTIP2, TBR1, FOXP2, SATB2, CUX1, and BRN2 were prepared in blocking buffer and incubated with the sample at 4 °C overnight. Fluorescently conjugated secondary antibodies were prepared in blocking buffer and incubated with the sample at room temperature for 90 min. Samples were counterstained with DAPI and stored in PBS until imaging.
High-Content Imaging and Analysis.
For neuronal identity markers, at least 25 randomly located images per well were acquired with a 20×/0.6 numerical aperture (NA) objective on an Operetta (PerkinElmer) and processed using custom code in Columbus (PerkinElmer). DAPI segmentation was used to identify cellular nuclei, and NeuN segmentation was used to identify regions of interest (ROIs) as neurons. NeuN immunoreactivity was not restricted to the nucleus during the neuronal subtype immunostaining, and thus both cytosolic and nuclear NeuN were used to demarcate neurons for those experiments. In experiments in which antigen retrieval was performed, NeuN segmentation in the nuclear compartment was used to identify NeuN+ ROIs. Neuronal subtype markers (GAD67 and TH) and cortical layer markers (BRN2, CTIP2, CUX1, FOXP2, SATB2, and TBR1) were identified based on colocalization with DAPI+NEUN+ ROIs. Quantification was performed on proportions of neurons displaying staining for each marker. Analysis was performed on final cell counts in R using full linear models (for neuron identity markers) or linear mixed-effects models (for NeuN).
RNA Isolation and qPCR.
Total RNA was isolated as described above, quantified using a NanoDrop spectrophotometer, and 100 ng was reverse-transcribed using SuperScript III. For neuron quality control, qPCR was performed using the Fluidigm Biomark System with TaqMan probes according to the manufacturer’s instructions. We utilized human-specific TaqMan probes that would not bind to rat sequences to validate gene expression in the human transcriptome, and rat-specific TaqMan probes that would not bind to the human sequence to validate gene expression in the rat transcriptome, and verified that these probes performed as expected by using controls from each species independently. Briefly, cDNA was amplified for 14 cycles of PCR using a combination of all included probes, and then the preamplified product was assayed in duplicate for each of 90 genes of interest, three housekeeping probes, and negative controls. The mean of the Ct values for the two technical replicates was computed and normalized against the mean of the Ct values for all three housekeeping genes.
RNA-Seq Data Generation and Processing.
Illumina RNA-seq libraries were constructed with Ribo-Zero Gold Kits across three batches and included External RNA Control Consortium (ERCC) spike-in sequences. The first batch—which only included hiPSC and NPC samples—had libraries prepared and sequenced at the Sequencing Core at the Lieber Institute for Brain Development on an Illumina HiSeq 3000. The second and third batches—which included all neuronal samples and five repeated NPC samples for comparability—were prepared and sequenced at Psomagen (formerly Macrogen) on an Illumina HiSeq 4000. A total of 304 samples were sequenced with paired-end 100-bp reads (2 × 100). Raw sequencing reads from all samples were aligned to a custom concatenated Gencode hg38 + rn6 reference genome using HISAT2 2.0.4 (53) (including NPC samples without any rodent RNA, for comparability with neuronal samples). Genes and exons were quantified with featureCounts v1.5.0-p3 (54) to a custom concatenated hg38+rn6 gtf file within each sample. ERCC spike-in sequences were quantified with kallisto (55), and we further calculated a bias factor for each sample using the observed versus expected abundances [∑((obs − exp)^2)]. A barcode of 738 exonic/coding SNPs were genotyped using the RNA-seq data to confirm sample identities using SAMtools mpileup (56). We used a previous RNA deconvolution method developed by our group to estimate the RNA fractions for 10 different cell classes in each sample from gene expression levels (9).
After processing, we performed quality control checks of samples for RNA quality, sequencing quality, and sample identities. We examined the distributions of read alignment and gene assignment rates to both human and rodent genomes, as well as the ERCC spike-in accuracy rates (sum of squared deviations from expected concentrations). We confirmed the sample identities within and across subclonal lines using the RNA-derived genotype barcodes (for donor confirmation) and RNA fractions (for time-point confirmation).
RNA-Seq Data Analysis.
After quality control checks, we performed a series of analyses on different subsets of samples.
-
1)
All samples (n = 304) for time-course/developmental changes.
-
2)
Neuronal samples (n = 94) for case–control modeling, including 51 CON samples (from 18 subclonal lines from 14 genomes) and 43 SCZ samples (from 14 lines from 12 genomes) across 23,119 expressed genes (based on subset-specific reads per kilobase million [RPKM]>0.2).
-
3)
NPC samples (n = 143) for case–control modeling, including 75 CON (from 17 subclones from 13 genomes) versus 68 SCZ samples (from 16 subclones from 14 genomes) across 23,974 expressed genes (based on subset-specific RPKM >0.2).
PCA of normalized gene expression levels (log2[RPKM + 1]) was used for exploratory data analysis within each subset of samples. The top component in each subset of samples (PC1) related to underlying RNA composition (which reflected developmental changes in the full dataset) and we therefore constructed the top PC of these 10 RNA fractions to capture their correlated effects (“cellPC”; SI Appendix, Fig. S4). We used linear mixed-effects modeling, treating each subclone line as a random intercept, using the limma voom approach (22). In neuronal samples, we specifically modeled trimmed mean of M-values (TMM)-normalized expression as a function of diagnosis, adjusting for main effects of ERCC bias (quantitative), cellPC (quantitative), DIV (binary, 8 or 10 wk), the human alignment fraction (quantitative), human gene assignment rate, and random intercept of the subclone line. In NPC samples, we used a related statistical model for diagnosis that adjusted for DIV, cellPC, chrM mapping rate (quantitative), human gene assignment rate, ERCC bias, and sequencing batch, with subclone again modeled as a random intercept. For both analyses, we calculated log2FCs for the SCZ term (relative to CON, such that positive logFCs indicated increased expression in patients), which were used to calculate empirical Bayes t statistics and corresponding P values [limma]. P values were adjusted for multiple testing using the Benjamini–Hochberg (B-H) procedure to control for an FDR of 0.1. Gene set enrichment analyses were performed with the clusterProfiler Bioconductor package (57), using the hypergeometric test to assess enrichment of marginally significant DEGs (at P < 0.005) that were stratified by case–control directionality, against a background of all expressed genes that had Entrez gene IDs. We again used the B-H FDR to control for multiple testing of gene sets.
Analysis of Association between Electrophysiological Measures and Diagnosis.
We performed linear regression analyses between 47 electrophysiology measures and SCZ diagnosis. The complete results are shown in Dataset S2. We adjusted for the researcher who differentiated the neurons (two people/levels/groups), the researcher who recorded from the neurons (three people/levels/groups), and the developmental time point of the neurons (binarized into early or late, two groups). We opted not to adjust for the donor/cell line of each neuron due to the design matrix being singular/collinear, which resulted in directional instabilities of the SCZ effect (e.g., different factor orderings of the Line term resulted in different directionalities of the SCZ effect on the ePhys measurements). Secondary analyses utilized Tukey post hoc tests to ensure directional/model consistency, as well as time point–specific sensitivity analyses to ensure results were not driven by a single time point. As many of the 47 variables were correlated, to calculate P values adjusted for the effective number of tests, we applied the Meff correction factor (58, 59), which was 34.91.
Analysis of Association between Electrophysiological Measures and Clinical and Cognitive Data.
In order to produce a single (genome-based) value for each subject for the seven electrophysiological phenotypes, we took the mean values across both 8- (DIV56) and 10-wk (DIV70) time points and across lines. Pearson correlations tested linear associations with clinical and cognitive variables separately for 13 SCZ and 15 CON. The complete results are shown in Datasets S3 (SCZ) and S4 (CON). These seven electrophysiological variables, themselves, were largely uncorrelated. The clinical (PANSS) and cognitive variables are each correlated, so we applied the Meff correction: For the 30 PANSS phenotypes, the correction factor was 24.24, and for the 8 cognitive variables, the correction factor was 4.88. The linear regression P values shown in Dataset S5 are uncorrected.
Virus Infection and Calcium Imaging and Analysis.
Neurons were infected at DIV23 with AAV1-hSyn1-mRuby2-P2A-GCaMP6s, a gift from Tobias Bonhoeffer, Mark Huebener, and Tobias Rose (Addgene viral prep 50942-AAV1; http://n2t.net/addgene:50942; RRID: Addgene_50942) (60). Imaging was performed in culture media at DIV42 and DIV63 on a Zeiss LSM 780 equipped with a 10×/0.45 NA objective and a temperature- and atmospheric-controlled enclosure. Analysis was performed as previously described (61). A reference image was acquired for each field of mRuby fluorescence, and then a time series was acquired at 4 Hz for 8 min. ROIs were identified from the red reference image, and fluorescence intensity over time was computed for each ROI and then normalized to ΔF/F. Peaks were identified and the number of events was calculated for each ROI. The synchronicity of these events was correlated on a per-image basis. In some cases, pharmacological inhibitors were then added to block glutamatergic synaptic transmission by application of dl-2-amino-5-phosphonopentanoic acid (dl-AP5) (100 μM) and 6,7-dinitroquinoxaline-2,3-dione (DNQX) (10 μM).
Whole-Cell Patch-Clamp Recordings.
Whole-cell electrophysiology recordings were performed using the following solutions. Extracellular recording buffer contained 128 mM NaCl, 30 mM glucose, 25 mM Hepes, 5 mM KCl, 2 mM CaCl2, and 1 mM MgCl2 (pH 7.3). Patch pipettes were fabricated from borosilicate glass (N51A; King Precision Glass) to a resistance of 2 to 5 MΩ. For the majority of current- and voltage-clamp measurements, pipettes were filled with 135 mM K-gluconate, 10 mM Tris-phosphocreatine, 10 mM Hepes, 5 mM ethylene glycol tetraacetic acid (EGTA), 4 mM adenosine 5′-triphosphate magnesium salt (MgATP), and 0.5 mM guanosine 5′-triphosphate sodium salt hydrate (Na2GTP) (pH 7.3). For Na+ channel kinetics experiments (Figs. 3 and 4 A–C), pipettes were filled with 10 mM NaCl, 140 mM CsF, 10 mM Hepes, 1 mM EGTA, 15 mM D-glucose, and 10 mM TEA-Cl (pH 7.3). Extracellular recording buffer consisted of 40 mM NaCl, 75 mM choline-Cl, 1 mM CaCl2, 1 mM MgCl2, 10 mM Hepes, 20 mM tetraethylammonium chloride (TEA-Cl), 4 mM 4-aminopyridine (4-AP), 0.1 mM CdCl2, and 10 mM D-glucose (pH 7.4). Current signals were recorded with either an Axopatch 200B (Molecular Devices) or a MultiClamp 700A amplifier (Molecular Devices) and were filtered at 2 kHz using a built-in Bessel filter and digitized at 10 kHz. Voltage signals were filtered at 2 kHz and digitized at 10 kHz. Data were acquired using AxoGraph on a Dell PC (Windows 7). For voltage-clamp recordings, cells were held at −70 mV for sodium/potassium currents and sEPSCs, and at 0 mV for sIPSCs. After recording a ramp protocol (−110 to 110 mV, 0.22 mV/s) for each neuron in voltage clamp, the recording configuration was switched to current clamp and a series of APs (20 episodes) was elicited at 0.5 Hz with a square current injection that was increased by 10 pA every episode. AP onset was defined when the slope exceeded 10 mV/ms. AP threshold and onset time were subsequently identified by AxoGraph. Baseline was set at onset time (i.e., threshold) to calculate AP peak amplitude. Width was calculated at 50% of peak amplitude. Calculation of dV/dt maximum and minimum was set as the maximum slope for the ascending and descending phases with three regression points per maximum slope. For LTI experiments, neurons were voltage-clamped at −90 mV and four consecutive voltage steps to −30 mV were applied for 16 ms with a 40-ms interpulse interval. AP-ISI was calculated from the maximum number of APs (threshold of 4 mV/ms) for neurons producing three or more APs.
Supplementary Material
Acknowledgments
We are grateful for the vision and generosity of the Lieber and Maltz families, who made this work possible. This project was supported by the Lieber Institute for Brain Development. The collection of the clinical and cognitive data and fibroblasts was supported by direct funding from the Intramural Research Program (IRP) of the NIMH to the Clinical Brain Disorders Branch (D.R.W., PI, Protocol 95-M-0150, NCT00001486, Annual Report no. ZIA MH002942053) with supplemental support from the Clinical and Translational Neuroscience Branch (K.F.B., PI). We thank all of the participants in the IRP study and their families. We thank Flora Vaccarino for helpful advice and review of the manuscript, and Pat Levitt for his insights about the data.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109395119/-/DCSupplemental.
Data Availability
All raw and processed sequencing data generated in this study are available from the National Center for Biotechnology Information Sequence Read Archive (https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP347253) (62). A web-based browser of the RNA expression time course of individual genes is available at https://stemcell.libd.org/schizophrenia/RNAseq/dataset001/ (63).
All study data are included in the article and/or supporting information.
References
- 1.Jin H., Mosweu I., The societal cost of schizophrenia: A systematic review. Pharmacoeconomics 35, 25–42 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Remington G., et al. , Treating negative symptoms in schizophrenia: An update. Curr. Treat. Options Psychiatry 3, 133–150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mallet J., Le Strat Y., Dubertret C., Gorwood P., Polygenic risk scores shed light on the relationship between schizophrenia and cognitive functioning: Review and meta-analysis. J. Clin. Med. 9, 341 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinberger D. R., Implications of normal brain development for the pathogenesis of schizophrenia. Arch. Gen. Psychiatry 44, 660–669 (1987). [DOI] [PubMed] [Google Scholar]
- 5.Schizophrenia Working Group of the Psychiatric Genomics Consortium, Ripke S., Walters J. T. R., O’Donovan M. C., Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. medRxiv [Preprint] (2020). 10.1101/2020.09.12.20192922 (Accessed 15 May 2021). [DOI]
- 6.Wray N. R., et al. , From basic science to clinical application of polygenic risk scores: A primer. JAMA Psychiatry 78, 101–109 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Janssens A. C. J. W., Validity of polygenic risk scores: Are we measuring what we think we are? Hum. Mol. Genet. 28, R143–R150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powell S. K., O’Shea C. P., Shannon S. R., Akbarian S., Brennand K. J., Investigation of schizophrenia with human induced pluripotent stem cells. Adv. Neurobiol. 25, 155–206 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burke E. E., et al. , Dissecting transcriptomic signatures of neuronal differentiation and maturation using iPSCs. Nat. Commun. 11, 462 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman G. E., et al. , Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat. Commun. 8, 2225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Topol A., et al. , Dysregulation of miRNA-9 in a subset of schizophrenia patient-derived neural progenitor cells. Cell Rep. 15, 1024–1036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen Z., et al. , Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 515, 414–418 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin M., et al. , Integrative transcriptome network analysis of iPSC-derived neurons from schizophrenia and schizoaffective disorder patients with 22q11.2 deletion. BMC Syst. Biol. 10, 105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon K.-J., et al. , Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell 15, 79–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brennand K., et al. , Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol. Psychiatry 20, 361–368 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao Z., et al. , Dysregulated protocadherin-pathway activity as an intrinsic defect in induced pluripotent stem cell-derived cortical interneurons from subjects with schizophrenia. Nat. Neurosci. 22, 229–242 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni P., et al. , iPSC-derived homogeneous populations of developing schizophrenia cortical interneurons have compromised mitochondrial function. Mol. Psychiatry 25, 2873–2888 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiihonen J., et al. , Sex-specific transcriptional and proteomic signatures in schizophrenia. Nat. Commun. 10, 3933 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sripathy S. R., et al. , Generation of 10 patient-specific induced pluripotent stem cells (iPSCs) to model Pitt-Hopkins syndrome. Stem Cell Res. (Amst.) 48, 102001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srikanth P., et al. , Genomic DISC1 disruption in hiPSCs alters Wnt signaling and neural cell fate. Cell Rep. 12, 1414–1429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mertens J., Marchetto M. C., Bardy C., Gage F. H., Evaluating cell reprogramming, differentiation and conversion technologies in neuroscience. Nat. Rev. Neurosci. 17, 424–437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Law C. W., Chen Y., Shi W., Smyth G. K., voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collado-Torres L., et al. , BrainSeq Consortium, Regional heterogeneity in gene expression, regulation, and coherence in the frontal cortex and hippocampus across development and schizophrenia. Neuron 103, 203–216.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Estacion M., Waxman S. G., The response of Na(V)1.3 sodium channels to ramp stimuli: Multiple components and mechanisms. J. Neurophysiol. 109, 306–314 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Dover K., Solinas S., D’Angelo E., Goldfarb M., Long-term inactivation particle for voltage-gated sodium channels. J. Physiol. 588, 3695–3711 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navarro M. A., et al. , Sodium channels implement a molecular leaky integrator that detects action potentials and regulates neuronal firing. eLife 9, e54940 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egan M. F., et al. , Relative risk of attention deficits in siblings of patients with schizophrenia. Am. J. Psychiatry 157, 1309–1316 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Callicott J. H., et al. , Abnormal fMRI response of the dorsolateral prefrontal cortex in cognitively intact siblings of patients with schizophrenia. Am. J. Psychiatry 160, 709–719 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Dickinson D., Goldberg T. E., Gold J. M., Elvevåg B., Weinberger D. R., Cognitive factor structure and invariance in people with schizophrenia, their unaffected siblings, and controls. Schizophr. Bull. 37, 1157–1167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickinson D., et al. , Attacking heterogeneity in schizophrenia by deriving clinical subgroups from widely available symptom data. Schizophr. Bull. 44, 101–113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weickert T. W., et al. , Cognitive impairments in patients with schizophrenia displaying preserved and compromised intellect. Arch. Gen. Psychiatry 57, 907–913 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Dickinson D., et al. , Distinct polygenic score profiles in schizophrenia subgroups with different trajectories of cognitive development. Am. J. Psychiatry 177, 298–307 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Temme S. J., Maher B. J., Christian K. M., Using induced pluripotent stem cells to investigate complex genetic psychiatric disorders. Curr. Behav. Neurosci. Rep. 3, 275–284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernando M. B., Ahfeldt T., Brennand K. J., Modeling the complex genetic architectures of brain disease. Nat. Genet. 52, 363–369 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christian K. M., Song H., Ming G.-L., Using two- and three-dimensional human iPSC culture systems to model psychiatric disorders. Adv. Neurobiol. 25, 237–257 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Radulescu E., et al. , Identification and prioritization of gene sets associated with schizophrenia risk by co-expression network analysis in human brain. Mol. Psychiatry 25, 791–804 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Jaffe A. E., et al. , BrainSeq Consortium, Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat. Neurosci. 21, 1117–1125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pers T. H., et al. , Schizophrenia Working Group of the Psychiatric Genomics Consortium, Comprehensive analysis of schizophrenia-associated loci highlights ion channel pathways and biologically plausible candidate causal genes. Hum. Mol. Genet. 25, 1247–1254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toritsuka M., et al. , Developmental dysregulation of excitatory-to-inhibitory GABA-polarity switch may underlie schizophrenia pathology: A monozygotic-twin discordant case analysis in human iPS cell-derived neurons. Neurochem. Int. 150, 105179 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Khan T. A., et al. , Neuronal defects in a human cellular model of 22q11.2 deletion syndrome. Nat. Med. 26, 1888–1898 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinberger D. R., Levitt P., “Neurodevelopmental origins of schizophrenia” in Schizophrenia, Weinberger D. R., Harrison P. J., Eds. (Wiley-Blackwell, 2010), pp. 393–412. [Google Scholar]
- 42.Uhlhaas P. J., Singer W., Abnormal neural oscillations and synchrony in schizophrenia. Nat. Rev. Neurosci. 11, 100–113 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Flaherty E., Maniatis T., The role of clustered protocadherins in neurodevelopment and neuropsychiatric diseases. Curr. Opin. Genet. Dev. 65, 144–150 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brennand K. J., et al. , Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evgrafov O. V., et al. , Gene expression in patient-derived neural progenitors implicates WNT5A signaling in the etiology of schizophrenia. Biol. Psychiatry 88, 236–247 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stertz L., et al. , Convergent genomic and pharmacological evidence of PI3K/GSK3 signaling alterations in neurons from schizophrenia patients. Neuropsychopharmacology 46, 673–682 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Potkin S. G., et al. , A PET study of the pathophysiology of negative symptoms in schizophrenia. Am. J. Psychiatry 159, 227–237 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Chen Q., et al. , Schizophrenia polygenic risk score predicts mnemonic hippocampal activity. Brain 141, 1218–1228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schizophrenia Working Group of the Psychiatric Genomics Consortium, Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fusaki N., Ban H., Nishiyama A., Saeki K., Hasegawa M., Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85, 348–362 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwartz J. P., Wilson D. J., Preparation and characterization of type 1 astrocytes cultured from adult rat cortex, cerebellum, and striatum. Glia 5, 75–80 (1992). [DOI] [PubMed] [Google Scholar]
- 52.McCarthy K. D., de Vellis J., Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 85, 890–902 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim D., Paggi J. M., Park C., Bennett C., Salzberg S. L., Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Li H., et al. , 1000 Genome Project Data Processing Subgroup, The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Derringer J., A simple correction for non-independent tests. PsyArXiv [Preprint] (2018). 10.31234/osf.io/f2tyw (Accessed 15 May 2021). [DOI]
- 59.Nyholt D. R., A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet. 74, 765–769 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rose T., Jaepel J., Hübener M., Bonhoeffer T., Cell-specific restoration of stimulus preference after monocular deprivation in the visual cortex. Science 352, 1319–1322 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Tippani M., et al. , CaPTure: Calcium Peak Toolbox for analysis of in vitro calcium imaging data. bioRxiv [Preprint] (2021). 10.1101/2021.09.08.458611 (Accessed 15 October 2021). [DOI]
- 62.S. C. Page et al., Electrophysiological measures from human iPSC-derived neurons are associated with schizophrenia clinical status and predict individual cognitive performance. NCBI Sequence Read Archive. https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP347253. Deposited 22 November 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.S. C. Page et al., Electrophysiological measures from human iPSC-derived neurons are associated with schizophrenia clinical status and predict individual cognitive performance. Schizophrenia Stem Cell Browser 1. https://stemcell.libd.org/schizophrenia/RNAseq/dataset001/. Deposited 22 November 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed sequencing data generated in this study are available from the National Center for Biotechnology Information Sequence Read Archive (https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP347253) (62). A web-based browser of the RNA expression time course of individual genes is available at https://stemcell.libd.org/schizophrenia/RNAseq/dataset001/ (63).
All study data are included in the article and/or supporting information.