Significance
Incipient species are at an intermediate stage of speciation where reproductive isolation is counteracted by the homogenizing effects of gene flow. Human activity sometimes leads such species to reunite, as seen in the Orange Sulphur butterfly, which forms large hybridizing populations with the Clouded Sulphur in alfalfa fields. Here we show that sex chromosomes maintain these species as distinct, while the rest of their genome is admixed. Sex chromosomes notably determine which males display to females a bright, iridescent UV signal on their wings. Genetic mapping, antibody stainings, and CRISPR knockouts collectively indicate that the gene bric a brac controls whether UV-iridescent nanostructures develop in each species, illustrating how a master switch gene modulates a male courtship signal.
Keywords: evo-devo, speciation, UV iridescence, large-Z effect, genetic coupling
Abstract
Mating cues evolve rapidly and can contribute to species formation and maintenance. However, little is known about how sexual signals diverge and how this variation integrates with other barrier loci to shape the genomic landscape of reproductive isolation. Here, we elucidate the genetic basis of ultraviolet (UV) iridescence, a courtship signal that differentiates the males of Colias eurytheme butterflies from a sister species, allowing females to avoid costly heterospecific matings. Anthropogenic range expansion of the two incipient species established a large zone of secondary contact across the eastern United States with strong signatures of genomic admixtures spanning all autosomes. In contrast, Z chromosomes are highly differentiated between the two species, supporting a disproportionate role of sex chromosomes in speciation known as the large-X (or large-Z) effect. Within this chromosome-wide reproductive barrier, linkage mapping indicates that cis-regulatory variation of bric a brac (bab) underlies the male UV-iridescence polymorphism between the two species. Bab is expressed in all non-UV scales, and butterflies of either species or sex acquire widespread ectopic iridescence following its CRISPR knockout, demonstrating that Bab functions as a suppressor of UV-scale differentiation that potentiates mating cue divergence. These results highlight how a genetic switch can regulate a premating signal and integrate with other reproductive barriers during intermediate phases of speciation.
Premating signals such as pheromones, calls, and displays often differ between sexes and species and, by helping animals to tell one another apart, they are integral to the formation of reproductive barriers during speciation itself (1, 2). Mating factors can diverge early in the speciation process due to local adaptation or later due to sexual selection that prevents the generation of unfit hybrids (3). While a coupling of premating and postmating isolation mechanisms is thought to be required for the completion of speciation (4), how mating cue variation actually coincides with other barrier loci to split lineages remains elusive in the empirical literature (5–7).
Previous work on the genetics of hybridization between the sulphur butterflies Colias eurytheme and Colias philodice highlights their potential for the study of intermediate phases of speciation with gene flow. Initially restricted to the western United States, the range of C. eurytheme expanded following both the spread of agricultural alfalfa and the reduction in forest cover in the past 200 y into regions once limited to C. philodice (8). As a result, the two species occur in secondary sympatry throughout an anthropogenic contact zone that includes the eastern United States and southern Canada. Both pre- and postzygotic reproductive barriers maintain species status in this system. However, heterospecific matings happen at increased frequency in dense populations (9, 10), partly because males can locate newly emerged females incapable of performing mate rejection behaviors [teneral mating (11)]. Hybrid female sterility forms an intrinsic postzygotic barrier that affects one of the two heterospecific crosses: oogenesis fails in female offspring that inherit a C. eurytheme W chromosome and a C. philodice Z chromosome (12, 13). This incompatibility is sex-linked and implies that to produce fully fertile progeny, C. eurytheme females must select males that are homozygous for a conspecific Z chromosome. An iridescent ultraviolet (UV) pattern acts as a visual mating cue in males and accurately displays their Z-chromosome status to females (9, 14) (Fig. 1A and SI Appendix, Fig. S1). UV occurs on the dorsal wing surfaces of C. eurytheme males only. The Mendelian U locus controls this interspecific variation and was previously mapped to the Z chromosome (15): C. eurytheme homozygous recessive males are UV-iridescent (u/u), advertising two compatible Z chromosomes to C. eurytheme females. Incompatible mates such as C. philodice males (U/U) and heterozygous hybrids (U/u) bear the dominant allele and lack UV. Finally, the female preference trait itself is also linked to the Z chromosome (14). This Z-linked inheritance of genetic incompatibility, mating signal, and mating preference supports an “indicator” model of speciation, which was previously theorized as a system where the mating cue can signal species identity and enable selection against hybrids (3, 16). In this study, we examine the genomic footprint of sex-linked reproductive barriers, and fine-map the allelic variation that switches on the male UV signal in C. eurytheme.
Fig. 1.
Large-Z architecture of species differentiation includes the U-locus candidate gene bab. (A) UV iridescence differentiates males from two incipient species. (B and B′) PCA (B) and distance-based phylogenetic network (B′) of 22 male whole-genome SNPs from the admixed Maryland population. (C and D) FST values for C. philodice vs. C. eurytheme plotted against recombination rate (C), and Manhattan plot (D). Red indicates windows with above-median recombination rate and in the 95th percentile of FST, including on the Z chromosome (asterisk). (E) Quantitative trait locus (QTL) analysis of the presence/absence of UV in 252 male offspring from F2 and BCs. (F) Genotype plot for the whole Z chromosome with resequencing data from 23 individuals. Each row is an individual, and each column is a color-coded SNP. The red bracket indicates a 2.5-Mb interval with high FST and above-median recombination rate. (G) Annotation of the U-locus zero-recombinant interval (box) and surrounding region.
Results
The Z Chromosomes Define Species Barriers in Secondary Sympatry.
To test a putative role of the Z chromosome as a barrier locus, we conducted a genome scan on 24 males from a sympatric population in Maryland, USA, where C. eurytheme settled in 1927 (17). Two individuals were identified as probable recent hybrids and excluded from further analyses. We retained 13 UV, orange males and 9 non-UV, yellow males which formed two discrete clusters based on genome-wide single-nucleotide polymorphism (SNP) clustering by principal-component analysis (PCA) (Fig. 1C and SI Appendix, Fig. S2 and Table S1). The Z chromosome showed a large increase in genetic differentiation when compared with autosomes (SI Appendix, Figs. S3 and S4 and Table S2), with a Z:A ratio of 12:1, the highest sex chromosome-to-autosome ratio of FST reported from a whole-genome dataset (7, 18). Heterogeneous landscapes of genomic differentiation can be explained by local barriers to gene flow or by linked selection in regions of low recombination (7, 19). To parse these two phenomena, we highlighted windows that were both in the top 5% FST and had an above-median recombination rate, thereby identifying three regions—two narrow autosomal FST peaks and a 2.5-Mb portion of the Z chromosome (Fig. 1 C and D). These data show that while only a restricted set of autosomal regions is likely under selection in each population, a large fraction of the Z chromosome is refractory to gene flow in a pattern consistent with a causal role in reproductive isolation. In other words, while Z chromosomes can recombine when hybrid F1 males are produced in the laboratory, this rarely persists in the field, presumably due to the aforementioned isolating mechanisms that select against hybrids.
In addition, nucleotide diversity (π) was depressed on the Z chromosome in both populations and divergence (dXY) was elevated, supporting the inference that the Z chromosome is highly differentiated (SI Appendix, Fig. S3 B and C′). In most scenarios, one expects π on the Z chromosome to be 75% of π on the autosomes (20). For C. eurytheme, πZ/πA was 0.751 ± 0.118, matching this expectation (SI Appendix, Fig. S3F). However, for C. philodice, πZ/πA was 0.532 ± 0.105, meaning πZ was lower than expected, which could reflect a recent selective sweep in C. philodice.
The Male Mating Signal Polymorphism Maps to bab.
The large-Z effect results in extended nonrecombining haplotypes in the natural population that prevent association mapping of trait variation (SI Appendix, Fig. S5). To gain further resolution on the genetic basis of the polymorphic UV signal, we turned to linkage mapping from controlled hybrid crosses. F2 and backcross (BC) broods showed Mendelian, recessive segregation of the UV state among male offspring (SI Appendix, Fig. S6). We genotyped 484 recombinant males and females using 2b-RAD sequencing, scored UV among the 252 genotyped males, and identified an LOD (logarithm of the odds) interval on the Z chromosome (Fig. 1E and SI Appendix, Fig. S6). We resequenced individuals with recombination events around the U locus and refined a 352-kb zero-recombinant window with 18 annotated genes (Fig. 1 F and G and SI Appendix, Tables S4 and S5). This mapping interval includes the 5′ intergenic region, promoter, and first exon of the gene bric a brac (bab), a salient candidate gene as it encodes a transcriptional repressor of male-limited traits in Drosophila such as abdominal pigmentation and sex combs (21–27).
Bab Expression Marks Non-UV Scale Cells.
To test a role in the regulation of Colias male UV, we characterized the expression and developmental functions of bab during color scale formation. Butterfly wing scales are macrochaete derivatives that each protrude from a single epidermal cell during pupal development (28). UV scales found in the subfamily Coliadinae are a derived scale type characterized by dense longitudinal ridges; each scale forms a multilayer of chitinous lamellae that selectively reflects UV light by the coherent scattering of incident light (29–32). UV scales are specific to the dorsal wing surface of C. eurytheme males, and cover the top of non-UV ground scales (Fig. 2 A and B). Immunofluorescence at the onset of scale emergence reveals Bab expression in the nuclei of all non–UV scale cell precursors (Fig. 2C, SI Appendix, Fig. S7, and Movie S1) regardless of the scale layer (ground, cover), pigment fate (yellow pterins, black melanin), wing surface, and sex. Remarkably, Bab is expressed in ground scales and is absent in the UV cover scales on the dorsal male wing surface. Thus, Bab is negatively correlated with the UV-scale type in C. eurytheme: it is consistently expressed in all scale cells fated as non-UV except in the wing cover scales of C. eurytheme males. Where it is not expressed, these scales develop layered nanostructures specifically capable of UV-iridescent reflectance. This inactivation in the sexually dichromatic pattern suggests a repressor function, analogous to the expression of Bab in the Drosophila abdominal epithelium (21, 23, 27).
Fig. 2.
Bab negatively correlates with UV-scale precursors in C. eurytheme male wings. (A) Pseudocolored SEM images highlighting the ultrastructural differentiation of the UV-iridescent dorsal cover scales (dcs; magenta), relative to non-UV ground scales (gs; orange) and non-UV ventral cover scales (vcs; green). (B) Microphotographs of adult C. eurytheme male wing surfaces in the visible and UV ranges. Line: damaged areas exposing non-UV ground scales. (C) Immunofluorescence detection of Bab (green) in all UV precursors at 46% pupal development. Magenta: DAPI (nuclei); orange: Dve; circles: cover-scale nuclei. (Scale bars, 2 μm [A], 100 μm [B], and 10 μm [C].)
CRISPR Knockouts Yield Ectopic UV Iridescence.
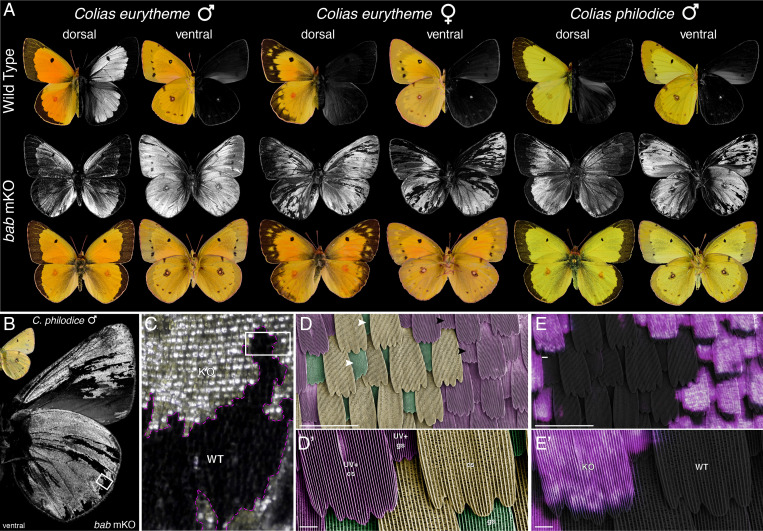
To directly test this model, we generated CRISPR-mediated loss-of-function mutations targeting the first exon of the bab coding sequence. We collected C. eurytheme and C. philodice females and microinjected eggs within 7 h postfertilization. G0 bab crispants showed mosaic phenotypes of high penetrance (51 out of 63 surviving adults), with a widespread gain of UV iridescence in both males and females of both species, including ventral surfaces (Fig. 3 and SI Appendix, Figs. S8–S13). Both pterin and melanin pigment scales and both cover and ground scales differentiated into UV scales following bab knockout (KO). Female-specific effects on pigmentation were also noted (SI Appendix, Fig. S11). These loss-of-function assays show that Bab represses the UV identity in all non-UV scale precursors regardless of wing surface, sex, or species. Male UV iridescence is widespread in the Colias genus (32), suggesting that the C. philodice absence of UV is due to a secondary loss of repression by Bab. The reappearance of UV in C. philodice bab crispants (an atavism) implies that the underlying network for producing UV scales is still present in this species.
Fig. 3.
Bab represses UV-scale fate. (A) G0 phenotypes resulting from CRISPR mosaic KOs (mKO) targeting the first exon of bab. Gain of UV iridescence was observed across both species and both sexes, including ventral wing surfaces. (B and C) Magnified views of male C. philodice wings featuring extensive ectopic iridescence following bab mKO. WT, wild type. (D and D′) False-colored SEM views of a mosaic region (box in C), with complete transformation of wild-type cover (cs; orange) and ground scales (green) into UV-iridescent scales with dense longitudinal ridges. Arrowheads indicate examples of ground scales in a wild-type state (white) and in a bab mutant clone. (E and E′) Correlative light and electron microscopy images featuring a superimposed view of UV-reflected light (magenta) on an SEM view. Both ground and cover scales from bab mutant clones are UV-iridescent. (Scale bars, 10 μm.)
bab Is the Causal Gene for UV-Iridescence Variation in Colias.
Linkage mapping of the U locus led to a zero-recombination interval of 19 genes (Fig. 1 F and G), and the combined evidence suggests that bab is the only gene in this interval that mediates the phenotypic variation. First, the expression pattern of Bab matches the pattern that would be predicted for a repressor of UV scales. This is consistent with the recessivity of the UV-iridescent allele, which would be difficult to explain if the causal gene was an activator of the UV state. Second, mosaic KOs of bab result in ectopic UV iridescence, meaning that no other gene in the genome blocks UV iridescence, and thus ruling out the existence of a second repressor at the U locus. No fixed amino acid variants of Bab were found between the two species (SI Appendix), suggesting that the mating signal difference involves a noncoding difference. Further experiments will be required to identify cis-regulatory elements blocking bab in UV-iridescent cover scales, and how it is derepressed in the alternative haplotype.
Discussion
Bab Is a Repressor of Male Dimorphic Traits.
Our linkage mapping, expression, and functional assays of UV iridescence show that 1) allelic variation of bab causes a male-specific mating signal difference between two incipient species; 2) Bab expression is sexually dimorphic in C. eurytheme; and 3) Bab functions as a repressor of the dimorphic trait. This repression of a male-specific feature is analogous to the expression and function of Bab paralogs in Drosophila sex-comb formation, abdominal pigmentation, and gonadal stem cell niches (21–23, 26, 27, 33, 34), with a male-specific repression in the presumptive cells forming the male feature, and loss of function resulting in gain or expansion of the male state. Bab is thus a major player in the development of sexually dimorphic features, and might have an ancestral function in the repression of male-specific development at the root of butterflies and flies. It will be critical to study the versatility of its functions comparatively in order to better understand how sexual forms evolve. As knockdowns and knockouts are increasingly amenable in new organisms, testing the repressive nature of Bab should be feasible and may yield a wide range of trait gain or masculinization phenotypes. Further work could also explore how the dimorphic (C. eurytheme) vs. monomorphic (C. philodice) expression of Bab is achieved. A potential regulator could be the transcription factor Doublesex, an integrator and master selector gene of somatic sexual identity in insects, including butterflies (35–38). In fruit flies, the DsxM male isoform of Doublesex directly represses Bab via a cis-regulatory element in the first intron of bab1, thus derepressing abdominal pigmentation, while the DsxF female isoform activates Bab, repressing pigmentation (24, 28). Extrapolating from this precedent, it will be interesting to test if DsxM similarly represses Bab in C. eurytheme cover scales, and if its binding sites have mutated in C. philodice.
bab Is a Genetic Hotspot of Sexual Phenotypic Variation.
There is replicated evidence that cis-regulatory evolution of bab directly causes sexual trait divergence in flies and Lepidoptera (butterflies and moths), making it a genetic hotspot of phenotypic variation (39). In Drosophila, linkage mapping studies have shown that regulatory alleles of the bab locus (bab1/2 recent paralogs) explain natural variation in dimorphic pigmentation and ovariole number (23, 26, 40). In corn borer moths (Ostrinia nubilalis), male response to a polymorphic female pheromone blend is driven by a Z-linked regulatory variation in the first intron of bab (41). This same intron is within the Colias U-locus interval. Thus, the 5′ portion of the lepidopteran bab locus likely underlies variation in a male olfactory preference in recently diverged Ostrinia species, as well as in a male visual signal in Colias. This leads us to propose that the bab locus is a hotspot for the evolution of reproductive isolation, driving species divergence and maintenance across Lepidoptera. Factors related to the function, regulation, and genomic location of bab may have predisposed it to the tuning of sexually selected traits. First, repressors have drastic effects on cis-regulatory regions and efficiently suppress transcription (42, 43). In this sense, evolving new Bab binding sites in DNA enhancers might be a path of least resistance for optimizing gene expression subtractively, rather than by adding activator binding sites for other selector genes. There is already precedent for the Drosophila Bab integrating into a preexisting network and sculpting sexual dichromatism (27, 44) consistent with this idea. Second, the published expression patterns of Bab show it integrates both spatial and sex-determination inputs (21, 24, 25, 34). Due to a hub-like position in gene-regulatory networks, Bab is potentially an input-output gene that facilitates tissue-specific change (45, 46). The large 5′ intergenic and intronic regions of bab in Colias and Ostrinia, and of its subfunctionalized duplicates in Drosophila, together suggest a complex cis-regulatory landscape bearing a multitude of enhancers or silencers (47–49), enabling the evolution of precise changes in specialized tissues (50). Last, the location of Bab on the lepidopteran Z chromosome is relevant to the divergence of premating traits under the assumption that sex chromosomes are more prone to the generation of reproductive isolation than autosomes (20, 51, 52). These possible generative biases will require further investigation among diverged Drosophila species, as well as in the Colias UV display and Ostrinia mate detection systems. Nonetheless, we speculate that the peculiar molecular function, regulation, and genomic location of Bab may collectively explain its propensity to fine-tune variation in sexual traits.
Extreme Large-Z Effect in an Anthropogenic Contact Zone.
Our genomic scan of differentiation focused on two incipient species that recently reunited due to human activity (alfalfa agriculture). We caught a remarkable signature of a large-Z effect (analogous to large-X) with widespread admixture across autosomes, and strong differentiation of the entire Z chromosome. This heterogeneous landscape of differentiation is the most pronounced identified so far from genome-wide data (18), supporting a role for sex chromosomes as key drivers of reproductive isolation (20, 51). Collectively with the evidence that both premating and postmating isolating mechanisms are sex-linked in this system (12, 14, 15), these data highlight that species status in this system is determined by Z chromosomes, while autosomes are largely exchangeable (though some divergence on autosomes was also observed). While linked selection (syn. divergence hitchhiking) in regions of low recombination can sometimes explain such extensive blocks of divergence (7, 19), our linkage map did not indicate that this is the case on the Z chromosome for the Colias sympatric pair. The U locus in particular overlapped with a 2.5-Mb region with above-median recombination in between-species crosses (red bracket in Fig. 1F). High FST indicates this region is refractory to gene flow in wild populations, suggesting it acts as a genomic barrier to gene flow rather than as a block of divergence hitchhiking, and might include further Z-linked barrier loci in addition to the male color signal variation (12–15). Overall, these data establish the anthropogenic contact zones of Colias butterflies as a promising system for the study of large-Z(X) effects in speciation with gene flow.
Coupling of Reproductive Barriers.
The data in hand suggest the U-locus variation causes premating isolation by accurately displaying sex-chromosome compatibility of courting males to C. eurytheme females. In summary, the Colias UV mating signal is polymorphic and recessive in areas of secondary contact, with both heterospecific and hybrid males lacking UV iridescence because they carry the dominant U allele of C. philodice. This allele drives uniform expression of Bab, preventing UV-scale development with a dominant effect. Conversely, the recessive C. eurytheme u allele represses expression of Bab in dorsal male cover scales, leading to UV fated scales. This Z-linked UV signal allows C. eurytheme females to choose Z-compatible mates, thereby prezygotically selecting against Z-linked hybrid sterility (3, 9, 10, 12–14). The recessivity of the trait allows the rejection of Z-heterozygous males and prevents a 25% cost in reproductive fitness (SI Appendix, Fig. S1), an effect that would not be possible if Bab were an activator of UV, which would likely be dominant. In this way, a large-Z effect that couples pre- and postmating barriers likely drives reproductive isolation in these butterflies (5, 18), akin to the genetic architectures of speciation in other species (16, 53), and previously conceptualized as an “indicator” mechanism that enables assortative mating (3). Further work is needed to decipher other putative barrier loci on this chromosome. In any case, these findings highlight how a simple genetic switch for a mating cue can influence the origin of species and the maintenance of biodiversity.
Experimental Procedures
Genomic Scans of Differentiation in Sympatric Males.
Samples originated from a large syntopic population at an alfalfa farm in Buckeystown, MD. Whole genomes from 24 males were resequenced at 14.3× mean coverage, aligned to the C. eurytheme reference genome assembly (54), and used in a population scan analysis pipeline (55).
Linkage Map.
Interspecific crosses consisting of an F2 and two BC broods generated 528 recombinant individuals of known pedigree, sex, and UV phenotype. 2b-RAD sequencing was used to genotype 484 males and females in a HiSeq 4000 SE50 run, using the BcgI enzyme and adapters, yielding a 16-fold representation reduction (56). Genotypes were used to build a linkage map as described elsewhere (54). Select individuals were resequenced at a 15× mean coverage to narrow the U-locus interval.
Bab Expression and Loss-of-Function Assays.
A custom rabbit polyclonal antibody was generated against the N-terminal 1 to 365 residues of the C. eurytheme Bab protein, and used with a guinea pig anti-Dve (57) for whole-mount immunofluorescence in pupal wings. Heteroduplex mixes of Cas9 recombinant protein and two equimolar sgRNAs (500:125:125 ng/μL) were injected into syncytial embryos 1 to 7 h after egg laying (AEL) for targeted mutagenesis of the first exon of bab.
CRISPR-Mediated KO of bab.
Two overlapping gRNAs were designed targeting the first exon of bab within the U locus. Heteroduplex mixes of Cas9:sgRNA1:sgRNA2 (500:125:125 ng/μL) were prepared and microinjected into butterfly syncytial embryos 1–7 h AEL. Eggs incubated for 2 d at 28 °C were then placed on vetch sprouts aged 7 to 14 d at an average greenhouse temperature of 24 °C. Two rounds of injections were successfully performed under these conditions resulting in 51 of 63 adults displaying crispant phenotypes.
UV Photography.
UV photography was performed using a full-spectrum converted Lumix G3 camera, mounted with Baader U-Venus filters and UV-transmitting lenses, under the illumination of blacklight bulbs or 365 nm light-emitting diodes.
Supplementary Material
Acknowledgments
We thank B. Wang, S. Barao, W. Watt, and R. Canalichio for butterfly rearing, access, and expertise; R. Rogers, S. Van Belleghem, and P. Rastas for bioinformatics assistance; C. Brantner and C. Day for scanning electron microscopy (SEM); M. Matz for 2b-RAD protocols; L. Livraghi for vector graphics; and M. Perry for sharing the Dve antibody. We also thank the core facilities at The George Washington University (GWU), University of Texas at Austin, and University of Maryland, Baltimore, for generating sequence data, and the GWU high-performance computing team for providing computing infrastructure (58). This work was supported by NSF Awards IOS-2108227 and IOS-1656553 and Swedish Research Council Award 2017-04386.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109255118/-/DCSupplemental.
Data Availability
Whole-genome sequencing data reported in this article have been deposited in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under BioProjects PRJNA663300, PRJNA719421, and PRJNA723900. SNP calling, genotyping data, and computer code have been deposited in the Dryad digital repository (10.5061/dryad.4b8gthtcc) (55).
References
- 1.Safran R. J., Scordato E. S., Symes L. B., Rodríguez R. L., Mendelson T. C., Contributions of natural and sexual selection to the evolution of premating reproductive isolation: A research agenda. Trends Ecol. Evol. 28, 643–650 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Servedio M. R., Boughman J. W., The role of sexual selection in local adaptation and speciation. Annu. Rev. Ecol. Evol. Syst. 48, 85–109 (2017). [Google Scholar]
- 3.Servedio M. R., The evolution of premating isolation: Local adaptation and natural and sexual selection against hybrids. Evolution 58, 913–924 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Kulmuni J., Butlin R. K., Lucek K., Savolainen V., Westram A. M., Towards the completion of speciation: The evolution of reproductive isolation beyond the first barriers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190528 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butlin R. K., Smadja C. M., Coupling, reinforcement, and speciation. Am. Nat. 191, 155–172 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Ortiz-Barrientos D., Grealy A., Nosil P., The genetics and ecology of reinforcement: Implications for the evolution of prezygotic isolation in sympatry and beyond. Ann. N. Y. Acad. Sci. 1168, 156–182 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Ravinet M., et al. , Interpreting the genomic landscape of speciation: A road map for finding barriers to gene flow. J. Evol. Biol. 30, 1450–1477 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Hovanitz W., The ecological significance of the color phases of Colias chrysotheme in North America. Ecology 25, 45–60 (1944). [Google Scholar]
- 9.Silberglied R. E., Taylor O. R., Ultraviolet reflection and its behavioral role in the courtship of the sulfur butterflies Colias eurytheme and C. philodice (Lepidoptera, Pieridae). Behav. Ecol. Sociobiol. 3, 203–243 (1978). [Google Scholar]
- 10.Taylor O. R. Jr., Random vs. non-random mating in the sulfur butterflies, Colias eurytheme and Colias philodice (Lepidoptera: Pieridae). Evolution 26, 344–356 (1972). [DOI] [PubMed] [Google Scholar]
- 11.Thurman T. J., Brodie E., Evans E., McMillan W. O., Facultative pupal mating in Heliconius erato: Implications for mate choice, female preference, and speciation. Ecol. Evol. 8, 1882–1889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grula J. W., Taylor O. R. Jr., Some characteristics of hybrids derived from the sulfur butterflies, Colias eurytheme and C. philodice: Phenotypic effects of the X-chromosome. Evolution 34, 673–687 (1980). [DOI] [PubMed] [Google Scholar]
- 13.Wang B., “Introgression and genomic differentiation in sympatric, hybridizing Colias butterflies,” PhD thesis, University of Massachusetts Amherst, Amherst, MA (2005).
- 14.Grula J. W., Taylor O. R. Jr., The effect of X-chromosome inheritance on mate-selection behavior in the sulfur butterflies, Colias eurytheme and C. philodice. Evolution 34, 688–695 (1980). [DOI] [PubMed] [Google Scholar]
- 15.Silberglied R. E., Taylor O. R., Ultraviolet differences between the sulphur butterflies, Colias eurytheme and C. philodice, and a possible isolating mechanism. Nature 241, 406–408 (1973). [DOI] [PubMed] [Google Scholar]
- 16.Saether S. A., et al. , Sex chromosome-linked species recognition and evolution of reproductive isolation in flycatchers. Science 318, 95–97 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Clark A. H., “The butterflies of the District of Columbia and vicinity” (Bulletin of the US National Museum, No. 157, US Government Printing Office, 1932).
- 18.Presgraves D. C., Evaluating genomic signatures of “the large X-effect” during complex speciation. Mol. Ecol. 27, 3822–3830 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burri R., et al. , Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 25, 1656–1665 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson Sayres M. A., Genetic diversity on the sex chromosomes. Genome Biol. Evol. 10, 1064–1078 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kopp A., Duncan I., Godt D., Carroll S. B., Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature 408, 553–559 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Couderc J.-L., et al. , The bric à brac locus consists of two paralogous genes encoding BTB/POZ domain proteins and acts as a homeotic and morphogenetic regulator of imaginal development in Drosophila. Development 129, 2419–2433 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Williams T. M., et al. , The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 134, 610–623 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers W. A., et al. , Recurrent modification of a conserved cis-regulatory element underlies fruit fly pigmentation diversity. PLoS Genet. 9, e1003740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salomone J. R., Rogers W. A., Rebeiz M., Williams T. M., The evolution of Bab paralog expression and abdominal pigmentation among Sophophora fruit fly species. Evol. Dev. 15, 442–457 (2013). [DOI] [PubMed] [Google Scholar]
- 26.De Castro S., Peronnet F., Gilles J.-F., Mouchel-Vielh E., Gibert J.-M., bric à brac (bab), a central player in the gene regulatory network that mediates thermal plasticity of pigmentation in Drosophila melanogaster. PLoS Genet. 14, e1007573 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roeske M. J., Camino E. M., Grover S., Rebeiz M., Williams T. M., cis-regulatory evolution integrated the Bric-à-brac transcription factors into a novel fruit fly gene regulatory network. eLife 7, e32273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dinwiddie A., et al. , Dynamics of F-actin prefigure the structure of butterfly wing scales. Dev. Biol. 392, 404–418 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Ghiradella H., Development of ultraviolet-reflecting butterfly scales: How to make an interference filter. J. Morphol. 142, 395–409 (1974). [DOI] [PubMed] [Google Scholar]
- 30.Rutowski R. L., Macedonia J. M., Morehouse N., Taylor-Taft L., Pterin pigments amplify iridescent ultraviolet signal in males of the orange sulphur butterfly, Colias eurytheme. Proc. Biol. Sci. 272, 2329–2335 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemp D. J., Vukusic P., Rutowski R. L., Stress-mediated covariance between nano-structural architecture and ultraviolet butterfly coloration. Funct. Ecol. 20, 282–289 (2006). [Google Scholar]
- 32.Stella D., Faltýnek Fric Z., Rindoš M., Kleisner K., Pechácek P., Distribution of ultraviolet ornaments in Colias butterflies (Lepidoptera: Pieridae). Environ. Entomol. 47, 1344–1354 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Camara N., Whitworth C., Dove A., Van Doren M., Doublesex controls specification and maintenance of the gonad stem cell niches in Drosophila. Development 146, dev170001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bourbon H.-M. G., et al. , Tissue-specific versus pleiotropic enhancers within the bric-a-brac tandem gene duplicates display differential regulatory activity and evolutionary conservation. bioRxiv [Preprint] (2021). 10.1101/2021.03.25.436949 (Accessed 20 December 2021). [DOI]
- 35.Kunte K., et al. , doublesex is a mimicry supergene. Nature 507, 229–232 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Nishikawa H., et al. , A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nat. Genet. 47, 405–409 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Caro F., et al. , Novel doublesex duplication associated with sexually dimorphic development of dogface butterfly wings. Mol. Biol. Evol. 38, 5021–5033 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prakash A., Monteiro A., Doublesex mediates the development of sex-specific pheromone organs in Bicyclus butterflies via multiple mechanisms. Mol. Biol. Evol. 37, 1694–1707 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin A., Orgogozo V., The loci of repeated evolution: A catalog of genetic hotspots of phenotypic variation. Evolution 67, 1235–1250 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Green D. A. II, Extavour C. G., Convergent evolution of a reproductive trait through distinct developmental mechanisms in Drosophila. Dev. Biol. 372, 120–130 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Unbehend M., et al. , bric à brac controls sex pheromone choice by male European corn borer moths. Nat. Commun. 12, 2818 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preger-Ben Noon E., Davis F. P., Stern D. L., Evolved repression overcomes enhancer robustness. Dev. Cell 39, 572–584 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Gisselbrecht S. S., et al. , Transcriptional silencers in Drosophila serve a dual role as transcriptional enhancers in alternate cellular contexts. Mol. Cell 77, 324–337.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hughes J. T., Williams M. E., Rebeiz M., Williams T. M., Widespread cis- and trans-regulatory evolution underlies the origin, diversification, and loss of a sexually dimorphic fruit fly pigmentation trait. J. Exp. Zoolog. B Mol. Dev. Evol., 10.1002/jez.b.23068 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Stern D. L., Orgogozo V., Is genetic evolution predictable? Science 323, 746–751 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orteu A., Jiggins C. D., The genomics of coloration provides insights into adaptive evolution. Nat. Rev. Genet. 21, 461–475 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Baudouin-Gonzalez L., et al. , Diverse cis-regulatory mechanisms contribute to expression evolution of tandem gene duplicates. Mol. Biol. Evol. 34, 3132–3147 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalay G., Lachowiec J., Rosas U., Dome M. R., Wittkopp P., Redundant and cryptic enhancer activities of the Drosophila yellow gene. Genetics 212, 343–360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borsari B., et al. , Enhancers with tissue-specific activity are enriched in intronic regions. Gen. Res. 31, 1325–1336 (2021). [DOI] [PMC free article] [PubMed]
- 50.Carroll S. B., Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134, 25–36 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Patten M. M., Selfish X chromosomes and speciation. Mol. Ecol. 27, 3772–3782 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Fraïsse C., Sachdeva H., The rates of introgression and barriers to genetic exchange between hybridizing species: Sex chromosomes vs autosomes. Genetics 217, iyaa025 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ortiz-Barrientos D., Counterman B. A., Noor M. A., The genetics of speciation by reinforcement. PLoS Biol. 2, e416 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tunström K., et al. , A complex interplay between balancing selection and introgression maintains a genus-wide alternative life-history strategy. bioRxiv [Preprint] (2021). 10.1101/2021.05.20.445023 (Accessed 20 December 2021). [DOI]
- 55.Ficarrotta V., et al. , Data from: A genetic switch for male UV-iridescence in an incipient species pair of sulphur butterflies (2021). Dryad. 10.5061/dryad.4b8gthtcc. Deposited 16 December 2021. [DOI] [PMC free article] [PubMed]
- 56.Wang S., Meyer E., McKay J. K., Matz M. V., 2b-RAD: A simple and flexible method for genome-wide genotyping. Nat. Methods 9, 808–810 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Perry M., et al. , Expanded color vision in butterflies: Molecular logic behind the three-way stochastic choices that expand butterfly colour vision. Nature 535, 280–284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.MacLachlan G., et al. , Building a shared resource HPC center across university schools and institutes: A case study. arXiv [Preprint] (2020). https://arxiv.org/abs/2003.13629 (Accessed 20 December 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole-genome sequencing data reported in this article have been deposited in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under BioProjects PRJNA663300, PRJNA719421, and PRJNA723900. SNP calling, genotyping data, and computer code have been deposited in the Dryad digital repository (10.5061/dryad.4b8gthtcc) (55).