Significance
Nitrogen fixation is economically important in agriculture. Enhancing nodulation in soybean can greatly enhance nitrogen fixation efficiency. Nodule formation in legumes starts with cortical cell divisions in the root. Despite the discovery of the Nod factors and plant hormones that are involved, it remains unclear how cortical cell division is spatiotemporally regulated. Here we provide evidence that soybean has acquired two SHORT-ROOT (SHR) paralogs during evolution, which are specifically induced in the early stage of nodule organogenesis. These SHR homologs can activate the expression of early nodulin genes and enhance cytokinin signaling and form part of a multilevel regulatory network to activate D-type cyclins during nodulation. Our results provide insight into the mechanisms of nodule organogenesis in soybean.
Keywords: nodule primordia, GmSHR4/5, cell division, cytokinin, GmCYCD6;1
Abstract
Nitrogen fixation in soybean takes place in root nodules that arise from de novo cell divisions in the root cortex. Although several early nodulin genes have been identified, the mechanism behind the stimulation of cortical cell division during nodulation has not been fully resolved. Here we provide evidence that two paralogs of soybean SHORT-ROOT (GmSHR) play vital roles in soybean nodulation. Expression of GmSHR4 and GmSHR5 (GmSHR4/5) is induced in cortical cells at the beginning of nodulation, when the first cell divisions occur. The expression level of GmSHR4/5 is positively associated with cortical cell division and nodulation. Knockdown of GmSHR5 inhibits cell division in outer cortical layers during nodulation. Knockdown of both paralogs disrupts the cell division throughout the cortex, resulting in poorly organized nodule primordia with delayed vascular tissue formation. GmSHR4/5 function by enhancing cytokinin signaling and activating early nodulin genes. Interestingly, D-type cyclins act downstream of GmSHR4/5, and GmSHR4/5 form a feedforward loop regulating D-type cyclins. Overexpression of D-type cyclins in soybean roots also enhanced nodulation. Collectively, we conclude that the GmSHR4/5-mediated pathway represents a vital module that triggers cytokinin signaling and activates D-type cyclins during nodulation in soybean.
Nodulation in legumes is coordinated by two coordinated processes: rhizobial infection and nodule formation (1, 2). Studies over the past decade have identified key components of the Nod factor-activated signaling pathway in the early symbiotic stage (3–6). Nodule organogenesis starts with new cell divisions in root cortical tissues (4). The root cortex in soybean consists of multiple layers, and the Nod factor-induced cell division occurs first in the outer cortical cells followed by divisions in the inner cortical tissues and the pericycle (7). A number of regulators of Nod factor perception have been identified, including NODULE INCEPTION (NIN) (8), an ERF family protein (ERN), and two GRAS family proteins called Nodulation Signaling Pathway (NSP1 and NSP2) (4–6). Overexpression of NIN in the absence of rhizobia is sufficient to induce cortical cell divisions and leads to spontaneous nodule-like structures (9). ENOD40, a marker gene for nodule primordium initiation, is up-regulated at the onset of nodulation (10–12). These Nod factor-inducible genes can be directly activated by NSP1 (13). The soybean orthologs of all these components have been identified (14).
In addition to the transcriptional regulators, the phytohormone cytokinin (CK) also plays a key role in promoting cortical cell division in the early symbiotic stages (15–25). Depletion of the endogenous CK by overexpressing a CK oxidase/dehydrogenase (CKX) or by repressing expression of the cytokinin biosynthetic gene LONELY GUY 1 (LOG1) dramatically decreases nodulation in Lotus japonicus and Medicago truncatula (25, 26). The crucial role of cytokinin is also evidenced by the effects of overexpressing ARABIDOPSIS THALIANA RESPONSE REGULATOR 5 (ARR5), MtRR4 type-ARR, or Two-Component Signaling Sensor (TCS), all of which increase cortical cell divisions and nodulation (19, 27). Interestingly, gain-of-function mutants of both LOTUS HISTIDINE KINASE 1 (LHK1) (L. japonicus lotus, named snf2) and Cytokinin Response 1 (MtCRE1) result in “spontaneous nodule formation” in the absence of rhizobia (24). Genetic analyses indicate that the spontaneous nodule formation in snf2 requires NIN and NSP2 (24).
As nodules form on roots, it has been hypothesized that nodule organogenesis is derived from the lateral root developmental program (5, 28–31). Recent studies in M. truncatula showed that several transcription factors, including WUS-RELATED HOMEOBOX 5 (MtWOX5) and MtPLETHORA, are expressed and function in the nodule meristem (32–34). In addition, RNA-seq of laser-microdissected nodules showed that close homologs for a series of key root meristem regulators are expressed in nodule primordia, including WOX5, ARABIDOPSIS CRINKLY 4 (ACR4), SHORT-ROOT (SHR), PLT, SCARECROW (SCR), and JACKDAW (JKD) (34). In Arabidopsis roots, SHR and SCR activate the expression of a D-type cyclin, CYCD6;1 to promote the periclinal division, which separates endodermis and cortex cell layers (35). Recently, it was revealed that MtSHR and MtSCR from M. truncatula form the module to drive the cell division in the cortex during nodule formation (36). These findings provided a hint that root developmental programs might be involved in nodulation. However, the function of important root meristem genes in nodule organogenesis has not been fully explored.
In this study, we obtained evidence that two distally evolved paralogs of the GRAS family protein SHR (GmSHR4 and GmSHR5) regulate cortical cell divisions and nodule organogenesis in soybean. After inoculation with rhizobia, GmSHR4/5 were activated mostly in outer cortical cells where the first cell divisions occur. Overexpression of GmSHR4/5 resulted in a substantial increase in nodulation, whereas GmSHR4/5-microRNA roots formed significantly fewer nodules. The expression of GmSHR4/5 in the cortex enhanced cytokinin signaling and up-regulated the early nodulin genes. Downstream of this cascade were a set of D-type cyclins, which were regulated by a classic feedforward mechanism mediated by GmSHR paralogs. Therefore, our results revealed a mechanism regulating nodule primordia formation in soybean.
Results
GmSHR Paralogs Are Induced in Cortical Tissues at the Early Stage of Nodulation.
In Glycine max, we identified six SHR homologs (named GmSHR1 to GmSHR6) by BLAST searches using AtSHR protein sequences as the query. The amino acid sequence of all six homologs had the five conserved domains of typical GRAS proteins, including LHR1, VHIID, LHRII, PFYRE, and SAW (SI Appendix, Fig. S1). Phylogenetic analysis revealed that GmSHR1, GmSHR2, GmSHR3, and GmSHR6 are closely related to Arabidopsis SHR (AtSHR) and Medicago SHRs (Medtr5g015490.1 and Medtr4g097080) (SI Appendix, Fig. S2A). Interestingly, two paralogs that arose during evolution of the soybean genome—GmSHR4 and GmSHR5—are only distally related to AtSHR (SI Appendix, Fig. S2A). These two GmSHR paralogs exhibited a tissue-specific expression pattern distinct from those of the other soybean SHRs (SI Appendix, Fig. S2B). In addition, the expression of GmSHR4 and GmSHR5 (GmSHR4/5 hereafter) in the roots was dramatically enhanced at 1 d after inoculation (dai) with rhizobia and then decreased with the progression of root nodulation (SI Appendix, Fig. S2C). By contrast, GmSHR1 did not appear to be up-regulated until 7 dai and its expression peaked in the mature nodule at 28 dai. Expression of GmSHR2 showed enhancement only in the mature nodule, while that of GmSHR3 and GmSHR6 changed very little during nodulation (SI Appendix, Fig. S2C). These results suggest that GmSHRs might participate in different stages of nodulation and GmSHR4/5 could be involved in the initiation stage.
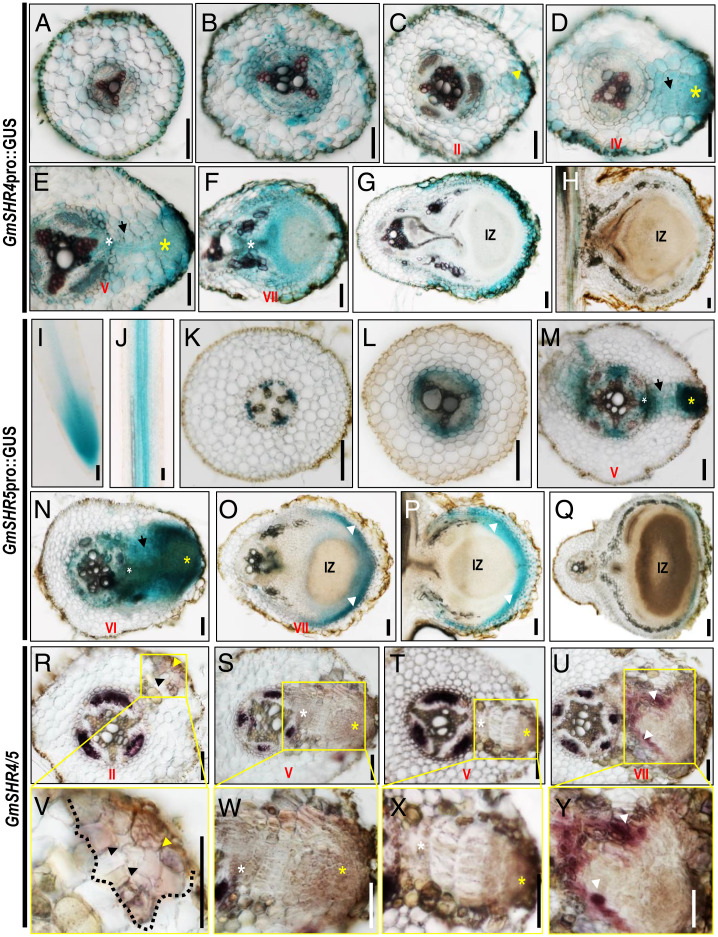
Within the 2-kb promoter regions upstream of GmSHR4/5, we identified many nodulin consensus sequence motifs (NODCON1GM and NODCON2GM) (SI Appendix, Fig. S2D). This implies that GmSHR4/5 expression could be triggered by the rhizobial infection. To understand the spatiotemporal expression pattern of GmSHR4/5 during nodulation, we first examined the nodule organogenesis in soybean roots. We performed histological visualization and divided the early nodule development into six stages (I to VI). We numbered the cortical layers from the outermost to the innermost (C1 to C5). In stages I and II, anticlinal divisions were initiated in the C1 layer and then occurred in C2 (SI Appendix, Fig. S3 A, B, G and H). In stage III, inner cortical cells started to divide (in C3 to C5), and more anticlinal divisions occurred in the C1 and C2 layers (SI Appendix, Fig. S3 C and I). In stage IV, divisions occurred in the endodermis and periclinal divisions started in the C1 layer (SI Appendix, Fig. S3 D and J). The pattern of cell divisions in the C1 layer also became more complex, leading to the formation of a primordial structure (SI Appendix, Fig. S3 D and J). In stage V, most of the cortical cells at the infection site had divided into smaller cells and cells in the pericycle started to divide (SI Appendix, Fig. S3 E and K). At stage VI, the young nodules distended (SI Appendix, Fig. S3 F and L). The newly formed nodules underwent steady growth and differentiation, in particular forming vascular bundles in stages VII through IX (SI Appendix, Fig. S3 M–O). To verify these histological observations, we performed in situ hybridization of the cell-cycle marker gene GmCyclB1.1. In stage V, the staining first became enriched mostly in the outer cortical cells (SI Appendix, Fig. S3P), followed by a high level of staining in the inner cortical cells, indicating a gradual expansion of mitotic activity during early nodulation (SI Appendix, Fig. S3Q). Consistent with the GmSHR4/5 expression pattern, GmCyclB1.1 expression gradually became undetectable in mature nodules (SI Appendix, Fig. S3 R and S). Further, we constructed promoter:GUS reporters of GmSHR4 and GmSHR5. In soybean roots, GmSHR4 had a weak expression and could occasionally be detected in the endodermal cells, vascular tissues, and cortex cells in the mature zone (Fig. 1A). GmSHR5 was expressed in the meristem (Fig. 1I) and confined only in vascular tissues in the mature zone (Fig. 1 J and K). Upon inoculation with rhizobia, the expression of GmSHR4/5 was greatly enhanced (Fig. 1 B, L, and M). GmSHR4/5 expression was enriched in the outer cortical tissues during the early stage of nodulation (stages II to VI) (Fig. 1 C–E, M, and N) and also maintained a weak expression in the inner cortex (Fig. 1 D, E, M, and N). GmSHR4/5 expression gradually disappeared as the nodules matured (Fig. 1 F–H and O–Q).
Fig. 1.
Spatial expression pattern of GmSHR genes during nodulation in soybean. (A–H) GmSHR4pro::GUS expression pattern during nodulation. The cross-sections of the roots without rhizobial infection (A) and the roots at 10 dai with rhizobia (B–G) and 20 dai (H) are shown. (I–Q) GmSHR5pro::GUS expression pattern during nodulation. The cross-sections of the roots at 0 dai (I–K), 10 dai (L–P), and 20 dai (Q) are shown. (R–U) In situ hybridization of GmSHR4/5 in roots at 10 dai. (V–Y) The areas in the yellow wireframe are the enlarged views of the boxed regions in R–U. IZ, infection zone. White arrows indicate the nodule parenchyma. Yellow and black arrows indicate the division of the outer cortical cells and inner cortical cells, respectively. White asterisks point to the division of the pericycle cells. Yellow asterisks point to the outer ground tissues. (Scale bars, 100 μm.)
The spatiotemporal expression pattern of GmSHR4/5 shown by the promoter GUS reporters was validated by in situ hybridization (Fig. 1 R–Y), further suggesting the potential involvement of these two SHR paralogs in the early stage of the nodulation. Unlike GmSHR4/5, GmSHR1/2 were expressed only in the vascular tissues (SI Appendix, Fig. S2E).
GmSHR4/5 Promote Cortical Cell Division and Positively Regulate Root Nodulation.
To examine GmSHR4/5 function, we manipulated their expression by overexpression (OX-GmSHR4/5 hereafter) and knockdown (GmSHR4-amiR, GmSHR5-amiR, and GmSHR4/5-amiR hereafter) in soybean roots (Fig. 2A and SI Appendix, Fig. S4 A–D). Interestingly, the expression of other GmSHRs appeared to be moderately reduced in the OX-GmSHR4/5 lines and increased in the GmSHR4/5-amiR roots, suggesting the existence of potential feedback inhibition among GmSHRs (SI Appendix, Fig. S4A).
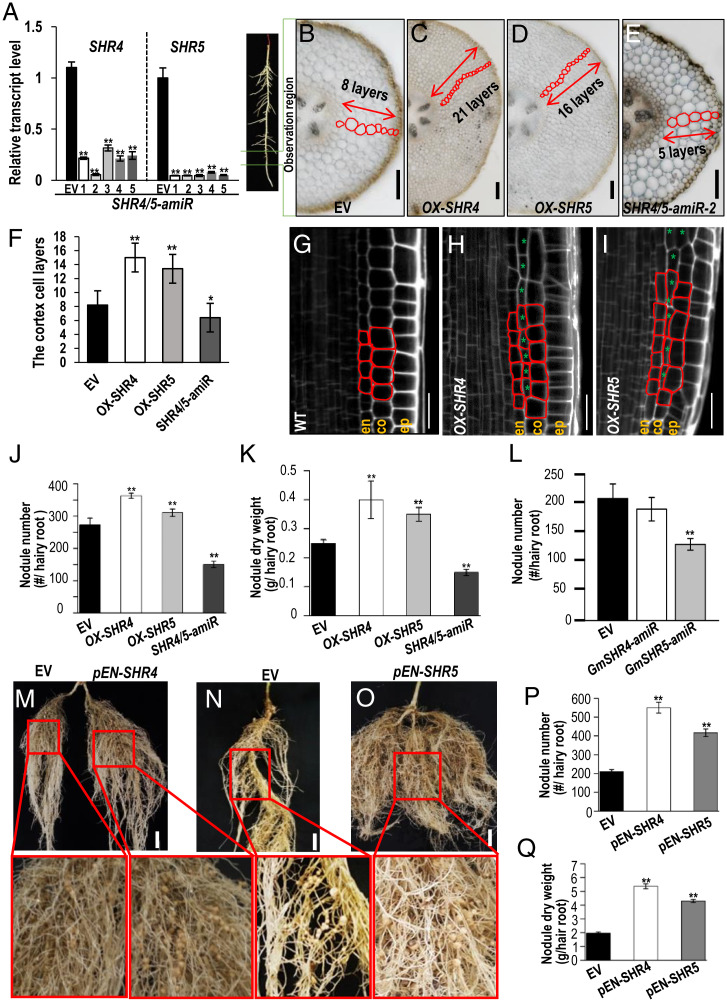
Fig. 2.
GmSHR4/5 expression level affects cortical cell division and root nodulation. (A) qRT-PCR shows that the expression of GmSHR4 and GmSHR5 is suppressed in the roots of artificial microRNA lines (GmSHR4/5-amiR). The RNA was extracted from the noninoculated roots. Error bars represent SD. GmEF1α (accession no. X56856) was used as the internal control. (B–E) The cortex cell layers in the root cross-section of GmSHR4/5 overexpression lines (OX-GmSHR4/5) and GmSHR4/5-amiR lines. All cortex sections were sampled from the same position of the hairy roots (n ≥ 15). (Scale bars, 100 μm.) (F) Quantification of the cortex cell layers shown in B–E (n ≥ 15). (G–I) Extra cortex cell division in the propidium iodide-stained Arabidopsis roots with ectopic expression of GmSHR4 and GmSHR5 (n ≥ 10). (Scale bars in G–I, 20 μm.) Green asterisks point to the extra cortex cells. (J and K) Nodule number (J) and nodule dry weight (K) in roots at 28 dai (n ≥ 25). (L) Nodule number per root at 28 DAI. (n ≥ 25). (M–Q) Nodulation in different backgrounds. (M) Split root showing the same root transformed with EV (Left) and pENOD40-GmSHR4 (Right). (N and O) EV and pENOD40-GmSHR5. Lower panels are the close-up views of the roots in M–O. (Scale bars, 2 cm.) (P) Nodule number. (Q) Nodule dry weight. EN, ENOD40 (n ≥ 20). Significant differences are observed between transgenic plants and EV plants. (**P < 0.01, *P < 0.05; Student’s t test). Error bars represent SD. n represents the number of independent biological samples. All experiments were repeated three times.
In Arabidopsis, SHR regulates periclinal cell division in cortical initial cells (35, 37). Therefore, we examined the cortical cell layers that are derived from periclinal cell divisions in soybean roots. As expected, the OX-GmSHR4/OX-GmSHR5 roots showed three to six additional layers of cortical tissues, while the GmSHR4/5-amiR roots had fewer (Fig. 2 B–F). In addition, we transformed Arabidopsis with GmSHR4 and GmSHR5 and observed extra cell divisions in the cortical tissues of the transgenic plants (Fig. 2 G–I). These results suggest that GmSHR4/5 can induce cortical cell divisions.
Next, we evaluated the nodulation of soybean roots with altered GmSHR4/5 expression at 28 dai with rhizobia. The total number and dry weight of nodules were significantly higher on OX-GmSHR4/5 roots and substantially lower on GmSHR4/5-amiR roots (Fig. 2 J and K and SI Appendix, Fig. S4E), indicating that GmSHR4/5 functioned as the positive regulators of soybean nodulation. Interestingly, unlike GmSHR5-amiR, GmSHR4-amiR did not change the nodulation (Fig. 2L). This finding suggests that GmSHR5 could be more involved than GmSHR4 in the initiation of nodule primordia.
To rule out the potential artifacts that can arise from overexpression, we specifically expressed GmSHR4/5 in nodule primordia under the promoter of GmENOD40. Both pENOD40-GmSHR4 and pENOD40-GmSHR5 lines resulted in a higher number of nodules (Fig. 2 M–P). The total dry weight of nodules of the pENOD40-GmSHR4 and pENOD40-GmSHR5 lines was 2.72 and 2.19 times that of the control, respectively (Fig. 2Q). These data further support that GmSHR4/5 function in nodule formation.
GmSHR4/5 Function in Cortical Tissues during the Nodule Initiation.
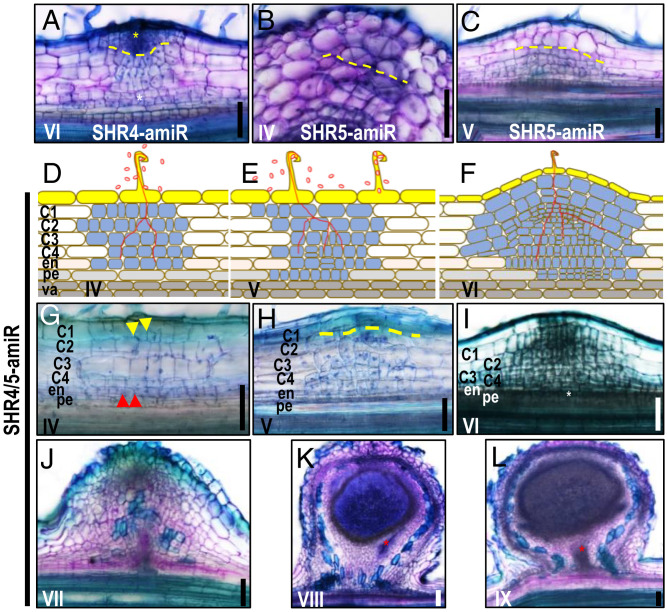
The spatial expression pattern of GmSHR4/5 in the cortical tissues suggests they could be involved in the division of cortical tissues. We next examined the effect of silencing GmSHR4/5 on the cortical cell division pattern. In GmSHR4-amiR roots, we detected no noticeable change in the nodulation process, suggesting the existence of functional redundancy (Fig. 3A and SI Appendix, Fig. S3 F and L). In GmSHR5-amiR roots, however, we observed fewer cell divisions in the outer cortical layers, along with a disordered pattern of division (Fig. 3 B and C and SI Appendix, Fig. S3 J and K). The simultaneous silencing of GmSHR4/5 disrupted the division pattern throughout the cortex (Fig. 3 D–I and SI Appendix, Fig. S3 J–L) and delayed the formation of the vascular tissues in the nodule primordia (Fig. 3 J–L and SI Appendix, Fig. S3 M–O). These results suggest that GmSHR5 is more important for the initial cell divisions in outer cortical layers, but both GmSHR4 and -5 redundantly promote ordered cortical cell divisions to form a nodule structure. In addition, divisions throughout the cortex contribute to the formation of nodules.
Fig. 3.
GmSHR4/5 function in cortical tissues during the nodule initiation and affect the formation of nodule vascular tissues. (A) Knockdown of GmSHR4 did not affect the nodule formation. (B and C) Knockdown of GmSHR5 leads to the disrupted cortical division in ground tissues. (D–I) Knockdown of both GmSHR4 and -5 leads to the disrupted cortical division in ground tissues. (J–L) Knockdown of both GmSHR4 and -5 delayed the formation of the vascular tissues in the nodule primordia. Red arrows and white asterisks point to the division of the endodermal cells and pericycle cells, respectively. Yellow dashed lines define the boundary of the division in outer ground tissues or inner ground tissues. Yellow asterisk points to the outer ground tissues. Red asterisks indicate the vascular tissues. Yellow arrows indicate the division of the outer cortical cells (n ≥ 20). n represents the number of independent biological samples. These experiments were repeated three times with similar results. (Scale bars, 100 μm.)
GmSHR4/5 Promote Nodule Formation by Activating Cytokinin Signaling.
It is known that cytokinin promotes cortical cell divisions during nodulation (15–24). To dissect the relationship between cytokinin and GmSHRs, we examined the expression of cytokinin-related genes in GmSHR4/5 transgenic systems. It was previously reported that CRE1/HK1 is the major cytokinin receptor involved in the nodulation of Medicago sativa and L. japonicus (17, 19). In soybean, we identified three HK1 homologs (named GmHK1-1, GmHK1-2, and GmHK1-3). Our qRT-PCR results showed that GmSHR4/5 had different activation targets, with GmSHR4 regulating GmHK1-1 and GmSHR5 targeting GmHK1-3 (Fig. 4A). In support of this, in situ hybridization showed that GmHK1-1 and GmHK1-3 were expressed in vascular and pericycle cells, as well as in dividing cortical cells (SI Appendix, Fig. S5A and Fig. 4B). In the mature nodule, GmHK1-3 expression was maintained and mostly restricted to the parenchyma and the infection zone, indicating other regulators might also be involved (Fig. 4B). In situ hybridization also showed that GmHK1-3 expression was noticeably decreased in GmSHR4/5-amiR roots (Fig. 4B). Many other genes involved in cytokinin biosynthesis also seemed to be activated by GmSHR4/5 (Fig. 4A). In GmSHR4/5-amiR roots, transcript levels for all these genes were significantly reduced (Fig. 4A). In addition, expression of CKX3, a cytokinin oxidase/dehydrogenase gene that negatively regulates cytokinin level, was decreased in OX-GmSHR4/5 roots and increased in GmSHR4/5-amiR roots at 8 dai (Fig. 4A). However, 6-Benzylaminopurine (6-BA) treatment seemed to have little effect on the expression of GmSHR4/5 (SI Appendix, Fig. S5B). To ensure the accuracy, we further examined the expression of SHR and cytokinin-related genes in individual roots of OX-GmSHR4, OX-GmSHR5, GmSHR4-amiR, GmSHR5-amiR, and GmSHR4/5-amiR (eight roots for each background). The result in Fig. 4A was verified in single-root expression analyses (SI Appendix, Fig. S5D), indicating that GmSHR4/5 promote nodule formation by activating cytokinin signaling.
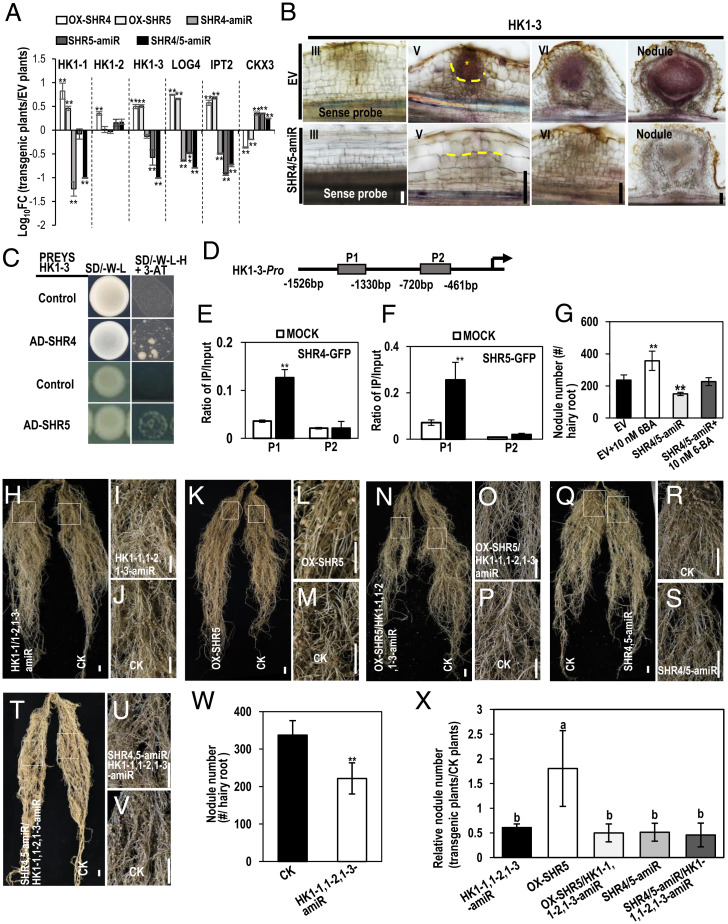
Fig. 4.
GmSHR4 and GmSHR5 function upstream of cytokinin signaling during nodulation. (A) The relative transcript of cytokinin genes in EV, OX-GmSHR4, OX-GmSHR5, GmSHR4-amiR, GmSHR5-amiR, and GmSHR4/5-amiR lines. The RNA was extracted from the noninoculated roots. GmEF1α (accession no. X56856) was used as the internal control. Significant differences are observed between transgenic plants and EV plants. (**P < 0.01; Student’s t test). Error bars represent SD. (B) In situ hybridization of GmHK1-3 in EV and GmSHR4/5-amiR lines. Yellow asterisk marks the division in outer ground tissues. Yellow dashed line defines the division in outer ground tissues (n ≥ 10). (C) Y1H assays showing the interaction between GmSHR4/5 and HK1-3 promoter. Promoters in pHIS 2 vector and pGADT7–GmSHR4/5 were cotransformed into yeast strain Y187. The empty pGADT7 vector (AD) was used as the negative control. The yeast clones were grown on SD/–Leu/–His/–Trp (–L–W–H) medium with 60 mM 3-AT (n = 3). (D) Regions of the GmCYCD6;1-6 promoter were used for ChIP (P1 to P2) assays. (E and F) The ratio of bound promoter fragments versus total input detected by qRT-PCR after immunoprecipitation of GFP-GmSHR4/5 by GFP antibodies. Data are means (± SE), n = 3. (G) The exogenous 6-BA can rescue the defective nodulation in GmSHR4/5-amiR roots. Shown is nodule number in roots with 10 nM 6-BA treatment (n ≥ 20). (H–V) Nodulation in GmHK1-1,1-2,1-3-amiR, OX-GmSHR5/GmHK1-1,1-2,1-3-amiR roots and GmSHR4/5-amiR/GmHK1-1,1-2,1-3-amiR roots. (I and J) Close-up view of the boxed areas in H. (L and M) Close-up view of the boxed areas in K. (O and P) Close-up view of the boxed areas in N. (R and S) Close-up view of the boxed areas in Q. (U and V) Close-up view of the boxed areas in T. (W and X) Nodule number. The roots were examined at 16 dai with rhizobia. Significant differences are observed between transgenic plants and CK plants (**P < 0.01, *P < 0.05; Student’s t test). Error bars represent SD. Different letters indicate significant differences between genotypes (P < 0.05 by Tukey’s test). (Scale bars, 1 cm.) CK, control check.
To examine how GmSHR4/5 regulate GmHK1-3 expression, we performed a yeast one-hybrid (Y1H) assay. We found that GmSHR4/5 could directly bind to the promoter of GmHK1-3 (Fig. 4C). To validate this result, we performed chromatin immunoprecipitation (ChIP)-qPCR using transgenic hairy roots expressing SHR4/5:GFP. Two primer pairs were designed to target two regions (P1 and P2) in the GmHK1-3 promoter (SI Appendix, Table S1). We detected strong enrichment of GmSHR4/5 in the P1 region of the GmHK1-3 promoter (Fig. 4 D–F).
Finally, we examined the effect of exogenous cytokinin (6-BA) on nodule formation. When the 6-BA concentration was increased from 5 to 10 nM, we started to see an increase in nodule number in the treated groups (SI Appendix, Fig. S6 A–D). When GmSHR4/5-amiR roots were treated with 10 nM 6-BA, their nodulation phenotype was fully rescued (Fig. 4G and SI Appendix, Fig. S6 E–H).
To further examine the function of HK1s, we knocked down their expression (GmHK1-1,1-2,1-3-amiR hereafter) in soybean roots (SI Appendix, Fig. S6I). As expected, the total number of nodules was significantly lower on GmHK1-1,1-2,1-3-amiR roots (Fig. 4 H–J and W).
To examine the genetic interactions between SHR and HK1, we combined GmHK1-1,1-2,1-3-amiR with both OX-SHR5 and SHR4/5-amiR roots in a transgenic hair root system (SI Appendix, Fig. S6 J and K). Compared with OX-SHR5 roots, OX-SHR5/GmHK1-1,1-2,1-3-amiR roots exhibited significantly decreased nodulation, showing the similar phenotypes to GmHK1-1,1-2,1-3-amiR roots (Fig. 4 H–V). Quantification of nodule numbers confirmed the similarity between OX-SHR5/GmHK1-1,1-2,1-3-amiR and GmHK1-1,1-2,1-3-amiR roots, suggesting the potential epistasis of HK1 over SHR5 (Fig. 4X). In addition, no further reduction of nodules was observed in GmSHR4/5-amiR/GmHK1-1,1-2,1-3-amiR lines, suggesting they presumably act in the same pathway.
Early Nodulin Genes Act Downstream of GmSHR4/5.
Cytokinin interacts with Nod factor signaling to control the expression of nodulin genes during nodulation. Therefore, we next examined how GmSHR4/5 interplay with these symbiotic marker genes (11, 27, 38, 39). We first verified that cytokinin indeed activated the early nodulin genes (SI Appendix, Fig. S4B). Most of the early nodulin genes, including GmNIN, GmNSP1, and GmENOD40, were up-regulated in OX-GmSHR4/5 roots while down-regulated in GmSHR4/5-amiR roots (SI Appendix, Fig. S7A). In situ hybridization showed that GmNIN was mostly expressed in the pericycle cells (SI Appendix, Fig. S7B), along with weak expression in the epidermis in soybean roots (SI Appendix, Fig. S7B). During nodulation, GmNIN was mainly expressed in the dividing cortical cells (SI Appendix, Fig. S7C). Notably, GmNIN expression was noticeably decreased in GmSHR4/5-amiR roots (SI Appendix, Fig. S7 E–G). Conversely, we also tested whether GmSHR4/5 expression was affected by the early nodulin genes. In the soybean roots overexpressing GmENOD40 or GmNSP, we detected no dramatic change in the expression of GmSHR4 (SI Appendix, Fig. S7H). However, GmSHR5 was surprisingly down-regulated in the presence of high levels of GmENOD40 or GmNSP1 (SI Appendix, Fig. S7H). These findings suggest that GmSHR4/5 likely function upstream of early nodulin genes but that GmSHR5 expression may be subject to feedback inhibition by these genes, which could partially explain why GmSHR5 transcript levels were dramatically reduced during the later stages of the nodulation. Furthermore, our Y1H results showed that GmSHR4 directly targeted the GmNSP1 promoter for activation (SI Appendix, Fig. S7I). Therefore, we concluded that these early nodulin genes are downstream components of the GmSHR4/5 and cytokinin signaling pathway.
GmCYCD6;1 Homologs Are Functional Components of GmSHR4/5-Mediated Nodulation.
In Arabidopsis, SHR regulates periclinal cell divisions by promoting the specific expression of a D-type cyclin, CYCD6;1 (35). Therefore, we asked whether this conserved module is part of the developmental program for nodulation in soybean. In the G. max genome, there are six GmCYCD6;1 homologs (named GmCYCD6;1-1 to GmCYCD6;1-6). Phylogenetic analysis revealed that GmCYCD6;1-1 to GmCYCD6;1-4 are closely related to Arabidopsis AtCYCD6;1 but GmCYCD6;1-5 and GmCYCD6;1-6 are distally related (SI Appendix, Fig. S8A). Transcriptional analysis showed that all GmCYCD6;1s were substantially up-regulated in OX-GmSHR4/5 roots and significantly down-regulated in GmSHR4/5-amiR roots (SI Appendix, Fig. S8B). We further examined the spatial expression patterns of GmCYCD6;1 using in situ hybridization. The results showed that GmCYCD6;1-1, GmCYCD6;1-2, GmCYCD6;1-3, GmCYCD6;1-4, and GmCYCD6;1-5 were all expressed in the vascular tissues (SI Appendix, Fig. S8 C–Q). In contrast, both in situ hybridization and the promoter:GUS reporter showed that GmCYCD6;1-6 expression was mostly enriched in the dividing cortical cells (SI Appendix, Fig. S8 S, U, and V). In mature nodules, GmCYCD6;1-6 was expressed in nodule parenchyma (SI Appendix, Fig. S8W).
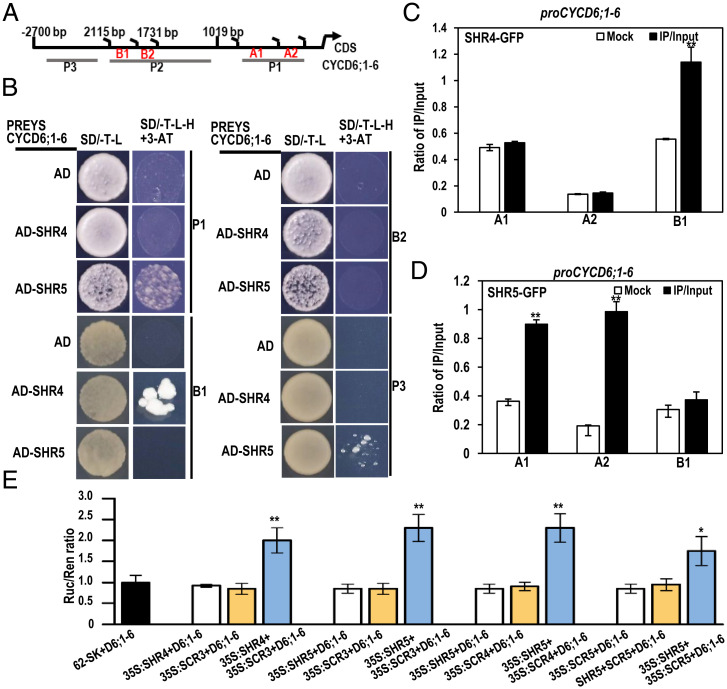
Next, we performed a Y1H assay to determine whether the regulation of GmCYCD6;1-6 by GmSHR4/5 is direct. We tested four regions of the GmCYCD6;1-6 promoter (P1, B1, B2, and P3), as shown in Fig. 5A. We found that GmSHR4 and GmSHR5 directly bound to the B1 and P1 regions of the GmCYCD6;1-6 promoter (Fig. 5B). To validate this result, we performed ChIP-qPCR using transgenic hairy roots expressing 35S:GmSHR4/5-GFP. To detect immunoprecipitated DNA fragments bound by GmSHR4/5-GFP, we designed primers targeting the GmCYCD6;1-6 promoter (regions A1, A2, and B1) (Fig. 5A). We observed a 2.05-fold enrichment of GmSHR4 in region B1 (Fig. 5C) and 2.48- and 5.15-fold enrichments of GmSHR5 in regions A1 and A2, respectively (Fig. 5D). Despite these results, we failed to detect the activation of GmCYCD6;1-6 by GmSHR4/5 in the LUC activation reporter system (Fig. 5E).
Fig. 5.
D-type cyclins act downstream of GmSHR4/5 to promote nodulation. (A) Regions of the GmCYCD6;1-6 promoter were used for ChIP and Y1H assays (A–D). (B) Y1H analyses of the interaction between GmSHR4/5 and the promoter of CYCD6;1-6 (n = 3). (C and D) The ratio of bound promoter fragments versus total input detected by qRT-PCR after immunoprecipitation of GFP-GmSHR4/5 by GFP antibodies. Data are means (± SE), n = 3. (E) LUC activation reporter assays. D6 represents the reporter of CYCD6;1-6 promoter. D6, CYCD6 (n = 3). Asterisks indicate significant differences using Student’s t test (*P < 0.05; **P < 0.01).
In Arabidopsis, AtSHR was reported to form a complex with AtSCR to activate CYCD6;1 (35). To see whether this mechanism is also conserved in soybean, we next examined the effect of GmSCRs on the expression of GmCYCD6;1-6. There are six GmSCR genes in the soybean genome, clustering into four groups in our phylogenetic analysis (SI Appendix, Fig. S9A). Using Y2H assays, we detected physical interactions between multiple GmSHR-GmSCR pairs (GmSHR4 and GmSCR3, GmSHR5 and GmSCR3, and GmSCR4 and GmSCR5) (SI Appendix, Fig. S9B). We then tested whether the various GmSCRs can help activate GmCYCD6;1-6. In a LUC reporter system, we indeed detected activation of GmCYCD6;1-6 when both GmSHR and GmSCR were added (Fig. 5E).
GmCYCD6;1s Function Downstream of Early Nodulin Genes.
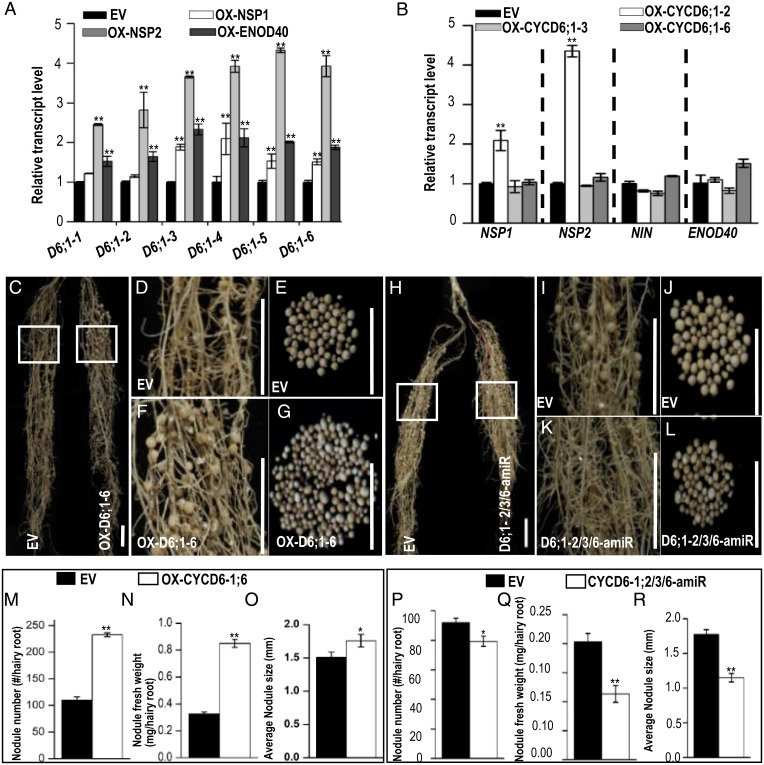
In OX-GmENOD40, OX-GmNSP1, and OX-GmNSP2 roots, the expression of GmCYCD6;1-3 to GmCYCD6;1-6 was markedly increased, while the expression of GmCYCD6;1-1 and GmCYCD6;1-2 seemed to be increased only in OX-GmENOD40 and OX-GmNSP2 roots (Fig. 6A). Although the transcriptional analysis supported that GmCYCD6;1s function downstream of early nodulin genes, GmCYCD6;1 seemed to have a feedback effect on nodulin gene expression. When GmCYCD6;1-2 was overexpressed in soybean roots (Fig. 6B), the expression of GmNSP1 and GmNSP2 was up-regulated (Fig. 6B). Interestingly, the spatial expression pattern of pGmENOD40:GUS was similar to that of pGmCYCD6;1-6:GUS (SI Appendix, Figs. S8 R–W and S9 C–F).
Fig. 6.
D-type cyclins act downstream of early nodulin genes. (A) The relative expression of GmCYCD6;1s in EV and the OX-GmNSP1, OX-GmNSP2, and OX-GmENOD40 lines. The RNA was extracted from the noninoculated roots. GmEF1α (accession no. X56856) was used as the internal control. Significant differences are observed between transgenic plants and EV plants (**P < 0.01, *P < 0.05; Student’s t test). Error bars represent SD. D6, CYCD6 (n = 3). (B) The relative expression of early nodulin genes in EV and the OX-GmCYCD6;1-2, OX-Gm CYCD6;1-3, and OX-CYCD6;1-6 lines. GmEF1α (accession no. X56856) was used as the internal control. Significant differences are observed between transgenic plants and EV plants (**P < 0.01, *P < 0.05; Student’s t test). Error bars represent SD (n = 3). (C–G) Nodulation in OX-GmCYCD6;1-6 roots. D6, CYCD6. (D and F) Close-up view of the boxed areas in C. (Scale bars: C, 2 cm; D and F, 5 mm.) D6, GmCYCD6. (H–L) Nodulation in CYCD6;1-2/3/6-amiR roots. (I and K) Close-up view of the boxed areas in H. (Scale bars: H, 2 cm; I and K, 5 mm.) D6, GmCYCD6. (M–R) The quantification of the nodulation in OX-CYCD6;1-6 roots and CYCD6-1;2/3/6-amiR. Shown are nodule number (M and P), nodule fresh weight (N and Q), and nodule size (O and R) (n ≥ 18). Significant differences are observed between transgenic plants and EV plants (**P < 0.01, *P < 0.05; Student’s t test). Error bars represent SD. n represents the number of independent biological samples. All experiments were repeated three times.
Cytokinin is a key regulator of root nodulation and the nodulin genes (23, 26). In soybean roots, cytokinin also markedly activated the expression of GmCYCD6;1-3/4/6 (SI Appendix, Fig. S5C). We next examined the role of the GmCYCD6;1 genes in soybean root nodulation. We manipulated the expression of three of the GmCYCD6;1 genes by overexpression and microRNA-mediated knockdown (SI Appendix, Fig. S10 A and B). As shown in SI Appendix, Figs. S7 C–R and S9 C–J, overexpression of GmCYCD6;1-2, GmCYCD6;1-3, and GmCYCD6;1-6 all significantly increased nodulation, as evidenced by increased nodule number, total nodule fresh weight, and nodule size. By contrast, knockdown of any of these three GmCYCD6;1 genes resulted in inhibition of nodulation. These results indicate that the GmCYCD6;1 genes function as positive regulators of soybean nodulation downstream of the early nodulation genes.
Spatially Separated Cortical Cell Divisions Account for the Formation of Nodule Primordia and Vascular Founder Cells.
The two distinct locations of cortical cell divisions in the early stage of nodulation prompted us to hypothesize that the cell divisions in the outer cortical layers are mainly responsible for the formation of nodule primordia and the divisions in the inner cortex produce the founder cells for the vascular tissues of the nodules. To test this hypothesis, we first examined the expression of GmSHR1 in nodule, a close homolog of AtSHR, which is expressed when the vascular founder cells are initiated in the globular stage of embryo development (39). Similar to AtSHR, GmSHR1 had a specific expression in the stele (SI Appendix, Figs. S2E and S11A). The GmSHR1 was not induced by the rhizobia inoculation, but showed patchy expression underlying the newly formed nodule primordia (SI Appendix, Fig. S11 B–D). This expression persisted and gradually became restricted to the newly formed vascular tissues of the nodule (SI Appendix, Fig. S11 E and F). In line with this hypothesis, knockdown of GmSHR4/5 considerably disrupted the cell divisions in inner cortex and affected the formation of nodule vascular tissues (Fig. 3 J–L).
In the outer cortex, repeated cell divisions drove the emergence of primordia-like structures from the soybean roots (SI Appendix, Fig. S3 E, K, and P). This bulging tissue was similar in morphology to the lateral root primordia. We therefore tested whether the primordia-like structures have stem cells by examining the expression of the marker gene GmWOX5. In situ hybridization showed that expression of GmWOX5 was initiated in the early stage of nodulation, when the bulging structures were formed (SI Appendix, Fig. S11G). The spatial expression of GmWOX5 in these structures was very similar to that in lateral roots, with enrichment in a few cells located at the top of the bulging tissue (SI Appendix, Fig. S11H). In the mature nodule, GmWOX5 expression was maintained, but was mostly restricted to the nodule parenchyma (SI Appendix, Fig. S11I).
Discussion
Several lines of evidence have shown that nodule organogenesis is evolutionarily related to root development (5, 28, 29). Genes important for regulating root stem cells, including WOX5 and PLETHORA (PLT1-4), play vital roles in the establishment of root nodule primordia (32–34). Nodule-specific knockdown of MtPLETHORA genes results in a reduced number of nodules or generates nodules with inactive meristems. Similar to lateral root development, nodule organogenesis starts from oriented cell divisions in root cortical tissues. However, we still have very limited understanding of the mechanism behind this cortical cell division during nodulation. In Arabidopsis roots, SHR-SCR-CYCD6;1 forms a regulatory module that spatiotemporally promotes periclinal cell divisions in cortex/endodermis initials (35). There is also evidence that AtSHR and rice SHR homologs can induce periclinal cell divisions in cortical tissues in root meristem (40, 41). The first cell division of a determinate nodule (such as those of soybean) occurs subepidermally in the outer cortex (42, 43). In this study, we provide evidence demonstrating that two SHR paralogs in soybean regulate nodulation by promoting cortical cell divisions in the cortex.
We identified six SHR homologs in soybean, which were differentially expressed in roots, stems, leaves, and nodules, implying that GmSHR could participate in a wide range of developmental processes in soybean. Four of the soybean SHR homologs are closely related to AtSHR and MtSHR1 (Medtr5g015490) (36) and exhibited a similar vascular expression pattern, while two distal paralogs (GmSHR4 and GmSHR5) had a distinct expression pattern. This implies that subfunctionalization of SHR homologs may have occurred during soybean evolution. Interestingly, GmSHR4 and GmSHR5 exhibited high levels of expression in the initial stage of nodule development, when nodule vascular tissues had not yet formed. In contrast, GmSHR1 and GmSHR2 were also induced during nodulation but were specifically expressed in the vascular tissues of the mature nodules. These results suggest that GmSHR family members may differentially regulate different stages of soybean nodulation. Upon the Rhizobial infection, the expression of GmSHR4/5 was activated in outer cortical cells, where nodule primordia were initiated. In the outer cortical cells, GmSHR5 seemed to play a more dominant role as the inhibition of GmSHR5 affected the formation of the nodule primordia. However, GmSHR4 and GmSHR5 could also be functionally redundant during nodulation. The overexpression of either of these two GmSHR genes substantially improved nodulation, suggesting that they could functionally replace each other.
Previous studies revealed that cytokinin signaling interacts with Nod factors to promote cortical cell division during nodulation (24). Although we know that a number of early nodulin genes are involved, the detailed mechanism of this stimulation of cortical cell division remains a mystery and it is not clear how the upstream signaling cascade is transduced to regulate cell division. Our results provide evidence that soybean GmSHR4/5 are a signaling component downstream of Nod factor perception but upstream of cytokinin as well as early nodulin genes, including GmNSP1, GmNIN, and GmENOD40. Interestingly, this regulation seems to involve a complex network rather than a linear cascade. A recent report showed increased MtSHR1 expression after 6-BA treatment in Medicago, and 6-BA effect seemed to be abolished in MtSHR1-SRDX hairy roots (36). However, in soybean roots, 6-BA treatment was insufficient to promote GmSHR4/5 expression and genetic analysis showed that GmHK1 presumably functions downstream of GmSHR4/5 during the nodulation in soybean. This difference might reflect the differential mechanism between indeterminate nodules and determinate nodules.
The expression of GmSHR5 could be suppressed when GmNSP1 and GmENOD40 were overexpressed. This potential feedback loop is also evidenced by the differential ability of GmSHR4 and GmSHR5 to induce nodules. Compared to the substantially increased nodule number induced by overexpression of GmSHR4, overexpression of GmSHR5 only moderately enhanced nodulation, which may reflect the feedback inhibition of GmNSP1 on GmSHR5. Both GmSHR4 and GmSHR5 seemed to play roles only in the early stage of nodulation, as their expression was hard to detect in mature nodules. Therefore, GmSHR4/5 could represent a key regulatory module of NFR-mediated early symbiotic events.
ENOD40 was previously shown to promote cortical cell division (44). However, the link between ENOD40 and downstream mitosis is not known. Here we showed that GmENOD40 induced the expression of D-type cyclins. Considering the role of the Arabidopsis D-type cyclin homolog (AtCYCD6;1) in promoting periclinal cell division, it is likely GmENOD40 promotes cortical cell division during nodulation by inducing GmCYCD6;1 expression. In line with this, GmCYCD6;1 expression in soybean roots was also enhanced by the application of cytokinin, which could induce spontaneous nodules without rhizobial infection.
In Arabidopsis, it was discovered that AtSHR regulates the development of vascular tissues by repressing cytokinin biosynthesis (45). Interestingly, in poplar, PtSHR1 seemed to increase cytokinin biosynthesis to promote both primary (height) and secondary (girth) growth (46). Soybean has six SHR homologs that can be clustered into two groups. GmSHR4/5 not only are distally related to the other GmSHR genes phylogenetically, but also exhibited expression patterns distinct from the other GmSHR genes. This suggests that the GmSHR genes have likely undergone subfunctionalization during their evolution. Through multiple assays, we showed that soybean SHRs promote nodulation by enhancing both cytokinin biosynthesis and signaling. In addition, GmSHR4/5 seemed to promote GmCYCD6;1-6 expression by directly binding to its promoter. This direct activation mechanism appears to be evolutionarily conserved as a similar mechanism also exists in Arabidopsis roots. Finally, GmSHR4/5 can also activate the expression of D-type cyclins via the early nodulins GmNIN and GmENOD40. The two SHR-mediated cascades that activate D-type cyclins form a classic feedforward loop to ensure the progression of nodule organogenesis (SI Appendix, Fig. S12).
Materials and Methods
Additional materials and methods are described at length in SI Appendix, SI Materials and Methods.
G. max cv. Williams-82 seeds and rhizobium strain Bradyrhizobium sp. BXYD3 were used throughout the experiments. Roots or nodules for histological analysis were embedded in a 5% agarose gel and 100-μm sections were obtained with a microtome (Leica VT1000S). Details are provided in SI Appendix, SI Materials and Methods.
Accession numbers are listed in SI Appendix, Table S2.
Supplementary Material
Acknowledgments
We thank Dr. Hong Liao for sharing research materials. This work is supported by the National Key Research and Development Program of China (2016YFD0100700, 2018YFD1000800) and the National Natural Science Foundation of China (31722006).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108641119/-/DCSupplemental.
Data Availability
qRT-PCR, GUS staining, construction of phylogenetic tree, genetics assay, in situ, and biochemistry assay data (Glyma.01G194200, Glyma.05G105600, Glyma.11G047700, Glyma.13G345700, Glyma.15G028600, Glyma.17G160800, Glyma.13G303000, Glyma.12G199300, Glyma.12G105600, Glyma.06G299700, Glyma.13G247600, Glyma.15G066200, Glyma.07G039400, Glyma.04G251900, Glyma.11G188100, Glyma.03G181300, Glyma.07G158800, Glyma.08G049000, Glyma.07G173700, Glyma.17G054500, Glyma.04G000600, Medtr8g020840.1, Glyma.01G028500, Glyma.03G123800, Glyma.02g42200, Glyma.09G270000, Glyma.11G096500, Glyma.12G022600, Glyma.10G038500) have been deposited in Phytozome. Construction of phylogenetic tree data (Solyc02g092370) have been deposited in Sol Genomics Network. Construction of phylogenetic tree data (AT4G37650 and AT4G03270) have been deposited in Tair. Construction of phylogenetic tree data (AJ832138, ABK35066.1, ABG49438.1, AAP06856.1, NP_192565.1, AJ832139, XP_012574062.1, XP_020232189.1) have been deposited in NCBI. All other study data are included in this article and/or SI Appendix.
References
- 1.Nishida H., Suzaki T., Nitrate-mediated control of root nodule symbiosis. Curr. Opin. Plant Biol. 44, 129–136 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Tsikou D., et al. , Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 362, 233–236 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Crespi M., Frugier F., De novo organ formation from differentiated cells: Root nodule organogenesis. Sci. Signal. 1, re11 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Ferguson B. J., et al. , Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 52, 61–76 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Desbrosses G. J., Stougaard J., Root nodulation: A paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 10, 348–358 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Oldroyd G. E., Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Calvert H. E., Pence M. K., Pierce M., Malik N. S., Bauer W. D., Anatomical analysis of the development and distribution of Rhizobium infections in soybean roots. Botany 62, 2375–2384 (1984). [Google Scholar]
- 8.Schauser L., Roussis A., Stiller J., Stougaard J., A plant regulator controlling development of symbiotic root nodules. Nature 402, 191–195 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Soyano T., Kouchi H., Hirota A., Hayashi M., Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 9, e1003352 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W. C., et al. , Characterization of GmENOD40, a gene showing novel patterns of cell-specific expression during soybean nodule development. Plant J. 3, 573–585 (1993). [DOI] [PubMed] [Google Scholar]
- 11.Crespi M. D., et al. , enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 13, 5099–5112 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Compaan B., Yang W. C., Bisseling T., Franssen H., ENOD40 expression in the pericycle precedes cortical cell division in Rhizobium-legume interaction and the highly conserved internal region of the gene does not encode a peptide. Plant Soil 230, 1–8 (2001). [Google Scholar]
- 13.Hirsch S., et al. , GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21, 545–557 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi S., et al. , Transient Nod factor-dependent gene expression in the nodulation-competent zone of soybean (Glycine max [L.] Merr.) roots. Plant Biotechnol. J. 10, 995–1010 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Miri M., Janakirama P., Held M., Ross L., Szczyglowski K., Into the root: How cytokinin controls rhizobial infection. Trends Plant Sci. 21, 178–186 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Liu H., Zhang C., Yang J., Yu N., Wang E., Hormone modulation of legume-rhizobial symbiosis. J. Integr. Plant Biol. 60, 632–648 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Plet J., et al. , MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 65, 622–633 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Heckmann A. B., et al. , Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Mol. Plant Microbe Interact. 24, 1385–1395 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Held M., et al. , Lotus japonicus cytokinin receptors work partially redundantly to mediate nodule formation. Plant Cell 26, 678–694 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferguson B. J., Mathesius U., Phytohormone regulation of legume-rhizobia interactions. J. Chem. Ecol. 40, 770–790 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Frugier F., Kosuta S., Murray J. D., Crespi M., Szczyglowski K., Cytokinin: Secret agent of symbiosis. Trends Plant Sci. 13, 115–120 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Boivin S., et al. , Different cytokinin histidine kinase receptors regulate nodule initiation as well as later nodule developmental stages in Medicago truncatula. Plant Cell Environ. 39, 2198–2209 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Gamas P., Brault M., Jardinaud M. F., Frugier F., Cytokinins in symbiotic nodulation: When, where, what for? Trends Plant Sci. 22, 792–802 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Tirichine L., et al. , A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315, 104–107 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Mortier V., et al. , Role of LONELY GUY genes in indeterminate nodulation on Medicago truncatula. New Phytol. 202, 582–593 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Lohar D. P., et al. , Cytokinins play opposite roles in lateral root formation, and nematode and Rhizobial symbioses. Plant J. 38, 203–214 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Vernié T., et al. , The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell 27, 3410–3424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roudier F., et al. , The Medicago species A2-type cyclin is auxin regulated and involved in meristem formation but dispensable for endoreduplication-associated developmental programs. Plant Physiol. 131, 1091–1103 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nutman P. S., Physiological studies on nodule formation: I. The relation between nodulation and lateral root formation in red clover. Ann. Bot. (Lond.) 12, 1–96 (1948). [Google Scholar]
- 30.Soyano T., Shimoda Y., Kawaguchi M., Hayashi M., A shared gene drives lateral root development and root nodule symbiosis pathways in Lotus. Science 366, 1021–1023 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Schiessl K., et al. , NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Curr. Biol. 29, 3657–3668.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franssen H. J., et al. , Root developmental programs shape the Medicago truncatula nodule meristem. Development 142, 2941–2950 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Osipova M. A., et al. , Wuschel-related homeobox5 gene expression and interaction of CLE peptides with components of the systemic control add two pieces to the puzzle of autoregulation of nodulation. Plant Physiol. 158, 1329–1341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roux B., et al. , An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77, 817–837 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Sozzani R., et al. , Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466, 128–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong W., et al. , An SHR-SCR module specifies legume cortical cell fate to enable nodulation. Nature 589, 586–590 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Cui H., Benfey P. N., Cortex proliferation: Simple phenotype, complex regulatory mechanisms. Plant Signal. Behav. 4, 551–553 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaló P., et al. , Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308, 1786–1789 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Smit P., et al. , NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308, 1789–1791 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Wu S., et al. , A plausible mechanism, based upon Short-Root movement, for regulating the number of cortex cell layers in roots. Proc. Natl. Acad. Sci. U.S.A. 111, 16184–16189 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu Q., et al. , Cell-fate specification in Arabidopsis roots requires coordinative action of lineage instruction and positional reprogramming. Plant Physiol. 175, 816–827 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szczyglowski K., et al. , Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol. Plant Microbe Interact. 11, 684–697 (1998). [Google Scholar]
- 43.van Spronsen P. C., Grønlund M., Pacios Bras C., Spaink H. P., Kijne J. W., Cell biological changes of outer cortical root cells in early determinate nodulation. Mol. Plant Microbe Interact. 14, 839–847 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Charon C., Johansson C., Kondorosi E., Kondorosi A., Crespi M., enod40 induces dedifferentiation and division of root cortical cells in legumes. Proc. Natl. Acad. Sci. U.S.A. 94, 8901–8906 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui H., et al. , Genome-wide direct target analysis reveals a role for SHORT-ROOT in root vascular patterning through cytokinin homeostasis. Plant Physiol. 157, 1221–1231 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miguel A., Milhinhos A., Novák O., Jones B., Miguel C. M., The SHORT-ROOT-like gene PtSHR2B is involved in Populus phellogen activity. J. Exp. Bot. 67, 1545–1555 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
qRT-PCR, GUS staining, construction of phylogenetic tree, genetics assay, in situ, and biochemistry assay data (Glyma.01G194200, Glyma.05G105600, Glyma.11G047700, Glyma.13G345700, Glyma.15G028600, Glyma.17G160800, Glyma.13G303000, Glyma.12G199300, Glyma.12G105600, Glyma.06G299700, Glyma.13G247600, Glyma.15G066200, Glyma.07G039400, Glyma.04G251900, Glyma.11G188100, Glyma.03G181300, Glyma.07G158800, Glyma.08G049000, Glyma.07G173700, Glyma.17G054500, Glyma.04G000600, Medtr8g020840.1, Glyma.01G028500, Glyma.03G123800, Glyma.02g42200, Glyma.09G270000, Glyma.11G096500, Glyma.12G022600, Glyma.10G038500) have been deposited in Phytozome. Construction of phylogenetic tree data (Solyc02g092370) have been deposited in Sol Genomics Network. Construction of phylogenetic tree data (AT4G37650 and AT4G03270) have been deposited in Tair. Construction of phylogenetic tree data (AJ832138, ABK35066.1, ABG49438.1, AAP06856.1, NP_192565.1, AJ832139, XP_012574062.1, XP_020232189.1) have been deposited in NCBI. All other study data are included in this article and/or SI Appendix.