Significance
Mutations in hTERT promoter are seen in over 19% of human cancers, irrespective of the cancer type. Understanding the molecular players that regulate Mut-hTERT promoters may help the design of effective targeting strategies to inhibit telomerase reactivation specifically in cancer cells. Our work uses genome-wide functional screens to identify 30 specific regulators of Mut-hTERT promoters. These candidates identified from the screening serve as an excellent resource to understand how telomerase is reactivated and as targets for making inhibitors to telomerase, a key driver of cancer.
Keywords: telomerase, TERT, cancer, chromatin
Abstract
Cancer-specific hTERT promoter mutations reported in 19% of cancers result in enhanced telomerase activity. Understanding the distinctions between transcriptional regulation of wild-type (WT) and mutant (Mut) hTERT promoters may open up avenues for development of inhibitors which specially block hTERT expression in cancer cells. To comprehensively identify physiological regulators of WT- or Mut-hTERT promoters, we generated several isogenic reporter cells driven by endogenous hTERT loci. Genome-wide CRISPR-Cas9 and small interfering RNA screens using these isogenic reporter lines identified specific regulators of Mut-hTERT promoters. We validate and characterize one of these hits, namely, MED12, a kinase subunit of mediator complex. We demonstrate that MED12 specifically drives expression of hTERT from the Mut-hTERT promoter by mediating long-range chromatin interaction between the proximal Mut-hTERT promoter and T-INT1 distal regulatory region 260 kb upstream. Several hits identified in our screens could serve as potential therapeutic targets, inhibition of which may specifically block Mut-hTERT promoter driven telomerase reactivation in cancers.
Telomerase reverse transcriptase (hTERT), the enzymatic component of telomerase holoenzyme, is expressed in stem cells and germ cells to maintain telomere length and chromosome integrity (1–3). However, hTERT is transcriptionally repressed in terminally differentiated somatic cells. Accumulation of driver mutations during oncogenesis and ensuing replicative stress in preneoplastic cells lead to reactivation of hTERT expression for the maintenance of critically short telomeres (4–6). Both canonical and noncanonical activities of hTERT are postulated to be essential drivers of various hallmarks of cancer (7). Reactivation of hTERT is hence considered the rate-limiting step in transformation (8). In the last decade, studies have identified that hTERT reactivation in different cancers may be achieved via distinct mechanisms which involve amplification of the hTERT gene itself, activation or overexpression of cancer cell–specific oncogenic transcription factors, chromosomal rearrangements, and cancer-specific promoter mutations (6, 9). Single residue mutations C250T (-124 C > T) and C228T (-146 C > T) in the proximal promoter of hTERT gene were first identified in melanomas (74%) (10, 11), and the cooccurrence of these mutations was found to predict worse outcomes in patients with concurrent RAS mutations (12). Subsequently, hTERT promoter mutations were shown to be highly prevalent in different cancer types, including glioblastoma (83%), urothelial bladder carcinoma (53.5%), and hepatocellular carcinoma (44%) (10, 11). These point mutations, located in the proximal region of hTERT promoter, create binding sites for the ETS family of transcription factors (10, 11, 13). These transcription factors enhance hTERT gene expression, and hence telomerase activity (10, 11, 13), by dimerization within the family or with other transcription factors such as NFκB (14, 15). It is now clear that hTERT reactivation requires chromatin elements way beyond the proximal promoter, and at least one long-range chromatin interaction with a distal region (T-INT1) located 260 kb upstream mediated by dimers of GA Binding Protein Transcription Factor Alpha (GABPA) transcription factors is required for productive transcriptional activation (6). Deletion of this region destabilizes GABPA on the proximal hTERT promoter and depletes H3K4Me3 and H3K9Ac marks, and POLII occupancy, leading to hTERT repression (6). These observations suggest that a complex three-dimensional (3D) hierarchy of chromatin organization is essential for functioning of at least the Mut-hTERT promoters. Given this new literature, it is important to ask, have we identified all the physiological regulators of hTERT activation?
In light of the observations that 3D chromatin architecture (16–19), long-range chromatin interactions (6, 19), and several chromatin modulating enzymes (6) are key for driving hTERT reactivation in vivo, a comprehensive discovery of the physiological regulators of hTERT would require high-throughput methods which truly report the expression of endogenous hTERT alleles via its proximal and distal regulatory elements. It is well known that endogenous hTERT messenger RNA (mRNA) levels are unreliably low for high-throughput quantification, and no currently available antibody faithfully reports endogenous hTERT protein levels (20). Therefore, herein, we use GFP and luciferase reporters which could be detected by sensitive methods in a high-throughput manner. These reporters are driven by endogenous hTERT promoters with or without C250T and C228T mutations. Unlike the plasmid-based reporter systems, we aimed to capture 3D regulatory events that might be significant for hTERT regulation in vivo. Previously, a targeted RNA interference (RNAi) screen in murine embryonic stem cells identified HIF1a as an important factor for hTERT expression and telomerase activity (21). Similarly, an RNAi screen directed toward the cellular kinases showed ERK8 as a therapeutic target for inhibiting telomerase activity (22). However, despite the advancements in small interfering RNA (siRNA) and CRISPR-Cas9–based genome-wide single guide RNA (sgRNA) screening methods, a whole-genome screen for regulators of Mut-hTERT promoter has been long overdue. Here we report genome-wide screens for factors governing Mut-hTERT expression using both RNAi and CRISPR libraries in isogenic cell lines. Using these isogenic reporter cell lines, we identify a cluster of mediator complex subunits, specifically MED12, as a key regulator for Mut-hTERT expression across cancers. Our study represents a comprehensive genome-wide screening to uncover the regulatory factors governing Mut-hTERT expression.
Results
Generation of hTERT Reporter Cell Lines.
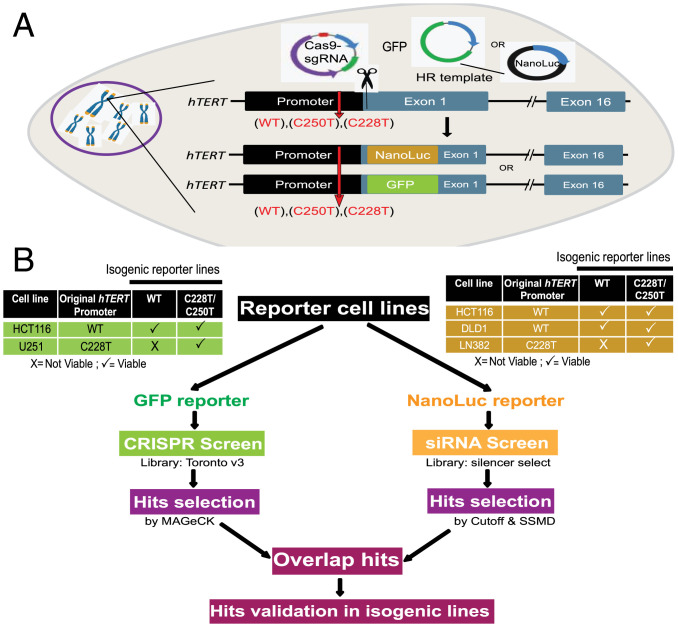
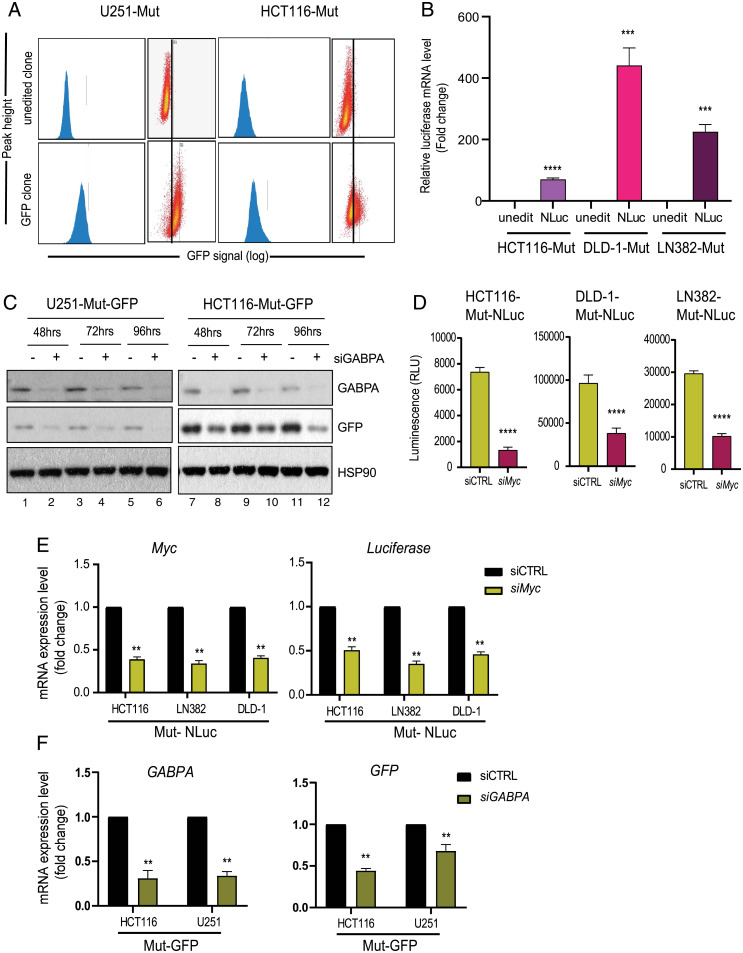
To comprehensively identify physiological regulators of wild-type (WT) and mutant (Mut) hTERT promoters, using CRISPR-Cas9–mediated genome editing, we generated several isogenic reporter cells that carry either WT or C228T/C250T Mut promoters in the endogenous hTERT locus (Fig. 1A). In these isogenic cells with three different versions of hTERT promoters (WT/C250T/C228T), we also knocked in either GFP or luciferase reporter genes to utilize them in genome-wide loss of function screens (Fig. 1B). Both reporter genes (GFP or luciferase) were inserted downstream of the hTERT transcription start site (TSS) (SI Appendix, Fig. S1A). To identify bona fide regulators of WT and Mut-hTERT promoters, irrespective of cancer type, we attempted to generate several colon cancer and glioblastoma lines as representatives of cancers that are from the opposite spectrum of hTERT promoter mutation frequencies (23). We could obtain viable engineered cells from four different cell lines representing these two cancer types (Fig. 1B). It is important to note that GFP and luciferase reporter genes were introduced only in one of the alleles, as it has been reported that 5′ tagging of hTERT gene results in decreased hTERT expression (24). It was evident that the remaining unedited copy was sufficient to maintain hTERT expression (SI Appendix, Fig. S1 B and C). Appropriate editing of the promoter mutations in the engineered lines was confirmed by Sanger sequencing (SI Appendix, Fig. S1D). In addition to confirming the precise editing of the sites, we also sequence verified the in-frame insertion of GFP and Nano luciferase genes in hTERT loci downstream of the promoter using Sanger sequencing (SI Appendix, Fig. S1E). We next checked for GFP and luciferase expression and found significant and detectable levels of GFP (Fig. 2A) or luciferase (Fig. 2B) in the reporter lines, indicating that in-frame inserted reporter genes are faithfully driven by the endogenous hTERT promoter. From here onward, we represent our engineered cell lines based on the following nomenclature that is, cell line (original name)-WT/Mut (hTERT promoter)-GFP/NLuc (reporter gene). To evaluate whether the reporter genes are functionally driven by the endogenous hTERT promoter and its relevant chromatin context, we transfected these reporter lines with siRNAs targeting known regulators of hTERT such as Myc and GABPA (6, 25–27). Myc binds to the two E-box sequences in hTERT proximal promoter and positively regulates hTERT (28). Cancer-specific mutations in the proximal hTERT promoter create binding sites for GABPA, binding of which enhances hTERT transcription (10, 11, 13, 29). Depletion of GABPA in the GFP reporter lines led to a significant reduction in GFP expression (Fig. 2C). Similarly, transient knockdown of Myc in luciferase reporter cell lines led to a significant reduction of luciferase signal (Fig. 2D). Furthermore, we also observed a significant reduction in the mRNA levels of Myc and luciferase genes upon Myc knockdown (Fig. 2E). Similarly, we also evaluated the functionality of GFP reporter cell lines, by transient knockdown of GABPA. Both reporter cells with siRNAs targeting GABPA showed reduced levels of GABPA and GFP (Fig. 2F). Collectively, these observations indicate the functionality and usability of our engineered reporter lines with promoter mutations for genome-wide loss/gain of function screens. These unique resources could be used for many chemical and genomic screens in the future.
Fig. 1.
Study design. (A) Schematic of the reporter cell lines created by CRISPR genome editing. Either GFP or NanoLuc gene is inserted under the control of endogenous hTERT promoter in exon 1 with in-frame fusion. GFP/NanoLuc insertion and promoter mutation corrections were done using a single homologous recombination (HR) template designed to span homology arms in hTERT promoter and downstream of exon 1. (B) Workflow of the genome-wide screens. The reporter lines and the algorithms that are used to identify hits from the CRISPR and siRNA screens are indicated. Hits from siRNA and CRISPR knockout screen were overlapped to identify commonality factors and were then chosen for the validation study.
Fig. 2.
Establishment and validation of reporter cell lines. (A) Representative plots of GFP sorting in U251-Mut and HCT116-Mut reporter cells. The x axis shows increased signal (log) of GFP in clones after CRISPR-mediated insertion of GFP into the endogenous hTERT locus. GFP-positive reporter lines are enriched using fluorescence-activated cell sorting (FACS) and named as U251-Mut-GFP and HCT116-Mut-GFP. (B) The qPCR data showing increased expression of the luciferase gene in clones after CRISPR-mediated insertion of NanoLuc into the endogenous hTERT locus (***P < 0.0005; ****P < 0.00005). (C) U251-Mut-GFP (Left) and HCT116-Mut-GFP (Right) cells were transfected with siRNA against GABPA. Cells were isolated for protein extraction 48, 72, and 96 h posttransfection. Protein expression analysis was performed for the indicated molecules by Western blot. HSP90 was used as a loading control. (D) HCT116-Mut-NLuc, DLD1-Mut-NLuc, and LN382-Mut-NLuc reporter lines were transfected with siControl (siCTRL) and siMYC. After 48 h, the luminescence signal of NanoLuc reporter was measured using Luciferase assay. The error bar indicates SD from at least three biological replicates (****P < 0.00005). (E) HCT116-Mut-NLuc, DLD1-Mut-NLuc, and LN382-Mut-NLuc reporter lines were transfected with siCTRL and siMYC. Gene expression analysis was performed for MYC and Luciferase by RT-qPCR. Actin was used as a control (**P < 0.01). (F) HCT116-Mut-GFP and U251-Mut-GFP reporter lines were transfected with siControl (siCTRL) and siGABPA. The qPCR results indicate the expression level of GABPA and GFP after transient knockdown of GABPA by siRNA. The error bars indicate SD from at least three biological replicates (**P < 0.01).
Whole-Genome siRNA Screen to Identify Regulators of hTERT Promoter.
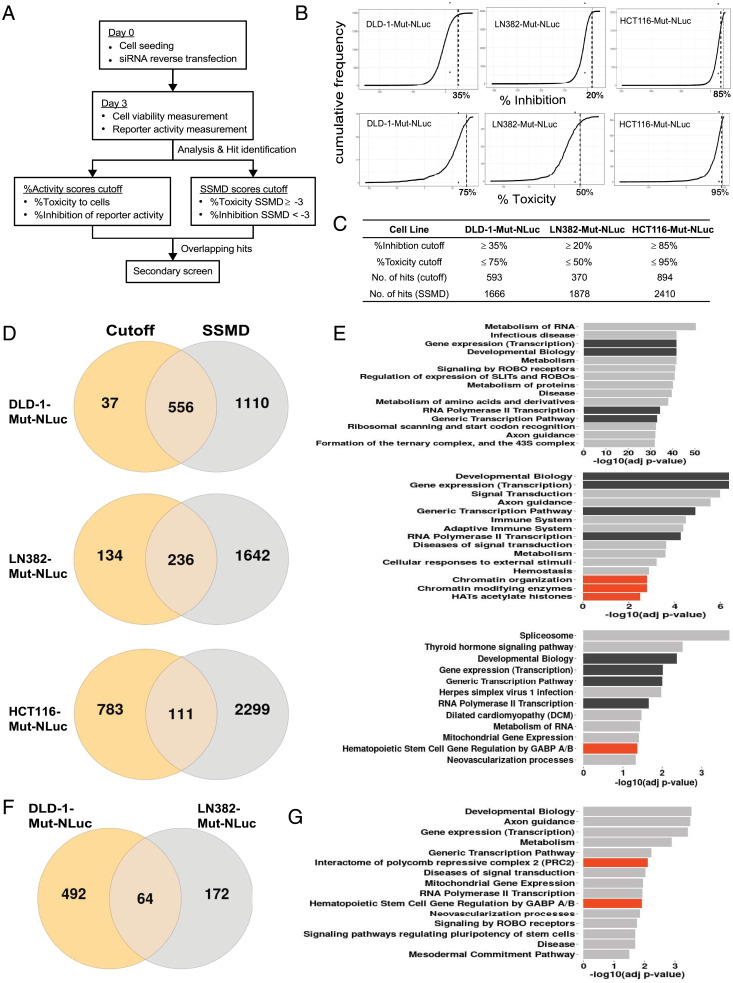
For the comprehensive and unbiased discovery of regulators of hTERT, we performed whole-genome siRNA screens in three cell lines which survived the editing process and could be expanded for large-scale screening. These lines, DLD-1-Mut-NLuc, LN382-Mut-NLuc, and HCT116-Mut-NLuc (Fig. 1B), are representative of different cancer types (i.e., DLD-1 vs. LN382) and different hTERT promoter mutation status (i.e., DLD-1 harbors the C228T mutations, while HCT116 harbors the C250T mutations), and we reasoned that common hits from these lines would likely help us narrow down the genuine regulators of Mut-hTERT promoter. The workflow for each siRNA screen is illustrated in Fig. 3A. Briefly, on day 0, cells were seeded and reverse transfected with arrayed siRNAs in 384-well plate format for 3 d. Signals obtained using PrestoBlue reagent had a poor correlation between replicates (SI Appendix, Fig. S2A). In contrast, luminescence-based NanoLuc readings (obtained using Nano-Glo luciferase assay system) to measure hTERT promoter activity displayed an excellent correlation between replicates (SI Appendix, Fig. S2B). Consequently, for the LN382-Mut-NLuc and HCT116-Mut-NLuc siRNA screens, we switched to using cytomegalovirus promoter-driven firefly luciferase (FLuc) expression to quantify and normalize the cell viability. This is also a luminescence-based readout similar to the NanoLuc reporter, except that the substrates used are different. Results from the subsequent screens using LN382-Mut-NLuc and HCT116-Mut-NLuc cells showed an excellent correlation between replicates with FLuc activity as a measure of cell viability (SI Appendix, Fig. S2 C–F). To identify hits, we used a combination of the percentage activity scores cutoff (cutoff method) and the strictly standardized means difference (SSMD) method and overlapped these results for common hit identification (30, 31). The main consideration in the selection of hits was that the siRNAs should have potent inhibition of the hTERT promoter-driven reporter activity (i.e., NanoLuc) while having minimal effects on the viability of the cells (i.e., low toxicity). This is to enable us to identify specific and direct regulators of hTERT promoter activity. In the first approach, we calculated the percentage toxicity (%Toxicity) and percentage hTERT promoter inhibition (%Inhibition) scores for each gene in the siRNA screen by normalizing their signals against the respective signals obtained from negative control siRNA–treated cells. A cumulative plot of all the %Inhibition scores for all the siRNAs investigated was then obtained for each cell line (Fig. 3B). Based on these cumulative plots, we observed cell line–specific differences in the effects of siRNA on hTERT promoter activity. This was expected, as each cell line screened has inherently different mutational and gene expression profiles due to their tissue of origin (32, 33). Based on the %Inhibition cumulative plots, we first decided on a cutoff for each cell line based on the point of inflection, as this point marks the boundary between specific and nonspecific effects (Fig. 3B). Genes for which the siRNAs resulted in %Inhibition scores greater than or equal to the cutoff (35% for DLD-1-Mut-NLuc, 20% for LN382-Mut-NLuc, and 85% for HCT116-Mut-NLuc) were plotted on a second cumulative plot based on their %Toxicity scores (Fig. 3B). From the second cumulative plot, a cutoff for the %Toxicity score was determined similarly as for the %Inhibition cutoff. Genes for which the siRNAs resulted in % Toxicity scores less than or equal to the cutoff (75% for DLD-1-Mut-NLuc, 50% for LN382-Mut-NLuc, and 95% for HCT116-Mut-NLuc) were selected. Consequently, based on this cutoff method, we identified 593, 370, and 894 hits for DLD-1-Mut-NLuc, LN382-Mut-NLuc, and HCT116-Mut-NLuc cells, respectively (Fig. 3C).
Fig. 3.
The siRNA screen in NanoLuc reporter cell lines. (A) Workflow of siRNA screen illustrating the performance and selection methods used to identify the hits. (B) Plots showing the inhibition and toxicity thresholds used in different lines to select hits from each siRNA screen. (C) Table listing the number of hits identified from each siRNA screen utilizing two different algorithms using threshold level from B. (D) Venn diagram showing the overlap of hits obtained from two algorithms in different cell lines (DLD-1-Mut-NLuc, Top; LN382-Mut-NLuc, Middle; HCT116-Mut-NLuc, Bottom). (E) Bar plots showing the enrichment of pathway terms (top 15, adj. P value < 0.05) for the overlapping hits (DLD-1-Mut-NLuc, Top; LN382-Mut-NLuc, Middle; HCT116-Mut-NLuc, Bottom) identified from D. The dark bars indicate common cell survival–related pathways seen among all the lines; orange bars indicate the pathway terms related to hTERT regulation seen in specific cell lines. (F) Overlap of hits identified from two different algorithms (Cutoff and SSMD) in DLD-1-Mut-NLuc and LN382-Mut-NLuc cell lines. The methods of Cutoff and SSMD are described in Methods. (G) Graph shows the pathway terms of the common genes between DLD-1-Mut-NLuc and LN382-Mut-NLuc reporter cell lines. The pathway terms were obtained from the gProfiler (g:GOst) using its inbuilt default functions.
Simultaneously, we also analyzed the siRNA screen results using GUItars, a siRNA screen analysis tool based on the SSMD method (30). For each cell line screened, we obtained two SSMD scores for each gene investigated: one SSMD score for toxicity (toxicity SSMD score) and one SSMD score for hTERT promoter activity inhibition (inhibition SSMD score). The use of SSMD as the criteria for hit selection has been reported as a means to control for false-positive and false-negative rates (34). The SSMD score obtained is also a measure for the strength of the effect with SSMD scores > 3, representing very strong activation or up-regulation effects and SSMD scores < −3, representing very strong inhibition or down-regulation effects (34). As mentioned above, our criteria for hit selection was that the siRNAs should have potent hTERT promoter activity inhibition with minimal toxicity. Since a toxicity SSMD score < −3 indicated strong toxic effects, we set a cutoff of ≥−3 for toxicity SSMD score. Conversely, for hTERT promoter activity inhibition, we selected siRNAs for which their inhibition SSMD score was <−3 (Fig. 3A). Based on these criteria, we identified 1,666, 1,878, and 2,410 hits for DLD-1-Mut-NLuc, LN382-Mut-NLuc, and HCT116-Mut-NLuc cells, respectively (Fig. 3D). Combining the hits identified from the cutoff method and the SSMD method, we found 556, 236, and 111 common hits for DLD-1-Mut-NLuc, LN382-Mut-NLuc, and HCT116-Mut-NLuc cells, respectively (Fig. 3D). Pathway enrichment analysis of the common hits for each cell line revealed similarities across the cell lines investigated. Among the top 15 enriched pathways for each cell line, pathways involved in transcription and development biology (Fig. 3E, black bars) are common across all three cell lines. Interestingly, in the LN382-Mut-NLuc cell line, we also identified chromatin organization and modification pathways (Fig. 3 E, Middle, red bars). Pathway enrichment of the hits identified in the HCT116-Mut-NLuc cell line also identified hematopoietic stem cell (HSC) gene regulation by GABPA/B (Fig. 3 E, Bottom, red bar) as one of the significantly enriched pathways. This was particularly reassuring, as this corroborates previous findings on the importance of GABPA in the regulation of hTERT expression specifically by Mut-hTERT promoters (5, 6). We also compared the hits identified from DLD-1-Mut-NLuc and LN382-Mut-NLuc siRNA screens and found 64 common hits (Fig. 3F). Pathway enrichment of the 64 common genes also identified HSC gene regulation by GABPA/B as one of the enriched pathways, as well as the interactome of the polycomb repressive complex 2 (Fig. 3G, red bars).
Whole-Genome CRISPR Knockout Screen to Identify Regulators of hTERT Expression.
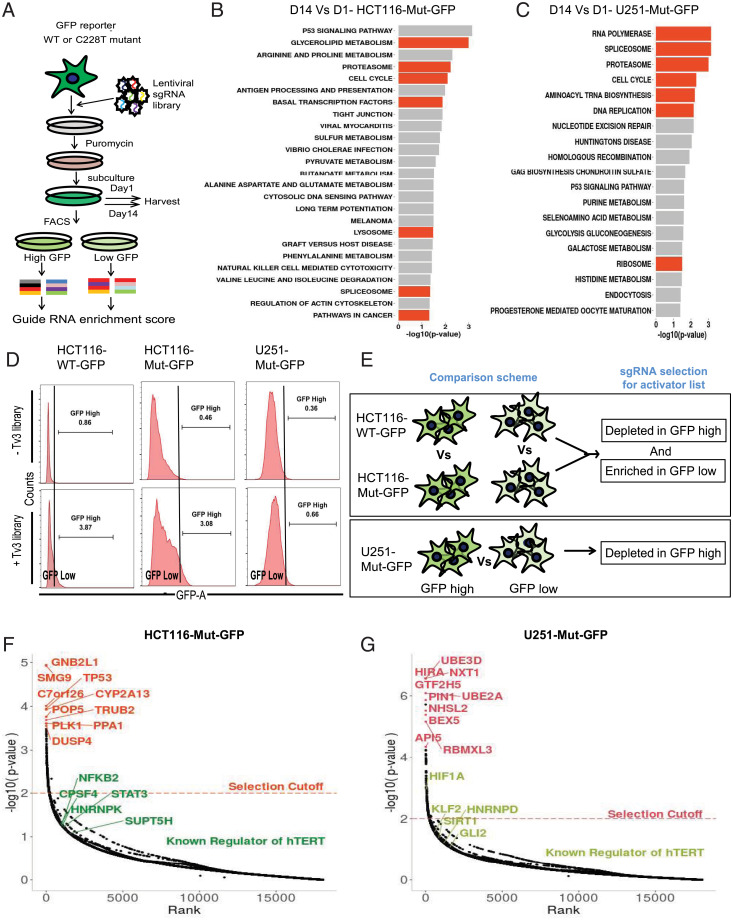
In addition to siRNA screens, which have their own limitations such as off-target effects and toxicity (35, 36), we utilized the Toronto v3 CRISPR knockout library containing a pool of 70,948 sgRNA targeting 18,053 genes in the human genome (37, 38). The advantage of this library is that it is an all-in-one system wherein the sgRNA and Cas9 are in the same plasmid, allowing single antibiotic selection after transduction, and it is hence superior to other CRISPR knockout libraries (37). As outlined in Fig. 4A, the screening process involves transducing the lentiviral pool of sgRNA+Cas9 in the target cells followed by puromycin selection to eliminate nontransduced cells. Sufficient cells were collected on day 1 and day 14 to compare the sgRNA representation of essential genes and pathways. On day 14, the cells were sorted, based on GFP expression, into GFP high and GFP low as compared to nontransduced cells. The sgRNA enrichment scores from each sample were plotted to identify genes essential for hTERT regulation (SI Appendix, Fig. S3 A and B). We observed consistent sgRNA scores across various samples (SI Appendix, Fig. S3 A and B). Besides, we also observed, in both HCT116-Mut-GFP and U251-Mut-GFP cells, from the pathway terms comparing day 14 with day 1, that sgRNA targeting essential pathways like cell cycle, spliceosome, proteasome and ribosomes were significantly depleted at day 14 (Fig. 4 B and C). These results uphold the functionality and usability of our CRISPR knockout screens for the identification of physiological regulators of Mut-hTERT promoter. We hence further analyzed the sgRNAs among GFP-high populations of HCT116-WT-GFP vs. HCT116-Mut-GFP cells and overlapped the hits with GFP-low population of HCT116-WT-GFP vs. HCT116-Mut-GFP cells (Fig. 4D). The sgRNAs depleted in GFP-high population most likely represent specific activators of Mut-hTERT promoter (Fig. 4E). CRISPR-Cas9–engineered U251 line harboring WT-hTERT promoter was not viable, due to dramatic loss of telomerase activity. Therefore, we compared sgRNAs that are enriched in the GFP-high compared to GFP-low population as activators (Fig. 4E). We have reported all the activators that are specific to each cell line, in SI Appendix, Table S1. We plotted the gene rank as indicative of the Mut-hTERT inhibition against the P value for each of the two cells lines (Fig. 4 F and G). In HCT116-GFP cells, we identified Mut-hTERT promoter-specific novel hits including PLK1, DUSP4, TP53, TRUB2, and PPA1 (Fig. 4F). In addition, we found previously reported regulators of hTERT expression, namely, Myc, GABPA, CPSF4, and hnRNPK, among significant top hits (P < 0.01) (Fig. 4F). Hits from U251-Mut-GFP cells identified genes like HIRA, GTF2H5, and PIN1 among the top 10 hits, while known regulators of hTERT expression like HIF1a, GLI2, KLF2, SIRT1, and hnRNPD came along as significant hits (P < 0.01) (Fig. 4G). We overlapped the hits from each screen and found varying degrees of overlap, depending on the cell lines and promoter mutation status (SI Appendix, Fig. S3 C and D).
Fig. 4.
CRISPR knockout screen in GFP reporter cell lines. (A) Workflow for CRISPR knockout screen. The GFP reporter lines were transduced with lentiviral encoded sgRNA library and selected with puromycin. GFP sorting was done on day 14 to segregate GFP high and low populations for calculating sgRNA enrichment score. (B and C) Enriched KEGG pathways for depleted (negative selected) sgRNA from reporter cells at day 14 compared to day 1. The sgRNA targeting important cell survival pathways (orange bars) were depleted in both HCT116-Mut-GFP (B) and U251-Mut-GFP (C) cells at day 14. (D) Representative FACS histograms of GFP sorted reporter cells (HCT116 WT-GFP, Left; HCT116-Mut-GFP, Middle; U251-Mut-GFP, Right) before and after Tv3 sgRNA library expression. The population with high GFP fluorescence was sorted as GFP high, while the remaining populations were sorted into GFP low for subsequent genomic DNA isolation to look for sgRNA enrichment in the populations. (E) Schematic showing the comparison scheme for sorted cells to find activators of hTERT. For activators in HCT116 isogenic reporter lines HCT116-WT-GFP and HCT116-Mut-GFP, the comparison was made between the same populations of either GFP high or GFP low to look for depleted or enriched sgRNA targeting genes, respectively. For the nonisogenic line (U251-Mut-GFP), sgRNA depleted in GFP high Vs. GFP low was derived to look for negatively selected sgRNA in GFP high population. (F) Plot showing the P value distribution of sgRNA targeted genes and top hits of activator genes (orange) and known regulators of hTERT among all hits (green) identified from the HCT116-Mut-GFP cell line. (G) Plot showing the P value distribution of sgRNA targeted genes and top hits of activator genes (orange) and known regulators of hTERT among all hits (green) identified from U251-Mut-GFP cells line.
Hit Comparison and Validation of MED12 as a Mutant-Specific hTERT Regulator.
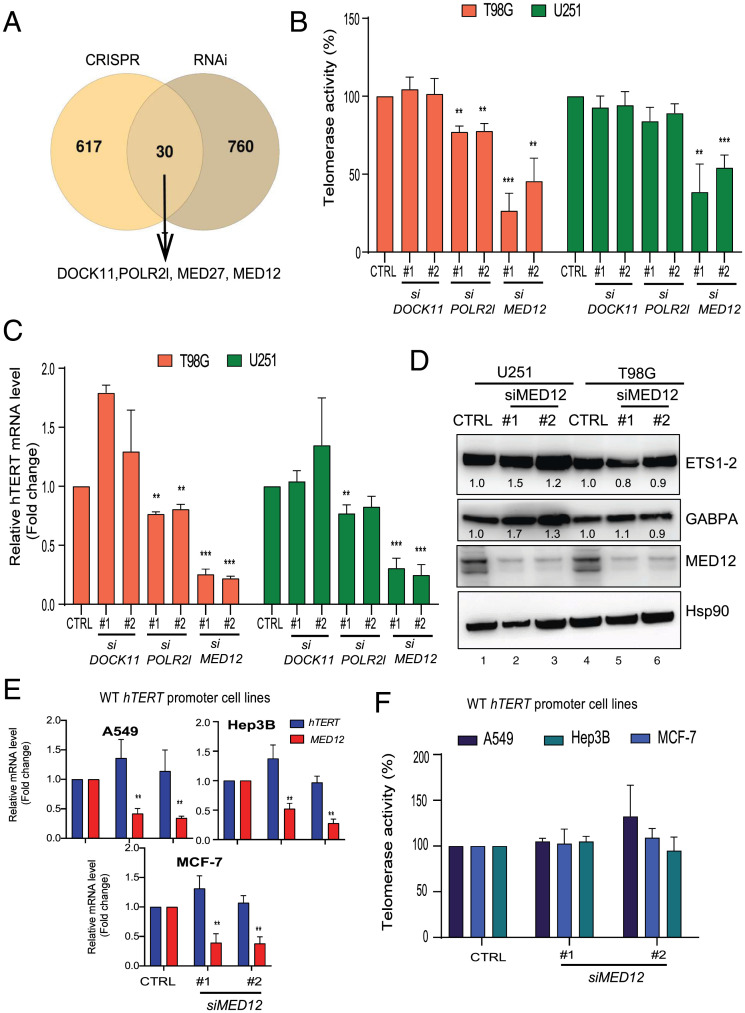
After creating a list of activators for each cell line used in our screens utilizing either CRISPR knockout library or siRNA library, we overlapped the hits to identify whether there are any common regulators. This analysis revealed 30 genes that surfaced as common hits between the two different screens performed in various isogenic cell lines (Fig. 5A). Among the common activators, we focused on genes related to transcription factors or DNA binding proteins, including mediator complex subunit (MED) genes, DOCK11, and POLR2l. Mediator complex subunits are important for gene transcription and have been shown to play a role in controlling expression of cell identity genes like Sox9 and Nanog. In recent years, the MED complex has been linked to the pathogenesis of cancer and is frequently mutated in different cancers (39–41). Particularly, MED12 has a high frequency of mutation in phylloid cancer, and the MED12 gene has been reported to play a role in the control of cell identity genes and superenhancer-associated gene expression (42). However, the mechanism by which MED12 functions in cancers, in general, and cancers with Mut-hTERT promoter remains elusive. With this backdrop on the identified common hits, we further analyzed three candidates, namely, DOCK11, POLR2l, and MED12, for their ability to control Mut-hTERT promoters. Transient knockdown of these candidate genes in parental (nonreporter) T98G and U251 cell lines containing inherent C250T and C228T mutations, respectively, showed a significant reduction of telomerase activity upon MED12 knockdown (Fig. 5B). POLR2l knockdown significantly reduced telomerase activity only in the T98G line (Fig. 5B). A similar trend was observed in expression of endogenous hTERT mRNA, indicating that MED12 exerts control of hTERT at gene expression level, supporting its reported role in the control of gene expression (Fig. 5C). As a proof of concept that our screens can identify bona fide regulators of Mut-hTERT promoter, our results prompted us to further validate MED12 as a candidate. To evaluate the mechanism of action of MED12, we first analyzed the influence of MED12 on the expression of well-known activators of Mut-hTERT promoters such as ETS1/2 and GABPA. Knocking down MED12 in both T98G and U251 lines did not show a reduction of either of these positive regulators of Mut-hTERT promoters (Fig. 5D and SI Appendix, Fig. S4A). Instead, we observed increased expression of GABPA in U251 lines with MED12 knockdown. While we have shown MED12 to regulate Mut-hTERT promoters in two different cell lines, we questioned its influence on WT-hTERT promoter. Depletion of MED12 by siRNAs in cell lines A549 (lung cancer), MCF-7 (breast cancer), and Hep3B (liver cancer), which harbor WT-hTERT promoter, showed no significant effect on hTERT expression and telomerase activity (Fig. 5 E and F). These observations further reiterate that MED12 is a specific positive regulator of Mut-hTERT promoters.
Fig. 5.
Comparison of screen hits and validation. (A) Venn diagram showing the overlap of all combined hits from various cell lines done by either CRISPR sgRNA library or siRNA library. The MED complex genes were identified as common among many different lines. (B) Telomerase activity assay was measured in nonreporter T98G and U251 cell lines after transient knockdown (with two siRNAs) of three selected hits from the overlap of screen data. (C) The qPCR data showing the expression of hTERT in nonreporter T98G and U251 cell lines after transient knockdown (with two siRNAs) of DOCK11, POLR2L, and MED12. Actin was used as a control. (D) Nonreporter T98G and U251 cell lines were transfected with siControl (siCTRL) and siMED12 (#1 and #2). Western blot image shows the protein levels of GABPA and ETS1-2 in T98G and U251 cell lines after transient knockdown of MED12 (with two siRNAs). HSP90 was used as a loading control. (E) The qPCR data showing the expression of hTERT and MED12 in different cell lines (A549, MCF-7, and Hep3B) with WT-hTERT promoter after transient knockdown (with two siRNAs) of MED12. (F) Telomerase activity assay was measured in WT-hTERT promoter lines after transient knockdown (with two siRNAs) of MED12. Error bars indicate mean ± SD of three independent experiments. P values were calculated by Student's t test method ( **P < 0.01; ***P < 0.001).
MED12 Specifically Regulates Mut-hTERT Promoter via Long-Range Chromatin Interaction.
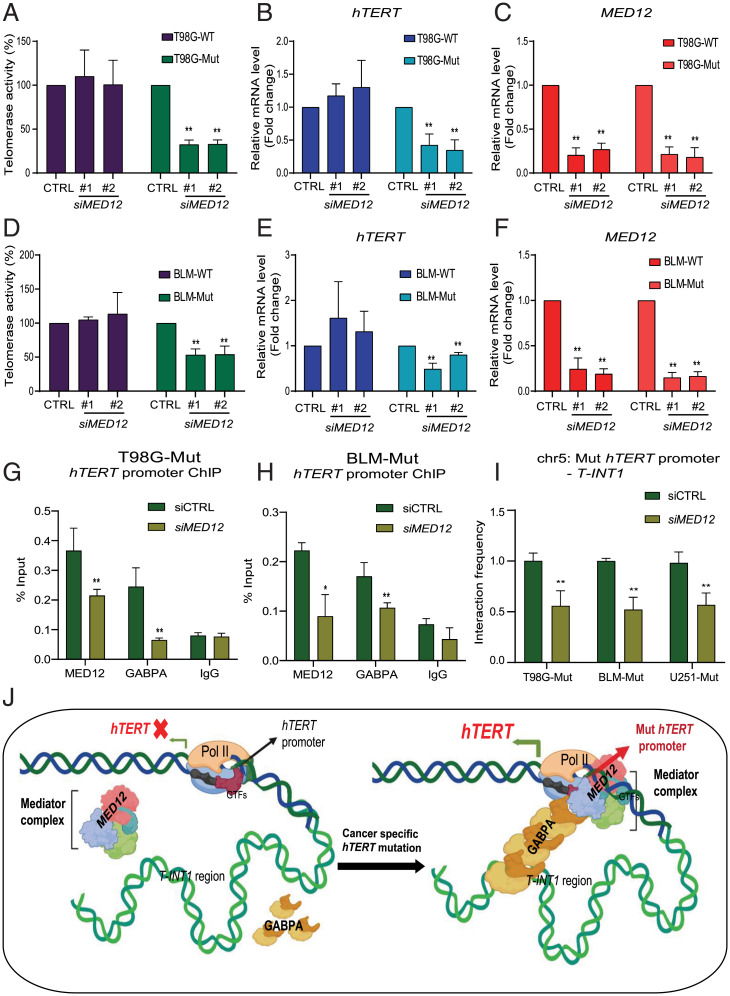
Having identified MED12 as a positive regulator that specifically activates Mut-hTERT promoters in parental lines with inherent C228T and C250T promoter mutations, we further evaluated the precise mechanism by which MED12 regulates Mut-hTERT promoter using nonreporter isogenic T98G (glioma) and BLM (melanoma) lines with WT or Mut-hTERT promoters. These isogenic lines had been instrumental in demonstrating the importance of the T-INT1 region in regulation of Mut-hTERT expression (6, 15). In these lines, depletion of MED12 using siRNAs inhibited telomerase activity specifically in Mut-hTERT promoter isogenic variants of both T98G and BLM cells (Fig. 6 A and D). We also observed a significant decrease of hTERT expression in these isogenic lines only with the Mut-hTERT promoter (Fig. 6 B and E). It is important to note that this specific effect of MED12 knockdown on the isogenic mutant line is not simply due to differences in knockdown efficiency of MED12 in the isogenic lines (Fig. 6 C and F). It is also not related to the immediate effect of MED12 knockdown on cell cycle (SI Appendix, Fig. S3E). Interestingly, this effect is attributed to reduced telomere length specifically in Mut-hTERT promoter cells (SI Appendix, Fig. S3F). Colony formation assays also showed that, upon MED12 knockdown, significant reduction in colony formation is observed, particularly in Mut-hTERT promoter cells, and this was rescued by restoration of hTERT activity by ectopic expression of MED12 (SI Appendix, Fig. S4 C–E). We next analyzed the role of MED12 in epigenetic control of mutant hTERT promoter by chromatin occupancy and chromatin interaction analyses. GABPA has been shown to bind to the de novo C250T and C228T sites to drive hTERT expression (29). GABPA dimers on the proximal hTERT promoter have been shown to interact with GABPA dimers located in a distal region (∼260 kb upstream of hTERT promoter) and form a long-range chromatin interaction to make a stable transcriptional hub (6). Therefore, we first tested whether MED12 influences GABPA occupancy on the proximal hTERT promoter in isogenic T98G and BLM cells. Chromatin immunoprecipitation (ChIP)-qPCR results demonstrated a significant reduction of GABPA occupancy on the proximal hTERT promoter upon MED12 depletion in both T98G-Mut and BLM-Mut cells (Fig. 6 G and H and SI Appendix, Fig. S5 A and B). Given that MED12 regulates GABPA occupancy, we checked for biochemical interaction between them by coimmunoprecipitation analysis. Using isogenic T98G cells, we showed that GABPA and MED12 interact irrespective of hTERT promoter status, suggesting that this complex might regulate other genes in the genome (SI Appendix, Fig. S5C). As these proteins interact, we tested the MED12 occupancy after siGABPA in T98G-Mut and BLM-Mut lines. MED12 occupancy is reduced in these lines after siGABPA treatment (SI Appendix, Fig. S5 D and E). Interestingly, we identified that MED12 binds to the proximal hTERT promoter, and its occupancy was reduced upon MED12 depletion (Fig. 6 G and H and SI Appendix, Fig. S5 A and B). Further, we observed that depletion of MED12 reduces interaction between the Mut-hTERT proximal promoter and the T-INT1 chromatin interaction region measured by chromatin conformation capture (3C) assay, as reduction of MED12 causes dissociation of GABPA subunits from the proximal hTERT promoter (Fig. 6I and SI Appendix, Fig. S5F). In the absence of GABPA recruitment to proximal Mut-hTERT promoter, the long-range interaction with the T-INT1 region is disrupted, and hence the productive 3D chromatin architecture along with the necessary chromatin modifiers required for activation of hTERT do not occur. Together, results from this study identify MED12 as a novel and specific regulator of Mut-hTERT promoter (Fig. 6J). These results also suggest that the isogenic reporter lines described here could serve as wonderful resources for discovering context and stimuli-specific genetic and chemical regulators of hTERT in cancers.
Fig. 6.
MED12 regulates Mut-hTERT promoter-driven gene expression. (A) Graph shows the relative telomerase activity of T98G-WT and T98G-Mut cells with (siMED12 #1 and siMED12 #2) and without (siCTRL) MED12 depletion by RT-qPCR. Relative telomerase activity was quantified as compared to siControl sample. (B and C) Gene expression analysis was performed for hTERT and MED12 genes by qPCR in T98G-WT and T98G-Mut cells transfected with siControl or siMED12. Cycle threshold values were normalized to actin gene. (D) Graph shows relative telomerase activity in BLM-WT and BLM-Mut cells transfected with siControl or siMED12. (E and F) Gene expression analysis was performed for hTERT and MED12 genes by qPCR in BLM-WT and BLM-Mut cells transfected with siControl or siMED12. (G and H) ChIP-qPCR was performed for MED12, GABPA, and IgG in T98G-Mut and BLM-Mut cells transfected with siControl and siMED12. Enrichment in the proximal hTERT promoter was calculated by using the percent input method. (I) Chromatin interaction frequency between Mut-hTERT promoter and T-INT1 region was analyzed by 3C-qPCR in T98G-Mut and BLM-Mut cells transfected with siControl and siMED12. Error bars indicate mean ± SD of three independent experiments. P values were calculated by Student's t test method (*P < 0.05; **P < 0.01). (J) Picture illustrates the hTERT promoter with its distal regulatory elements. In the absence of hTERT promoter mutation (Left), hTERT promoter and T-INT1 interaction does not form, as GABPA and MED12 are not bound to the WT-hTERT promoter. Upon acquiring cancer-specific hTERT promoter mutation, GABPA and MED12 bind to the Mut-hTERT promoter and mediate long-range chromatin interaction between Mut-hTERT promoter and the T-INT1 region which recruits Pol II to the promoter to transcribe hTERT expression.
Discussion
Cancer-specific mutations are unique features that help us understand tumorigenesis and also serve as tools for making cancer-specific therapeutic targets. As cancer cells continuously divide, they gain additional mutations that provide growth advantages and enable them to bypass host surveillance mechanisms. In general, these mutations alter protein structure or RNA stability, or rewire transcriptional regulation by creating de novo transcription binding sites. Cancer-specific recurrent hTERT promoter mutations (C228T and C250T) in the proximal promoter region were reported to increase hTERT expression (43). These mutations create a de novo ETS binding motif that drives transcription of hTERT in a unique mechanism compared to cells with WT-hTERT promoter. Therefore, in this study, we aimed to identify transcriptional modulators that specifically drive Mut-hTERT promoter that could potentially allow targeting of cancer cells harboring the mutant promoters. While siRNA screens have been commonly used for knockdown screens in many models for decades, the recent emergence of CRISPR technology to perform similar large-scale knockout-based loss-of-function screens allows us to overcome limitations of the siRNA technology and identify important regulator mechanisms. Here, we utilized both these complementary genome-scale technologies to discover physiological activators of the endogenous hTERT gene. We generated isogenic lines including colorectal cancer (DLD-1 and HCT116), glioblastoma (U251 and LN382), and melanoma (BLM) that differ by only a single nucleotide C250T or C228T to exclude the effect from other genomic differences. We simultaneously inserted GFP or luciferase reporter genes after the ATG start codon to quantify the activity of WT- and Mut-hTERT promoter. However, only the isogenic lines from colorectal cancers (edited from WT to mutant) survived the screening procedure, as other cell lines were highly susceptible to apoptosis by alteration of hTERT promoter status (edited from mutant to WT), indicating the essentiality of hTERT expression via mutant promoter for cell survival. Secondly, it is important to note that our study using an engineered, endogenous reporter system provides a significant advantage over previous hTERT screens that were performed using plasmid-based reporters. Specifically, our reporter models allow us to capture and functionally interrogate 3D chromatin regulation, which is a key mechanism for the regulation of Mut-hTERT promoters.
Our CRISPR knockout screen in the isogenic line (HCT116-WT-GFP and HCT116-Mut-GFP) revealed interesting insights. Similar screens have been conducted in various genomic contexts, leading to the identification of susceptibility factors (44). Our screen identified many MED complex subunits in different cell lines. The mediator complex regulates enhancer–promoter interactions to modulate gene expression (45–47). Using WT and Mut isogenic BLM and T98G (melanoma and glioma) cells among which the only difference is the single nucleotide (C250T), we illustrated that depletion of MED12 specifically represses hTERT expression and inhibits telomerase activity in cells harboring Mut-hTERT promoter (Fig. 6 A–E). Our results showed that depletion of MED12 significantly reduced GABPA occupancy in BLM-Mut and T98G-Mut cells (Fig. 6 G and H). MED12 also interacts with GABPA biochemically and forms a stable complex on the proximal hTERT promoter. Previously, MED12 was shown to regulate transcriptional activation of specific genes associated with enhancers and superenhancer regions (42, 48, 49). These regions bind to transcription factors and bridge the transcription of genes located several kilobases apart in the genome. It has been shown that Mut-hTERT promoter is regulated by long-range chromatin interaction where GABPA dimers form between the Mut-hTERT promoter and the distal region T-INT1 located ∼260 kb upstream of the TSS of hTERT gene. Our 3C analysis showed that MED12 regulates the formation of Mut-hTERT promoter-specific long-range chromatin interaction through stabilizing GABPA on the hTERT promoter (Fig. 6I). These results further confirm the efficiency of our screening and the validity of our reporter lines generated for the hTERT promoter activity quantification. MED12 gain of function mutations are observed in human cancers (50, 51). Targeting MED12 could be an advantage in cancers like phyllodes tumors where MED12 and hTERT promoter mutations cooccur (52, 53). Additionally, other oncogenic targets of MED12–GABPA complex should be further investigated, as these genes can be inhibited by new drugs which can dissociate MED12–GABPA interaction. Aside from MED12, we identified 29 other candidates which require further validation. Similar to MED12, some of the hits might inhibit Mut-hTERT promoter across different cancer types or might be specific to a particular oncogenic signal that is linked to hTERT expression regulation.
While there are multiple points to showcase the utility of our study for any follow-up work, it also has some limitations. For example, DOCK11, which was identified as a common hit in both screens, did not show any influence on Mut-hTERT expression in validation studies. On the other hand, specific regulators of Mut-hTERT like GABPA came out as a significant hit only in the RNAi screen but not in the CRISPR knockout screen. As CRISPR knockout screens rely on the number of sgRNA and their targeting efficiency, it could be that sgRNA targeting GABPA in our CRISPR knockout library is not efficient or the region of sgRNA incorporation in the genome forms a heterochromatic structure, which therefore does not allow its efficient expression. It may also be explained by the technicality that CRISPR editing results in true loss of function in contrast to RNAi that generally causes hypomorphic effects. Complete deletion of GABPA by CRISPR could be lethal to cells, thereby causing a dropout of this hit from the screen. Another limitation we found was the successful establishment of isogenic lines with the reporter gene. While, previously, we made T98G and BLM cell lines with an isogenic mutation in the hTERT promoter locus, creating those cell lines with a reporter gene was unsuccessful. This could be because the insertion of a reporter gene copy replaces the endogenous hTERT, thereby reducing the copy numbers of hTERT gene in the cells. For cell lines like T98G and BLM, it may be necessary to have a sufficient copy number of hTERT to survive, and its editing leads to cell death. Despite these limitations, our study represents one of the few reports combining large-scale genomic loss of function screens (CRISPR and RNAi) to uncover essential genes for cancer cell survival. Previously, two such observations were independently reported by Evers et al. (54) and Morgens et al. (55) to show the outperformance of CRISPR knockout screen over RNAi (56). The novelty in our study stems from the fact that we used unique engineered cell lines with a reporter gene under the hTERT locus to understand Mut-hTERT promoter-specific regulator, unlike the other two studies which used cancer cells to perform the screen for essential genes. Our study also opens up a few intriguing questions: Do 3D chromatin structure and long-range interactions drive expression from WT-hTERT promoters? The hTERT is driven by WT promoters in stem cells and immune cells. If, indeed, the WT- and Mut-hTERT promoters’ activation involves a differential mechanistic approach, what are the endogenous factors that distinctly drive these promoters in their native chromatin contexts? In conclusion, future functional validation and mechanistic dissection of the Mut-hTERT–specific modulators identified in our study could be highly beneficial for the identification of the next generation of drugs that can inhibit cancer-specific Mut-hTERT promoter with minimal to no cytotoxicity on the stem cell compartment.
Methods
Generation of Reporter Cell Lines.
Generation of reporter lines are described in SI Appendix.
Whole-Genome CRISPR and siRNA Screen.
Silencer Select Human whole-genome siRNA library and Toronto KnockOut Library v3 (TKOv3) were used for siRNA and CRISPR screens, respectively. Details are described in SI Appendix.
Telomerase Activity Assay.
Telomerase activity assay (TRAP) assay was performed from cells 36 h posttransfection as described previously (20).
The 3C Chromatin Interaction Assay.
The 3C assay was performed as described previously (57). Details are described in SI Appendix.
Statistical Analysis.
The two-tailed Student’s t test was used to analyze groups for the qPCR, 3C-qPCR, ChIP-qPCR, and TRAP assay. The mean ± SD of each gene expression, ChIP, chromatin interaction, and TRAP assays were obtained from at least three independent experiments, as indicated in the figure legends.
Supplementary Material
Acknowledgments
The V.T. laboratory is supported by the National Research Foundation-Competitive Research Programme (NRF-CRP17-2017-02) and core funding from IMCB A*STAR. This research is supported by the Singapore Ministry of Health’s National Medical Research Council-Young Individual Research Grant (NMRC/OFYIRG/18MAY-0008 to S.C.A.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105171119/-/DCSupplemental.
Data Availability
All study data are included in the article, SI Appendix, and/or Datasets S1 and S2.
References
- 1.Shay J. W., Wright W. E., Senescence and immortalization: Role of telomeres and telomerase. Carcinogenesis 26, 867–874 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Wu L., Fidan K., Um J. Y., Ahn K. S., Telomerase: Key regulator of inflammation and cancer. Pharmacol. Res. 155, 104726 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Akincilar S. C., Chan C. H. T., Ng Q. F., Fidan K., Tergaonkar V., Non-canonical roles of canonical telomere binding proteins in cancers. Cell. Mol. Life Sci. 78, 4235–4257 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macheret M., Halazonetis T. D., DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 10, 425–448 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Yuan X., Larsson C., Xu D., Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: Old actors and new players. Oncogene 38, 6172–6183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akıncılar S. C., et al. , Long-range chromatin interactions drive mutant TERT promoter activation. Cancer Discov. 6, 1276–1291 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Low K. C., Tergaonkar V., Telomerase: Central regulator of all of the hallmarks of cancer. Trends Biochem. Sci. 38, 426–434 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Counter C. M., et al. , Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene 16, 1217–1222 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Stern J. L., et al. , Allele-specific DNA methylation and its interplay with repressive histone marks at promoter-mutant TERT genes. Cell Rep. 21, 3700–3707 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horn S., et al. , TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Huang F. W., et al. , Highly recurrent TERT promoter mutations in human melanoma. Science 339, 957–959 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y., Cheng H. S., Chng W. J., Tergaonkar V., Activation of mutant TERT promoter by RAS-ERK signaling is a key step in malignant progression of BRAF-mutant human melanomas. Proc. Natl. Acad. Sci. U.S.A. 113, 14402–14407 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinagre J., et al. , Frequency of TERT promoter mutations in human cancers. Nat. Commun. 4, 2185 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Xu X., et al. , Structural basis for reactivating the mutant TERT promoter by cooperative binding of p52 and ETS1. Nat. Commun. 9, 3183 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., et al. , Non-canonical NF-κB signalling and ETS1/2 cooperatively drive C250T mutant TERT promoter activation. Nat. Cell Biol. 17, 1327–1338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robin J. D., et al. , Telomere position effect: Regulation of gene expression with progressive telomere shortening over long distances. Genes Dev. 28, 2464–2476 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Min J., Shay J. W., TERT promoter mutations enhance telomerase activation by long-range chromatin interactions. Cancer Discov. 6, 1212–1214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim W., Shay J. W., Long-range telomere regulation of gene expression: Telomere looping and telomere position effect over long distances (TPE-OLD). Differentiation 99, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim W., et al. , Regulation of the human telomerase gene TERT by telomere position effect-over long distances (TPE-OLD): Implications for aging and cancer. PLoS Biol. 14, e2000016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akıncılar S. C., et al. , Quantitative assessment of telomerase components in cancer cell lines. FEBS Lett. 589, 974–984 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Coussens M., et al. , RNAi screen for telomerase reverse transcriptase transcriptional regulators identifies HIF1alpha as critical for telomerase function in murine embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 107, 13842–13847 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerone M. A., Burgess D. J., Naceur-Lombardelli C., Lord C. J., Ashworth A., High-throughput RNAi screening reveals novel regulators of telomerase. Cancer Res. 71, 3328–3340 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Killela P. J., et al. , TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. U.S.A. 110, 6021–6026 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiba K., et al. , Endogenous telomerase reverse transcriptase N-terminal tagging affects human telomerase function at telomeres in vivo. Mol. Cell. Biol. 37, e00541-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deshane J., et al. , Sp1 regulates chromatin looping between an intronic enhancer and distal promoter of the human heme oxygenase-1 gene in renal cells. J. Biol. Chem. 285, 16476–16486 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nolis I. K., et al. , Transcription factors mediate long-range enhancer-promoter interactions. Proc. Natl. Acad. Sci. U.S.A. 106, 20222–20227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kieffer-Kwon K. R., et al. , Myc regulates chromatin decompaction and nuclear architecture during B cell activation. Mol. Cell 67, 566–578.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyo S., et al. , Sp1 cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT). Nucleic Acids Res. 28, 669–677 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bell R. J., et al. , Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 348, 1036–1039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goktug A. N., Ong S. S., Chen T., GUItars: A GUI tool for analysis of high-throughput RNA interference screening data. PLoS One 7, e49386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birmingham A., et al. , Statistical methods for analysis of high-throughput RNA interference screens. Nat. Methods 6, 569–575 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Djebali S., et al. , Landscape of transcription in human cells. Nature 489, 101–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghandi M., et al. , Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X. D., et al. , The use of strictly standardized mean difference for hit selection in primary RNA interference high-throughput screening experiments. J. Biomol. Screen. 12, 497–509 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Mohr S. E., Smith J. A., Shamu C. E., Neumüller R. A., Perrimon N., RNAi screening comes of age: Improved techniques and complementary approaches. Nat. Rev. Mol. Cell Biol. 15, 591–600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campeau E., Gobeil S., RNA interference in mammals: Behind the screen. Brief. Funct. Genomics 10, 215–226 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Hart T., et al. , Evaluation and design of genome-wide CRISPR/SpCas9 knockout screens. G3 (Bethesda) 7, 2719–2727 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mair B., et al. , Essential gene profiles for human pluripotent stem cells identify uncharacterized genes and substrate dependencies. Cell Rep. 27, 599–615.e12 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Laé M., et al. , MED12 mutations in breast phyllodes tumors: Evidence of temporal tumoral heterogeneity and identification of associated critical signaling pathways. Oncotarget 7, 84428–84438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siraj A. K., et al. , MED12 is recurrently mutated in Middle Eastern colorectal cancer. Gut 67, 663–671 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kämpjärvi K., et al. , Somatic MED12 mutations in uterine leiomyosarcoma and colorectal cancer. Br. J. Cancer 107, 1761–1765 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aranda-Orgilles B., et al. , MED12 regulates HSC-specific enhancers independently of mediator kinase activity to control hematopoiesis. Cell Stem Cell 19, 784–799 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borah S., et al. , Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science 347, 1006–1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hinze L., et al. , Synthetic lethality of Wnt pathway activation and asparaginase in drug-resistant acute leukemias. Cancer Cell 35, 664–676.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poss Z. C., Ebmeier C. C., Taatjes D. J., The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 48, 575–608 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malik S., Roeder R. G., The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11, 761–772 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borggrefe T., Yue X., Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin. Cell Dev. Biol. 22, 759–768 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Kuuluvainen E., Domènech-Moreno E., Niemelä E. H., Mäkelä T. P., Depletion of Mediator kinase module subunits represses superenhancer-associated genes in colon cancer cells. Mol. Cell. Biol. 38, e00573-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin J. W., Wang G., The Mediator complex: A master coordinator of transcription and cell lineage development. Development 141, 977–987 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Zhang S., O’Regan R., Xu W., The emerging role of mediator complex subunit 12 in tumorigenesis and response to chemotherapeutics. Cancer 126, 939–948 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mittal P., et al. , Med12 gain-of-function mutation causes leiomyomas and genomic instability. J. Clin. Invest. 125, 3280–3284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia-Dios D. A., et al. , MED12, TERT promoter and RBM15 mutations in primary and recurrent phyllodes tumours. Br. J. Cancer 118, 277–284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida M., et al. , TERT promoter mutations are frequent and show association with MED12 mutations in phyllodes tumors of the breast. Br. J. Cancer 113, 1244–1248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evers B., et al. , CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat. Biotechnol. 34, 631–633 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Morgens D. W., Deans R. M., Li A., Bassik M. C., Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 34, 634–636 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Housden B. E., Perrimon N., Comparing CRISPR and RNAi-based screening technologies. Nat. Biotechnol. 34, 621–623 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Hagège H., et al. , Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2, 1722–1733 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article, SI Appendix, and/or Datasets S1 and S2.