Abstract
Besides molecular and phenotypic variations observed in cancer cells, intratumoral heterogeneity also occurs in the tumor microenvironment. Correlative stiffness maps of different intratumor locations in breast tumor biopsies show that stiffness increases from core to periphery. However, how different local ECM stiffness regulates key functions of cancer cells in tumor progression remains unclear. Although increased tissue stiffness is an established driver of breast cancer progression, conclusions from 2D cultures do not correspond with newer data from cancer cells in 3D environments. Many past studies of breast cancer in 3D culture fail to recapitulate the stiffness of a real breast tumor or the various local stiffnesses present in a tumor microenvironment. In this study, we developed a series of collagen/alginate hybrid hydrogels with adjustable stiffness to match the core, middle, and peripheral zones of a breast tumor. We used this hydrogel system to investigate effects of different local stiffness on morphology, proliferation, and migration of breast cancer cells. RNA sequencing of cells in hydrogels with different stiffness revealed changes in multiple cellular processes underlying cancer progression, including angiogenesis and metabolism. We discovered that tumor cells in a soft environment enriched YAP1 and AP1 signaling related genes, whereas tumor cells in a stiff environment became more pro-angiogenic by upregulating fibronectin 1 (FN1) and matrix metalloproteinase 9 (MMP9) expression. This systematic study defines how the range of environmental stiffnesses present in a breast tumor regulates cancer cells, providing new insights into tumorigenesis and disease progression at the tumor-stroma interface.
Keywords: tumor microenvironment, ECM stiffness, hybrid hydrogel, 3D tumor model, intratumor heterogeneity
Introduction
Breast cancer, like most other malignancies, is marked by heterogeneity at multiple levels. Breast tumors fall into multiple molecular subtypes with different receptors that inform selection of therapies [1, 2]. With increasing use of single-cell sequencing, researchers now realize that heterogeneity of breast cancer exists not only among different tumors but also within a single tumor[3, 4]. Besides molecular and phenotypic variations observed in cancer cells, intratumoral heterogeneity also extends to the tumor microenvironment[5, 6]. While studies have focused on composition and heterogeneity of stromal cells in tumor microenvironments in vitro and in vivo[7–9], heterogeneity of the non-cellular component, namely physical and mechanical changes in the extracellular matrix (ECM) that alter parameters such as stiffness[10] has largely been overlooked. Correlative stiffness maps obtained from different locations in breast tumor biopsies show that stiffness varies from core to periphery. The core is the softest with stiffness usually lower than 2 kPa[11], while stiffness in the periphery can go as high as 20 kPa[12]. The diversity of biomechanical profiles underscores the importance of understanding heterogeneity in the extracellular microenvironment and how it regulates disease progression in breast cancer.
Tumor initiation and progression generate complex structural changes in the ECM and cytoskeletal architecture[13–15]. Differences in local ECM stiffness, therefore, may elicit varying mechanical responses among cancer cells. Investigating how such biomechanical heterogeneities affect pro-metastatic functions of cancer cells requires the ability to measure cellular behaviors within the complicated native tumor microenvironment in vivo[16] while separating effects caused by changes in systemic physiology. Specifically, dissecting effects of biomechanical heterogeneity in tumor microenvironments on functions of cancer cells necessitates an in vitro tumor model with adequate spatial/structural resolution to study regional variations of local ECM stiffness.
Long-standing ambiguities exist about biomechanics in cancer because of fundamental differences between 2D versus 3D environments. Numerous studies have investigated isolated cells on 2D substrates with various stiffness and shown that increased stiffness exerted profound effects on cancer progression[17], inducing oncogenic intracellular signaling to aid tumorigenesis by activating pathways such as FAK[18, 19], AKT[20, 21], and PI3K[22, 23]. By comparison, 3D culture has been reported to be crucially important when modeling breast cancer[24–27]. Cell morphology and signaling alter significantly in 3D culture[28–31], producing sometimes contradictory outcomes from 2D settings[32–34]. In addition, most existing studies of breast cancer in 3D culture fail to recapitulate the stiffness of a real breast tumor[21, 35–37]. As such, effects of varied ECM stiffness on breast cancer cells in 3D culture and subsequent cellular responses still remain unclear.
In this study, we developed a series of collagen/alginate hybrid hydrogels with adjustable stiffness to match the core, middle, and peripheral zones of a breast tumor. We used these gels to investigate effects of varying mechanical stiffness on multiple responses of MDA-MB-231 breast cancer cells. We previously established that our collagen/alginate hybrid hydrogel possesses a well-organized, homogenous microstructure with adjustable mechanical stiffness and permeability of nutrients from culture medium. We also established a 3D tumor invasion model showing follow-the-leader migration with fibroblasts leading invasion of cancer cells similar to in vivo[38]. Furthermore, we demonstrated that human mammary fibroblasts embedded in these 3D matrices remodel the collagen network to oriented, thicker fibrillar tracks, facilitating invasion of tumor cells[39]. Based on these results, we further explore effects of 3D hydrogel stiffness, which mimics heterogeneous local stiffness of tumor ECM, on breast cancer cell morphology, proliferation, and migration. Using RNA sequencing, we also identified critical cellular processes regulated by tissue mechanics, including metabolism and angiogenesis. This systematic study reveals key processes regulated by heterogeneous mechanical tumor microenvironments in breast cancer, providing links of mechanical stiffness with tumor formation and progression.
Methods
Hydrogel preparation.
We prepared hydrogels as previously described. Briefly, we mixed collagen type I (Ibidi) with low viscosity alginic acid sodium salt stock solution (Sigma Aldrich), 10× DMEM (Sigma Aldrich), and cell culture medium (with or without live cells and spheroids) to achieve a final concentration of 3 mg/ml collagen and 5 mg/ml alginate for all gels. We then adjusted to neutral pH and crosslinked gels with different molar concentrations of CaCl2 (Sigma Aldrich) (3.75 mM for the soft gels, 7.5 mM for the middle gels, and 30 mM for the stiff gels). After mixing with CaCl2, we cured gels in a humidified tissue culture incubator with 5% CO2 for 30 minutes at 37 °C and then added fresh cell culture medium on top.
Stiffness measurements.
We measured compressive elastic moduli of gels as previously described. Briefly, we prepared cylindrical gel samples with a diameter of 3 mm and thickness of 1 mm and compressed the hydrogels with a MicroSquisher (CellScale) in a water bath. We determined the compressive modulus from the slope of the linear region on the stress-strain curve (n = 5 samples per condition).
Cell culture.
We cultured all cells in DMEM (Thermo Fisher) with 10% fetal bovine serum (Thermo Fisher) and 1% added penicillin/streptomycin/glutamine (Thermo Fisher) in a humidified 37 °C tissue culture incubator with 5% CO2. MDA-MB-231 and MCF-7 human breast cancer cells (obtained from the ATCC and verified by short tandem repeat profiling) with LifeAct-GFP protein or unfused GFP have been described previously[40]. We previously described human mammary fibroblasts (HMFs) (gift of Dr. Daniel Hayes, University of Michigan) stably immortalized with a viral vector co-expressing telomerase and GFP. We added mCherry to these HMFs to distinguish them by dual-color fluorescence (GFP and mCherry)[41]. We previously described cells stably expressing FUCCI cell cycle reporters[42].
Preparation of tumor spheroids.
We prepared spheroids with HMFs, HS5 cells, and MDA-MB-231 breast cancer cells as described[40]. Briefly, we added 3×103 cells in 20 μl culture medium to 384-well low volume, non-adhesive, round bottom plates (Corning) and cultured cells for 48 hours before embedding in gels.
Fluorescence microscopy.
We captured all microscopic images of cells and spheroids with an upright Olympus FVMPE-RS two photon microscope with Insight-DS+ laser (Spectra-Physics) and 25× NIR corrected objective (XLPLN25XWMP, NA=1.05, Olympus). We used 920 nm excitation for GFP and 1040 nm excitation for red fluorescent proteins, collecting emitted light in green (495–540 nm) and red (575–630 nm) channels, respectively. To monitor actin and YAP1 expression in hydrogel containing cells, we used actin-tracker red-rhodamine (C2207S, Beyotime) and YAP1 (D8H1X, Cell Signaling Pathway). To limit signal attenuation throughout 300 μm stacks (5 μm step size), we adjusted laser transmission to the sample and detector gain to maximize signal per slice. To capture invasion from spheroids, we stitched images with a resolution of 1024×1024 to show the entire field. We used the same acquisition parameters for all cells and spheroids compared within a single experiment. We performed SHG imaging using 880 nm for excitation and collected emitted light at 460 nm. We exported SHG images to Imaris software for image segmentation and data analysis.
Quantification of cell proliferation.
We used the FUCCI fluorescent reporter system to detect cell cycle phases. We did not use any experimental manipulations to synchronize cell cycles of breast cancer cells prior to introducing them into hydrogels. We took fluorescent images as described above and analyzed images using Imaris software. The fluorescence intensity of each cell nucleus was quantified in red and green. Following thresholding, the numbers of nuclei of in green and red channels as well as the total number of cells were quantified. We defined cells that showed both fluorescence in red and green as yellow, which indicated a transition phase from G1 to S. For each group, we quantified 6 (3×2) stitched 1024×1024 images with 300 μm stacks (5 μm step size), and we performed three independent replicates to achieve statistical significance.
Quantification of spheroid invasion distance.
We defined the margin of invasion as the circular outer edge of the radial migration of cells from spheroids. To quantify invasion distance, we took the invasive margin of spheroids in gels as a circle and then measured the diameter using CellSens software (Olympus). Invasion distances were calculated as the diameter of the invasion margin on each day minus that on day 0. For each condition, we measured 3 spheroids and repeated experiments with 3 independent replicates of cells to achieve statistical significance.
Cell line RT-qPCR assay.
We washed gels from different conditions using PBS for three times and then incubated the gels with 50 mg/ml sodium citrate for 5–10 minutes, following by 1 mg/ml collagenase lysis. We then extracted total RNA using TRIzol reagent (Invitrogen, 15596018) according to the manufacture’s protocol and examined RNA quality by Nanodrop (Thermo) analysis. Total RNA was reversed transcribed into cDNA using a reverse transcription kit (Takara, RR047A) according to manufacturer’s protocol. The relative quantity of mRNA was determined by qPCR using ABI Stepone system with SYBR Green reagent (Takara TB Green, RR420A). The expression levels of genes were quantified using the comparative Ct method. The expression level of each mRNA was normalized to the expression of GAPDH mRNA or β-actin mRNA and calculated as the fold difference relative to the control.
The human primer sequences used in qPCR are shown as below:
GAPDH: GGAGCGAGATCCCTCCAAAAT; GGCTGTTGTCATACTTCTCATGG
DHFR: ACTCAAGGAACCTCCACAAGG; GCCACCAACTATCCAGACCA
NDUFB3: GCTGGCTGCAAAAGGGCTA; CTCCTACAGCTACCACAAATGC
MFN2: CTCTCGATGCAACTCTATCGTC; TCCTGTACGTGTCTTCAAGGAA
DNM1L: TCACCCGGAGACCTCTCATTC; GGTTCAGGGCTTACTCCCTTAT
PGK1: TGGACGTTAAAGGGAAGCGG; GCTCATAAGGACTACCGACTTGG
LDHA: ATGGCAACTCTAAAGGATCAGC; CCAACCCCAACAACTGTAATCT
SLC2A1: GGCCAAGAGTGTGCTAAAGAA; ACAGCGTTGATGCCAGACAG
YAP1: TAGCCCTGCGTAGCCAGTTA; TCATGCTTAGTCCACTGTCTGT
FOS: GGGGCAAGGTGGAACAGTTAT; CCGCTTGGAGTGTATCAGTCA
FOSB: GCTGCAAGATCCCCTACGAAG; ACGAAGAAGTGTACGAAGGGTT
FOSL: CAGGCGGAGACTGACAAACTG; TCCTTCCGGGATTTTGCAGAT
MMP9: AGACCTGGGCAGATTCCAAAC; CGGCAAGTCTTCCGAGTAGT
FN1: CGGTGGCTGTCAGTCAAAG; AAACCTCGGCTTCCTCCATAA
β-actin : TGACGTGGACATCCGCAAAG; CTGGAAGGTGGACAGCGAGG
The murine primer sequences used in qPCR are shown as below:
FN1: ATGTGGACCCCTCCTGATAGT; GCCCAGTGATTTCAGCAAAGG
MMP9: CTGGACAGCCAGACACTAAAG; CTCGCGGCAAGTCTTCAGAG
ANG: CCAGGCCCGTTGTTCTTGAT; GGAAGGGAGACTTGCTCATTC
β-actin : GAGACCTTCAACACCCCAGC; ATGTCACGCACGATTTCCC
RNA-seq mRNA library construction.
Oligo(dT)-attached magnetic beads were used to purify mRNA, followed by fragmentation into small pieces with fragment buffer. Then first-strand cDNA was generated using random hexamer-primed reverse transcription, followed by a second-strand cDNA synthesis. After that, A-Tailing Mix and RNA Index Adapters were added to end repair. The cDNA fragments obtained from the previous step were amplified by PCR, and products were purified by Ampure XB Beads and then dissolved in EB solution. The product’s quality control was validated on the Agilent Technologies 2100 bioanalyzer. The double stranded PCR products from the previous step were heated, denatured, and circularized by the splint oligo sequence to get the final library, and the single strand circle DNA (ssCir DNA) was formatted as the final library. The copies of one molecular, DNBs were loaded into the patterned nanoarray and single end 50 bases reads were generated on BGIseq500 platform (BGI-Shenzhen, China).
Bioinformatic analysis of RNA-seq data.
For RNA-seq data analysis, data were filtered with SOAPnuke (v1.5.2) by removing reads containing sequencing adapter; removing reads whose low-quality base ratio (base quality less than or equal to 5) exceeded 20%; and removing reads whose unknown base (‘N’ base) ratio exceeded 5%. Resulting clean reads were obtained and stored in FASTQ format. The clean reads were mapped to the reference genome using HISAT2 (v2.0.4). Bowtie2 (v2.2.5) was applied to align the clean reads to the reference coding geneset, then expression levels of genes were calculated by RSEM (v1.2.12). The heatmap was drawn by pheatmap (R language package) according to the gene expression in different samples. Essentially, differential expression analysis was performed using the DESeq2 with Q value ≤ 0.05.
GO (http://www.geneontology.org/) and KEGG (https://www.kegg.jp/) enrichment analysis of annotated different expressed gene was performed by Phyper (https://en.wikipedia.org/wiki/Hypergeometric_distribution) based on Hypergeometric test. Significant levels of terms and pathways were corrected by Q value with a rigorous threshold (Q value ≤ 0.05) by Bonferroni correction.
Hydrogel assay in vivo analysis.
All animal experiments were approved by the Animal Ethics Committee of Sun Yat-sen University (Approval No. SYSU-IACUC-2020–000425) and performed in accordance with the Animal Care and Use guidelines of Sun Yat-sen University. 8-week-old female BALB/c nude mice were subcutaneously injected with 2×106 MDA-MB-231 cells in 100 μl hydrogel solution. The gels were still liquid during injection, and the body temperature of mice then initiated crosslinking. Hydrogels with tumors were harvested at end point as indicated. Hydrogels were fixed in 4% paraformaldehyde at room temperature for at least 48 hours.
Immunohistochemistry (IHC) and microvessel density (MVD).
The hydrogels were fixed in 4% paraformaldehyde at 4°C for 48 hours, embedded in paraffin and cut into 5 μm sections. The paraffin sections were dehydrated in an alcohol gradient, dewaxed in xylene, and then incubated with 3% H2O2 and super-blocking reagent for 10 minutes at room temperature. The sections were subsequently incubated with the following primary antibodies at 4°C: CD31 (Arigobio, ARG52748). IHC staining was detected using a secondary antibody detection kit (ABsin, abs957) according to the manufacturer’s instructions. After staining with DAB, sections were counterstained with hematoxylin and sealed with neutral gum. The sample slices were imaged by light microscopy. Six randomly selected high-power fields (400×) were evaluated, and CD31+ microvessls were counted and averaged in the most vascular regions of hydrogel sections. Mean MVD was assessed as the number of microvessels/mm2.
Statistical analysis.
For Fig 1–3 we report all statistics as means ± standard deviation (SD). We used analysis of variance (ANOVA) tests followed by the Tukey test for post-hoc pairwise comparisons with p<0.05 defining significance. Statistical analysis of relative gene mRNA levels and MVD difference were performed using GraphPad Prism 8 software (San Diego, California). We analyzed data with a two tailed t-test or two-way ANOVA with p<0.05 defining significant differences. All quantitative data were reported as mean ± standard error of the mean (SEM) deviation from at least three independent experiments.
Fig. 1. Representative stiffnesses of 3D hydrogel environment mimicking the local mechanical properties of human breast cancer.

a The corresponding stiffness range of breast cancer changes from core to periphery[11, 12] (data reproduced with combination of human and mouse tissue analysis). b The stiffness of 3D hydrogel environment can be adjusted to match the core, middle, and peripheral areas of a breast tumor. *: > soft, p < 0.01; #: > middle, p < 0.01.
Fig. 3. Cell migration and interaction with ECM in 3D hydrogel environment mimicking the local mechanical stiffness of human breast cancer.
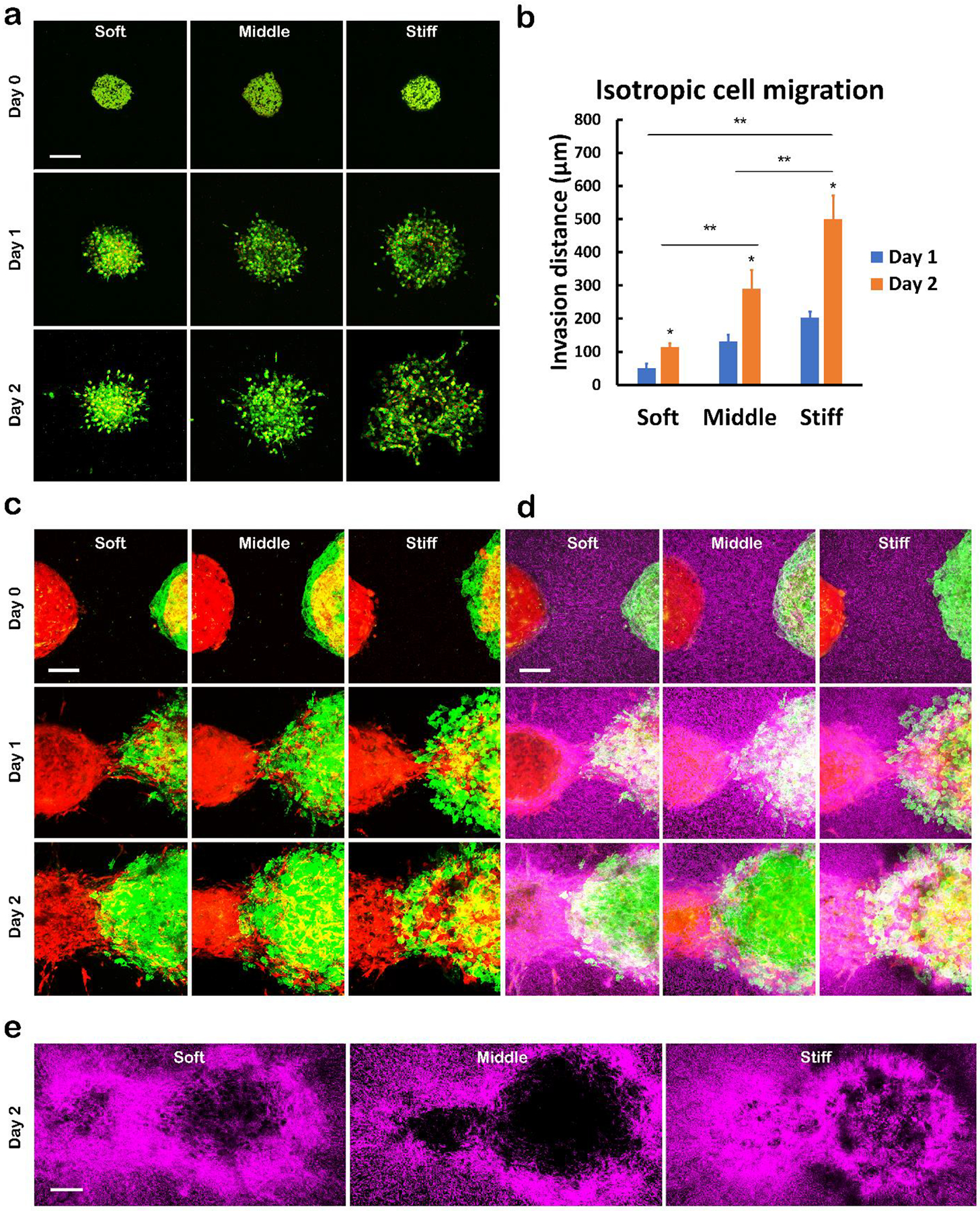
a Isotropic cell migration in hydrogels of different stiffnesses. Invasion of cells from a tumor spheroid made of MDA-MB-231 breast cancer cells (GFP labelled cytoplasm and mCherry labelled nucleus) and HS5 cells was imaged using 2-photon microscopy after culturing in gels on day 0, 1, and 2. Scale bar indicates 200 μm. b Quantification of the invasion distances in the isotropic cell migration assay shown in a *: > day 1, p <0.01; **: p <0.01. c Anisotropic cell migration in hydrogels of different stiffness. A spheroid of mCherry-labelled HMFs and spheroid of LifeAct MDA-MB-231 cells and mCherry-labelled HMFs were placed in gels with an original separation of about 200 μm. Cell migration into the gap was imaged using 2-photon microscopy on day 0, 1, and 2. Scale bar indicates 100 μm. d Overlay of second harmonic images of collagen network (pseudocolor in purple) with the images of two-spheroid migration shown in c. Scale bar indicates 100 μm. e Whole field view of collagen networks in anisotropic two-spheroid migration on day 2. Scale bar indicates 100 μm.
Results
Hydrogel system mimicking the heterogeneous stiffness of a tumor microenvironment.
We prepared collagen/alginate hybrid hydrogels with adjustable stiffness to mimic the local stiffnesses in human breast tumors. Based on previous published data from both human and mouse tissues (both showed similar trends and stiffness values), Fig 1a shows a schematic map of stiffness distributions in a breast tumor, where the ranges of stiffness in core, middle, and periphery were 0–2, 2–5, and 4–20 kPa, respectively[11, 12]. Correspondingly, we adjusted the gel stiffness by varying the degree of calcium crosslinking to match the three local stiffnesses of a breast tumor microenvironment. Fig 1b shows the gel stiffnesses we chose in this study, where soft (2.27±0.46kPa), middle (3.94±0.27kPa), and stiff (16.85±1.12kPa) gels were in accordance with stiffness ranges of the core, middle, and peripheral areas of a breast tumor. We used separate gels of three different stiffnesses instead of one gel with a stiffness range from soft to stiff. This strategy allows us to focus on the local microenvironment and dissect effects of pure local stiffness from a stiffness gradient, which may induce cell migration and other crosstalk between cells in different locations. We believe the current design provides essential knowledge for future studies on behaviors of cells in 3D environments with gradients of stiffness.
Prior investigations showed that mechanical rigidity regulates invasion of cancer cells from spheroids. However, gels used in prior studies typically failed to recapitulate the stiffness of real tumors[43, 44]. In addition, methods used to change gel stiffness most often involved varying concentrations of major extracellular matrix components, which also altered gel microstructure and cell binding motifs[45, 46]. The hydrogel we used in this study is a well-defined system that allows us to control stiffness while maintaining constant amounts of alginate and collagen. The amounts of calcium we used in this protocol ranged from 3.75 to 30 mM, which is a relatively low concentration as compared with previous studies where investigators used up to several hundred mM CaCl2 to crosslink alginate[47–49]. In the soft gel the calcium concentration is very close to the calcium level in DMEM cell culture medium (1.8 mM). Hence, the leaching of calcium ions from the gel and the free calcium in the medium can reach a balance, and the cells inside the gel will not be stimulated by a burst release of calcium ions. In addition, the alginate chains that are not crosslinked by calcium will not diffuse out of the network due to hydrogen bonding with collagen fibers. We tested the stiffness with regular culture medium changes over 24 hours and 7 days after gelation for the stiff gels and did not find a significant change over time (Sup Fig 1). Our previous studies have thoroughly defined material properties of this hydrogel system, including mechanism of gelation, porosimetry, and the remodeling of fibrillar collagen microstructure, demonstrating this gel system as a versatile in vitro platform to study tumor invasion[38, 39]. This gel system offers a purified mechanical microenvironment with stiffness as the only variable, while minimizing confounding variables such as cell binding motifs and gel porosity. Since type I collagen is the major component of tumor ECM and the crosslinked alginate provides good mechanical support to achieve the designed stiffness, this biomimetic gel system is capable of recapitulating the mechanical microenvironment of a breast tumor.
Cellular morphology and proliferation in environments with different local stiffness
We first explored changes in cell morphology and proliferation in heterogeneous tumor microenvironments of different stiffness. We cultured MDA-MB-231 human breast cancer cells, a highly invasive basal subtype of breast cancer, in the hydrogel for 4 days. We seeded MDA-MB-231 cells stably expressing LifeAct, a marker of polymerized actin, fused to green fluorescent protein at the same density (50,000 cells/ml) for all three conditions. Fig 2a displays cell morphologies after 4 days of culture. Presented images represent maximum intensity in z-projection over a depth of 200 μm. As compared with soft gels where MDA-MB-231 cells showed both elongated and round morphologies (Fig 2a first column), cells in the middle and stiff gels extended long fine protrusions that formed connections between cells. We also observed small actin protrusions (tiny brighter projections demarked by white arrows in the zoomed images in Fig 2a) on the cell bodies. Actin filaments are known to play an important role in mechano-sensing[50, 51]. We take these actin protrusions as an indication of breast cancer cells interacting with the mechanical microenvironment through mechano-sensing, where cells actively sense the microenvironment through protrusion and respond to the stiffness of extracellular matrix. Similarly, we also found the morphology of MCF-7 cells behaved comparably to MDA-MB-231 cells, where cells formed small round aggregates in the soft gel and showed a spindle shape in the stiff gel (Sup Fig 2).
Fig. 2. Morphology and proliferation of breast cancer cells in heterogeneous 3D microenvironments.

a Cell morphology represented by LifeAct MDA-MB-231 cells (green fluorescence tagged F-actin) in 3D hydrogels of different stiffnesses. White arrows denote protrusions of cytoskeleton into surrounding environment. Scale bar indicates 50 μm. b Overall proliferation curves of cells in heterogeneous environments. *: p<0.01. c Proliferation and cell cycle in heterogeneous environments represented by cells with the FUCCI reporter, where cell nuclei showing green fluorescence indicates S, G2, and M phases, red fluorescence indicates G1, and yellow fluorescence indicates transition from G1 to S. Scale bar indicates 100 μm. d Quantification of FUCCI reporter fluorescence in cells after culturing in 3D hydrogel environment of different stiffnesses for 2 and 4 days.
We quantified overall proliferation of cells in different microenvironments in Fig 2b. We counted the numbers of cells on day 0, 2, and 4 and normalized to the data of day 0 to define fold change in proliferation. Breast cancer cells in the soft and middle gels showed significantly higher proliferation than the stiff gel on day 4 (Fig 2b). We also investigated changes in cell cycle in microenvironments of different stiffnesses using cells expressing the FUCCI system[52]. Fig 2c shows the growth of FUCCI cells in heterogeneous gel environments, where the fluorescent color in cell nuclei indicates different phases of the cell cycle. Green nuclei indicate cells in S, G2, and M phases, while red nuclei indicate the cells in G1. There is also a transitional phase from G1 to S, where nuclei were shown as yellow (overlay of red and green) fluorescence. We quantified cells in each phase plotted as percentages over time and culture conditions in Fig 2d. Results showed cells in soft environments proliferated more than in a stiff environment on day 2, indicated by 35% cells in S, G2, and M phases in soft gels as compared with 15% in stiff gels. Moreover, the day 4 results also showed higher proliferation in a soft environment with less cells (54%) in G1 phase in soft gels as compared with in stiff gels (77%). To summarize, MDA-MB-231 cells showed higher proliferation in a soft versus stiff microenvironment. Cells in a stiff microenvironment showed more actin protrusions, which are essential for cell migration.
Cell invasion and remodeling of ECM responding to different local mechanical stiffness
Building on our observation that breast cancer cells in a stiff microenvironment had more actin protrusions, we next investigated effects of stiffness on cell migration. We have previously shown invasion with multi-cellular tumor spheroids made of MDA-MB-231 breast cancer cells and human mammary fibroblasts (HMFs) in collagen/alginate hydrogels, where HMFs led the way for breast cancer cells as has been reported in vivo[38]. To avoid ‘follow-the leader’ migration and just focus on migration of cancer cells, we substituted the HMFs with another type of stromal cell line (HS5 bone marrow stromal cells) known to be non-invasive[53]. Fig 3a shows invasion of tumor spheroids made of MDA-MB-231 and HS5 cells in hydrogel environments of different stiffness. MDA-MB-231 breast cancer cells were labeled with GFP in cytoplasm and mCherry in the nucleus, while the HS5 cells had no fluorescence labeling. The tumor spheroids were initially approximately 350 μm diameter (top row in Fig 3a), and the size grew after culturing in hydrogels due to cell proliferation and migration. From Fig 3a, cancer cells proliferated more in the soft gel as indicated by highly compacted cells showing higher intensity on day 2. In the stiff gel, the cancer cells migrated more, as shown by the large sparsely occupied fluorescent area from non-migratory stromal cells remaining alone in the core (shown as black area in the center). These results demonstrated isotropic invasion of cancer cells into the surrounding environment without assistance from stromal cells on days 1 and 2. We quantified the invasion distance of tumor spheroids as previously described[38] and compared the cell migration distance on each day in different gels as shown in Fig 3b. The isotropic cell migration distance increased significantly with the stiffness of tumor microenvironment, indicating mechanics of a heterogeneous tumor microenvironment regulates cancer cell migration.
We also investigated cell migration in co-cultures with HMFs, which are well-known as important regulators of tumor invasion[54, 55]. In Fig 3c we placed two spheroids in the same gel environment separated by a short distance (approximately 200 μm), where spheroids on the left consisted of only HMFs (mCherry) and the right contained both MDA-MB-231 cells (GFP) and HMFs (mCherry), respectively. This 2-spheroid model allowed us to monitor migration aided by stromal cells. As shown in Fig 3c the HMFs on both sides extended tracks to build a bridge connecting the two spheroids on day 1, where cancer cells migrated along the bridge from right to left. This migration behavior differs from the isotropic migration in Fig 3a, since the cancer cells preferentially migrated to the left side in an anisotropic pattern. We did not observe any notable difference in cancer cell migration distance on day 1 among all the three conditions, while on day 2 bridging cells merged the two spheroids. The cancer cells in the stiff gel extended a further leading edge onto the HMF spheroid while leaving some empty space between leading edge and the spheroid on the right (less green fluorescence and more red fluorescence) due to a lower proliferation rate. In the soft and middle gels, the cells proliferated more and filled all the gap (brighter green fluorescence intensity and overlay with red). These results show interaction with stromal cells promotes migration of cancer cells, establishing that both cellular components (stromal cells) and acellular components (ECM) regulate tumor invasion. The two migration models we used here also mimic different scenarios in vivo, where in the tumor stroma fibroblasts assist and lead the way for cancer cell invasion, while bone marrow stromal cells regulate several steps in bone metastasis[56].
We further explored remodeling of ECM in the same 2-spheroid model. In Fig 3d and 3e, we detected the collagen network (pseudo-colored in purple) with second harmonic imaging (SHG) using two-photon microscopy. Fig 3d showed overlaid images of the collagen network with spheroids, where we found the collagen network initially homogeneously surrounded spheroids on day 0. On day 1 there was a dense collagen ring wrapping around the HMF spheroid and along the bridge area in all three conditions, which became denser with greater spreading with both HMFs and cancer cells. Fig 3e shows the full areas of Fig 3d without overlays, where the collagen rings and bridges clearly showed the locations of spheroids. These thick collagen rings indicated remodeling of the surrounding collagen network by stromal cells, which is in accordance with our previous finding[39]. Note the remodeling of the gel was purely done by cells rather than decomposition of gels since blank gels without cells remain stable with regular medium changes over 6 months (Sup Fig 1). To summarize, the heterogeneous stiffness in tumor microenvironments affects migration of cancer cells, while the existence of stromal cells also plays a vital role in regulating cancer cell migration. In addition, HMFs can remodel the collagen network of tumor microenvironment.
Transcriptomic profile changes responding to matrix stiffness
To elucidate alterations in transcriptomes of tumor cells in environments with different stiffnesses, we performed transcriptomic profiling by RNA sequencing. We cultured MDA-MB-231 cells in soft, middle and stiff hydrogels for 48 hours, then extracted total RNA and performed transcriptomic profiling by RNA-seq. To provide a better comparison with most previous studies, we also used cells on 2D tissue culture plastic as a control for RNA-seq. The heatmap in Fig 4a shows distinct differences in total gene expression for cells in 2D, soft, middle and stiff microenvironments. According to Gene Ontology (GO) function annotation, we found that the altered genes mostly involved binding, catalytic activity and transcription regulator activity. The Venn diagram in Fig 4b shows that 15,966 genes were co-expressed in four groups with 243, 284, 304 and 120 genes exclusively expressed in 2D, soft, middle and stiff groups. Then we analyzed Differentially Expressed Genes (DEGs) between the control group (2D culture) and other three hydrogel groups (soft, middle and stiff) in supplemental figure (Sup Fig 3a–d). We also analyzed the DEGs in these groups in Fig 4c by Venn diagram. The unique DEGs in soft, middle and stiff hydrogels compared to 2D control were 113, 920 and 1246, respectively. Consistent differential expression of 904 genes occurred in these groups with enrichment computed by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis. Fig 4d shows the top 20 enriched pathways of the 904 DEGs set, including breast cancer, cell cycle, ECM-receptor interaction and metabolism. Changes in genes associated with cell cycle, ECM interactions, and metabolism parallel prior observations in expression when comparing cells in 2D versus 3D cultures[57–60]. We further focused on transcriptional changes induced by heterogeneous hydrogel stiffness. Sup Fig 3e–g shows the expression heatmaps of soft-vs-middle, middle-vs-stiff and soft-vs-stiff groups respectively. We showed the DEGs between three groups in Fig 4e by Venn diagram. There were 151, 458 and 92 genes that differed in soft-vs-middle, middle-vs- stiff and soft-vs-stiff groups, respectively. The GO function enrichment of the DEGs showed the top 3 pathways including binding, catalytic activity and molecular function regulator (Fig 4f). We next performed the KEGG pathway enrichment analysis in Fig 4g–h. MAPK and PI3K-Akt pathways regulate multiple processes in breast cancer and other malignancies, including proliferation, migration, invasion, and survival[61–63]. Enrichment of these pathways may reflect capabilities of cells in middle stiffness environments to proliferate efficiently while still retaining the ability to migrate. Nevertheless, the definite and specific signaling processes involved in responses to stiffness requires more research. IL-17 drives invasion in cancer cells, consistent with greater migration of breast cancer cells in middle versus soft stiffness environments[64]. While MAPK and Ras signaling may drive both proliferation and invasion in cancer, preferential expression of these pathways in cells cultured in soft environments suggests greater functions in proliferation, a behavior upregulated in soft versus stiff cultures. Similarly, Notch signaling promotes proliferation in a variety of physiologic and pathologic settings[65]
Fig. 4. Microenvironmental stiffness alters transcriptomic profiles of breast cancer cells.

a Gene expression of breast cancer cells in 2D, soft, middle, stiff rigidities groups. b Total gene expression in 2D, soft, middle and stiff hydrogel groups. 243, 284, 304 and 120 genes were exclusively expressed in 2D, soft, middle and stiff hydrogel groups, respectively. 15,966 genes were co-expressed in four groups. c DEGs between 2D culture and hydrogels of different stiffness conditions. 113, 920 and 1,246 genes were exclusively expressed in 2D-vs-soft, 2D-vs-middle and 2D-vs-stiff hydrogel groups, respectively. 904 genes were co-expressed as indicated. d KEGG enrichment analysis of DEGs indicated DNA repair, ECM receptor interaction and cell cycle pathways. e DEGs between hydrogel culture conditions of different stiffnesses. 151, 458 and 92 genes were exclusively expressed in soft-vs-middle, middle-vs-stiff and soft-vs-stiff hydrogel groups, respectively. 13 DEGs genes were co-expressed as indicated. f GO analysis revealed enrichment for pathways of binding, catalytic activity and molecular function regulator. g-h KEGG pathway analysis revealed pathway enrichment of soft-vs-middle (g), soft-vs-stiff (h), respectively.
Cancer cells shift from glycolysis to OXPHOS and FA metabolism responding to a stiff microenvironment
To establish how matrix rigidity affects cellular activities, we performed overall Geneset Enrichment Analysis (GSEA) of sequencing data. Interestingly, the results showed enrichment of glycolysis in a soft microenvironment (Fig 5a), whereas a stiff microenvironment made tumor cells upregulate oxidative phosphorylation (OXPHOS) and fatty acid metabolism (FA) processes (Fig 5b–c). The clustering heatmap illustrates genes involved in glycolysis, OXPHOS and FA (Sup Fig 4a–c). In Fig 5d we listed the top ranked genes for glycolysis. The top ranked 10 genes of OXPHOS and FA metabolism profiles were listed in Fig 5e and Fig 5f, respectively. Next, since OXPHOS correlates with mitochondrial respiration, we also enriched the mitochondrial respiration chain complex assembly transcriptomic profile in Fig 5g–h. Compared to the soft stiffness microenvironment, breast tumor cells elevated expression of genes in the mitochondrial respiratory chain in the stiff microenvironment. NDUFB and NDUFS, components of mitochondrial respiratory complex I, were upregulated in tumor cells by the stiffer microenvironment. To confirm the RNA-seq results, we detected mitochondrial respiratory and dynamics related genes, including NDUFB3, DNM1L (DRP-1) and MFN2 in cells of different conditions using RT-qPCR (Fig 5i). Consistent with the sequencing data, we confirmed that these significant genes were increased in breast cancer cells responding to stiffness. Alterations in genes involved in glycolysis and OXPHOS pathways, such as DHFR, SLC2A1, PGK1 and LDHA, also were detected by RT-qPCR (Fig 5j and Sup Fig 6g). With the significance upregulation of DHFR and downregulation of PGK1 in MCF-7 and MDA-MB-231 cells (Sup Fig 6g), these results demonstrate that heterogeneous tumor mechanics alter genes related to cellular metabolism.
Fig. 5. Breast cancer cells shift from glycolysis to OXPHOS and fatty acid metabolism responding to stiff matrix microenvironment.

a Geneset Enrichment Analysis (GSEA) of overall gene expression of breast cancer cells in soft rigidity environment. Hallmark genes in glycolysis were enriched in cells cultured in the soft rigidity environment. b-c GSEA enrichment of oxidative phosphorylation (b) and fatty acid metabolism (c) hallmarks in breast cancer cells cultured in the stiff rigidity environment. d-f Clustering of top ranked genes involved in glycolysis (d), oxidative phosphorylation (e) and fatty acid metabolism pathways (f). Red and blue represent up- and down-regulated genes, respectively. Heatmap was normalized by z-score (row direction), and the expression level was the mean of three biological repeats as indicated. g Mitochondrial respiratory chain complex assembly genes were enriched in cells cultured in stiff microenvironment. h Clustering of leading genes involved in mitochondrial respiratory chain complex assembly geneset. Red represented the upregulated, blue represented the downregulated. Heatmap was normalized by z-score (row direction). i MDA-MB-231 cells were cultured in hydrogels of different stiffnesses for 24 hours before extraction of total RNA. Relative NDUFB3, DNM1L and MFN2 expression levels were detected by RT-qPCR. j Relative DHFR, SLC2A1, PGK1 and LDHA expression levels were detected by RT-qPCR in MDA-MB-231 cells. Error bars represent standard deviations of three independent experiments. p values were shown as indicated in graph. Significance was indicated as p<0.05.
Early vascular angiogenesis responding to heterogeneous local stiffness
Next, we performed several Key Driver-genes Analyses (KDA) in angiogenesis (Fig 6a), OXPHOS, ECM modeling and EMT pathways (Sup Fig 5a–c), respectively. Comparing key genes in the normal and invasive breast carcinoma databases, we found that angiogenesis pathway was obviously different between normal and patients. Consistently, we found enrichment of pro-angiogenic genes in increased rigidity microenvironments (Fig 6b). We confirmed by RT-qPCR that the expression of MMP9 and FN1 in MDA-MB-231 and MCF-7 cells increased with stiffness (Fig 6c and Sup Fig 6a–b). Next, to examine to what extent stiffness of a hydrogel microenvironment can regulate angiogenesis in vivo, we established subcutaneous tumor xenografts by injecting MDA-MB-231 cells mixed with hydrogels of different stiffness. To monitor early angiogenesis, we euthanized mice 10 days after injection according to previous research[66], when the hydrogel had not been totally absorbed in vivo. The in situ xenografts of hydrogels were photographed in Sup Fig 7a–b. More obvious vascular formation was shown in middle and stiff hydrogel groups by CD31 staining and MVD analysis in Fig 6d. To further evaluate key genes involved in rigidity-related modulation of angiogenesis, we detected murine FN1, ANG and MMP9 from hydrogel-containing cells from xenograft models, which indicated that breast cancer cells in stiffer rigidity promoted angiogenesis in vivo (Fig 6e). Next, we analyzed additional signaling pathways or transcriptional programs regulated by stiffness. By GSEA analysis, mostly E2F-related genes were enriched in the stiff group (Fig 6f). Besides E2F, we also found enrichment of KRAS, STAT3 and mTORC1 activation in our transcriptional profiles by KEGG enrichment and analysis (Fig 6g).
Fig. 6. Breast cancer cells upregulate angiogenesis in stiff microenvironmental niche.

a KDA revealed the most 10 significant genes involved in angiogenesis pathways participating in matrix stiffness environments. b GSEA of overall gene expression of breast cancer cells in middle rigidity environment compared to soft environment. Hallmarks of angiogenesis were enriched in cells cultured in microenvironment of middle stiffness. c MDA-MB-231 were cultured in different hydrogels for 24 hours and extracted total RNA. Relative MMP9 and FN1 mRNA expression were detected by RT-qPCR. d Hydrogels of different stiffnesses were mixed with 2×106 MDA-MB-231 cells, followed by subcutaneous injection into BALB/c nude mice (each group contained 5 mice). The hydrogels containing cells were harvested after 10 days for analysis of CD31 staining and MVD analysis. Scale bar indicates 50 μm. e Total RNA was extracted from hydrogels recovered from mice. Relative FN1, ANG, and MMP9 expression were detected by RT-qPCR. Error bars represent standard deviations of three independent experiments. p values were shown as indicated in graph. Significance was indicated as p<0.05. f-g GSEA (f) and GO (g) analysis of overall gene expression of breast cancer cells in a microenvironment of stiff rigidity compared to soft environment. E2F signaling was enriched in cells cultured in a stiff environment.
YAP1 and AP1 signaling pathways were downregulated by increased local stiffness
To reveal signaling pathways downregulated by microenvironmental stiffness in cancer cells, we performed GSEA analysis and found enrichment of YAP1 target gene set in the soft microenvironment (Fig 7a). The YAP1/Hippo pathway is known to contribute to tumorigenesis[67]. We confirmed YAP1 expression by RT-qPCR in Fig 7b and Sup Fig 6c, which showed that YAP1 expression decreased in MDA-MB-231 and MCF-7 cells with increasing stiffness. As MDA-MB-231 cells exhibit a mesenchymal phenotype, we confirmed the findings from MDA-MB-231 cells using a second cell line, MCF-7, with an epithelial phenotype. Fig 7c showed that more YAP1 protein located in nuclei, a marker of transcription by this protein, when cells were cultured in soft hydrogels compared with a stiffer environment (Fig 7c). These results agreed with a recent study establishing that 2D YAP1 mechano-transduction studies do not recapitulate conditions seen in clinical samples and 3D models, as the high activation of YAP1 seen in 2D culture did not occur in 3D cultures. Notably, expression of YAP1 target genes decreased with greater stiffness in 3D culture[32], but the consequences remain unclear. Besides YAP1, another important transcriptional factor, AP1 was also enriched in breast cancer cells in a softer microenvironment (Fig 7d). Expression of the AP1 subunits, FOS, FOSL and FOSB, decreased with increased stiffness (Fig 7e–g and Sup Fig 6d–f).
Fig. 7. Breast cancer cells down regulate YAP and AP1 signaling pathways responding to the microenvironmental stiffness.

a GSEA of YAP1 signaling expression of breast cancer cells in soft rigidity environment compared to stiff environment. b MDA-MB-231 cells were cultured in different hydrogels for 24 hours and extracted total RNA. Relative YAP1 mRNA expression was detected in two cell lines by RT-qPCR. c Immunofluorescence of YAP1 (red) and DAPI (blue) expression in MDA-MB-231 cells cultured in different rigidity groups were detected and imaged. Scale bar indicates 20 μm. d GSEA of AP1 signaling expression of breast cancer cells in soft rigidity environment compared to stiff environment. e-g MDA-MB-231 cells were cultured in different hydrogels for 24 hours and extracted total RNA. Relative FOS (e), FOSL (f) and FOSB (g) mRNA expression were detected by RT-qPCR. Error bars represent standard deviations of three independent experiments. p values were shown as indicated in graph. Significance was indicated as p<0.05. h Hydrogels of different stiffnesses were mixed with 2×106 MDA-MB-231 cells, followed by subcutaneous injection into BALB/c nude mice for 30 days. Tumors were dissected from xenograft mice (soft group: n=8, middle group: n=7, stiff group: n=3). Scale bar indicates 10 mm. The tumor volumes were calculated by measuring length and width (mean ±SEM).
To quantify long-term tumorigenesis and proliferation, we euthanized mice 30-days after subcutaneous injection. Percentages of tumors formed in soft, middle, and stiff hydrogel xenograft groups were 100%, 87.5%, and 37.5%, respectively. Although the soft gel degraded the fastest as compared with the middle and stiff groups (as shown in the 10-day tumor formation data where the soft group showed the smallest volume), tumors in the soft hydrogels had the highest tumor formation percentage. There were no gels left in the 30-day tumor tissue, showing that all hydrogels eventually degraded over time. These data indicate that hydrogels mediated greatest effects on tumor formation and growth at earlier times after implantation. Tumors formed in middle hydrogels had the largest volume among the three groups, suggesting that tumor cells cultured in middle stiffness balanced metabolism, angiogenesis, and signaling to proliferate. However, tumors formed from soft and stiff gels showed comparable, small volumes (Fig 7h), which may due to the downregulation in key genes of angiogenesis. Overall, the middle stiffness hydrogel provided the most suitable overall environment for tumor growth with the stiff environment being least conducive.
Discussion
In this study, we applied a mature 3D hydrogel culture system to model the heterogeneous mechanics in different parts of a breast tumor and investigate changes in both cellular behaviors and transcriptomic profiles regulated by distinct microenvironmental stiffnesses. Our results demonstrate how heterogeneous stiffnesses of a tumor microenvironment affect breast cancer cells. The three locations we chose correspond with different functional requirements for tumor progression. Stiffnesses in the core and periphery promote proliferation or invasion of cancer cells, while the middle stiffness supports both proliferation and invasion. As MDA-MB-231 cells already exhibit a very ‘mesenchymal’ phenotype, we focused more on cell growth, changes in metabolism and early angiogenesis in our research.
Various biomaterials have been applied to grow 3D tumor models in vitro, although there is still not a gold standard formulation that optimally mimics the tumor microenvironment. Investigators select different materials to best satisfy the experimental needs and questions. Synthetic polymers such as PEG or polyesters are frequently used with chemically functionalized cell-binding motifs, which present adjustable mechanical properties and well-defined molecular structures[68, 69]. Natural materials such as proteins (collagen, gelatin)[70, 71], polysaccharides (hyaluronic acid, alginate)[72, 73], and ECM extracts (decellularized ECM, Matrigel)[74, 75], on the other hand, present better cellular responses as compared with synthetic polymers, albeit with less flexibility in manipulating material properties. Here we chose the collagen/alginate gel as a well-defined system to eliminate confounding effects due to unknown factors. We note that prior publications using hydrogels for 3D cell culture have used many different formulations, which may account for inconsistences not only with our results but also across tissue-engineered models of cancer. Differences in material components of hydrogels, culture conditions, and mechanics contribute to divergent outcomes in various model systems. The hydrogel system described in this manuscript, which uses readily available components and reproducible design parameters, positions us to investigate mechanisms accounting for these differences across studies in breast cancer and other malignancies. As both collagen and alginate are commonly used natural materials[76, 77], our gel system thus can provide comparable results with previous and future studies. Since the concentrations of collagen and alginate were fixed, we only varied millimolar concentrations of CaCl2 to control crosslinking and stiffness. As gels were all saturated in cell culture medium, the water uptake and surface charge also remained consistent with negligible changes. The varied crosslinking level of alginate may affect the overall sterically available functional groups within the gel, which requires further in-depth study on molecular conformations of hydrogels. We only focused on cell binding sites on the collagen fibers, which remained constant with the unchanged collagen concentration. Our previous results showed that while hydrated, the gel behaved in a manner of rubber-like materials where the linear region of stress-strain curve occurred at 0–25% strain. Microstructure illustrated the secondary network of crosslinked alginate over a network of collagen, where the collagen formed D-banded fibrils at neutral pH that can be captured by SHG imaging using two-photon microscopy. The gels were always kept under a saturated condition, so the microenvironment remained stable with no additional swelling or contracting.
We present the new observation that 3D matrix stiffness regulates metabolic pathways related to OXPHOS versus glycolysis. A stiff microenvironment altered genes related to mitochondrial respiration, enhancing processes associated with OXPHOS and FA metabolism. Prior research demonstrated that invasive cancer cells preferentially utilized OXPHOS[78]. By comparison, cancer cells in soft hydrogels representing the core of a tumor upregulate genes related to glycolysis and high YAP1 expression. Cancer cells commonly rely on glycolysis to meet energetic and anabolic requirements of proliferation, consistent with our results showing greater proliferation of cancer cells in soft and middle stiffness environments in vitro[79]. Our system controls mechanical properties of ECM without changing composition of the hydrogel itself, which may account for differences between studies. Further research will investigate mechanics controlling intersections of ECM mechanics and metabolism in cancer.
Substrate stiffness may promote migration and angiogenesis capacities of lung cancer through overexpression of MMPs and angiogenesis-related growth factors[80]. Consistently, short-term xenograft experiments in our research showed formation of blood vessels consistent with upregulation of FN1, ANG and MMP9 in breast cancer cells in a stiff microenvironment, whereas other angiogenesis-related genes (VEGFA, IL-18) remained unchanged or inconsistent in two cell lines (MDA-MB-231 and MCF-7). GSEA analysis showed enrichment of Rb-E2F signaling in cells cultured in a stiff microenvironment. The Rb-E2F pathway regulates genes involved in angiogenesis and invasion such as MMPs or FN1 in retinoblastoma and human circulating tumor cells (CTCs)[81, 82]. E2F also plays a regulatory role in energy metabolism of cancer cells[83]. FN1 promotes angiogenesis in prostate cancer[84] and hepatocellular carcinoma[85], regulated by MAPK signaling and the TFCP2 transcriptional factor. In accordance with pro-angiogenic functions, FN1 also promoted aggressive metastatic cancer cells. Secreted FN1 supports vascular mimicry contributing to tumor angiogenesis [86] through activation of ZEB. MMP9 in our experiments was increased by increasing stiffness, contributing to ECM degradation and basement membrane remodeling during angiogenesis. Tumor cells sense mechanics through multiple interconnected systems, including the actin cytoskeleton, integrins, cell-cell adhesion receptors, receptor tyrosine kinases, and other membrane proteins including ion channels and G-protein-coupled receptors[87]. Interestingly, in our Fig4 analysis, GO analysis showed enrichment of binding proteins, opening further studies in the future.
Effects of hydrogel stiffness likely diminish with time in vivo as regulation shifts toward ECM materials deposited by endogenous mouse stroma and contraction of ECM by fibroblasts in the tumor environment. In addition, these data indicate that interactions between environmental and cancer cell states regulate overall tumor formation. Long-term xenograft experiments showed higher proliferation of tumor cells in soft and middle microenvironments. Both the soft and middle stiffness hydrogels promoted comparable engraftment and proliferation in vitro and in mouse xenografts. Reduced tumor size in soft hydrogels may be due to dissolution of the soft hydrogel in vivo, resulting in loss of the expected tissue mechanics. Different calcium crosslinking ratios used to produce gel networks with desired stiffness led to different degradation kinetics in vivo, which affected tumor growth in the early time points. GSEA demonstrated an increase in YAP1 and AP1 signaling pathways in cells cultured in soft hydrogels compared to stiff hydrogels. Tumors require angiogenesis and proliferation to continue growth beyond millimeter size because of limits on diffusion of oxygen and nutrients[88]. The ability of middle stiffness hydrogels to support both proliferation and angiogenesis of cancer cells may account for greater tumor size and overall growth. Although the soft hydrogel has 100% engraftment in mice, the middle hydrogel was the most suitable environment for later tumor growth and proliferation. Previous studies indicate that plasticity of cancer cells and the ability to shift between proliferative and invasive states promotes disease progression in breast cancer and other malignancies[89–91]. Our results support the need for plasticity in proliferative and invasive cell states in tumor formation.
Our system utilized common biomaterials for construction of a simplified 3D tumor microenvironment, leaving opportunities for future improvements. While the simplified components provided well-controlled mechanical stiffness, the environment lacks other ECM components, such as laminin, present in tumor microenvironments. In addition, we simplified the cellular components and only studied the cancer cells and fibroblasts as stromal cells. Future advances in the system should include other cells, including subsets of immune cells and vascular endothelium. We recognize that a limitation of this study arises from using MDA-MB-231 cells, an established breast cancer cell line adapted to standard 2D cell culture conditions. Established cell lines may have genetic and/or epigenetic changes not representative of cancer cells in patients. We also acknowledge that relying on cell lines known to exhibit mesenchymal features may reduce the extent to which data generalize to all breast cancers. Extending these studies to patient-derived cells represents an exciting future direction for our hydrogel technology. While our hydrogel system controls mechanical properties of the environment, other variables in 3D cultures, including hypoxia and restricted diffusion of nutrients, also may affect behaviors of cells and results of RNA sequencing.
Heterogeneity in a tumor microenvironment involves complex effects arising not only from multiple cells but also from non-cellular ECM components. To investigate effects of ECM on cancer cells, we first started with a model focused on local stiffness as a single regulator and demonstrated effects of various ECM stiffnesses on cell behaviors. We believe our results will provide important basic knowledge for future studies on heterogeneous tumor microenvironments.
Conclusion
In this study, we employed a well-defined biomimetic ECM material to model different stiffnesses of a tumor microenvironment and investigated effects of microenvironmental stiffness on cell proliferation, migration, metabolism, angiogenesis and other transcriptomics of breast cancer cells. Our results illustrate functional and transcriptomics effects of different ECM microenvironmental stiffnesses on cancer cells. We believe this study will provide an essential foundation for future investigations of ECM mechanics in breast cancer and spark new approaches for therapy.
Supplementary Material
Acknowledgments
The authors acknowledge funding from United States National Institutes of Health grants R01CA238042, R01CA196018, U01CA210152, R01CA238023, R33CA225549, and R37CA222563 (GDL). This work was supported in part by the National Natural Science Foundation of China (grant no. 81970191); Guangdong Basic and Applied Basic Research Foundation (2019A1515110005); and the Fundamental program Funds of Shenzhen Science and Technology Innovation Commission (JCYJ20190807160209294; JCYJ20190807160813467).
Conflict of Interest
GDL receives research funding from Polyphor.
References
- [1].Alizadeh AA, Aranda V, Bardelli A, Blanpain C, Bock C, Borowski C, Caldas C, Califano A, Doherty M, Elsner M, Esteller M, Fitzgerald R, Korbel JO, Lichter P, Mason CE, Navin N, Pe’er D, Polyak K, Roberts CW, Siu L, Snyder A, Stower H, Swanton C, Verhaak RG, Zenklusen JC, Zuber J, Zucman-Rossi J, Toward understanding and exploiting tumor heterogeneity, Nat Med 21(8) (2015) 846–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fidler IJ, Tumor heterogeneity and the biology of cancer invasion and metastasis, Cancer Res 38(9) (1978) 2651–60. [PubMed] [Google Scholar]
- [3].Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C, Intratumor heterogeneity and branched evolution revealed by multiregion sequencing, N Engl J Med 366(10) (2012) 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, Halushka MK, Sukumar S, Parker LM, Anderson KS, Harris LN, Garber JE, Richardson AL, Schnitt SJ, Nikolsky Y, Gelman RS, Polyak K, Molecular definition of breast tumor heterogeneity, Cancer Cell 11(3) (2007) 259–73. [DOI] [PubMed] [Google Scholar]
- [5].Hida K, Ishii G, Editorial: Targeting tumor microenvironment heterogeneity, Adv Drug Deliv Rev 99(Pt B) (2016) 139. [DOI] [PubMed] [Google Scholar]
- [6].Hirata E, Sahai E, Tumor Microenvironment and Differential Responses to Therapy, Cold Spring Harb Perspect Med 7(7) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Keller L, Pantel K, Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells, Nat Rev Cancer 19(10) (2019) 553–567. [DOI] [PubMed] [Google Scholar]
- [8].McGranahan N, Swanton C, Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future, Cell 168(4) (2017) 613–628. [DOI] [PubMed] [Google Scholar]
- [9].Park CC, Bissell MJ, Barcellos-Hoff MH, The influence of the microenvironment on the malignant phenotype, Mol Med Today 6(8) (2000) 324–9. [DOI] [PubMed] [Google Scholar]
- [10].Pickup MW, Mouw JK, Weaver VM, The extracellular matrix modulates the hallmarks of cancer, EMBO Rep 15(12) (2014) 1243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM, Tensional homeostasis and the malignant phenotype, Cancer Cell 8(3) (2005) 241–254. [DOI] [PubMed] [Google Scholar]
- [12].Plodinec M, Loparic M, Monnier CA, Obermann EC, Zanetti-Dallenbach R, Oertle P, Hyotyla JT, Aebi U, Bentires-Alj M, Lim RY, Schoenenberger CA, The nanomechanical signature of breast cancer, Nat Nanotechnol 7(11) (2012) 757–65. [DOI] [PubMed] [Google Scholar]
- [13].Baker BM, Trappmann B, Wang WY, Sakar MS, Kim IL, Shenoy VB, Burdick JA, Chen CS, Cell-mediated fibre recruitment drives extracellular matrix mechanosensing in engineered fibrillar microenvironments, Nat Mater 14(12) (2015) 1262–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Berger AJ, Linsmeier KM, Kreeger PK, Masters KS, Decoupling the effects of stiffness and fiber density on cellular behaviors via an interpenetrating network of gelatin-methacrylate and collagen, Biomaterials 141 (2017) 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lu P, Weaver VM, Werb Z, The extracellular matrix: a dynamic niche in cancer progression, J Cell Biol 196(4) (2012) 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li Y, Kumacheva E, Hydrogel microenvironments for cancer spheroid growth and drug screening, Sci Adv 4(4) (2018) eaas8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ulrich TA, de Juan Pardo EM, Kumar S, The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells, Cancer Res 69(10) (2009) 4167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gilbert PM, Havenstrite KL, Magnusson KE, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM, Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture, Science 329(5995) (2010) 1078–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM, Matrix crosslinking forces tumor progression by enhancing integrin signaling, Cell 139(5) (2009) 891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Larue L, Bellacosa A, Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3’ kinase/AKT pathways, Oncogene 24(50) (2005) 7443–54. [DOI] [PubMed] [Google Scholar]
- [21].Reid SE, Kay EJ, Neilson LJ, Henze AT, Serneels J, McGhee EJ, Dhayade S, Nixon C, Mackey JB, Santi A, Swaminathan K, Athineos D, Papalazarou V, Patella F, Roman-Fernandez A, ElMaghloob Y, Hernandez-Fernaud JR, Adams RH, Ismail S, Bryant DM, Salmeron-Sanchez M, Machesky LM, Carlin LM, Blyth K, Mazzone M, Zanivan S, Tumor matrix stiffness promotes metastatic cancer cell interaction with the endothelium, EMBO J 36(16) (2017) 2373–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Broders-Bondon F, Nguyen Ho-Bouldoires TH, Fernandez-Sanchez ME, Farge E, Mechanotransduction in tumor progression: The dark side of the force, J Cell Biol 217(5) (2018) 1571–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chaudhuri O, Koshy ST, Branco da Cunha C, Shin JW, Verbeke CS, Allison KH, Mooney DJ, Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium, Nat Mater 13(10) (2014) 970–8. [DOI] [PubMed] [Google Scholar]
- [24].Hong H, Stegemann JP, 2D and 3D collagen and fibrin biopolymers promote specific ECM and integrin gene expression by vascular smooth muscle cells, J Biomater Sci Polym Ed 19(10) (2008) 1279–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hutmacher DW, Biomaterials offer cancer research the third dimension, Nat Mater 9(2) (2010) 90–3. [DOI] [PubMed] [Google Scholar]
- [26].Thakuri PS, Liu C, Luker GD, Tavana H, Biomaterials-Based Approaches to Tumor Spheroid and Organoid Modeling, Adv Healthc Mater 7(6) (2018) e1700980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Han K, Pierce SE, Li A, Spees K, Anderson GR, Seoane JA, Lo Y-H, Dubreuil M, Olivas M, Kamber RA, Wainberg M, Kostyrko K, Kelly MR, Yousefi M, Simpkins SW, Yao D, Lee K, Kuo CJ, Jackson PK, Sweet-Cordero A, Kundaje A, Gentles AJ, Curtis C, Winslow MM, Bassik MC, CRISPR screens in cancer spheroids identify 3D growth-specific vulnerabilities, Nature 580(7801) (2020) 136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Baker BM, Chen CS, Deconstructing the third dimension – how 3D culture microenvironments alter cellular cues, Journal of Cell Science 125(13) (2012) 3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Brancato V, Gioiella F, Imparato G, Guarnieri D, Urciuolo F, Netti PA, 3D breast cancer microtissue reveals the role of tumor microenvironment on the transport and efficacy of free-doxorubicin in vitro, Acta Biomater 75 (2018) 200–212. [DOI] [PubMed] [Google Scholar]
- [30].Lee JY, Chaudhuri O, Regulation of Breast Cancer Progression by Extracellular Matrix Mechanics: Insights from 3D Culture Models, ACS Biomaterials Science & Engineering 4(2) (2018) 302–313. [DOI] [PubMed] [Google Scholar]
- [31].Serebriiskii I, Castello-Cros R, Lamb A, Golemis EA, Cukierman E, Fibroblast-derived 3D matrix differentially regulates the growth and drug-responsiveness of human cancer cells, Matrix Biol 27(6) (2008) 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lee JY, Chang JK, Dominguez AA, Lee HP, Nam S, Chang J, Varma S, Qi LS, West RB, Chaudhuri O, YAP-independent mechanotransduction drives breast cancer progression, Nat Commun 10(1) (2019) 1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pickl M, Ries CH, Comparison of 3D and 2D tumor models reveals enhanced HER2 activation in 3D associated with an increased response to trastuzumab, Oncogene 28(3) (2009) 461–8. [DOI] [PubMed] [Google Scholar]
- [34].Riedl A, Schlederer M, Pudelko K, Stadler M, Walter S, Unterleuthner D, Unger C, Kramer N, Hengstschlager M, Kenner L, Pfeiffer D, Krupitza G, Dolznig H, Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses, J Cell Sci 130(1) (2017) 203–218. [DOI] [PubMed] [Google Scholar]
- [35].Khavari A, Nydén M, Weitz DA, Ehrlicher AJ, Composite alginate gels for tunable cellular microenvironment mechanics, Scientific Reports 6(1) (2016) 30854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cavo M, Caria M, Pulsoni I, Beltrame F, Fato M, Scaglione S, A new cell-laden 3D Alginate-Matrigel hydrogel resembles human breast cancer cell malignant morphology, spread and invasion capability observed “in vivo”, Scientific Reports 8(1) (2018) 5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Huang D, Nakamura Y, Ogata A, Kidoaki S, Characterization of 3D matrix conditions for cancer cell migration with elasticity/porosity-independent tunable microfiber gels, Polymer Journal 52(3) (2020) 333–344. [Google Scholar]
- [38].Liu C, Lewin Mejia D, Chiang B, Luker KE, Luker GD, Hybrid collagen alginate hydrogel as a platform for 3D tumor spheroid invasion, Acta Biomaterialia 75 (2018) 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Liu C, Chiang B, Lewin Mejia D, Luker KE, Luker GD, Lee A, Mammary fibroblasts remodel fibrillar collagen microstructure in a biomimetic nanocomposite hydrogel, Acta Biomater 83 (2019) 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Humphries BA, Buschhaus JM, Chen YC, Haley HR, Qyli T, Chiang B, Shen N, Rajendran S, Cutter A, Cheng YH, Chen YT, Cong J, Spinosa PC, Yoon E, Luker KE, Luker GD, Plasminogen Activator Inhibitor 1 (PAI1) Promotes Actin Cytoskeleton Reorganization and Glycolytic Metabolism in Triple-Negative Breast Cancer, Mol Cancer Res 17(5) (2019) 1142–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cavnar SP, Xiao A, Gibbons AE, Rickelmann AD, Neely T, Luker KE, Takayama S, Luker GD, Imaging Sensitivity of Quiescent Cancer Cells to Metabolic Perturbations in Bone Marrow Spheroids, Tomography : a journal for imaging research 2(2) (2016) 146–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cavnar SP, Rickelmann AD, Meguiar KF, Xiao A, Dosch J, Leung BM, Cai Lesher-Perez S, Chitta S, Luker KE, Takayama S, Luker GD, Modeling selective elimination of quiescent cancer cells from bone marrow, Neoplasia 17(8) (2015) 625–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Taubenberger AV, Bray LJ, Haller B, Shaposhnykov A, Binner M, Freudenberg U, Guck J, Werner C, 3D extracellular matrix interactions modulate tumour cell growth, invasion and angiogenesis in engineered tumour microenvironments, Acta Biomater 36 (2016) 73–85. [DOI] [PubMed] [Google Scholar]
- [44].Ulrich TA, de Juan Pardo EM, Kumar S, The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells, Cancer Res 69(10) (2009) 4167–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Liang Y, Jeong J, DeVolder RJ, Cha C, Wang F, Tong YW, Kong H, A cell-instructive hydrogel to regulate malignancy of 3D tumor spheroids with matrix rigidity, Biomaterials 32(35) (2011) 9308–15. [DOI] [PubMed] [Google Scholar]
- [46].Szot CS, Buchanan CF, Freeman JW, Rylander MN, 3D in vitro bioengineered tumors based on collagen I hydrogels, Biomaterials 32(31) (2011) 7905–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Jang J, Seol Y-J, Kim HJ, Kundu J, Kim SW, Cho D-W, Effects of alginate hydrogel crosslinking density on mechanical and biological behaviors for tissue engineering, J Mech Behav Biomed 37 (2014) 69–77. [DOI] [PubMed] [Google Scholar]
- [48].Cao N, Chen XB, Schreyer DJ, Influence of Calcium Ions on Cell Survival and Proliferation in the Context of an Alginate Hydrogel, ISRN Chemical Engineering 2012 (2012) 516461. [Google Scholar]
- [49].Fajardo AR, Silva MB, Lopes LC, Piai JF, Rubira AF, Muniz EC, Hydrogel based on an alginate–Ca2+/chondroitin sulfate matrix as a potential colon-specific drug delivery system, Rsc Adv 2(29) (2012) 11095–11103. [Google Scholar]
- [50].Hayakawa K, Tatsumi H, Sokabe M, Mechano-sensing by actin filaments and focal adhesion proteins, Commun Integr Biol 5(6) (2012) 572–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Mei L, Espinosa de los Reyes S, Reynolds MJ, Leicher R, Liu S, Alushin GM, Molecular mechanism for direct actin force-sensing by α-catenin, eLife 9 (2020) e62514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, Imamura T, Ogawa M, Masai H, Miyawaki A, Visualizing spatiotemporal dynamics of multicellular cell-cycle progression, Cell 132(3) (2008) 487–98. [DOI] [PubMed] [Google Scholar]
- [53].Windus LC, Glover TT, Avery VM, Bone-stromal cells up-regulate tumourigenic markers in a tumour-stromal 3D model of prostate cancer, Mol Cancer 12(1) (2013) 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dvorak KM, Pettee KM, Rubinic-Minotti K, Su R, Nestor-Kalinoski A, Eisenmann KM, Carcinoma associated fibroblasts (CAFs) promote breast cancer motility by suppressing mammalian Diaphanous-related formin-2 (mDia2), Plos One 13(3) (2018) e0195278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bischel LL, Beebe DJ, Sung KE, Microfluidic model of ductal carcinoma in situ with 3D, organotypic structure, BMC Cancer 15 (2015) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Graham N, Qian BZ, Mesenchymal Stromal Cells: Emerging Roles in Bone Metastasis, Int J Mol Sci 19(4) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Riedl A, Schlederer M, Pudelko K, Stadler M, Walter S, Unterleuthner D, Unger C, Kramer N, Hengstschläger M, Kenner L, Pfeiffer D, Krupitza G, Dolznig H, Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT–mTOR–S6K signaling and drug responses, Journal of Cell Science 130(1) (2017) 203. [DOI] [PubMed] [Google Scholar]
- [58].Lagies S, Schlimpert M, Neumann S, Wäldin A, Kammerer B, Borner C, Peintner L, Cells grown in three-dimensional spheroids mirror in vivo metabolic response of epithelial cells, Communications Biology 3(1) (2020) 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kapałczyńska M, Kolenda T, Przybyła W, Zajączkowska M, Teresiak A, Filas V, Ibbs M, Bliźniak R, Łuczewski Ł, Lamperska K, 2D and 3D cell cultures - a comparison of different types of cancer cell cultures, Arch Med Sci 14(4) (2018) 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Duval K, Grover H, Han L-H, Mou Y, Pegoraro AF, Fredberg J, Chen Z, Modeling Physiological Events in 2D vs. 3D Cell Culture, Physiology (Bethesda) 32(4) (2017) 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [61].Yu JSL, Cui W, Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination, Development 143(17) (2016) 3050. [DOI] [PubMed] [Google Scholar]
- [62].Chin YR, Toker A, Function of Akt/PKB signaling to cell motility, invasion and the tumor stroma in cancer, Cell Signal 21(4) (2009) 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Schevzov G, Kee AJ, Wang B, Sequeira VB, Hook J, Coombes JD, Lucas CA, Stehn JR, Musgrove EA, Cretu A, Assoian R, Fath T, Hanoch T, Seger R, Pleines I, Kile BT, Hardeman EC, Gunning PW, Regulation of cell proliferation by ERK and signal-dependent nuclear translocation of ERK is dependent on Tm5NM1-containing actin filaments, Molecular biology of the cell 26(13) (2015) 2475–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Guo N, Shen G, Zhang Y, Moustafa AA, Ge D, You Z, Interleukin-17 Promotes Migration and Invasion of Human Cancer Cells Through Upregulation of MTA1 Expression, Frontiers in oncology 9 (2019) 546–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Baonza A, Garcia-Bellido A, Notch signaling directly controls cell proliferation in the Drosophila wing disc, P Natl Acad Sci USA 97(6) (2000) 2609–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Low-Marchelli JM, Ardi VC, Vizcarra EA, van Rooijen N, Quigley JP, Yang J, Twist1 Induces CCL2 and Recruits Macrophages to Promote Angiogenesis, Cancer Res 73(2) (2013) 662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Tremblay AM, Missiaglia E, Galli GG, Hettmer S, Urcia R, Carrara M, Judson RN, Thway K, Nadal G, Selfe JL, Murray G, Calogero RA, De Bari C, Zammit PS, Delorenzi M, Wagers AJ, Shipley J, Wackerhage H, Camargo FD, The Hippo transducer YAP1 transforms activated satellite cells and is a potent effector of embryonal rhabdomyosarcoma formation, Cancer Cell 26(2) (2014) 273–87. [DOI] [PubMed] [Google Scholar]
- [68].Del Bufalo F, Manzo T, Hoyos V, Yagyu S, Caruana I, Jacot J, Benavides O, Rosen D, Brenner MK, 3D modeling of human cancer: A PEG-fibrin hydrogel system to study the role of tumor microenvironment and recapitulate the in vivo effect of oncolytic adenovirus, Biomaterials 84 (2016) 76–85. [DOI] [PubMed] [Google Scholar]
- [69].Rijal G, Bathula C, Li W, Application of Synthetic Polymeric Scaffolds in Breast Cancer 3D Tissue Cultures and Animal Tumor Models, Int J Biomater 2017 (2017) 8074890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Qiu Y, Qiu S, Deng L, Nie L, Gong L, Liao X, Zheng X, Jin K, Li J, Tu X, Liu L, Liu Z, Bao Y, Ai J, Lin T, Yang L, Wei Q, Biomaterial 3D collagen I gel culture model: A novel approach to investigate tumorigenesis and dormancy of bladder cancer cells induced by tumor microenvironment, Biomaterials 256 (2020) 120217. [DOI] [PubMed] [Google Scholar]
- [71].Zhu J, Zheng S, Liu H, Wang Y, Jiao Z, Nie Y, Wang H, Liu T, Song K, Evaluation of anti-tumor effects of crocin on a novel 3D tissue-engineered tumor model based on sodium alginate/gelatin microbead, Int J Biol Macromol 174 (2021) 339–351. [DOI] [PubMed] [Google Scholar]
- [72].Amorim S, Soares da Costa D, Pashkuleva I, Reis CA, Reis RL, Pires RA, 3D hydrogel mimics of the tumor microenvironment: the interplay among hyaluronic acid, stem cells and cancer cells, Biomater Sci 9(1) (2021) 252–260. [DOI] [PubMed] [Google Scholar]
- [73].Bonnesoeur S, Morin-Grognet S, Thoumire O, Le Cerf D, Boyer O, Vannier JP, Labat B, Hyaluronan-based hydrogels as versatile tumor-like models: Tunable ECM and stiffness with genipin-crosslinking, J Biomed Mater Res A 108(5) (2020) 1256–1268. [DOI] [PubMed] [Google Scholar]
- [74].Kwak TJ, Lee E, In vitro modeling of solid tumor interactions with perfused blood vessels, Sci Rep 10(1) (2020) 20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Alabi BR, LaRanger R, Shay JW, Decellularized mice colons as models to study the contribution of the extracellular matrix to cell behavior and colon cancer progression, Acta Biomater 100 (2019) 213–222. [DOI] [PubMed] [Google Scholar]
- [76].Szot CS, Buchanan CF, Freeman JW, Rylander MN, 3D in vitro bioengineered tumors based on collagen I hydrogels, Biomaterials 32(31) (2011) 7905–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Qiao SP, Zhao YF, Li CF, Yin YB, Meng QY, Lin FH, Liu Y, Hou XL, Guo K, Chen XB, Tian WM, An alginate-based platform for cancer stem cell research, Acta Biomater 37 (2016) 83–92. [DOI] [PubMed] [Google Scholar]
- [78].Caino MC, Ghosh JC, Chae YC, Vaira V, Rivadeneira DB, Faversani A, Rampini P, Kossenkov AV, Aird KM, Zhang R, Webster MR, Weeraratna AT, Bosari S, Languino LR, Altieri DC, PI3K therapy reprograms mitochondrial trafficking to fuel tumor cell invasion, Proc Natl Acad Sci U S A 112(28) (2015) 8638–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB, Brick by brick: metabolism and tumor cell growth, Curr Opin Genet Dev 18(1) (2008) 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zhao D, Xue C, Li Q, Liu M, Ma W, Zhou T, Lin Y, Substrate stiffness regulated migration and angiogenesis potential of A549 cells and HUVECs, J Cell Physiol 233(4) (2018) 3407–3417. [DOI] [PubMed] [Google Scholar]
- [81].Meier C, Spitschak A, Abshagen K, Gupta S, Mor JM, Wolkenhauer O, Haier J, Vollmar B, Alla V, Pützer BM, Association of RHAMM with E2F1 promotes tumour cell extravasation by transcriptional up-regulation of fibronectin, The Journal of Pathology 234(3) (2014) 351–364. [DOI] [PubMed] [Google Scholar]
- [82].Schaal C, Pillai S, Chellappan SP, The Rb-E2F transcriptional regulatory pathway in tumor angiogenesis and metastasis, Adv Cancer Res 121 (2014) 147–182. [DOI] [PubMed] [Google Scholar]
- [83].Schaal C, Pillai S, Chellappan SP, Chapter Four - The Rb–E2F Transcriptional Regulatory Pathway in Tumor Angiogenesis and Metastasis, in: Tew KD, Fisher PB (Eds.), Advances in Cancer Research, Academic Press; 2014, pp. 147–182. [DOI] [PubMed] [Google Scholar]
- [84].Hybel TE, Dietrichs D, Sahana J, Corydon TJ, Nassef MZ, Wehland M, Kruger M, Magnusson NE, Bauer J, Utpatel K, Infanger M, Grimm D, Kopp S, Simulated Microgravity Influences VEGF, MAPK, and PAM Signaling in Prostate Cancer Cells, Int J Mol Sci 21(4) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Xu X, Liu Z, Zhou L, Xie H, Cheng J, Ling Q, Wang J, Guo H, Wei X, Zheng S, Characterization of genome-wide TFCP2 targets in hepatocellular carcinoma: implication of targets FN1 and TJP1 in metastasis, Journal of Experimental & Clinical Cancer Research 34(1) (2015) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Arslan AD, Sassano A, Saleiro D, Lisowski P, Kosciuczuk EM, Fischietti M, Eckerdt F, Fish EN, Platanias LC, Human SLFN5 is a transcriptional co-repressor of STAT1-mediated interferon responses and promotes the malignant phenotype in glioblastoma, Oncogene 36(43) (2017) 6006–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Zanotelli MR, Reinhart-King CA, Mechanical Forces in Tumor Angiogenesis, in: Dong C, Zahir N, Konstantopoulos K (Eds.), Biomechanics in Oncology, Springer International Publishing, Cham, 2018, pp. 91–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Folkman J, Role of angiogenesis in tumor growth and metastasis, Semin Oncol 29(6 Suppl 16) (2002) 15–8. [DOI] [PubMed] [Google Scholar]
- [89].Luo M, Brooks M, Wicha MS, Epithelial-mesenchymal plasticity of breast cancer stem cells: implications for metastasis and therapeutic resistance, Curr Pharm Des 21(10) (2015) 1301–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Kong D, Hughes CJ, Ford HL, Cellular Plasticity in Breast Cancer Progression and Therapy, Frontiers in Molecular Biosciences 7(72) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Lu W, Kang Y, Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis, Dev Cell 49(3) (2019) 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


